Significance
Syncytins are “captured” genes of retroviral origin, corresponding to the fusogenic envelope gene of endogenized retroviruses. They are present in a series of eutherian mammals, including humans and mice where they play an essential role in placentation. Here we show that marsupials—which diverged from eutherian mammals ∼190 Mya but still possess a primitive, short-lived placenta (rapidly left by the embryo for development in an external pouch)—have also captured such genes. The present characterization of the syncytin-Opo1 gene in the opossum placenta, together with the identification of two additional endogenous retroviral envelope gene captures, allow a recapitulation of the natural history of these unusual genes and definitely extends their “symbiotic niche” to all clades of placental mammals.
Keywords: endogenous retrovirus, envelope protein, fusogenic activity, syncytiotrophoblast, marsupials
Abstract
Syncytins are genes of retroviral origin captured by eutherian mammals, with a role in placentation. Here we show that some marsupials—which are the closest living relatives to eutherian mammals, although they diverged from the latter ∼190 Mya—also possess a syncytin gene. The gene identified in the South American marsupial opossum and dubbed syncytin-Opo1 has all of the characteristic features of a bona fide syncytin gene: It is fusogenic in an ex vivo cell–cell fusion assay; it is specifically expressed in the short-lived placenta at the level of the syncytial feto–maternal interface; and it is conserved in a functional state in a series of Monodelphis species. We further identify a nonfusogenic retroviral envelope gene that has been conserved for >80 My of evolution among all marsupials (including the opossum and the Australian tammar wallaby), with evidence for purifying selection and conservation of a canonical immunosuppressive domain, but with only limited expression in the placenta. This unusual captured gene, together with a third class of envelope genes from recently endogenized retroviruses—displaying strong expression in the uterine glands where retroviral particles can be detected—plausibly correspond to the different evolutionary statuses of a captured retroviral envelope gene, with only syncytin-Opo1 being the present-day bona fide syncytin active in the opossum and related species. This study would accordingly recapitulate the natural history of syncytin exaptation and evolution in a single species, and definitely extends the presence of such genes to all major placental mammalian clades.
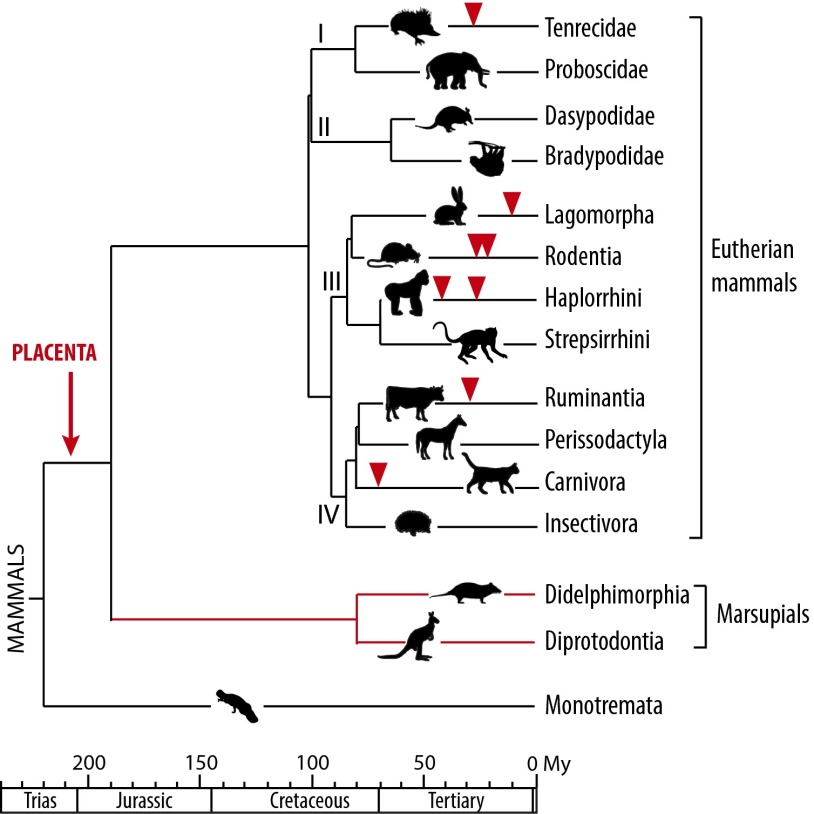
Marsupial mammals, such as kangaroos and opossums, are the closest living relatives to placental eutherian mammals (Fig. 1), from which, however, they diverged ∼190 million years ago (Mya) (1, 2). The latter comprise four major clades—namely, the Laurasiatheria (e.g., the ruminants and carnivorans), the Euarchontoglires (e.g., primates, rodents and lagomorphs), the Xenarthra, and the Afrotheria. All are viviparous animals, which gestate their young internally with a specialized organ of fetal origin—the placenta—allowing prolonged nutrient and gas exchanges between the mother and fetus. The structure of the feto-maternal interface displays strong variations, from simply apposed fetal and maternal membranes (the epitheliochorial placenta) to highly invasive placental tissues bathed by the maternal blood (the hemochorial placenta). In marsupials, a short-lived placenta is also formed, but the period of intimate contact between this transient organ and the maternal endometrium is very short—from 4 to 10 days—compared with that for eutherian mammals—up to 22 months for the elephant—with the marsupial fetus being rapidly released, in most species in an external pouch, where it will develop via lactation (3).
Fig. 1.
Phylogeny of mammals and previously identified syncytin genes. Mammals comprise the monotremes, the marsupials, and the eutherians, the latter comprising the four major clades: Afrotheria (I), Xenarthra (II), Euarchontoglires (III), and Laurasiatheria (IV) (data from ref. 1). Branch length is proportional to time (in My), and the time of insertion of the different syncytins identified to date is indicated with arrowheads.
In eutherian mammals, previous studies have identified envelope (env) genes of retroviral origin that have been independently captured and “co-opted” by their host, most probably for a function in placentation, and which have been named syncytins (reviewed in refs. 4 and 5). In simians, syncytin-1 (6–9) and syncytin-2 (10, 11), as bona fide syncytins, entered the primate genome 25 and >40 Mya, respectively, retained their coding capacity in all of the subsequent lineages, display placenta-specific expression, and are fusogenic in ex vivo cell–cell fusion assays. Furthermore, one of them displays immunosuppressive activity (12). A pair of env genes from endogenous retroviruses (ERVs) was also identified in the Muroidea, named syncytin-A and -B, which share closely related functional properties, although they have a completely distinct origin, showing a divergent sequence and a different genomic location compared with the primate syncytins (13, 14). Via the generation of syncytin knockout mice, we have unambiguously demonstrated that these genes are indeed essential for placentation, with a lack of cell–cell fusion observed in vivo at the level of the syncytiotrophoblast interhemal layer of the mutant placenta, resulting in impaired feto-maternal exchanges and embryo development and survival (15, 16). Recently, syncytin genes have been identified in four other clades among eutherian mammals—namely, in lagomorphs, carnivorans, ruminants, and the primitive afrotherian tenrecs (refs. 17–20; see also ref. 21). They all are unrelated to the simian and murine genes, but most probably share with them a function in placentation.
Here, we asked whether marsupials—despite their specific short-lived placenta and their ancestral divergence from the eutherian mammals—have also acquired syncytin genes of retroviral origin, as the latter. By combining (i) an in silico search for candidate genes in sequenced marsupial genomes; (ii) assays for their in vivo transcriptional activity by RT-PCR among a large panel of tissues that were recovered from the gray short-tailed opossum (Monodelphis domestica), including the placenta; (iii) cloning of the candidate genes from this species; and (iv) in situ hybridization (ISH) of placenta sections with appropriate probes, we identified a placenta-specific ERV env gene, displaying all of the characteristic features of a bona fide syncytin gene, that we accordingly named syncytin-Opo1. It is specifically expressed within syncytial structures at the feto-maternal interface; it is fusogenic in an ex vivo cell–cell fusion assay; and it is conserved in the Monodelphis genus with evidence for purifying selection. Moreover, we identified an ancestrally captured retroviral env gene, conserved within all marsupials and displaying strong purifying selection as demonstrated by sequencing the orthologous copies in 26 marsupial species from both the Australian (e.g., wallaby) and American (e.g., opossum) lineages, thus dating its capture between 80 and 190 Mya. This gene is nonfusogenic because it does not possess a transmembrane anchoring domain and thus cannot be involved in syncytiotrophoblast formation. Although it is not expressed at a significant level in the placenta—at least at the postimplantation stages—it possesses a canonical immunosuppressive domain (ISD) and, as such, could still be involved in placenta formation via this latter function. Finally, a third retroviral env gene was found to be expressed in the maternal uterine glands, belonging to recently acquired endogenous proviral sequences possibly responsible for the formation of the viral-like particles that we observed by electron microscopy at the same sites. All together, the present study clearly extends the range of mammals in which retroviral env gene captures and, in some instances, bona fide syncytin exaptations have taken place in the course of evolution, now including the marsupials in addition to the eutherian mammals. It is consistent with syncytins playing a role in the emergence of placentation from primitive egg-laying species, with the monotremes—e.g., the mammalian oviparous platypus—still existing as their rare descendants today.
Results
In Silico Search for Retroviral env Genes Within the Opossum (M. domestica) Genome.
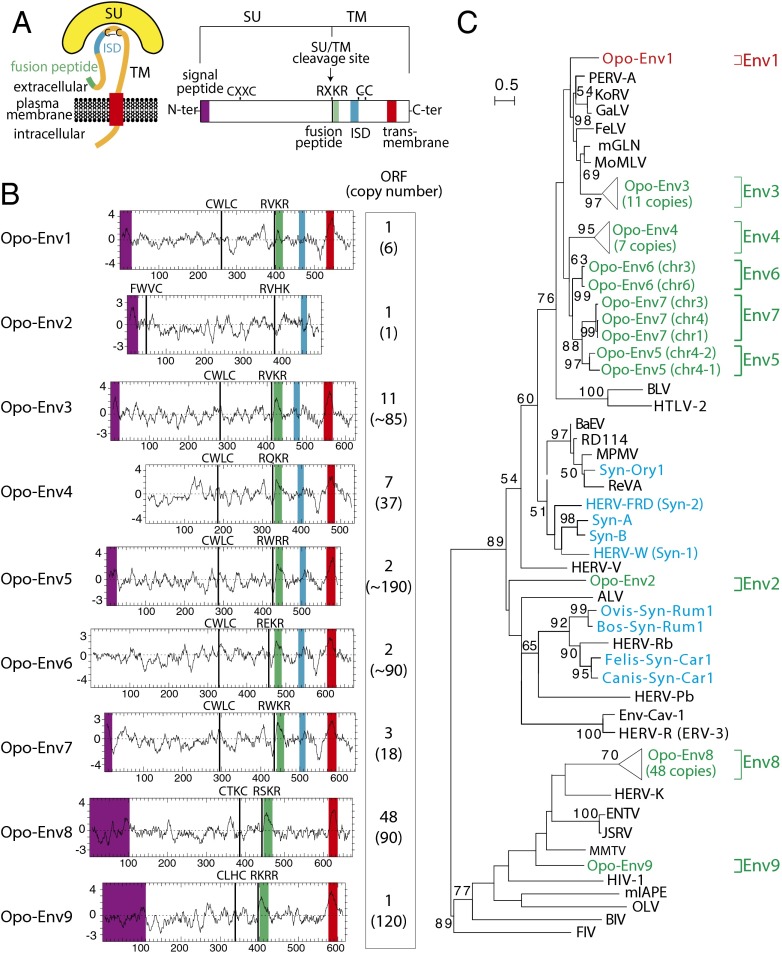
To identify putative env-derived syncytin genes, we made use of the opossum genome assembly [6.8× coverage assembly of the M. domestica genome; University of California, Santa Cruz (UCSC) Broad Institute MonDom5; October 2006]. A BLAST search for ORFs (from the Met start codon to the stop codon) >450 aa was performed by using a selected series of Env sequences representative of both infectious and ERV families, including all identified syncytins (Methods). It yielded 76 sequences, incorporated into the phylogenetic Env tree shown in Fig. 2C. Some of the sequences can be grouped into single families, resulting finally in nine families that we named Opo-Env1 to -Env9 (Fig. 2B). Analysis of the overall structure of the nine identified Env families (Fig. 2) strongly suggests that they indeed correspond to bona fide retroviral Env proteins, with some of their characteristic features, including a putative furin cleavage site delineating a surface (SU) and a transmembrane (TM) subunit and a CXXC motif in the SU subunit corresponding to a binding domain between the two subunits. Hydrophobicity plots identify a putative hydrophobic fusion peptide at the N terminus of the TM subunit, as well as the hydrophobic transmembrane domain within the TM subunits required for anchoring the Env protein within the plasma membrane, with the exception of Opo-Env2 (Fig. 2 and Fig. S1) that contains a premature stop codon before this transmembrane domain. A putative signal peptide can be predicted at the N terminus of most genes, with the exception of Opo-Env4 and -Env6. Opo-Env1 to -Env7 contain a canonical ISD (12). Only the opo-env1, -env2, and -env9 genes are present as a single copy encoding a full-length ORF. Finally, a BLAST search disclosed that only the opo-env1 and -env2 gene families are present at a low copy number (six and one, respectively), whereas the seven other env gene families display a much higher copy number (between 18 and 190; Fig. 2B).
Fig. 2.
Structure of a canonical retroviral Env protein and characterization of the identified opossum candidates. (A) Schematic representation of a retroviral Env protein, delineating the SU and TM subunits. The furin cleavage site (consensus: R/K-X-R/K-R) between the two subunits, the C-X-X-C motif involved in SU–TM interaction, the hydrophobic signal peptide (purple), the fusion peptide (green), the transmembrane domain (red), and the putative ISD (blue) along with the conserved C-X5/6/7-C motif are indicated. (B) Characterization of the candidate opossum (M. domestica) Env proteins. (B, Left) The hydrophobicity profile of each candidate is shown with the canonical structural features highlighted in A positioned, when present (same color code). (B, Right) Number of full-length env gene ORFs within each family of element and total number of genomic copies (in parentheses). (C) Retroviral Env protein-based phylogenetic tree with the identified Opo-Env protein candidates. Env proteins that were grouped into single families of elements are distinguished by indication of their chromosomal position (Table S1). A maximum-likelihood tree using TM subunit amino acid sequences from syncytins and a series of endogenous and infectious retroviruses is shown. The horizontal branch length is proportional to the percentage of amino acid substitutions from the node (scale bar on the left), and the percent bootstrap values >50% (obtained from 1,000 replicates) are indicated at the nodes. ALV, avian leukemia virus; MPMV, Mason–Pfizer monkey virus; Env-Cav1, “syncytin-like” Cavia porcellus Env1 protein; KoRV, koala retrovirus; GaLV, gibbon ape leukemia virus; PERV, porcine endogenous retrovirus; BaEV, baboon endogenous virus; HIV1, HIV type 1; HERV, human endogenous retrovirus; MoMLV, Moloney murine leukemia virus; HTLV-2, human T-lymphotropic virus type 2; BLV, bovine leukemia virus; JSRV, Jaagsiekte retrovirus; MMTV, murine mammary tumor virus; mIAPE, M. musculus intracisternal A-type particle with an Env gene; FeLV, feline leukemia virus; FIV, feline immunodeficiency virus; RD114, feline endogenous type-C retrovirus.
Transcription Profile Analyses for the Identification of Placenta-Specific env Genes.
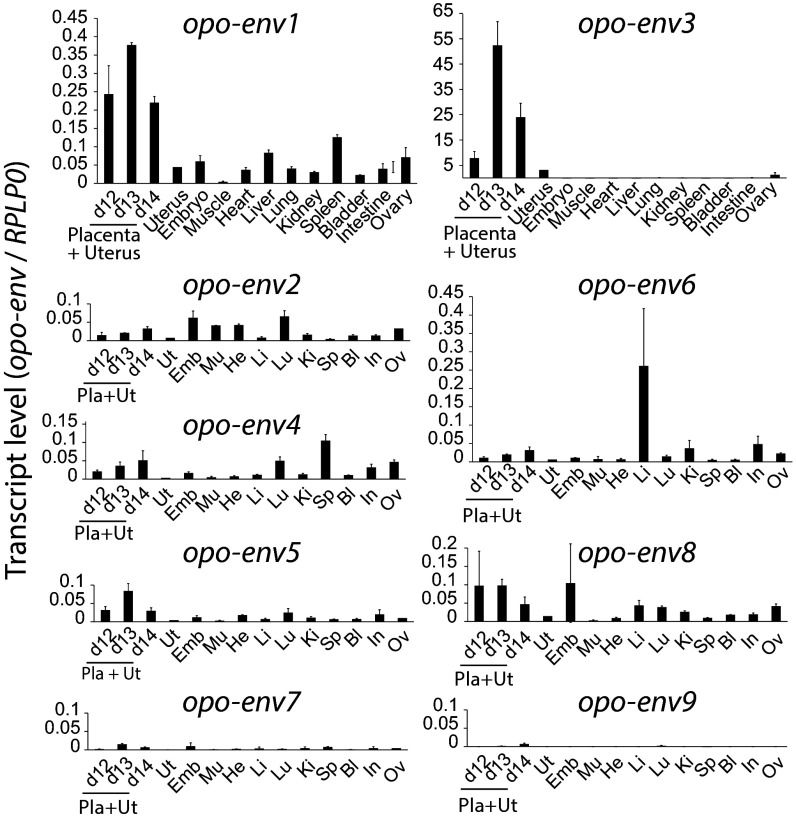
We then examined the transcript levels of the nine candidate env gene families in the opossum placenta and in a panel of other tissues. Quantitative RT-PCR (qRT-PCR) analyses were performed by using primers designed to be specific for the ORF-containing sequences within each family of elements (Table S2). In the opossum (M. domestica), gestation time is 15 days, with a transitory placenta being established shortly before parturition, between days 12 and 15. Because of the interpenetration of the two tissues, the uterus and the attached placenta were collected as a whole, at days 12, 13, and 14. As illustrated in the qRT-PCR analysis in Fig. 3, two genes—namely, opo-env1 and -env3—are expressed at a significant level in the placenta and uterus, as expected for candidate syncytin genes, with reduced expression in the other organs (including the uterus from nonpregnant females). Opo-env3 is the most highly expressed gene, up to 60-fold higher than the housekeeping ribosomal protein RPLP0 gene, and with >100-fold higher expression at day 13 of gestation than env1 (but it is present at a >10-fold higher copy number than opo-env1; Fig. 2). The other env genes, with the exception of opo-env6 in the liver, showed no or only limited expression in all organs. To go further into the analysis of the expression of both opo-env1 and -env3 in the placenta, and more precisely to determine—especially in the case of the multicopy opo-env3 ORF—whether the specific expression observed in the placenta originates from definite and/or single copies, RT-PCR analyses were performed to sort out both the Env1 and Env3 protein-encoding transcripts in the placenta, using primers designed to amplify all Env protein-coding copies. Sequencing of the bulk of the RT-PCR products disclosed only one copy with a full-length ORF (no polymorphism detectable in the amplified transcripts) for both opo-env1 (consistent with the in silico analysis) and, more unexpectedly, for opo-env3. It strongly suggests that in both cases, only one Env protein-coding copy (among the 11 copies identified in silico for opo-env3) is actively transcribed, as further confirmed by subcloning the bulk of the RT-PCR products and sequencing a series of molecular clones.
Fig. 3.
Real-time qRT-PCR analysis of the candidate opo-env gene transcripts from the opossum (M. domestica). Transcript levels are expressed as the ratio of the expression level of each opo-env gene to that of the RPLP0 control gene (SI Methods). Because of the high interpenetration of maternal and fetal tissues, placental and uterine tissues are analyzed as a whole at three gestational dates (days 12, 13, and 14). The results obtained with the same series of tissues for the nine env gene candidates are shown. Values are the means of duplicates from three samples ± SEM.
Altogether, the in silico analyses combined with the RT- and qRT-PCR assays for the opossum retroviral env genes clearly identify opo-env1 and -env3 as candidate syncytin genes (aa sequences provided in Fig. S1).
In situ hybridization on Placenta Sections.
The opossum placenta is a transitory organ, lasting for only 4 days between the shell coat rupture at 11 days postcoitus (dpc) and birth at 15 dpc (22). At 14 dpc, the definitive placenta is vascularized, with fetal vessels coming in between the inner endodermal layer and the outer trophoblastic layer of the placenta, forming the so-called “trilaminar” placenta. On the maternal side, three layers can also be observed, with, from mother to placenta: (i) the uterine myometrium, (ii) the trabecular endometrial uterine glands that secrete nutrients into the uterine lumen, and (iii) the maternal uterine epithelium with underlying maternal capillaries (Fig. 4A). During gestation, the uterine epithelium proliferates, forming villi and crypts that will be further colonized by the growing fetal placenta. At the feto-maternal interface, syncytial structures can be observed, formed by the cellular fusion of fetal trophoblast cells (22). Nuclei of the syncytia are grouped together, forming “syncytial knots” of two to five nuclei, separated by thin projections of the syncytia cytoplasm (Fig. 4A). The maternal epithelium is invaded at regular intervals by the syncytiotrophoblast that will displace the uterine cells without degrading them (Fig. 4 A and B). At these sites of invasion, the syncytiotrophoblast will directly contact the maternal capillaries, forming regions locally similar to that of the endotheliochorial placenta of eutherian mammals, where the syncytiotrophoblast layer surrounds the maternal vessels.
Fig. 4.
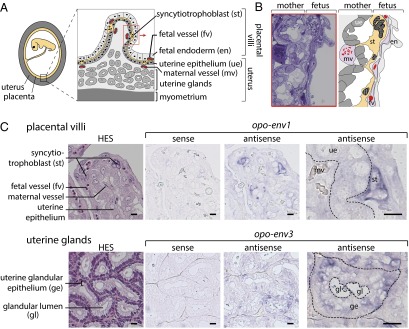
Structure of the opossum (M. domestica) placenta and ISH for opo-env1 and -env3 expression on placental sections. (A) Schematic representation of the opossum placenta. (A, Left) Overview of a gravid uterus displaying the apposed maternal and fetal tissues. The yellow and gray areas represent the fetus and mother tissues, respectively. (A, Right) Detailed scheme of the definitive placental structure. The trilaminar placenta (fetal endoderm, fetal vessels, and syncytiotropholast layer) is apposed to the maternal uterine epithelium. Locally, the maternal epithelium is invaded by projections from the syncytiotrophoblast layer that directly contact the maternal vessels. A large trabecular meshwork of uterine glands can be seen underneath the maternal uterine epithelium. (B) Semithin section of the feto-maternal interface, corresponding to the area boxed in A, with the various constituents delineated on the right. The multinucleate syncytiotrophoblast (st, yellow) penetrates the uterine epithelium (ue, dark gray) and contacts the maternal vessel (mv, pink). (C) hematoxylin eosin saffron (HES)-stained sections of placenta and ISH on serial sections for opo-env1 (Upper) or opo-env3 (Lower) placental transcripts using digoxigenin-labeled antisense or sense riboprobes revealed with an alkaline phosphatase-conjugated antidigoxigenin antibody. (C, Upper) Placental villi. (C, Lower) Uterine glands. Specific staining is observed at the level of the feto-maternal interface of placental villi for opo-env1 and at the level of the maternal uterine glands for opo-env3 (enlarged views in Right). (Scale bar: 10 µm.)
To assess the physiological relevance of opo-env1 and -env3 expression, we performed ISH experiments on paraffin sections of opossum placenta. Specific digoxigenin-labeled antisense riboprobes were synthesized for the detection of the opo-env1 and -env3 transcripts, as well as the corresponding sense riboprobes to be used as negative controls. As illustrated in Fig. 4C, specific labeling was observed only with the antisense probes, and not with the control probes for both genes. Specific hybridization with the opo-env1 antisense probe was observed at the feto-maternal interface. More precisely, opo-env1 is expressed at the level of the multinucleate syncytiotrophoblast, with the observation of a strong labeling of the syncytial knots, where syncytiotrophoblast nuclei are grouped. This labeling is consistent with a role for opo-env1 in the formation of the syncytiotrophoblast. In contrast to opo-env1, specific labeling with the opo-env3 antisense probe was not observed at the level of the feto-maternal interface (Fig. S2), but at the level of the uterine endometrial glands (Fig. 4C). Interestingly, as illustrated in Fig. S3, electron microscopy analysis of these uterine glands revealed the presence of viral particles budding into the glandular lumen. In the extracellular medium, these particles display a dense—icosahedral—core, as classically observed for mature retroviral particles.
Given the relative proportion of the uterine glands to the fetal syncytiotrophoblast, this pattern of expression—with opo-env3 expressed in the uterine glands and opo-env1 expressed in the syncytiotrophoblast layer—is compatible with the higher levels of expression of opo-env3 compared with opo-env1 in the mixed placenta and uterus tissues observed above in the qRT-PCR experiment. All together, these data strongly argue for opo-env1, but not opo-env3, to be a candidate syncytin gene in the opossum, provided that it has fusogenic activity.
Env1 Is a Fusogenic Retroviral Env Protein.
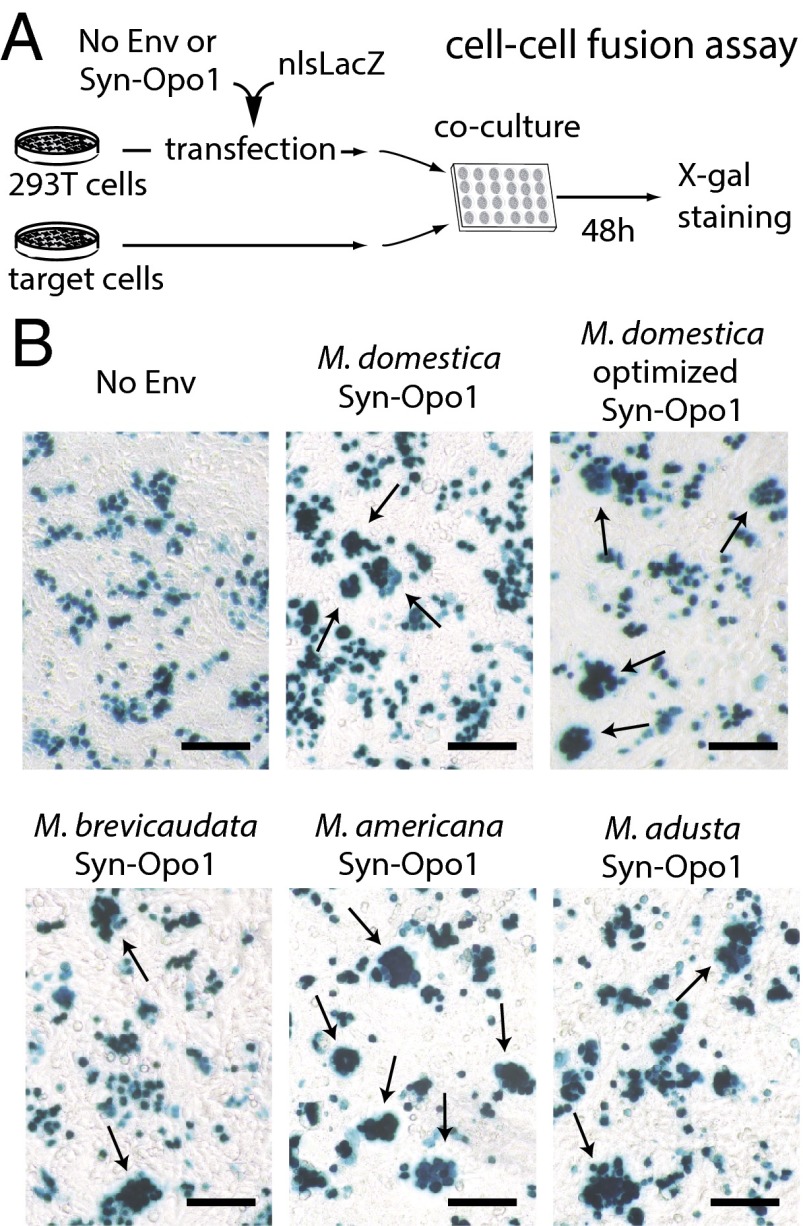
The functionality of Opo-Env1 as a retrovirally derived, fusogenic Env protein was assayed ex vivo as described in Fig. 5. Basically, we tested whether the opossum Env protein could induce the formation of syncytia in a cell–cell fusion assay. An expression vector for env1 was constructed (phCMV-env1; SI Methods), which was introduced (together with an nlsLacZ vector; Fig. 5) into the highly transfectable human 293T cell line. The transfected cells were then cocultured with target cells from different species (SI Methods), and cell–cell fusion was detected after 48 h of coculture, upon X-gal staining for visualization of the syncytia formed between the Env-expressing and target cells. As shown in Fig. 5, expression of opo-env1 in 293T cells resulted in cell–cell fusion when using the A23 hamster cells as the target, leading to the formation of multinucleated syncytial structures. The effect was not observed with an empty “none” vector as a control. Interestingly, the same opo-env1 expression vector construct, but modified to contain optimized codons for expression in human cells (synthetic gene; SI Methods), slightly increased fusion efficiency, as expected (Table S3). Of note, no significant syncytium formation could be detected for any of the other target cell lines used in the assay, including the opossum kidney (OK) cells (Table S3), a situation reminiscent of what had been observed for the fusogenic mouse Syncytin-B, which induced syncytium formation in vivo (16) but was found to be fusogenic ex vivo with only one cell line among all those tested (the dog MDCK cells; ref. 13). These unusual situations could be accounted for by assuming that the—still-to-be-identified—receptors for the corresponding Env proteins are not—or are only poorly—expressed in most of the tested cell lines and/or, in the case of the nonmarsupial cells, that the Env–receptor interactions have been impaired due to species-specific mutations. Although a complete understanding of the “fusion pattern” of Opo-Env1 would require the identification of its cognate receptor, the present experiments establish that Opo-Env1 is a fusogenic Env protein, which will now be named Syncytin-Opo1.
Fig. 5.
Syncytin-Opo1 is a fusogenic retroviral Env protein. (A) Schematic representation of the coculture assay for cell–cell fusion with Syncytin-Opo1. Human 293T cells were transfected with an expression vector for Syncytin-Opo1 (or an empty “No Env” vector as a control) and a plasmid expressing a nuclear beta-galactosidase (nlsLacZ). After transfection, the 293T cells were cocultured with target cells and X-gal–stained 48 h later. (B) Syncytium formation (see arrows) with the indicated Syncytin-Opo1, using A23 cells as the target (no syncytium with the No Env control). (Scale bars: 200 µm.)
Characterization of the syncytin-Opo1 Genomic Locus.
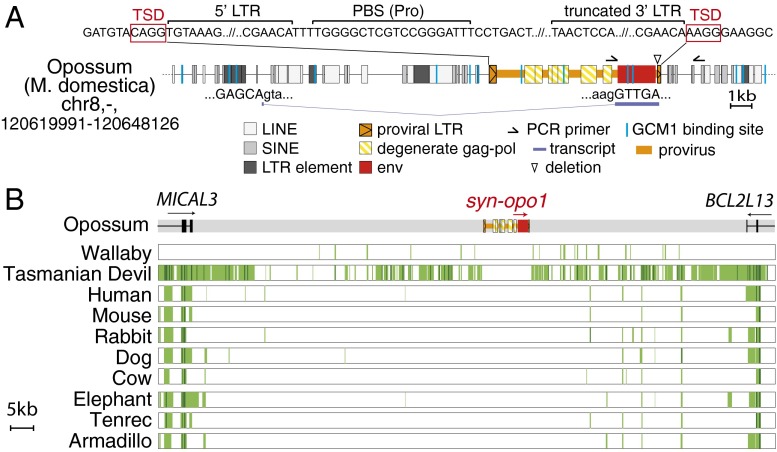
As illustrated in Fig. 6, syncytin-Opo1 is part of a proviral sequence with degenerate, but still identifiable, long terminal repeats (LTR) and gag–pol gene sequences (Fig. 6A). Its 3′ LTR is 5′-truncated, but the provirus flanking sequences disclose a degenerate target site duplication (TSD) of four nucleotides, consistent with proviral integration (Fig. 6A). A primer binding site (PBS) sequence can also be identified downstream to the 5′ LTR, as commonly observed for retroviruses, in which it is used for priming reverse transcription, and is found in the present case to be complementary to proline tRNA (Fig. 6A). The retroviral origin of the syncytin-Opo1 gene is further supported by the occurrence of a retroviral-like mode of generation of the subgenomic env transcripts, with a putative acceptor splice site located just upstream of the Env initiation codon. Its position and functionality were ascertained by RACE-PCR analysis of Syncytin-Opo1–encoding transcripts in the placenta, using appropriate primers (Fig. 6A). Interestingly, at variance with most syncytin genes, the promoter responsible for placental expression of syncytin-Opo1 does not seem to be located within the 5′ LTR of the proviral sequence, but, rather, within the genomic flanking sequence, ∼10 kb 5′ to the provirus. Of note, several putative binding sites for glial cell missing 1 (GCM1), a placental transcription factor known to regulate expression of some human and murine syncytins (reviewed in ref. 5), can be identified close to this placental promoter, which may account for the placental expression of syncytin-Opo1. As shown in Fig. 6B, the syncytin-Opo1–containing provirus is located in an intergenic region, between the MICAL3 and BCL2L13 cellular genes. In silico analysis of the syntenic locus in the human, mouse, rabbit, dog, cow, elephant, tenrec, and armadillo genomes, by using the MultiPipMaker synteny building tool, revealed that both the provirus and the syncytin-Opo1 orthologous gene are absent in all these species. Interestingly, it is also not found in the more closely related tammar wallaby and Tasmanian devil marsupial species using the same approach.
Fig. 6.
Characterization of the opossum (M. domestica) syncytin-Opo1 gene and its proviral integration site. (A) Structure of the syncytin-Opo1 proviral sequence and integration site. Repeated mobile elements as identified by the RepeatMasker web program are positioned. Of note, the provirus shows signs of degeneration, with a deletion within the 3′ LTR and degenerate gag-pol sequences, with the noticeable exception of the syncytin-opo1 gene. PCR primers used to identify the syncytin-Opo1 orthologous copy in other Monodelphis species are indicated (black half arrows). The spliced env subgenomic transcript as determined by RACE-PCR of opossum placental RNA is indicated (GenBank accession no. KM235357). Putative GCM1 binding sites are indicated as blue vertical lines. Sequence of the integration site of syncytin-Opo1, showing evidence for retroviral integration, is provided above the schematized provirus: characteristic sequences are shown, including LTRs (with TG . . . CA borders), PBS for a proline tRNA, and TSD (red boxes). (B) Absence of Syncytin-Opo1 in the genomes of representative species of mammalian lineages. The genomic locus of syncytin-Opo1 (with the env gene in red), along with the surrounding MICAL3 and BCL2L13 genes were recovered from the UCSC Genome Browser (genome.ucsc.edu/), together with the syntenic loci of marsupials (i.e., wallaby, and Tasmanian devil) and eutherian mammals genomes; exons (vertical lines) of the MICAL3 and BCL2L13 genes and the sense of transcription (arrows) are indicated. Homology of the syntenic loci was analyzed by using the MultiPipMaker alignment building tool. Homologous regions are shown as pale green boxes, and highly conserved regions (>100 bp without a gap displaying at least 70% identity) are shown as dark green boxes.
Insertion Date and Conservation of syncytin-Opo1 in Marsupial Evolution.
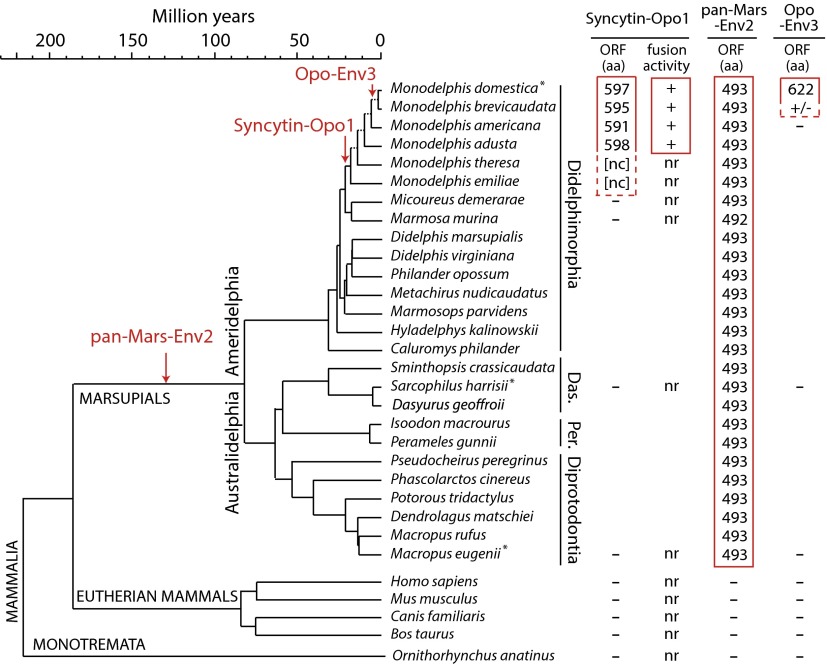
To further characterize syncytin-Opo1 and determine its date of genome insertion and status in evolution, we searched for the orthologous gene in representative species of the Monodelphis genus and in related branches of Didelphidae, as well as in other marsupial species (Fig. 7). Locus-specific pairs of PCR primers (forward primer upstream of syncytin-Opo1 and reverse primer downstream of the provirus in the 3′ flanking sequence; Table S2) were used to amplify genomic DNA from representative species. In Monodelphis species, PCR amplification showed the expected amplification product with a conserved size (except for the most divergent Monodelphis emiliae species, for which the 3′ primer was internal to the provirus), thus strongly suggesting the presence of the orthologous syncytin-Opo1 throughout the Monodelphis lineage. This finding was confirmed by sequencing the PCR products (sequences deposited in GenBank; accession nos. KM235324–29), which revealed the presence of a syncytin-Opo1 ortholog in the species tested (Fig. 7). Analysis of the gene sequences revealed that syncytin-Opo1 encodes a full-length ORF in the three species closely related to M. domestica (namely, in Monodelphis brevicaudata, Monodelphis americana, and Monodelphis adusta; 591–598 aa long). However, the gene has no coding capacity in the two distant Monodelphis theresa and M. emiliae species. Finally, PCRs using genomic DNA from other marsupial species (belonging to the Didelphimorphia order, or to more distant species of the Australidelphia superorder) were found to be negative. The absence of syncytin-Opo1 was confirmed by using PCR primers internal to the syncytin-Opo1 ORF, placed at positions where all previously sequenced genes showed a strictly identical nucleotide sequence. Although we cannot formally exclude that sequences may be too divergent to allow primer annealing and PCR amplification, and despite the fact that we could not amplify the “empty locus” in the Micoureus demerarae and Marmosa murina species, the data strongly suggest that syncytin-Opo1 has inserted into the identified locus after the divergence between Monodelphidae and the latter two species—i.e., ∼20 Mya.
Fig. 7.
Insertion date and conservation of syncytin-Opo1, pan-Mars-env2, and opo-env3 during marsupial evolution. (Left) Marsupial phylogenetic tree (data from refs. 1 and 37–40). Horizontal branch length is proportional to time (scale bar on top), with the exception of the Monodelphis lineage where no information is available to date. The names of the 26 marsupial species tested are indicated, together with the names of the Ameridelphia and Australidelphia corresponding orders (Das., Dasyuromorphia; Per., Peramelemorphia). Asterisks indicate species whose genome is available in genomic databases. (Right) The length (in amino acids) of the different env genes that could be identified is indicated. [nc], presence of a full-length noncoding syncytin-Opo1 gene; +/−, multiple homologous mixed sequences; −, no env homologous sequence identified, by either PCR-amplification or database search. The fusion activity for each syncytin-Opo1 cloned gene, as determined by the cell–cell fusion assay, is indicated. nr, not relevant. Arrows indicate the respective insertion dates of the different env genes.
Finally, as illustrated in Fig. S4, close examination of the full-length syncytin-Opo1 ORFs identified above demonstrates high similarities, ranging from 88.6% to 98.7% amino acid identity, and shows signs of purifying selection, with nonsynonymous to synonymous mutation ratios (dN/dS) between all pairs of species lower than unity (i.e., 0.40 < dN/dS < 0.61), as expected for a bona fide cellular gene. Of note, the phylogenetic tree generated from an alignment of these syncytin-Opo1 sequences is congruent with the Monodelphidae phylogenetic tree. To further assess the functional conservation of the syncytin-Opo1 gene, an ex vivo assay for its fusogenic activity, as already documented in Fig. 5 for M. domestica, was performed, with the PCR-amplified syncytin-Opo1 genes cloned into the same eukaryotic expression vector. The cell–cell fusion assay, as shown in Fig. 5 and Table S3, disclosed positive results for all of the species tested, demonstrating functional conservation of syncytin-Opo1.
Together, the data indicate that syncytin-Opo1 integrated into the genome of the ancestor of Monodelphidae and that it has been co-opted for a functional role in placentation in a subgroup of species not comprising the M. emiliae and M. theresa species. In this respect, it would be of interest to determine whether the latter two species lack a syncytial structure at the feto-maternal interface.
Search for More Ancestrally Captured Marsupial Env Genes: env2 Is Present and Conserved in the Entire Marsupial Lineage.
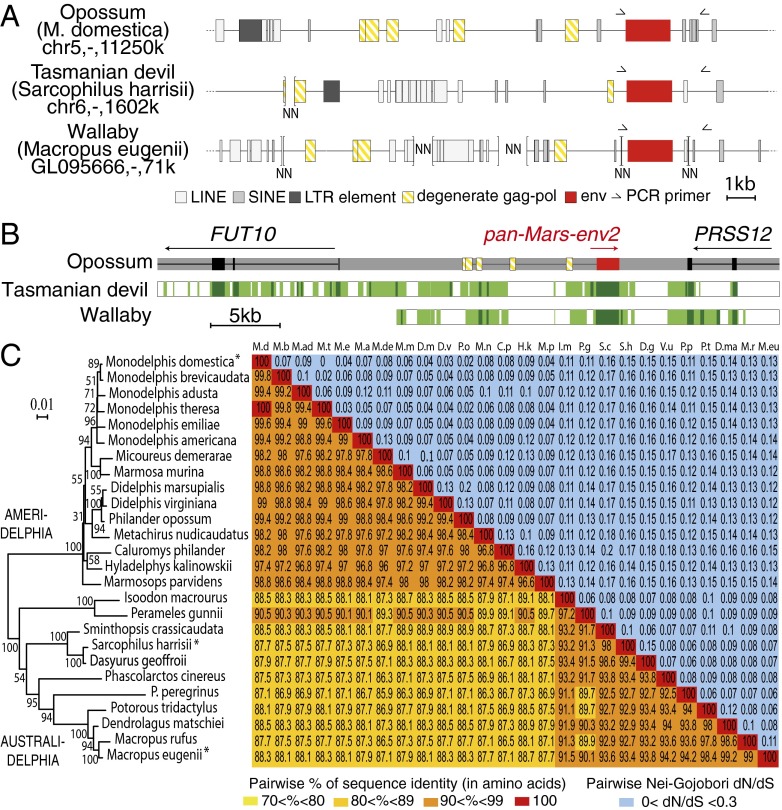
Although the insertion date of syncytin-Opo1 in the marsupial clade, as well as its topical expression, is consistent with the emergence of a syncytialized placental feto-maternal interface in Monodelphis species, the question of the existence of other captured env sequences which could predate syncytin-Opo1 entry can be assessed—to some extent—via the search of sequences common to all sequenced marsupial genomes, including that of the most distantly related tammar wallaby. Actually, a BLAST in silico search for sequences among the wallaby and Tasmanian devil genomes common with those identified in the opossum revealed that env2—and only env2—can be found in all three species, with a high level of sequence conservation (see below). In all three species, env2 is part of a highly degenerate proviral sequence, with no identifiable LTR and only few remnants of gag and pol genes (Fig. 8A). Analysis of the site of integration of env2 reveals that this gene is integrated at the same genomic orthologous locus, within an intergenic region between the FUT10 and PRSS12 cellular genes, in antisense orientation compared with the sense of transcription of these genes, as commonly observed for ERVs (Fig. 8B). This result indicates that the env2-containing provirus had integrated into the ancestor of all marsupial species—i.e., between 80 and 190 Mya. This finding was confirmed by PCR analysis of a large series of marsupial genomic DNAs, comprising Ameridelphia and Australidelphia species, using primers indicated in Fig. 8A. It yielded in all cases an orthologous gene that we further named pan-Mars-env2 (Fig. 7). Its sequencing revealed high conservation in all cases of the coding sequence (86–100% identity in amino acids; sequences deposited in GenBank, accession nos. KM235331–56) (Fig. 8C), with the presence of a premature stop codon before the transmembrane part of the TM subunit, at a position conserved for all genes, and 3′ to the preserved ISD sequence. It results in a truncated, most probably soluble and unambiguously nonfusogenic protein (see Fig. S1 for the opossum pan-Mars-Env2 protein), as previously described for the primate HERV-R Env (ERV3) protein (Discussion). However, because of the ancestry and the high sequence conservation of pan-Mars-env2, and despite the lack of a transmembrane domain for this Env protein, it was further analyzed for the selective pressure to which it might have been subjected in evolution. Interestingly, the phylogenetic tree generated from an alignment of these sequences (Fig. 8C) is congruent with the marsupial phylogenetic tree in Fig. 7, with the Ameridelphia and Australidelphia sequences branching into two distinct monophyletic groups. Closer examination only revealed minor differences within these groups, which actually correspond to nodes with low bootstrap values. As for syncytin-Opo1, conservation through evolution of the pan-Mars-env2 gene among marsupials was further analyzed by measuring the dN/dS ratio between all pairs of species. Accordingly, the entire env gene displayed very strong purifying selection, with dN/dS ratios all <0.2 (Fig. 8C). To further characterize the conservation of pan-Mars-env2, we performed a refined analysis of the sequences, using methods based on the rate of nonsynonymous vs. synonymous substitutions within the complete set of sequences and allowing differences in selection pressure between different domains of the proteins to be revealed (site-specific selection). Such an analysis, using the PAML package (23), provided support for a model (model M7) in which all of the codons are under purifying selection (dN/dS < 0.7) (Fig. S5). There was no significant support for a positive selection model (model M8 vs. M7: Chi2 = 2.8 × 10−4, df = 2, P > 0.99), suggesting that no sites are under positive selection. Analyses using the HyPhy package (24) with slightly different site-specific models [fixed effect likelihood (FEL) and random effect likelihood (REL)] led to similar conclusions (Fig. S5). This finding strongly suggests that pan-Mars-env2 is a genuine cellular gene most probably endowed with a physiological function. Taking into account that we could not demonstrate—by qRT-PCR on a series of opossum tissues and ISH on placental sections—that the gene is expressed at a significant level (similar negative results were obtained with tammar wallaby placental samples), it remains difficult to assess its possible role. Because its ISD is highly conserved in evolution, one could hypothesize that this gene is expressed at preimplantation stages and is involved in maternal tolerance during the early stages of pregnancy, but this hypothesis remains to be demonstrated via the—difficult to perform—recovery of embryos at the corresponding stages.
Fig. 8.
Characterization of the marsupial pan-Mars-env2 gene and of its proviral integration site. (A) Structure of the opossum (M. domestica), Tasmanian devil (S. harrisii), and wallaby (M. eugenii) env2 proviral sequences and integration sites. Repeated mobile elements as identified by the RepeatMasker Web program are positioned. Of note, the proviruses show signs of degeneration, with no LTR identifiable and highly degenerate gag-pol sequences. PCR primers used to identify the env2 orthologous copies in marsupial species are indicated (black half arrows). (B) Demonstration of the orthology of env2 (dubbed pan-Mars-env2) in the genomes of sequenced marsupial species. The genomic locus of the opossum pan-Mars-env2 (in red), along with the surrounding FUT10 and PRSS12 genes, was recovered from the UCSC Genome Browser (genome.ucsc.edu/), together with the syntenic loci of the wallaby and the Tasmanian devil genomes. The genome of wallaby is incompletely assembled, and the FUT10 and PRSS12 genes are located on two distinct unassembled scaffolds, with pan-Mars-env2 being located on the scaffold containing the PRSS12 gene. The FUT10 and PRSS12 genes were not located on the same chromosome in other eutherian mammalian species, preventing syntenic comparisons. Same color code and symbols are used as in Fig. 6. (C) Sequence conservation and evidence for purifying selection pan-Mars-Env2 in marsupials. (C, Left) pan-Mars-Env2 phylogenetic tree determined using amino acid alignment of the encoded proteins identified in Fig. 7, by the maximum-likelihood method. The horizontal branch length and scale indicate the percentage of amino acid substitutions. Percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. (C, Right) Double-entry table for the pairwise percentage of amino acid sequence identity between the pan-Mars-env2 gene among the indicated species (lower triangle), and the pairwise Nei–Gojobori nonsynonymous to synonymous mutation rate ratio (dN/dS; upper triangle). A color code is provided for both series of values.
Opo-env3 Is Part of a Recently Acquired ERV with Evidence for Polymorphic Sites of Insertion in the Opossum.
Opo-env3 is present at a high copy number in the opossum genome, with >85 copies, among which 11 copies have full-length ORFs. Some of them are part of proviruses containing still identifiable retroviral gag, pro, pol, and env genes, together with LTR sequences. A consensus sequence (Fig. S6A) indicates that this ERV (dubbed Opo-Env3-ERV) has a mosaic structure, with a betaretrovirus pol gene, whereas its env gene is that of a gammaretrovirus, as was similarly observed for the infectious Mason–Pfizer monkey virus primate retrovirus (Fig. S6B). Although only one Opo-Env3–encoding copy is expressed in the uterine tissue, with no polymorphisms detectable in the amplified transcripts, this placental transcript, rather unexpectedly, does not match exactly with any of the genomic coding sequences present in the opossum genomic database. The best matching copy, located on Chr2, only display 99.6% of identity with the placental transcript. Because the mismatch between the placental transcript of the opossum individual from which RNA was extracted and the genomic copy of the opossum database could be due to specific polymorphisms between our subject animal and the one used as a reference for the genomic sequencing (as an alternative to incomplete genome coverage), we tentatively amplified the corresponding opo-env3 copy on Chr2 in the genomic DNA extracted from our individual (using a forward primer located within the provirus at the 5′ end of the env gene and a reverse primer located downstream of the provirus in the 3′ flanking sequence). However, no amplification product could be obtained by using this combination of primers, suggesting the absence of the proviral sequence at this position in our subject individual. This absence was confirmed by using another forward primer located in the 5′ flanking sequence of the putatively inserted provirus. A PCR assay using this combination of primers resulted in a single amplification product with the expected size for an empty “provirus-free” locus, and sequencing confirmed the absence of any proviral sequence at this locus, thus demonstrating an insertional polymorphism between the opossum individual used here and the animal used as a reference for the genomic sequence. Such an insertional polymorphism suggests (i) that some opo-env3–containing proviruses may still be replicative and infectious in the opossum, and (ii) that the endogenization process might be recent. Actually, a qRT-PCR analysis of the opo-env3 associated gag gene expression displayed specific expression within the placenta and uterine tissues, as observed for opo-env3 (Fig. S6), whereas a RACE-PCR analysis of the corresponding transcripts disclosed the presence of protein-encoding gag-pro and env transcripts (GenBank accession nos. KM235358–59), consistent with the observation in Fig. S3 of mature viral particles within the uterine gland tissues (although we could not formally demonstrate that they are actually encoded by the identified ERV). Finally, consistent with a recent endogenization, search for opo-env3 copies within marsupial species, using pairs of PCR primers internal to the provirus env gene, demonstrated the presence of env3 genes only within the two most closely related M. domestica and M. brevicaudata species (Fig. 7).
Discussion
Here we have identified syncytin-Opo1, the env gene from an ERV that has integrated into the genome of an opossum ancestor, ∼20 Mya, and has been maintained as a functional env gene in a definite group of Monodelphis species. This gene displays all of the canonical characteristics of a syncytin gene: (i) it exhibits fusogenic activity, in an ex vivo cell–cell fusion assay; (ii) it has been subject to purifying selection in the course of evolution, displaying low rates of nonsynonymous to synonymous substitutions and conservation of its fusogenic property; and (iii) it is specifically expressed in the placenta, as determined by both RT-PCR analyses and ISH of placental tissue sections. ISH experiments using syncytin-Opo1 sequences as a probe clearly show that expression takes place at the level of the syncytial layer of the feto-maternal interface, consistent with a direct role of this fusogenic syncytin gene in syncytiotrophoblast formation. Clearly, syncytin-Opo1 adds to the syncytin genes previously identified in the eutherian mammals. In the case of the murine syncytin-A and -B genes, knockout mice unambiguously demonstrated that they are absolutely required for placentation, with evidence for a defect in syncytiotrophoblast formation, resulting in decreased feto-maternal exchange and impaired embryo survival (15, 16). It can, therefore, be proposed that all of the identified syncytins, including the newly discovered syncytin-Opo1, are likely to play a similar role in placentation by being involved in syncytiotrophoblast formation.
An important outcome of the present investigation is that the discovery of syncytin-Opo1 extends the presence of syncytins outside the eutherian mammals, to now include the marsupials, at least the American lineage. These species diverged from eutherian mammals ∼190 Mya and display a very specific mode of reproduction, with an only short-lived placenta, with the fetus being rapidly released in most cases in an external pouch, where it will develop via lactation. In the presently analyzed species, implantation lasts only from day 12 to 14, still with evidence for syncytium formation (22), and syncytin-Opo1 was precisely found to be expressed at these stages and at the location expected for a role in syncytium formation. Altogether the present data—combined with the fact that syncytin-Opo1 is distinct from all previously identified syncytins—indicate that syncytin capture has been a widespread process, which finally turns out to have taken place in several widely separate lineages during the mammalian radiation. It could thus be hypothesized that the remarkable variability in placental structures observed among mammals might in part result from the diversity of the syncytin genes that have been stochastically captured in the course of mammalian evolution (5).
However, an important question remains to be answered, if one takes into account the relatively recent occurrence of syncytin-Opo1 capture, in comparison with the time of emergence of placental mammals (including marsupials) from egg-laying animals, ∼200 Mya. We had previously proposed (reviewed in refs. 4 and 5) that this evolutionary transition was most probably favored by the primitive capture of an ancestral “founding” retroviral env gene that allowed the retention of the growing embryo within the mother, despite her immune system, most probably thanks to the immunosuppressive activity of retroviral Env proteins, allowing establishment of a primitive feto-maternal tolerance. In this scenario, it was further proposed that subsequent env gene exaptation could naturally take place, because ERV capture is an ongoing process, thus resulting in “new” syncytins being retained due to an increased benefit to the host compared with the primitive one, that would then simply be replaced and vanish. An interesting outcome of the present investigation is that this hypothetical scenario can be substantiated by the discovery of pan-Mars-env2. Indeed, as illustrated here, we found that pan-Mars-env2 capture predates the divergence of the American and Australasian marsupials, because we could demonstrate that this gene of retroviral origin is present in all of the marsupials in which we searched for it. Furthermore, this gene is (i) located at the same orthologous locus, (ii) discloses high sequence conservation, and (iii) is subjected to purifying selection, with a very low rate of nonsynonymous-to-synonymous substitutions, as expected for a bona fide gene and as observed—although on a more restricted time scale—for syncytin-Opo1. However, this env gene has an uncommon structural characteristic, because it has a conserved stop codon just upstream of the transmembrane domain of the TM subunit, thus resulting in a soluble nontransmembrane protein. Although rare, this structural feature is not unique. A primate ancestrally captured retroviral env, namely ERV3, has a closely related structure, being similarly truncated, and this truncation is also “ancient,” because it is found in all primates at the same position (25). Although this ERV3 Env is highly expressed in the placenta (26–28), and has further been shown to be immunosuppressive (12), it is not found in the gorilla (25) and a natural knockout of the gene (via an additional more premature stop codon) in the homozygous state has been identified in 1% of the human population (29), thus excluding any pivotal role in placentation, at least in humans. We had previously proposed that this gene was most probably a “decaying” syncytin, now replaced by syncytin-1 and -2 in higher primates (30). Along these lines, it can be hypothesized that pan-Mars-env2 in marsupials plays a role close to that played by ERV3 in primates before it was functionally replaced by bona fide syncytins and that both the pan-Mars-Env2 and ERV3 proteins are—or have been—involved in the establishment of feto-maternal tolerance, via their common immunosuppressive function. Such a function would account for the high sequence conservation of pan-Mars-Env2—including the ISD—together with the aforementioned strong purifying selection applying to it. The pan-Mars-Env2 truncation—that de facto renders it nonfusogenic—is consistent with the fact that the placenta of the tammar wallaby displays no evidence of syncytium formation (31) and that occurrence of a syncytium in the opossum is linked to the more recent exaptation of the fusogenic syncytin-Opo1. Accordingly, it turns out that the diversity of syncytin captures is likely to be responsible for the diversity observed at the level of the refined structure of the corresponding placentae, as already strongly suggested in the case of eutherian mammals (5).
Finally, a third interesting outcome of the present investigation is the discovery within the opossum of viral particles that are produced in the placenta and, more precisely, within the maternal uterine glands. Occurrence of placental viral particles has long been described in a series of mammalian species—including humans, mice, and ruminants (e.g., refs 32 and 33)—and can actually be minimally attributed to the presence of an active retroviral gag gene, which is sufficient to produce such virus-like particles. In most cases, these particles are defective for replication (no active pol gene, no functional env gene) but this is most probably not the case for the presently identified ERV. Indeed, the present in silico search identified a full-length protein-coding env gene (env3) that seems to have entered the opossum genome quite recently because it is only found within the closely related M. domestica and M. brevicaudata species. In addition, we found a polymorphism in the locus of insertion of one of the proviral copies, suggesting that the corresponding ERVs are relatively recent and might still be active. Consistently, in silico analyses disclose env3-associated ERV copies with full-length retroviral gag-pro-pol coding sequences, and at least Gag and Pro encoding transcripts proved to be expressed in the placenta. These findings could account for the presence and morphology of the viral particles—yet not unambiguously identified—detected in this tissue, with evidence for “core” maturation and condensation as classically observed for functional retroviruses. Interestingly, a similar situation has been described in the sheep for the endogenous Jaagsiekte retrovirus (enJSRV) ERVs, which are also very abundantly expressed in the uterine glands and genital tract and which can be transmitted to the conceptus and are thus still active retroviral elements, with evidence for polymorphism in their genomic position among species (33–35). In any case, the presence of retroviral particles within the placenta is likely to be due to recently captured ERVs, which might in turn supply new syncytin or placental genes to come.
Conclusively, the present investigation would recapitulate the complete lifestyle of captured retroviruses, with as a starting point the endogenization process per se, resulting in possibly still active proviruses in the case of the env3-associated elements, followed by env gene exaptation in the case of env1 and env2, with the former being a bona fide syncytin responsible for the emergence of a syncytialized organization of the opossum placenta and the latter being possibly involved in the emergence of the marsupial ancestral placenta >80 Mya.
Methods
Database Screening and Sequence Analyses.
Retroviral endogenous env gene sequences were searched for by BLAST on the opossum genome (6.8× coverage assembly of the M. domestica genome; Broad monDom5; October 2006): Sequences containing an ORF >450 aa (from start to stop codons) were extracted from the monDom5 genomic database by using the getorf program of the EMBOSS package (emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) and translated into amino acid sequences. These sequences were BLASTed against the TM subunit amino acid sequences of 35 retroviral Env glycoproteins (from representative ERVs—among which known syncytins—and infectious retroviruses) by using the BLASTP program of the NCBI (www.ncbi.nlm.nih.gov/BLAST). Putative Env protein sequences were then selected based on the presence of a hydrophobic domain (transmembrane domain) located 3′ to a highly conserved C-X5,6,7-C motif. The identified Env-encoding sequence coordinates are listed in Table S1.
The opossum genome was secondarily screened with the identified Env glycoprotein sequences by using the BLAST programs from the NCBI (www.ncbi.nlm.nih.gov/BLAST). Multiple alignments of amino acid sequences were carried out by using the Seaview program under ClustalW protocol. Maximum-likelihood phylogenetic trees were constructed with RaxML (Version 7.3.2; ref. 36), with bootstrap percentages computed after 1,000 replicates by using the GAMMA + GTR model for the rapid bootstrapping algorithm. PAML4 (23) was used to run site-specific selection tests and obtain dN/dS ratios for all syncytin-Opo1 sequences. PAML models analyzed assumed no molecular clock (clock = 0) and a single dN/dS for all tree branches (model = 0), and we used likelihood-ratio tests to compare the improvement in likelihood for a model (M8) allowing for positive selection compared with a model (M7) (NS site = 7–8) that does not. Each analysis ran until convergence (Small_Diff = 0.5e-6), and the control file is available upon request. HyPhy (24) was used on the datamonkey web server (www.datamonkey.org) to run the site-specific FEL and REL models.
The tammar wallaby (Macropus eugenii; UCSC/Baylor Institute macEug2; September 2009; 2× coverage), the Tasmanian devil (Sarcophilus harrisii; UCSC/NCBI; sarHar1; February 2011), the human (Homo sapiens; UCSC GRCh37/hg19; 2009), the mouse (Mus musculus; UCSC GRCm38; 2011), the rabbit (Oryctolagus cuniculus; UCSC/Broad oryCun2.0; 2009; 7.48× coverage), the dog (Canis lupus familiaris; UCSC/Broad CanFam3.1; 2011), the cow (Bos taurus; UCSC/Baylor bosTau7; 2011), the elephant (Loxodonta africana; UCSC/Broad loxAfr3; 2009; 7× coverage), the tenrec (Echinops telfairi; UCSC/Broad echTel2; 2012), and the armadillo (Dasypus novemcinctus; UCSC/Baylor dasNov3; 2011) were also screened for the presence of the identified syncytin-Opo1–containing provirus sequence, using syntenic genomic regions from the UCSC genome browser (genome.ucsc.edu/). Analyses of the syntenic conserved sequences in each genome were performed by using the MultiPipMaker alignment tool (pipmaker.bx.psu.edu) with the opossum genome sequence as a reference.
Search for syncytin-Opo1, pan-Mars-env2, and Opo-env3 in Other Species.
PCRs were performed on 100 ng of genomic DNA, using Accuprime Taq DNA Polymerase (Invitrogen). A highly sensitive touchdown PCR protocol was performed (elongation time at 68 °C, with 30-s hybridization at temperatures ranging from 60 °C to 50 °C, −1 °C per cycle for 10 cycles, followed by 40 cycles at 55 °C). Genomic DNAs were tentatively amplified as indicated in Results with primers either external to the provirus (locus primers; Table S2), close to the start and stop codons of the syncytin ORF (ORF primers; Table S2), or internal to the ORF and conserved among all sequenced genes (internal primers; Table S2). PCR products were directly sequenced without cloning to avoid low-level mutations introduced by PCR.
Materials, Real-Time RT-PCR, In Situ Hybridization, Electron Microscopy, and Syncytin-Opo1 Oxpression Vector and Fusion Assay.
See SI Methods. Maintenance of and experiments on animals were performed under registration of the responsible State Office of Health and Social Affairs Berlin (LaGeSo), Germany.
Supplementary Material
Acknowledgments
We thank J. Zeller, Humboldt University, for technical assistance; A. Billepp and P. Grimm, Humboldt University, for assistance in care and breeding of M. domestica; A. Janke, LOEWE Biodiversity and Climate Research Centre, for providing some marsupial genomic DNAs; C. Conroy, Museum of Vertebrate Zoology, for the gift of several Monodelphis tissues; A. Leskowicz and V. Marquis, UMR5503, for the gift of OK cells; Dr. R. Potier, ZooParc de Beauval, for blood samples; O. Bawa, Institut Gustave Roussy, for contribution to the histological analyses; and C. Lavialle, Institut Gustave Roussy, for discussion and critical reading of the manuscript. This work was supported by the CNRS and by grants from the Ligue Nationale Contre Le Cancer (Equipe Labellisée) (to T.H.) and Agence Nationale de la Recherche (Retro-Placenta) (to T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KM235324–KM235359).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417000112/-/DCSupplemental.
References
- 1.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 2.Luo Z-X, Yuan C-X, Meng Q-J, Ji Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476(7361):442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 3.Freyer C, Zeller U, Renfree MB. The marsupial placenta: A phylogenetic analysis. J Exp Zoolog A Comp Exp Biol. 2003;299(1):59–77. doi: 10.1002/jez.a.10291. [DOI] [PubMed] [Google Scholar]
- 4.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta. 2012;33(9):663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Lavialle C, et al. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 2013;368(1626):20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74(7):3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 8.Mallet F, et al. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA. 2004;101(6):1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cáceres M, Thomas JW. NISC Comparative Sequencing Program The gene of retroviral origin Syncytin 1 is specific to hominoids and is inactive in Old World monkeys. J Hered. 2006;97(2):100–106. doi: 10.1093/jhered/esj011. [DOI] [PubMed] [Google Scholar]
- 10.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100(22):13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaise S, Ruggieri A, Dewannieux M, Cosset F-L, Heidmann T. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J Virol. 2004;78(2):1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangeney M, et al. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104(51):20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102(3):725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernochet C, et al. The captured retroviral envelope Syncytin-A and Syncytin-B genes are conserved in the Spalacidae together with hemotrichorial placentation. Biol Reprod. 2014 doi: 10.1095/biolreprod.114.124818. [DOI] [PubMed] [Google Scholar]
- 15.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106(29):12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupressoir A, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA. 2011;108(46):E1164–E1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis G, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci USA. 2012;109(7):E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornelis G, et al. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc Natl Acad Sci USA. 2013;110(9):E828–E837. doi: 10.1073/pnas.1215787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornelis G, et al. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc Natl Acad Sci USA. 2014;111(41):E4332–E4341. doi: 10.1073/pnas.1412268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunlap KA, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103(39):14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeller U, Freyer C. Early ontogeny and placentation of the grey short ‐ tailed opossum, Monodelphis domestica (Didelphidae: Marsupialia): Contribution to the reconstruction of the marsupial morphotype. J Zoological Syst Evol Res. 2001;39(3):137–158. [Google Scholar]
- 23.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 24.Kosakovsky Pond SL, Frost SDW. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 25.Hervé CA, Forrest G, Löwer R, Griffiths DJ, Venables PJW. Conservation and loss of the ERV3 open reading frame in primates. Genomics. 2004;83(5):940–943. doi: 10.1016/j.ygeno.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Kato N, et al. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: Two of the three ERV3 mRNAs contain human cellular sequences. J Virol. 1987;61(7):2182–2191. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd MT, Bax CM, Bax BE, Bloxam DL, Weiss RA. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology. 1993;196(2):905–909. doi: 10.1006/viro.1993.1556. [DOI] [PubMed] [Google Scholar]
- 28.Venables PJ, Brookes SM, Griffiths D, Weiss RA, Boyd MT. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology. 1995;211(2):589–592. doi: 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- 29.de Parseval N, Heidmann T. Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J Virol. 1998;72(4):3442–3445. doi: 10.1128/jvi.72.4.3442-3445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esnault C, Cornelis G, Heidmann O, Heidmann T. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 2013;9(3):e1003400. doi: 10.1371/journal.pgen.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freyer C, Zeller U, Renfree MB. Ultrastructure of the placenta of the tammar wallaby, Macropus eugenii: Comparison with the grey short-tailed opossum, Monodelphis domestica. J Anat. 2002;201(2):101–119. doi: 10.1046/j.1469-7580.2002.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris JR. Placental endogenous retrovirus (ERV): Structural, functional, and evolutionary significance. BioEssays. 1998;20(4):307–316. doi: 10.1002/(SICI)1521-1878(199804)20:4<307::AID-BIES7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Black SG, et al. Viral particles of endogenous betaretroviruses are released in the sheep uterus and infect the conceptus trophectoderm in a transspecies embryo transfer model. J Virol. 2010;84(18):9078–9085. doi: 10.1128/JVI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chessa B, et al. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324(5926):532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmarini M, et al. Expression of endogenous betaretroviruses in the ovine uterus: Effects of neonatal age, estrous cycle, pregnancy, and progesterone. J Virol. 2001;75(23):11319–11327. doi: 10.1128/JVI.75.23.11319-11327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 37.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446(7135):507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson MA, et al. Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 2010;8(7):e1000436. doi: 10.1371/journal.pbio.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de A Carvalho B, et al. Phylogenetic relationships and phylogeographic patterns in Monodelphis (Didelphimorphia: Didelphidae) J Mammal. 2011;92(1):121–133. [Google Scholar]
- 40.Solari S. A molecular perspective on the diversification of short-tailed opossums. Mastozoología Neotropical. 2010;17(2):317–333. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.