Educated by his deep appreciation of nature, Darwin observed that “from so simple a beginning endless forms most beautiful” have arisen throughout the evolutionary history of life on earth (1). The spectacular diversity of orchids (2) and beetles (3) has long fascinated naturalists and casual observers alike. More recently, the adaptive radiations of Hawaiian drosophilids (4), Caribbean Anolis lizards (5), and African cichlid fishes (6) have become prime examples for understanding the mechanisms that enable diversification. Gene duplication and deletion are generally considered important evolutionary mechanisms that give rise to phenotypic diversity (7). Following gene duplication and loss, adaptation and speciation appear to proceed through a combination of both structural and cis-regulatory changes in one or more paralogous genes (8). Recent advances in sequencing technology have enabled researchers to make significant progress in understanding the molecular evolution that has facilitated diversification. In PNAS, Cortesi et al. (9) examine the evolution of vertebrate opsin genes as a spectacular example of how gene duplication and deletion events that affect spectral sensitivity have driven adaptation to diverse light environments and visual displays.
Gene families comprise several to many genes of similar nucleotide or amino acid sequences; they share similar cellular functions and commonly arise as a result of gene or genome duplication events. The expansion or contraction of gene families over evolutionary time in different lineages can be random or the result of natural selection, although demonstrating the latter can be difficult (10). Several mechanisms, such as tandem duplications, segmental duplications, or even whole-genome duplications can lead to the expansion of gene families. Importantly, during the evolution of chordates, the ancestral deuterostome genome (likely in a cephalochordate ancestor) experienced two rounds of whole-genome duplication followed by a genome duplication in actinopterygian (ray-finned) fishes, but not in the sarcopterygian (lobe-finned) fishes, the lineage that includes land vertebrates (11). Even though most duplicated genes were secondarily lost, many evolved new functions, in support of the notion that gene and genome duplications might provide a major mechanism for generating phenotypic diversity in evolution (7).
Opsin Gene Family Expansion and Light Sensitivity
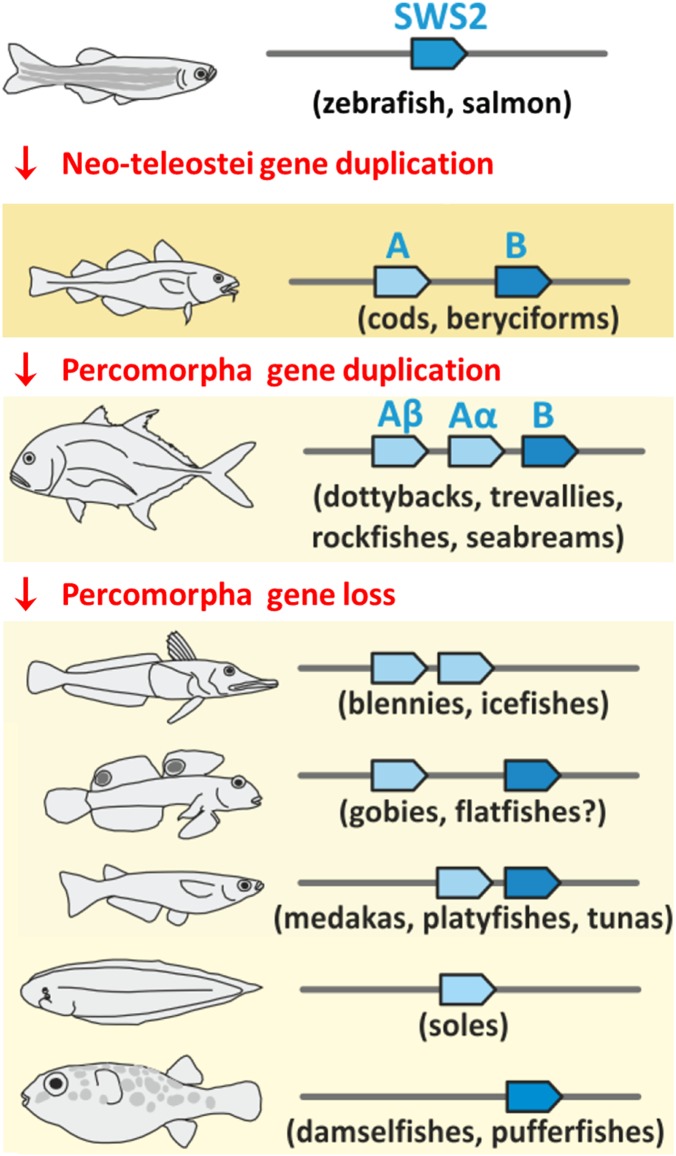
Opsin genes expressed in photoreceptor cells are fundamental to animal vision and are a major force underlying the evolutionary adaptation to variable photic environments (12). The diversity of these genes is achieved by gene duplication followed by changes in amino acid sequence at key tuning sites. Opsins have been crucial to the successful colonization of diverse habitats, especially in teleost fishes, the most species-rich lineage of vertebrates. In a tour de force comparative genomics analysis, Cortesi et al. (9) describe a newly discovered violet/blue short wavelength-sensitive 2 (SWS2) opsin, which arose alongside the radiation of the highly diverse percomorph fishes (which include cichlid fishes, wrasses, and other diverse and colorful families). Specifically, the authors compared almost 100 fish genomes to examine the complex evolutionary history of SWS2, including numerous duplication, deletion, and pseudogenization events, and possibly even the “resurrection” of functional genes from pseudogenes (Fig. 1). Several amino acid substitutions are described that likely facilitated the adaptive differentiation between SWS2 gene copies, probably by conferring sensitivities to different wavelengths of light or by being differentially expressed as organisms move through their ontogenetic and life history stages. The study by Cortesi et al. (9) illustrates the complexity that results from gene duplication and loss, and which in turn enables the evolution of animal diversity.
Fig. 1.
Abbreviated history of violet-blue–sensitive (SWS2) genes in teleost fishes. The SWS2 gene was present in a single copy before the neo-teleostei gene duplication, which gave rise to SWS2A (light blue) and SWS2B paralogs (dark blue). In the percomorpha lineage, a subsequent duplication event gave rise to SWS2Aα and SWS2β paralogs. Although these three paralogs have been retained in many species, one or more paralogs have been lost in percomorph fishes. The syntenic relationship is shown to illustrate that these were tandem duplications occurring on the same chromosome, with example species listed below in parentheses. Modified from ref. 9.
Gene Family Evolution and Phenotypic Diversification
The evolution of opsin genes has received ample scrutiny, yet several other gene families deserve mention because of their likely role in phenotypic diversification. One of the most spectacular examples comprises the large and diverse family of olfactory receptor genes, which vary widely in number across vertebrate genomes (13). For example, although rodents have approximately 1,000 olfactory receptor genes, humans have only around 400 (a reduction likely caused by massive loss and pseudogenization of olfactory receptor genes in the human lineage). Compared with mammals, the olfactory receptor gene family is considerably more diverse in fishes (eight subfamilies are present), yet the total number of olfactory receptor genes is much smaller, suggesting that the mammalian olfactory receptor gene family is much less complex compared with that in the ancestor of vertebrates. Even though it is often assumed that gene families evolve adaptively, signatures of selection are often difficult to demonstrate. In this regard olfactory receptor genes also serve as a warning against adaptive scenarios because the number and types of olfactory receptor genes apparently have evolved only in part in response to environmental needs (13).
Voltage-gated sodium channels provide another compelling example, as they form the basis for electrical excitability in animals (14). These sodium channels evolved from calcium channels and were present in the last common ancestor of choanoflagellates and metazoa (animals), thus they already existed when neurons first evolved. A motif that evolved early in chordate evolution allows voltage-gated sodium channels to cluster where action potentials are generated to greatly enhance conduction velocity. After the late Devonian extinction, when teleosts and tetrapods each diversified in their respective habitats and the complexity of their brains increased concomitantly, the voltage-gated sodium channel gene family expanded in parallel in tetrapods and teleosts, possibly allowing more complex neural computations along with energy savings. In addition, these channels have been selected to encode diverse communication signals in weakly electric fish (15) and to protect against lethal sodium channel toxins (e.g., in snakes, newts, pufferfish, insects), providing unprecedented opportunities for drug design and therapeutic applications (16).
Gene families involved in cell-to-cell signaling appear to expand less in the course of evolution, possibly because the genes that encode the receptors and ligands need to evolve in a coordinated manner. Examples include steroid hormones, which classically bind to receptors that belong to the nuclear receptor family of transcription factors. These genes have coevolved with those that encode the enzymes that synthesize steroids at key transitions in the evolution of vertebrates and during gene family expansion (17), likely contributing to the diversification of vertebrates through their fundamental roles in reproduction, development, homeostasis, and stress response.
Integration of Functional and Evolutionary Genomics
The study by Cortesi et al. (9) does not describe any analyses of regulatory sequence evolution, nor do the authors investigate tissue- or temporally specific gene-expression patterns that may have arisen following regulatory changes. Coyne and Hoekstra (8) argue that adaptation and speciation proceed through a combination of both structural and cis-regulatory changes in one or more paralogous genes. Regrettably, much of our knowledge regarding the influence of structural and regulatory contribution to phenotypic diversity comes from studies examining these two mechanisms in isolation rather than through concurrent examination in the same system. However, a recent study by Harris et al. (18) combined structural, functional, and regulatory analyses to examine the evolution of the pro-opiomelanocortin gene family. By integrating temporal and spatial expression measurements with sequence variation and regulatory interactions, these authors were able to shed new light on the mechanisms of phenotypic diversification. It would be interesting to see future studies on the opsin gene family that incorporate both an analysis of gene expression across time or cell type with a bioinformatic analysis of regulatory sequence evolution.
The Power of the Comparative Approach
All these studies underscore the power of the comparative approach for understanding gene family evolution and, ultimately, the origins of animal diversity. Aristotle (in his book Peri Zoon Morion) already championed the promise of comparing different species for achieving a deep understanding about nature. If conducted within a phylogenetic framework, comparative analyses can provide inference similar to that obtainable with experimental approaches (19). Furthermore, a deeper understanding of the detailed relationship between orthologous and paralogous genes is crucial if we want to fully capitalize on the wealth of data generated with comparative omics approaches. In an era where biologists use fewer and fewer model systems, to the detriment of the entire biomedical research enterprise (20), the Cortesi et al. (9) paper provides a timely reminder for this notion, as it convincingly demonstrates a likely role for opsin gene evolution in the ability of animals to conquer new niches and acquire new modes of communication.
Acknowledgments
Research in our laboratory is supported by National Science Foundation Grants IOS-0843712 and IOS-1354942 (to H.A.H.) and DBI-0939454 for the BEACON Center for the Study of Evolution in Action.
Footnotes
The authors declare no conflict of interest.
See companion article on page 1493.
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection. J Murray; London: 1859. [Google Scholar]
- 2.Dressler RL. Phylogeny and Classification of the Orchid Family. Timber Press; Portland, OR: 1993. [Google Scholar]
- 3.Farrell BD. “Inordinate Fondness” explained: Why are there so many beetles? Science. 1998;281(5376):555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- 4.Kambysellis MP, Craddock EM. Ecological and reproductive shifts in the diversification of the endemic Hawaiian Drosophila. In: Givnish TJ, Sytsma KJ, editors. Molecular Evolution and Adaptive Radiation. Cambridge Univ Press; Cambridge, UK: 1997. pp. 475–509. [Google Scholar]
- 5.Losos JB. Evolution: A lizard’s tale. Sci Am. 2001;284(3):64–69. doi: 10.1038/scientificamerican0301-64. [DOI] [PubMed] [Google Scholar]
- 6.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513(7518):375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno S. Evolution by Gene Duplication. Springer; New York: 1970. [Google Scholar]
- 8.Coyne JA, Hoekstra HE. Evolution of protein expression: New genes for a new diet. Curr Biol. 2007;17(23):R1014–R1016. doi: 10.1016/j.cub.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Cortesi F, et al. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc Natl Acad Sci USA. 2015;112:1493–1498. doi: 10.1073/pnas.1417803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartl DL, Clark AG. Principles of Population Genetics. 4th Ed Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- 11.Meyer A, Van de Peer Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27(9):937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 12.Carleton K. Cichlid fish visual systems: Mechanisms of spectral tuning. Integr Zool. 2009;4(1):75–86. doi: 10.1111/j.1749-4877.2008.00137.x. [DOI] [PubMed] [Google Scholar]
- 13.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51(6):505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zakon HH. Adaptive evolution of voltage-gated sodium channels: The first 800 million years. Proc Natl Acad Sci USA. 2012;109(Suppl 1):10619–10625. doi: 10.1073/pnas.1201884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zakon HH, Lu Y, Zwickl DJ, Hillis DM. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: Convergent molecular evolution. Proc Natl Acad Sci USA. 2006;103(10):3675–3680. doi: 10.1073/pnas.0600160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Billen B, Bosmans F, Tytgat J. Animal peptides targeting voltage-activated sodium channels. Curr Pharm Des. 2008;14(24):2492–2502. doi: 10.2174/138161208785777423. [DOI] [PubMed] [Google Scholar]
- 17.Baker ME, Nelson DR, Studer RA. Origin of the response to adrenal and sex steroids: Roles of promiscuity and co-evolution of enzymes and steroid receptors. J Steroid Biochem Mol Biol. 2014 doi: 10.1016/j.jsbmb.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Harris RM, Dijkstra PD, Hofmann HA. Complex structural and regulatory evolution of the pro-opiomelanocortin gene family. Gen Comp Endocrinol. 2014;195:107–115. doi: 10.1016/j.ygcen.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Maynard-Smith J, Holliday R, editors. Evolution of Adaptation by Natural Selection. The Royal Society; London: 1979. Preface to Evolution of Adaptation by Natural Selection. [Google Scholar]
- 20.Bolker J. Model organisms: There’s more to life than rats and flies. Nature. 2012;491(7422):31–33. doi: 10.1038/491031a. [DOI] [PubMed] [Google Scholar]