Significance
Heterotrimeric G proteins are activated by exchange of a bound GDP for GTP at the active site of the alpha subunit (Gα), typically mediated by a transmembrane G protein-coupled receptor (GPCR). Resistance to inhibitors of cholinesterase 8A (Ric-8A), a soluble intracellular protein that is essential for embryonic development, can also catalyze nucleotide exchange and activate G proteins. Here, in a mechanistic study of a nonreceptor exchange factor, site-directed spin labeling and dipolar EPR spectroscopy are used to show that Ric-8A triggers the separation of the Ras-like and helical domains, also observed in GPCR-G protein complexes. This separation and induction of structural plasticity around the nucleotide-binding cavity of Gα promote nucleotide release. Ric-8A and GPCRs, although structurally unrelated, exhibit distinct but related mechanisms of action.
Keywords: G protein, guanine nucleotide exchange factor, double electron electron resonance spectroscopy, tertiary structure, protein dynamics
Abstract
Heterotrimeric G proteins are activated by exchange of GDP for GTP at the G protein alpha subunit (Gα), most notably by G protein-coupled transmembrane receptors. Ric-8A is a soluble cytoplasmic protein essential for embryonic development that acts as both a guanine nucleotide exchange factor (GEF) and a chaperone for Gα subunits of the i, q, and 12/13 classes. Previous studies demonstrated that Ric-8A stabilizes a dynamically disordered state of nucleotide-free Gα as the catalytic intermediate for nucleotide exchange, but no information was obtained on the structures involved or the magnitude of the structural fluctuations. In the present study, site-directed spin labeling (SDSL) together with double electron-electron resonance (DEER) spectroscopy is used to provide global distance constraints that identify discrete members of a conformational ensemble in the Gαi1:Ric-8A complex and the magnitude of structural differences between them. In the complex, the helical and Ras-like nucleotide-binding domains of Gαi1 pivot apart to occupy multiple resolved states with displacements as large as 25 Å. The domain displacement appears to be distinct from that observed in Gαs upon binding of Gs to the β2 adrenergic receptor. Moreover, the Ras-like domain exhibits structural plasticity within and around the nucleotide-binding cavity, and the switch I and switch II regions, which are known to adopt different conformations in the GDP- and GTP-bound states of Gα, undergo structural rearrangements. Collectively, the data show that Ric-8A induces a conformationally heterogeneous state of Gαi and provide insight into the mechanism of action of a nonreceptor Gα GEF.
Heterotrimeric G proteins are activated by exchange of GDP for GTP at the alpha subunit (Gα), a reaction with a high-activation energy barrier (1). Guanine dinucleotides and trinucleotides bind tightly to Gα with affinities in the low nanomolar range (2, 3), and contribute substantially to the overall stability of Gα tertiary structure. Indeed, nucleotide-free Gα exhibits properties characteristic of a molten globule (4). In cells, agonist-stimulated 7-transmembrane helical G protein-coupled receptors (GPCRs) catalyze nucleotide exchange from G protein heterotrimers, in which Gα•GDP is bound to a heterodimer of Gβ and Gγ subunits (5). The cytosolic proteins Ric-8A and Ric-8B, which are structurally unrelated to GPCRs, have been shown to have guanine nucleotide exchange (GEF) activity toward Gα•GDP subunits in the absence of Gβγ (6, 7), thus functionally activating the subunit. In Caenorhabditis elegans, Drosophila, and mouse, Ric-8 homologs have been shown to be essential for asymmetric cell division, where they are assumed to function as GEFs (8–12). Ric-8 proteins also promote efficient folding and membrane localization of certain Gα subunits (13, 14), and inhibit their ubiquitination and degradation (15, 16). With respect to these activities, Ric-8A acts specifically on Gα subunits of the i, q, and 12/13 classes, whereas Ric-8B is active toward Gαs (17).
Gα subunits are composed of two structural domains (3). The Ras-like domain is homologous to guanine nucleotide-binding domains of the Ras superfamily. Within the Ras-like domain are three so-called switch segments that integrate catalytic (guanine nucleotide-binding and GTP hydrolysis) with regulatory function (effector regulation). The conformations of these peptide segments differ between the GTP and GDP-bound states of Gα (3). In crystals of Gαi1–nucleotide complexes, switch I and switch II are well-ordered in the GTP-bound state, but partially (switch I) or fully (switch II) disordered when GDP is bound (18, 19). Inserted into switch I of the Ras domain is a helical domain that is unique to the family of heterotrimeric G proteins. The helical domain flanks the guanine nucleotide-binding site and, while it makes few direct contacts with the nucleotide, shields it from solvent and may affect the rate of its dissociation (20, 21).
We have shown that in the complex of nucleotide-free Gαi1 and Ric-8A, an intermediate in the nucleotide exchange reaction (6, 22), Gαi1 is conformationally heterogeneous and dynamic, but the structures involved and the magnitude of the structural fluctuations were not determined (4). The nucleotide-free Gαi1:Ric-8A complex is stable and can be readily isolated. In the present study, we used site-directed spin labeling (SDSL) and both continuous wave (CW) and double electron-electron resonance (DEER) (23, 24) spectroscopy to map sequence-specific structural and dynamical changes in Gαi1 upon complex formation with Ric-8A. The data reveal that binding of Ric-8A to Gαi1 induces structural heterogeneity due to new conformations in which the helical domain has pivoted away from the Ras-like domain, exposing the nucleotide-binding site to solvent, thus providing an escape (and entry) pathway for the nucleotide. A similar change is induced in Gαi1 upon formation of the nucleotide-free complex with the activated GPCR rhodopsin (20), but is distinctly different from that in the crystal structure of Gαs in the complex with β2 adrenergic receptor (β2R) (25). In addition to the global changes in tertiary structure, binding of Ric-8A also triggers deformation within the Ras-like domain, particularly of structural elements that surround the nucleotide-binding pocket. Together, these changes reveal salient features of a mechanism underlying the GEF activity of Ric-8A.
Results
In the background of Hexa I Gαi1, in which all native surface-exposed and reactive cysteine residues were mutated (26), SDSL methods (27) were used to introduce pairs of nitroxide side chains (R1) at selected sites in nonmyristoylated GDP-bound Gαi1; sites were selected pairwise from the set shown in Fig. 1. Spin-label pairs were designed to detect changes in the distance, monitored by DEER (24), between the helical and Ras-like domains and within these domains upon binding of Ric-8A to Gαi1•GDP. All Hexa I Gαi1 mutants and their R1-labeled forms exhibited GTPγS-binding rates within a factor of 2 of that for wild-type Gαi1. In all cases, rates were enhanced at least 10-fold by Ric-8A (Table 1), comparable to its action on wild-type Gαi1. Heterodimeric, nucleotide-free complexes between Ric-8A and mutants of HexaI-Gαi1 and their R1-labeled forms were homogeneous by HPLC analysis (Fig. S1). DEER data on the spin labeled mutants were obtained from samples flash-frozen in liquid nitrogen (Materials and Methods). The basic DEER data are a dipolar evolution function from which an interspin distance distribution is derived. Background-corrected dipolar evolution functions for all samples are provided in Fig. S2.
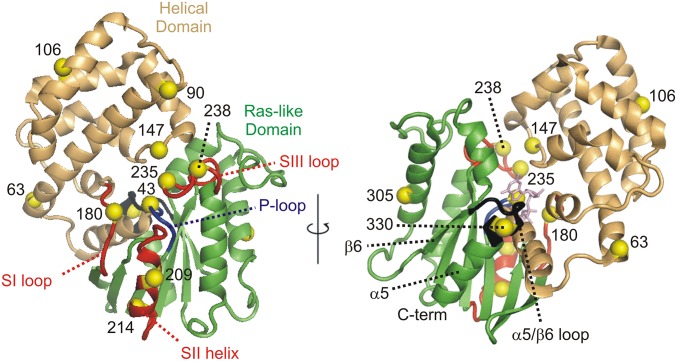
Fig. 1.
The set of sites selected for introduction of R1 in Gαi1 (yellow alpha carbon spheres). The helical domain and Ras-like domains are shown as light orange and green ribbons, respectively. The ribbon corresponding to the switch I loop, the switch II helix, and the switch III loop are all colored red, whereas the phosphate binding P-loop is shown in blue. Other structural elements mentioned in the text are labeled. The structure of Gαi1:GTPγS (PDB ID code 1GIA) is shown because switch II and III are ordered in that complex, but not in Gαi1:GDP (PDB ID code 1GDD). A stick figure of GTPγS (light magenta) is shown at the nucleotide-binding site, Right.
Table 1.
Rates of GTPγS binding to Hexa I Gαi1 and Hexa I R1-Gαi1
| Mutant* | R1 sites† | GTPγS binding,‡ min−1 | ||
| Nat | R1 | R1 + Ric-8A | ||
| Gαi1 | — | 0.043 | — | 0.56 (43) |
| Hexa I Gαi1 | — | 0.021 | — | 0.28 (32) |
| 90C,106C | H | 0.018 | 0.019 | 0.22 (25) |
| 90C,214C | H-R | 0.024 | 0.018 | 0.27 (24) |
| 90C,238C | H-R | 0.025 | 0.015 | 0.32 (32) |
| 106C-214C | H-R | 0.019 | 0.025 | 0.33 (35) |
| 106C-238C | H-R | 0.033 | 0.029 | 0.35 (32) |
| 147C-235C | H-R | 0.025 | 0.02 | 0.29 (32) |
| 63C-238C | H-R | 0.016 | 0.016 | 0.11 (28) |
| 63C-209C | H-R | 0.022 | 0.022 | 0.29 (29) |
| 43C-330C | R | 0.042 | 0.04 | 0.50 (34) |
| 180C-305C | R | 0.023 | 0.02 | 0.28 (29) |
| 209C-330C | R | 0.033 | 0.031 | 0.36 (30) |
| 305C-330C | R | 0.041 | 0.032 | 0.49 (35) |
All R1 pairs are in the Hexa I Gαi1 background.
H, both R1 residues in Helical domain; H-R, one R1 residue in helical and one R1 in Ras domain; R, both R1 residues in Ras domain.
Nat, unlabeled cysteine mutant in Hexa I background; R1, R1-derivatized cysteine mutant in Hexa I background; R1 + Ric-8A, R1-derivatized mutant in the presence of Ric-8A (see Materials and Methods for assay conditions). Values of individual measurements are within 5% of the average value. Overall percent change in tryptophan fluorescence after 10 min of incubation with GTPγS is shown in parenthesis.
Ric-8A–Induced Gαi1 Interdomain Motions.
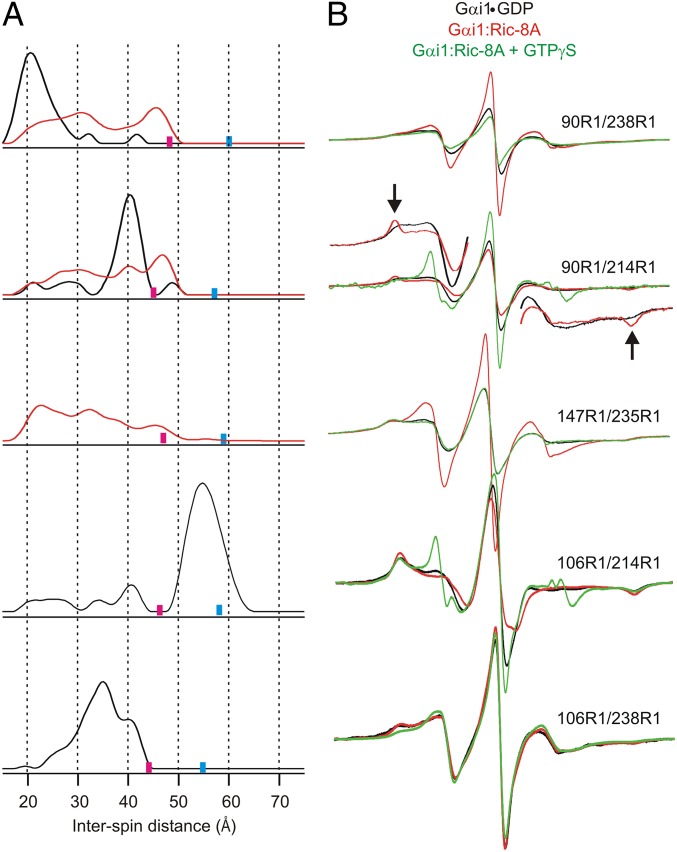
To detect and quantify the action of Ric-8A on the interdomain geometry of Gαi1 using DEER distance mapping, we made use of spin label pairs with one R1 in each domain. The pairs were selected from a set consisting of three sites in the helical domain (residues 90, 106, and 147) and three functionally significant sites in the Ras-like domain (residue 214 in switch II and 235 and 238 in switch III) that are well-ordered in the activated GTP-bound state but flexible with GDP bound (19, 28) (Fig. 1). The five pairs investigated are 90R1/238R1, 90R1/214R1, 147R1/235R1, 106R1/214R1, and 106R1/238R1. In principle, the experimental DEER distances can be compared with predictions based on modeling of R1 in the appropriate crystal structure. However, for residues 209, 214, 235, and 238, electron density is missing in the Gαi1•GDP crystal structure (PDB ID code 1GDD), and this comparison cannot be made for any of the above R1 pairs.
Model-free DEER analysis for 90R1/238R1 and 90R1/214R1 in Gαi1•GDP yields interdomain distance probability distributions in which single maxima account for the majority of the populations (Fig. 2A, black traces). The widths of the distributions for the major populations are on the order of those expected for known rotamers of R1 (29). However, the minor populations in 90R1/214R1 at both shorter and longer distances are outside the range for rotamers of the R1 side chain, suggesting large-amplitude but low-probability fluctuations in the structure that involve relative movements of the C-terminal portion of the flexible switch II sequence and residue 90R1 in the helical domain in the GDP-bound state of Gαi1. The smaller populations in 90R1/238R1 may not be reliably determined by the data. When bound to Ric-8A, there are striking changes in the distance distributions of 90R1/238R1 and 90R1/214R1 relative to Gαi1•GDP. For example, the distance distribution for 90R1/238R1 in the Gαi1:Ric-8A complex broadens strongly toward longer distances and is multimodal, suggesting that the Ras and helical domains separate to occupy a manifold of substates, one of which (∼20 Å) is apparently the same as in Gαi1•GDP. For 90R1/214R1, the major population corresponding to Gαi1•GDP (∼40 Å) drops sharply, whereas shorter and longer distances, similar to those of the minor states in Gαi1•GDP, are strongly populated.
Fig. 2.
Distance distributions for spin pairs that span the Ras-like and helical domains of Gαi1•GDP and the corresponding Gαi1:Ric-8A complex. (A) Distance distributions for the indicated pairs are shown for Gαi1•GDP (black traces) and for the complex with Ric-8A (red traces). Based on the DEER data collection time, upper limits for reliable distance and width determination are shown as cyan and magenta bars, respectively, on the distance axis. (B) CW EPR spectra for the corresponding doubly labeled proteins in the Gαi1•GDP (black), Gαi1:Ric-8A complex (red), and after the addition of GTPγS (green). The scan width in magnetic field is 100 G. Insets for 90R1/214R1 provide vertically expanded views of the low and high field regions in the GDP and Ric-8A-bound states to more clearly reveal features corresponding to immobilized states (arrows). For 147R1/235R1 broadening extending beyond 100 G reveals magnetic dipolar interaction in Gαi1•GDP but not in the complex with Ric-8A. Slight dipolar broadening is also seen in 90R1/238R1.
The nitroxides of the interdomain 147R1/235R1 pair are sufficiently close in Gαi1•GDP (≤15 Å) for the CW EPR spectra to show magnetic dipolar broadening (30, 31) (Fig. 2B), consistent with expectations based on any of the crystal structures of the Gαi1 subunit (Fig. 1). The interspin distance is too short for determination by DEER (2). However, upon complex formation with Ric-8A, the dipolar broadening disappears, as evidenced by the sharp increase in spectral intensity that accompanies the narrowing of the resonance lines in the CW spectra, and DEER data reveal a broad distribution extending beyond 40 Å, consistent with the domain separation and conformational heterogeneity identified by the other spin pairs.
The interdomain pairs 106R1/214R1 and 106R1/238R1 have resolved DEER data for Gαi1•GDP that give the broad distance distributions shown in Fig. 2A. For 106R1/214R1, the width of the main population at ∼54 Å is uncertain because of the limited data duration (Fig. S2 and Fig. 2), but the most probable distance is well-determined (Materials and Methods). The width of the distance distribution for 106R1/238R1 can be accounted for by R1 rotamers. The key point is that upon complex formation with Ric-8A, DEER signals are lost in both cases (Fig. S2), indicating that the interspin distances have increased beyond ∼60 Å for 106R1/214R1 and beyond ∼50 Å for 106R1/238R1, further supporting domain separation in the Ric-8A complex.
Interpretation of the distance distributions in terms of a specific trajectory for the interdomain separation would require analysis of more pairs, but the data presented above are sufficient to unequivocally demonstrate the main conclusion—that Ric-8A binding induces domain separation and conformational heterogeneity in Gαi1.
Ric-8A–Induced Structural Heterogeneity Within the Gαi1 Domains.
To examine internal structural changes in the Ras-like domain upon complex formation, R1 pairs were selected so that at least one is placed within a functional sequence. Included in this set are the following: 180, 209, and 238 in switches I, II, and III, respectively, residue 330, located at the N terminus of helix α5 near the juncture of the β6-α5 loop that contacts the guanine ring of the nucleotide (3), and residue 43, proximal to the nucleotide phosphate binding loop (P-loop). Residues 43, 180, and 330 are in structural elements involved in nucleotide binding and hydrolysis (3).
To monitor movement of the above residues in the Ras-like domain because of Ric-8A binding, sites 63 and 305 were selected as reference sites. Residue 63R1 is formally in the helical domain, but located at an apparent hinge point between the Ras and helical domains (Fig. 1) and is not anticipated to undergo large amplitude motions during domain separation (25). Residue 305 in helix α4 is in a structurally invariant region in the crystal structures of Gαi1•GDP, Gαi1•GTPγS, and the complex with Gβγ. Moreover, the corresponding sequence in Gαs does not change position upon complex formation with β2R (25). Thus, residue 305 is assumed to not undergo significant changes upon complex formation with Ric-8A. With 63R1 and 305R1 as references, changes in distance distributions for pairs involving these residues are tentatively assigned to movement of the partner.
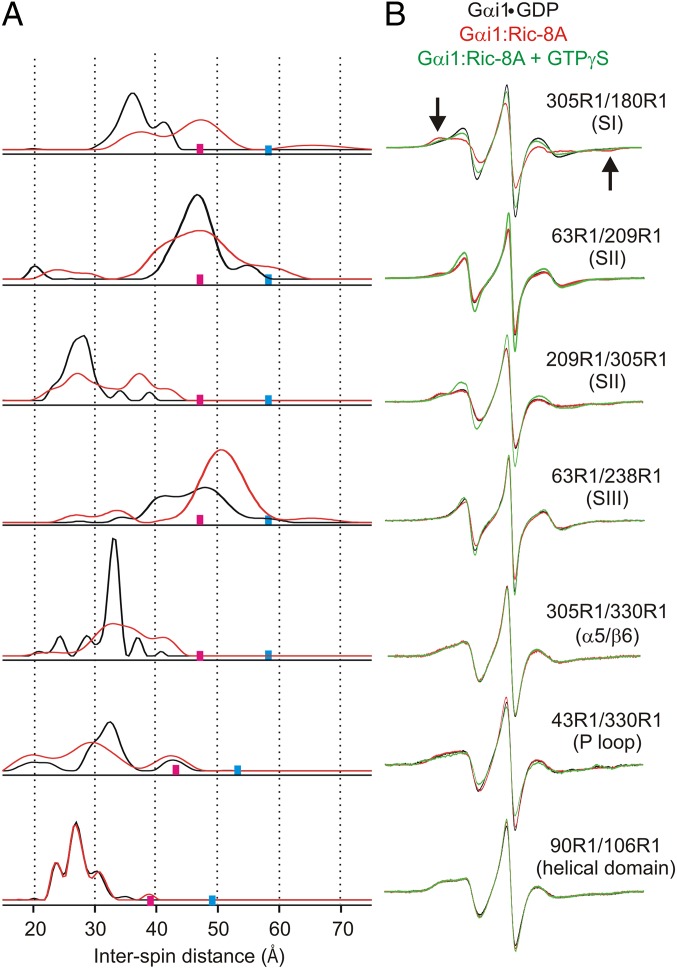
Fig. 3 shows DEER distance distributions for five R1 pairs involving these reference sites. As indicated in the figure, four of the pairs monitor the positions of the switch regions (SI–SIII), whereas 330R1/305R1 monitors the position of the β6-α5 junction, in each case relative to the indicated reference. In addition, 43R1/330R1 is included to measure movements of the P loop relative to β6-α5 junction, and the single pair 90R1/106R1 to monitor internal changes in the helical domain. In the crystal structure of Gαi1•GDP, electron density is resolved for all relevant residues except for 209 and 238, and the experimental DEER distributions for the four pairs not involving these residues can be compared with predictions from modeling based on multiscale modeling of macromolecules (MMM) (32). In each case, the experimental and modeled distributions based on the Gαi1•GDP structure are in reasonable agreement, except for 43R1/330R1, where the width of the broad trimodal distribution considerably exceeds that from MMM modeling (Fig. S3), suggesting that the distribution is determined by protein flexibility rather than R1 rotamers. The origin of the distribution could be motion of either 43R1 or 330R1 or both, but the narrow dominant population of 305R1/330R1 suggests that it is due to 43R1 in the P loop that defines a boundary of the nucleotide-binding site.
Fig. 3.
Distance distributions for spin pairs within the Ras-like domain in Gαi1•GDP and the corresponding Gαi1:Ric-8A complex. (A) Distance distributions for the indicated pairs are shown for Gαi1•GDP (black traces) and for the complex with Ric-8A (red traces). Upper limits for reliable distance and width determination are shown as cyan and magenta bars, respectively, on the distance axis. (B) CW EPR spectra for the corresponding doubly labeled proteins in the Gαi1•GDP (black), Gαi1:Ric-8A complex (red), and after the addition of GTPγS (green).
Upon formation of the Gαi1:Ric-8A complex, there are changes in the distance distributions for all pairs. The general nature of these changes is moderate to strong broadening of the overall distribution, leading to the important conclusion that complex formation with Ric-8A results in an increase in conformational heterogeneity in the Ras-like domain, presumably corresponding to fluctuations in solution at ambient temperatures. In addition to the overall broadening, there are large shifts in distance distributions for some R1 residues relative to the selected reference. In particular, the distribution for switch I (180R1) shows the appearance of a new population at a displacement of ∼10 Å relative to 305R1. Distributions for switch II (209) relative to both 63R1 and 305R1 undergo strong broadening, and new populations appear at longer distances for 209R1/305R1. The broad multimodal nature of the distributions for the 180/305R1 and 209R1/305R1 pairs in the complex indicate fluctuations of switch I and II between conformational substates, one of which is apparently similar to the Gαi1•GDP state. The position of switch III, monitored by 238R1 relative to 63R1, shifts to longer distances corresponding to one of the two states present in the Gαi1•GDP state, suggesting the possibility of conformational selection. The distance distribution for 43R1/330R1 shows shifts in relative populations upon complex formation, but is also broadened, consistent with fluctuations along the P-loop/purine binding site axis. In contrast, the distance distribution for 90R1/106R1, which monitors the internal structure of the helical domain, shows essentially no change, despite its large-scale separation from the Ras domain.
The interresidue distance changes observed upon Ric-8A binding are likely to originate from the combined displacements of multiple structural elements in Gαi1. Under any circumstance, the collective data are consistent with substantial fluctuations in the dimensions of the nucleotide-binding site in the complex.
CW EPR Spectra Reflect Structural Changes Due to Ric-8A and GTPγS Binding.
Changes in the CW EPR spectra upon complex formation with Ric-8A at room temperature reflect changes in Gαi1 conformation and dynamics in multiple ways. For example, in 147R1/235R1, the nitroxides are sufficiently close to show distant-dependent magnetic dipolar broadening in the CW spectra for Gαi1•GDP (30) as mentioned above; there is also a weak dipolar broadening in 90R1/238R1 (Fig. 2) This broadening disappears upon formation of the complex (dramatically in the case of 147R1/235R1), signaling separation of the spins beyond approximately 15 Å (compare black and red traces; Fig. 2B) and supporting the conclusions from DEER distance measurements made at cryogenic temperatures. Addition of GTPγS to the nucleotide-free complex (green traces) leads to complete recovery of the dipolar broadening, demonstrating reversible closing of the domains.
In the absence of dipolar broadening, the CW EPR spectral lineshapes of R1 reflect the local structure (33) and backbone dynamics (34, 35), and changes in lineshape can be interpreted in those terms. For example, the spectra of the 90R1/214R1 and 305R1/180R1 doubles show an increase in the population of a more immobilized state upon Ric-8A binding (arrows, Figs. 2B and 3B). Although these spectra are composites of those for the individual sites, the absence of spectral changes for other pairs involving the same partners (i.e., 90R1/106R1, 305R1/330R1, and 305R1/209R1) suggests that 214R1 and 180R1 are the source of the change. CW spectra of single 214R1 and 180R1 mutants confirm this conclusion (Fig. S4). Residue 214R1 is located in switch II, and an interesting possibility to account for the strong immobilization is a direct contact of the switch II sequence with Ric-8A in the complex. The same model could account for the immobilization of 180R1 in the nearby switch I.
Collectively, the CW EPR lineshape changes observed upon formation of the Ric-8A complex reverse after the addition of GTPγS to 305R1/180R1, consistent with the dissociation of the complex (Fig. 3B, green traces). The dramatic change of the 90R1/214R1 spectrum upon GTPγS addition to this mutant is a hallmark of the dynamic transition in switch II upon moving from the GDP to the GTPγS-bound state (28).
Discussion
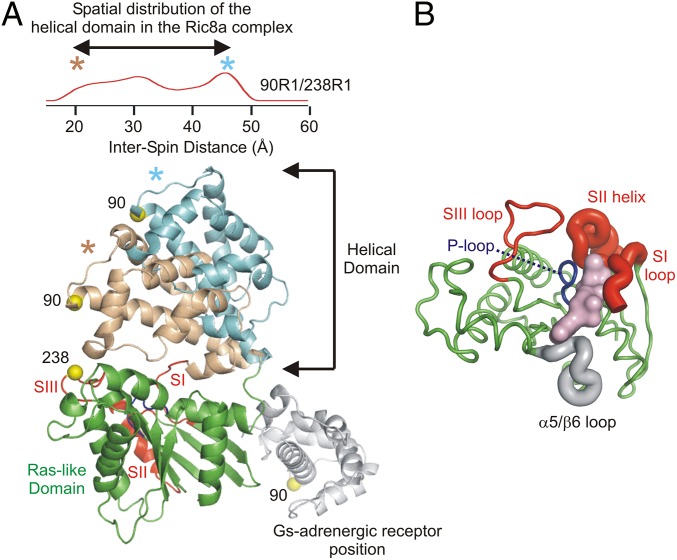
The results presented here afford insight into mechanistic features of nucleotide exchange that are specific to Ric-8A activation of monomeric Gα in contrast to GPCR activation of Gα in a heterotrimeric context, and those that appear to be common to both. Fig. 4 summarizes graphically the main findings of this study regarding the nature and topography of changes in Gαi1 that occur upon binding to Ric-8A. Although they are structurally unrelated, GPCRs and Ric-8A appear at least superficially to use similar mechanisms to induce nucleotide release. Thus, both GEFs bind to the C terminus (4, 25, 36–39) of Gα and trigger changes that destabilize the contacts between the helical and Ras-like domains of Gα. In addition to the appearance of an open state in which residue 90R1 in the helical domain is separated by ∼48 Å from 214R1 in switch II and 238R1 in switch III, there appears to be essentially a continuum of intermediate states that approach the fully closed Gαi1•GDP conformation, suggesting a ratchet-like separation that may facilitate nucleotide release while clearly providing an entry route to GTP (20, 25) (Fig. 4A). However, a model of the Ric-8A–bound open state for Gαi1 based on the crystal structure of the Gs:β2R complex (25) is not consistent with the DEER data presented here. For example, in the crystal structure, the helical domain occupies a position relative to the nucleotide domain that is ∼20 Å further than that measured by the maximum 90R1-238R1 distance in the distribution (Fig. 4A). However, the 106R1-214R1 distance is found to be beyond the detection limit of the DEER experiment (Fig. S2) and, thus, must be substantially longer then would be expected for a Gs:β2R-like complex (∼50 Å) in which the distance would be resolved. To the extent that they can be compared, the displacement and broad distribution in position of the Giα1 helical domain in the Ric-8A complex is similar to that observed in the complex with activated rhodopsin (20). A distribution of helical domain positions is also observed in cryo-EM studies of the β2R:Gs complex (40). The large displacement in the crystal structure of the Gs:β2R complex may be due to lattice interactions.
Fig. 4.
Models summarizing Gαi1 structural heterogeneity in the Gαi1:Ric-8A complex as determined by DEER. (A, Upper) The distance distributions between 90R1 (helical domain) and 238R1 (nucleotide domain) in the Gαi1:Ric-8A complex (red trace) reproduced from Fig. 2; the asterisks mark the short and long distance extremes in the nearly continuous distribution. Lower shows a ribbon model of Gαi1 with the helical domain occupying the two distance extremes of the distribution; the ribbons are color-coded to match the asterisks in Upper, and the nucleotide domain is shown in green with a red ribbon for SI, SII, and SIII. For reference, the position of the helical domain modeled from the Gs:β2R crystal structure (PDB ID code 3SN6) is shown as a gray ribbon. (B) Structural heterogeneity around the nucleotide-binding site. With the helical domain removed, a top view of the Ras-like domain is displayed. The thickness of the backbone segment illustrates increased heterogeneity in the Gαi1:Ric-8A complex versus Gαi1•GDP. The GTPγS nucleotide is shown as a surface rendering in light magenta.
In addition to the separation of the helical and Ras-like domains in the nucleotide-free state of Gα, both GPCRs and Ric-8A induce internal perturbations within the Ras-like domain. For example, in the crystal structure of the Gs:β2R complexes, the P-loop, β6-α5 connector and the N-terminal segment of switch II that encompass the GTP binding site, all undergo receptor-induced conformational changes (25). However, while β6-α5, the P-loop, and flanking segments of secondary structure are resolved in the crystal structure, hydrogen–deuterium exchange experiments show that these segments become conformationally dynamic (41). These changes are allosterically induced, because the receptor forms no direct contacts with the switch regions, or elements that form the nucleotide-binding site. The extent of conformational dynamics appears to be localized to discrete regions of secondary structure.
Ric-8A likewise induces conformational dynamics within the Ras domain. The DEER results presented here suggest that the amplitude of these dynamic changes are large and global in scope. Thus, the centroid distribution of 305R1-330R1 distances increases in breadth by 10 Å at its half maximum. The CW spectra show that the local environments at the probe positions do not change upon complex formation, showing that there is no direct contact with Ric-8A, indicating that Ric-8A acts allosterically to induce these structural changes. Similarly, we observe increases in structural heterogeneity across the nucleotide-binding site, indicated by the loss of substructure in the major peak of the 43R1/330R1 distance distribution relative to that observed for Gαi1•GDP. The centroid of this distribution is down-shifted by 3 Å, indicative of a contraction in the nucleotide-binding site along the P-loop—β6-α5 axis in Ric-8A–bound nucleotide-free Gαi1, a change that is not observed in the Gs:β2R complex. The large excursions in switch I and switch II, as revealed in the 180R1/330R1 and 209R1/330R1 distributions (and also in the increase in the width of the 63R1/209R1 distribution) are accompanied by immobilization of 180R1, and quite dramatically, of 214R1, suggesting that Ric-8A may induce these structural displacements by making direct contact with the two switch segments. Thus, the magnitude of Ras domain heterogeneity induced by Ric-8A appear to be markedly greater than those caused by Gα–GPCR interactions as inferred from the crystal structure.
That Ric-8A binds to monomeric Gαi1•GDP, whereas GPCRs preferentially recognize intact G protein heterotrimers—thereby preserving contacts between Gα switch II and Gβγ—may, in part, explain the differences in the amplitude of changes induced by the two exchange factors on the Ras domain of Gα. The presence of Gβγ in GPCR complexes with G protein heterotrimers may provide some degree of global stabilization of nucleotide-free Gα that is not afforded in the complex with Ric-8A, and this stabilization may be reflected in the kinetics of nucleotide exchange. Detergent-solubilized rhodopsin catalyzes rapid (50 min−1) nucleotide exchange from heterotrimeric transducin (42). In contrast, the slow kinetics (8 min−1) of Ric-8A–catalyzed exchange (22) may reflect the high activation energies associated with induction of unfolding events in monomeric Gα, manifested in the formation of an ensemble of structurally heterogeneous nucleotide-free states observed in the experiments described here.
Materials and Methods
Molecular Cloning, Protein Expression, and Spin Labeling.
A cDNA construct encoding bovine Gαi1 with six amino acid substitutions at solvent-exposed cysteine residues (C3S, C66A, C214S, C305S, C325A, C351I; Gαi1 Hexa I) and a hexahistidine sequence inserted between residues M119 and T120, was amplified by PCR using an attB-modified forward primer encoding a tobacco etch virus (TEV) protease site N-terminal to the Gαi1 sequence, and subcloned into a pDEST-15 destination vector for expression as a GST fusion protein using the Gateway cloning system (Invitrogen). This vector was used for construction of double-cysteine mutants by QuikChange mutagenesis (Agilent). The C147/C235 mutant was constructed as described (20). Hexa I Gαi1 harboring cysteine pairs were expressed and purified as described by Thomas et al. (4). A fully active fragment of rat Ric-8A composed of residues 1–492 was purified as described (4) and used for all experiments.
R1 adducts of cysteine double mutants of Hexa I Gαi1 were generated by incubation with the sulfhydryl spin label S-(1-oxy-2,2,5,5,-tetramethylpyrroline-3-methyl)-methanethiosulfonate and purified as described by Van Eps et al. (28). Intrinsic and Ric-8A–catalyzed rates of GTPγS binding was assayed for the double cysteine mutants and the R1-labeled species as indicated in Table 1. Complexes of doubly R1-labeled Hexa I Gαi1 with Ric-8A (1–492) were prepared and purified as described by Thomas et al (4)., and subjected to HPLC analysis as indicated in Fig. S1. To check labeling, preliminary CW spectra were taken on a Bruker EMX Plus EPR spectrometer by using a 200-G field scan at a microwave power of 0.6325 mW.
Rates of GTPγS Binding to Hexa I and R1-Labeled Hexa I Gαi1.
Exchange of GTPγS for GDP bound to Gαi1, Hexa I, their cysteine mutants, and R1 adducts was followed by monitoring the change in the tryptophan fluorescence of Gαi1. Gαi1•GDP (1 µM) in assay buffer (20 mM Hepes, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 0.1 mM DTT, 2 µM GDP, and 0.05% C12E10) in a reaction volume of 400 µL was allowed to equilibrate for 10–15 min at 20 °C in a quartz fluorescence cuvette. GTPγS (final concentration, 10 µM) was added to the reaction mixture in the absence or presence of 1.5 µM Ric-8A, and the increase in fluorescence at 340 nm was monitored upon excitation at 290 nm. Fluorescence measurements were conducted by using an LS55 spectrofluorometer (PerkinElmer Life Sciences) attached to a circulating water bath to maintain a steady sample temperature of 20 °C. Excitation and emission slit widths were set at 2.5 nm. All exciting light was eliminated by use of a 290-nm cutoff filter positioned in front of the emission photomultiplier. All kinetic rate parameters are numerical averages obtained by fitting triplicate datasets to a single exponential (y = y0 + a(1 − e-kt)) equation in Sigma Plot 8.0 or Graph Pad Prism 5.0. Values of individual measurements are within 5% of the average value. Incubation of R1-labeled Gαi1 proteins in assay buffer for 2 h at 25 °C, followed by acquisition of CW spectra, demonstrated that R1-side chains are not cleaved from the protein to a significant extent by disulfide exchange in the presence of 0.1–0.2 mM DTT.
HPLC analysis of R1-Gαi1:Ric-8A complexes were performed on a TSKgel SuperSW S3000 with 50 mM Tris, pH 8.0, 150 mM NaCl, and 0.2 mM DTT as the running buffer using an Agilent 1200 HPLC. R1-labeled Gαi1•GDP samples (containing 50 μM Gαi1•GDP, 10 μM GDP in 100 μL) were mixed in equal molar ratio of Ric-8A, incubated for 2 h and concentrated by using a Millipore micro-Centricon to remove excess GDP. After centrifugation at 18,300 × g on a benchtop microcentrifuge, samples (25 μL) were injected onto the column, eluted at 0.3 mL/min and monitored at 280 nm. Bio-Rad gel filtration standards (15 μL) were injected and eluted at 0.3 mL/min in the running buffer to provide molecular weight markers, and check column flow characteristics and peak shapes.
CW EPR and DEER Measurements.
CW EPR spectra over a 100-G range were recorded at room temperature on a Bruker E580 spectrometer by using a high-sensitivity resonator (HS0118) at X-band microwave frequencies with an observe microwave power of 20 mW. Field-modulation amplitudes were selected to give maximal signal intensity without line-shape distortion. The data were typically averages of 20–30 scans.
For DEER measurements, the spin-labeled proteins were flash-frozen in quartz capillaries (1.5 mm inner diameter and 1.8 mm outer diameter) in a liquid nitrogen bath. Data were collected on a Bruker Elexsys 580 spectrometer with a Super Q-FTu Bridge by using a Bruker EN 5107D2 resonator at 80 K. A 36-ns π-pump pulse was applied to the low field peak of the nitroxide absorption spectrum, and the observer π/2 (16 ns) and π (32 ns) pulses were positioned 50 MHz (17.8 G) upfield, which corresponds to the nitroxide center resonance. Model-free distance distributions were obtained from the raw dipolar evolution data by using the program “LongDistances” developed by Christian Altenbach. The program is freely available online at www.biochemistry.ucla.edu/biochem/Faculty/Hubbell. Upper distance limits for reliable distance and width determination were calculated as described (24).
Supplementary Material
Acknowledgments
We thank Gwendolyn Nix for outstanding technical assistance, Ian M. Chrisman for collecting CW EPR spectra at the University of Montana, Heidi E. Hamm (Vanderbilt University) and Suzanne Scarlata (State University of New York at Stony Brook) for providing Hexa I Gαi1 template DNA and mutants, and Anita M. Preininger (Vanderbilt University) for advice on Hexa I Gαi1 expression and purification. This work was supported by the Jules Stein Professor Endowment (to W.L.H.) and National Institute of Health Grants R01GM105993 (to S.R.S.), R01EY05216 (to W.L.H.), and P30EY00331 for research core support to the Jules Stein Eye Institute. The Bruker EMXplus EPR spectrometer and MALDI TOF instrument at the University of Montana was purchased with funds from National Science Foundation Awards CHE-1048623 and CHE-1039814, respectively, and operated with support from Center for Biomolecular Structure and Dynamics NIH Centers of Biomedical Research Excellence Award P20GM103546.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423878112/-/DCSupplemental.
References
- 1.Kohl B, Hofmann KP. Temperature dependence of G-protein activation in photoreceptor membranes. Transient extra metarhodopsin II on bovine disk membranes. Biophys J. 1987;52(2):271–277. doi: 10.1016/S0006-3495(87)83214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. Effects of Mg2+ and the beta gamma-subunit complex on the interactions of guanine nucleotides with G proteins. J Biol Chem. 1987;262(2):762–766. [PubMed] [Google Scholar]
- 3.Sprang SR, Chen Z, Du X. Structural basis of effector regulation and signal termination in heterotrimeric Galpha proteins. Adv Protein Chem. 2007;74:1–65. doi: 10.1016/S0065-3233(07)74001-9. [DOI] [PubMed] [Google Scholar]
- 4.Thomas CJ, et al. The nucleotide exchange factor Ric-8A is a chaperone for the conformationally dynamic nucleotide-free state of Gαi1. PLoS ONE. 2011;6(8):e23197. doi: 10.1371/journal.pone.0023197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J Biol Chem. 2003;278(10):8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- 7.Chan P, Gabay M, Wright FA, Tall GG. Ric-8B is a GTP-dependent G protein alphas guanine nucleotide exchange factor. J Biol Chem. 2011;286(22):19932–19942. doi: 10.1074/jbc.M110.163675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David NB, et al. Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol. 2005;7(11):1083–1090. doi: 10.1038/ncb1319. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, et al. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat Cell Biol. 2005;7(11):1091–1098. doi: 10.1038/ncb1317. [DOI] [PubMed] [Google Scholar]
- 10.Afshar K, Willard FS, Colombo K, Siderovski DP, Gönczy P. Cortical localization of the Galpha protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 2005;132(20):4449–4459. doi: 10.1242/dev.02039. [DOI] [PubMed] [Google Scholar]
- 11.Afshar K, et al. RIC-8 is required for GPR-1/2-dependent Galpha function during asymmetric division of C. elegans embryos. Cell. 2004;119(2):219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 12.Tõnissoo T, et al. Nucleotide exchange factor RIC-8 is indispensable in mammalian early development. Dev Dyn. 2010;239(12):3404–3415. doi: 10.1002/dvdy.22480. [DOI] [PubMed] [Google Scholar]
- 13.Gabay M, et al. Ric-8 proteins are molecular chaperones that direct nascent G protein α subunit membrane association. Sci Signal. 2011;4(200):ra79. doi: 10.1126/scisignal.2002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan P, Thomas CJ, Sprang SR, Tall GG. Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein α subunits. Proc Natl Acad Sci USA. 2013;110(10):3794–3799. doi: 10.1073/pnas.1220943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai Y, Nishimura A, Tago K, Mizuno N, Itoh H. Ric-8B stabilizes the alpha subunit of stimulatory G protein by inhibiting its ubiquitination. J Biol Chem. 2010;285(15):11114–11120. doi: 10.1074/jbc.M109.063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chishiki K, Kamakura S, Yuzawa S, Hayase J, Sumimoto H. Ubiquitination of the heterotrimeric G protein α subunits Gαi2 and Gαq is prevented by the guanine nucleotide exchange factor Ric-8A. Biochem Biophys Res Commun. 2013;435(3):414–419. doi: 10.1016/j.bbrc.2013.04.103. [DOI] [PubMed] [Google Scholar]
- 17.Tall GG. Ric-8 regulation of heterotrimeric G proteins. J Recept Signal Transduct Res. 2013;33(3):139–143. doi: 10.3109/10799893.2013.763828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman DE, et al. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265(5177):1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 19.Mixon MB, et al. Tertiary and quaternary structural changes in Giα1 induced by GTP hydrolysis. Science. 1995;270(5238):954–960. doi: 10.1126/science.270.5238.954. [DOI] [PubMed] [Google Scholar]
- 20.Van Eps N, et al. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci USA. 2011;108(23):9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science. 1993;262(5141):1895–1901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 22.Thomas CJ, Tall GG, Adhikari A, Sprang SR. Ric-8A catalyzes guanine nucleotide exchange on G alphai1 bound to the GPR/GoLoco exchange inhibitor AGS3. J Biol Chem. 2008;283(34):23150–23160. doi: 10.1074/jbc.M802422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen RG, Singel DJ. Double electron-electron resonance spin-echo modulation - spectroscopic measurement of electron-spin pair separations in orientationally disordered solids. J Chem Phys. 1993;98(7):5134–5146. [Google Scholar]
- 24.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medkova M, Preininger AM, Yu NJ, Hubbell WL, Hamm HE. Conformational changes in the amino-terminal helix of the G protein alpha(i1) following dissociation from Gbetagamma subunit and activation. Biochemistry. 2002;41(31):9962–9972. doi: 10.1021/bi0255726. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell WL, López CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 2013;23(5):725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Eps N, Oldham WM, Hamm HE, Hubbell WL. Structural and dynamical changes in an alpha-subunit of a heterotrimeric G protein along the activation pathway. Proc Natl Acad Sci USA. 2006;103(44):16194–16199. doi: 10.1073/pnas.0607972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klose D, et al. Simulation vs. reality: A comparison of in silico distance predictions with DEER and FRET measurements. PLoS ONE. 2012;7(6):e39492. doi: 10.1371/journal.pone.0039492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci USA. 1995;92(18):8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry. 2001;40(51):15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 32.Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13(6):2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 33.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 34.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends in Biochemical Sci. 2002;27(6):288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 35.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51(33):6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamm HE, et al. Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science. 1988;241(4867):832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 37.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 38.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13(9):772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 39.Vellano CP, et al. Activation of the regulator of G protein signaling 14-Gαi1-GDP signaling complex is regulated by resistance to inhibitors of cholinesterase-8A. Biochemistry. 2011;50(5):752–762. doi: 10.1021/bi101910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westfield GH, et al. Structural flexibility of the G α s α-helical domain in the β2-adrenoceptor Gs complex. Proc Natl Acad Sci USA. 2011;108(38):16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung KY, et al. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477(7366):611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci USA. 2007;104(26):10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.