Significance
A crucial factor for drug efficacy is not just the binding affinity, but also the mean residence time in the binding pocket, usually quantified by its inverse, koff. This is an important parameter that regulates the time during which the drug is active. Whereas the calculation of the binding affinity is by now routine, the calculation of koff has proven more challenging because the timescales involved far exceed the limits of standard molecular dynamics simulation. We propose a metadynamics-based strategy that allows reaching timescales of seconds, and estimate koff along with unbinding pathways and associated dynamical bottlenecks. The protocol is exemplified for trypsin–benzamidine unbinding. This work is a step towards a more effective computer-based drug design.
Keywords: protein–ligand unbinding, kinetics, enhanced sampling, drug design
Abstract
The ability to predict the mechanisms and the associated rate constants of protein–ligand unbinding is of great practical importance in drug design. In this work we demonstrate how a recently introduced metadynamics-based approach allows exploration of the unbinding pathways, estimation of the rates, and determination of the rate-limiting steps in the paradigmatic case of the trypsin–benzamidine system. Protein, ligand, and solvent are described with full atomic resolution. Using metadynamics, multiple unbinding trajectories that start with the ligand in the crystallographic binding pose and end with the ligand in the fully solvated state are generated. The unbinding rate is computed from the mean residence time of the ligand. Using our previously computed binding affinity we also obtain the binding rate . Both rates are in agreement with reported experimental values. We uncover the complex pathways of unbinding trajectories and describe the critical rate-limiting steps with unprecedented detail. Our findings illuminate the role played by the coupling between subtle protein backbone fluctuations and the solvation by water molecules that enter the binding pocket and assist in the breaking of the shielded hydrogen bonds. We expect our approach to be useful in calculating rates for general protein–ligand systems and a valid support for drug design.
Understanding the thermodynamics and kinetics of protein–ligand interactions is of paramount relevance in the early stages of drug discovery (1–3). So far the major emphasis has been placed on predicting the most likely binding pose as determined by the highest binding affinity (4, 5). In contrast, it has not been possible to predict the pathways for unbinding and the associated rates. However, it is by now well-recognized that one of the most pertinent factors for sustained drug efficacy and safety is not just its affinity, but possibly even more so, the mean lifetime of the protein–ligand complex (1–3). The latter property is strictly related to the time during which the ligand remains in the binding site (1, 2), and is typically expressed by its inverse, the dissociation rate (2). In principle should be amenable to calculations through all-atom molecular dynamics (MD) simulations. These simulations could give detailed and useful insights into the atomic interactions at work during unbinding, especially in the ephemeral but kinetically most relevant transition state ensemble (TSE) (6, 7). Such information is of great value in designing modifications of the ligand that might improve its pharmaceutical properties.
However, despite the potential of MD simulations no such calculation has yet been reported. This is a consequence of the limited timescales of MD simulations. Even with the most modern purpose-built supercomputers or massive distributed computing, one can barely reach the timescale of milliseconds (3). Unfortunately most of the reported ligand–protein dissociation times far exceed this timescale (2). These timescales can be reached either by transition path sampling methods (8, 9), quasi-classical approximations (10), by the construction of Markov state models (11, 12), or through carefully designed enhanced sampling methods (8, 13–30) that make accessible the timescale of seconds and beyond in a controlled and accurate way. The enhanced sampling method we use in this work is based on metadynamics (13–15), which has been widely and successfully applied to a variety of systems including complex protein–ligand systems (25–30), and has been rigorously proven to converge to the correct free-energy surface (31, 32).
Recently, we have extended the scope of metadynamics by showing that it can also be used to recover kinetic information (15). Furthermore, we showed that by using an a posteriori statistical analysis (33) one can also establish the reliability of the kinetics thus generated. The use of metadynamics for obtaining kinetic information is still in its infancy, however its usefulness has been tested by us and other groups in a range of systems (15, 33–36).
In this work, we demonstrate that the scope of the method reported in ref. 15 can be extended to study protein–ligand dissociation pathways and to determine in an accurate way the ligand unbinding rates. We reach well into the hundreds of milliseconds regime and longer, maintaining at the same time full atomic resolution for protein, ligand, and solvent. Specifically, we study the unbinding of the inhibitor benzamidine from trypsin, a serine protease protein (27, 37, 38) using classical force fields (39, 40). Using our acceleration method (15, 33) we are able to harness 21 independent successful unbinding trajectories in which the ligand goes from the bound to the fully unbound state. We find that one of the most distinctive features of the unbinding process is the role played by the water molecules (41, 42). In particular, the solvent promotes unbinding by assisting in the breakage of shielded hydrogen bonds through the formation of water bridge interactions (41).
From the analysis of the unbinding trajectories we find that along the unbinding pathways the ligand rests for times ranging from nanoseconds to milliseconds in a number of intermediate structures. We calculate the rates for all possible transitions between these intermediates and construct a Markov model for the unbinding process (11, 43, 44). The overall escape rate computed from this Markov model is in good agreement with the direct estimation of the mean unbinding time that comes from the metadynamics runs. Reassured by this agreement we use the Markov model to determine the dominant unbinding pathways and rate-limiting steps. To this end, starting from the metadynamics reactive trajectories, we perform a committor analysis and determine the TSE (6). Using the recently computed value of the binding affinity (27) we also estimate the binding rate constant . Our calculated unbinding and binding rates compare reasonably well with the known experimental measurement (37), especially taking into account the margin of error in the experiment and the inaccuracy of the force field used in the simulations (42). Unprecedented structural features of the target are also disclosed. In particular, we find that in its apo state trypsin can exist in two forms. In the first form, loop Val207–Tyr224 (hereafter labeled loop L) oscillates around the crystallographic state. In the other form, a small distortion of this loop is stabilized. The mean lifetime of this distorted state is nearly 0.7 ms and during this time the ligand cannot reach the binding site.
We believe that this metadynamics-based strategy is, to our knowledge, the first direct approach for calculating from MD simulations of unbinding. Previous studies have focused on the calculation of and the magnitude of was only indirectly obtained (12, 38). Our strategy should be easily applicable for calculating unbinding pathways and rates for generic protein–ligand systems, thus complementing and extending the role of enhanced sampling-based simulations in drug discovery.
Methods
Metadynamics.
Metadynamics is by now a well-established method whose details can be found in many review papers; the interested reader is referred to the growing literature (14, 22, 26, 27). Here we underline only some features that are relevant to the present discussion and provide details in SI Appendix. In metadynamics, one first identifies a small subset of the difficult to sample but relevant degrees of freedom, called collective variables (CVs) (22). A history-dependent biasing potential is then constructed on the fly as a function of these CVs. By gradually enhancing the fluctuations in the CVs, the system is discouraged from getting trapped in the low free-energy basins of phase space. Thus, using metadynamics one can observe processes that would be far beyond the timescales accessible to normal MD, while still maintaining complete atomic resolution. Metadynamics fully takes into account the dynamical ever-fluctuating nature of the protein and the complex role played by the molecular solvent. At the end of a metadynamics run, the probability distribution of any observable can be either computed directly (14, 32) or through a reweighting procedure (32, 45).
Unbiased Kinetics from Biased Metadynamics.
Inspired by previous work (16, 17), we recently extended the scope of metadynamics and showed how to extract unbiased rates from biased ones with minimal extra computational burden (15). By kinetic information, we specifically mean pathways, the associated rates, and rate-limiting steps. The key assumptions for our approach to work are (i) the process being investigated is characterized by movements from one stable state to another via dynamical bottlenecks that are rarely but quickly crossed, or in other words, there exists a separation of timescales, and (ii) although there is no need to know beforehand the nature or location of such bottlenecks, one should have CVs that can distinguish between stable basins. Under these two key assumptions, by making the bias deposition slower than the time spent in dynamical bottlenecks, one can keep these bottlenecks bias-free throughout the course of the metadynamics run. This preserves the unbiased sequence of state-to-state transitions and allows one to access the acceleration of transition rates achieved through biasing, by appealing to a generalized version of transition state theory (15, 46, 47). This acceleration is provided by the following running average accumulated through the course of metadynamics (15–17):
| [1] |
where s is the collective variable being biased, β is the inverse temperature, and is the bias experienced at time t. The above expression is valid even if there are multiple intermediate states and numerous alternative reactive pathways (15, 33).
In a successive work we proposed a way to assess the reliability of the two assumptions above (33). This relies on the fact that the escape times from a long-lived metastable state obey a time-homogeneous Poisson statistics (48). A statistical analysis based on the Kolmogorov–Smirnov (KS) test can quantitatively assess how precisely our assumptions have been met (33).
CVs from Preliminary Free-Energy Surface Exploration.
The identification of appropriate CVs is a challenging problem that in recent years has witnessed much progress (49, 50). For the problem at hand there exists a practical way to build CVs, namely path-based CVs (51, 52). These are extremely powerful and versatile when one wants to study transitions between two states A and B. For protein–ligand unbinding these states would be the docked pose and the fully solvated unbound state.
In the path CV formalism one defines a reference path that takes the system from A to B, and a distance s defining the position of the system along the chosen starting path. However, such a variable might not be discriminating enough, and several possible pathways can collapse into one (53). By biasing together the values of s and a second CV that lifts the degeneracy in s (SI Appendix), metadynamics naturally generates an ensemble of pathways.
As a first step toward the construction of the path we have to characterize the space in which it is defined. Specifically, this requires identifying the relevant protein–ligand interactions. These can be found through a preliminary funnel metadynamics run (27, 54) using for instance a protein–ligand distance as CV. At this stage we do not need to obtain a converged free-energy surface (FES) (32); however, funnel metadynamics allows us to explore a range of relevant intermediate states in a limited computer time by facilitating repeated binding and unbinding. The residues that are found to play a role in the unbinding trajectory thus generated are included in the list of interactions used to build the path CV.
If performing metadynamics using the trial path leads to visiting intermediates that exhibit stable interactions not included in the original list, these are added to it and the process iterated until no further relevant interaction is found. For trypsin–benzamidine just one iteration was necessary to obtain good CVs.
Calculation of .
With CVs optimized as described in the previous section, we then calculate . This simply amounts to performing several independent infrequently biased metadynamics runs starting in the bound X-ray pose [Protein Data Bank (PDB) ID code 3atl] and stopping when the ligand is unbound and fully solvated. We analyze the distribution of unbinding times using the approach of ref. 33 and make sure that it passes the KS test (SI Appendix). From this analysis we get a direct estimate of . See SI Appendix for more details on the metadynamics parameters including the details of infrequent biasing.
Further Analysis of the Unbinding Mechanisms Through Construction of a Markov Model.
Over the last few years, Markov state modeling has proven to be a useful tool in the analysis of data generated from MD simulations (11, 12, 43, 44). Inspired by this approach, we build a kinetic equation for transitions between various intermediate states in the unbinding of benzamidine from trypsin. We stress that this step is not needed if all one seeks is the magnitude of that can be directly obtained from the simpler protocol described in the previous section. This second step needs to be taken when one seeks detailed insight into the roles of the different intermediate states observed in the metadynamics runs and to determine the dynamical bottlenecks.
As in all Markov state models, also in our analysis the most delicate step is the enumeration of intermediates. Here we are assisted by the metadynamics runs which directly provide an estimate of where the system spends most of its time (13, 32). Our criterion was to include only those states in which during the full unbinding runs the system typically spends a much longer time than the interval between successive bias depositions. These states can be identified with the main free-energy basins even if a converged FES is neither achieved nor required in our calculations. These states are described in detail in the following sections.
We then do a second set of metadynamics runs with infrequent bias deposition starting in each of these states. These are done with absorbing boundary conditions (48), in which the simulation is stopped as soon as that state is exited for the first time. Around 25 such runs were performed starting from each of the identified stable states.
From these metadynamics runs, again making use of Eq. 1 we calculate mean lifetime of each state along with respective KS tests (SI Appendix). We then build a matrix of state-to-state transition rates by taking into account the mean lifetimes and the number of transitions from one state into another as described for instance in ref. 55. Solving for the slowest eigenvalue of this matrix gives an alternative estimate of and an analysis based on the eigenvalues provides the rate-limiting steps and fluxes through them (56, 57) (SI Appendix).
One of the main features of infrequent metadynamics is that corresponding to these rate-limiting steps, the segments of trajectories that cross the barrier between successive bias depositions are representatives of the unbiased transition path ensemble. On this set of transition paths, we perform detailed committor analysis (SI Appendix) and identify the respective TSEs (6, 8).
Results
Unbinding and Binding Rates from Infrequent Metadynamics.
The metadynamics runs start with the ligand in the docked pose and end in the fully unbound state. Using 21 such independent simulations we find a of 9.1 ± 2.5 s−1 with a very good metric on the KS test (SI Appendix). The values are in agreement with the reported experimental value of 600 ± 300 s−1 (37), especially taking into account the conservative error of 2 kcal/mol in force fields and other factors detailed in Discussion (39, 40, 58). In SI Appendix we show that around 12 independent metadynamics runs are sufficient to obtain a reliable estimate of .
We then indirectly compute the binding rate by using the value previously computed with funnel metadynamics at equilibrium concentration of 1 M. To calculate we use the relation , where is the standard concentration. We thus obtain , in excellent agreement with the experimental value of .
Kinetically Relevant States.
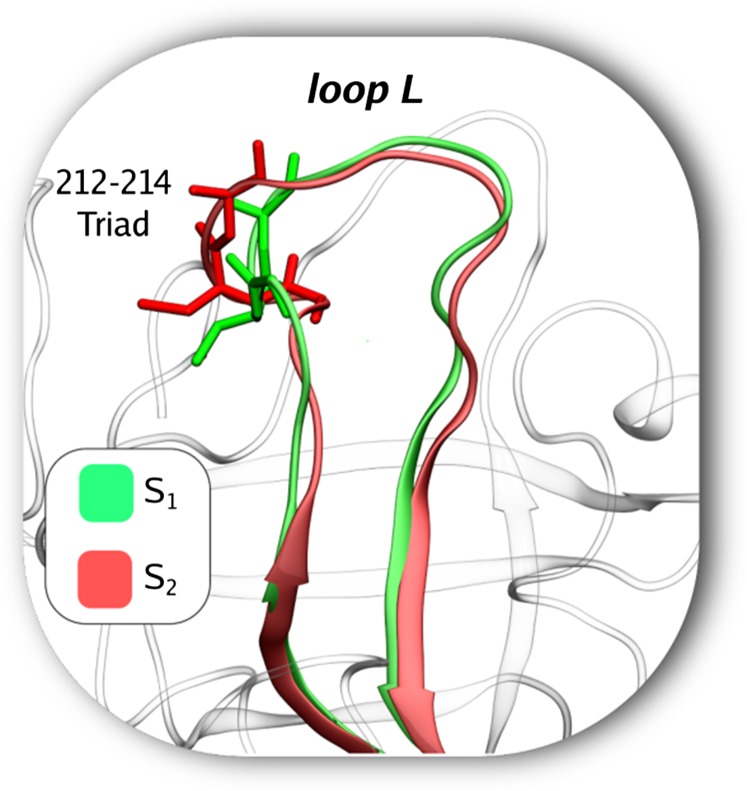
From our metadynamics trajectories, we find that there are several kinetically relevant stable states during the unbinding of benzamidine from trypsin (Fig. 1). These include the X-ray binding pose (A), another bound state (B) in which the ligand is slightly rotated with respect to A, a presolvated state (P) in which the ligand’s diamino group points toward the solvent, and two solvated unbound states ( and ) which differ in the arrangement of the loop L (Fig. 2). In the protein is in its undistorted crystallographic pose and is available for further binding. In the loop L is distorted and the protein is temporarily inactive.
Fig. 1.
States relevant for the unbinding of trypsin–benzamidine complex. The specific interactions that stabilize these states are indicated. Also indicated are the water molecules that play defining roles. See text for further details and mean lifetimes.
Fig. 2.
Trypsin in its apo state can exist in substates S1 (green) and S2 (red). The key difference between these two states is in the loop L. In S1 the loop is as in the X-ray pose and the protein is available for binding. In S2 the loop has undergone a distortion initiated primarily by glycine–serine residues (S213 and G214) that engage in hydrogen bond interaction with other residues (D216 and Q217). In this state, the protein is temporarily unavailable for binding. See text for further details and mean lifetimes.
Our statistical analyses and the agreement of the results obtained using multiple metadynamics protocols, comprising more than 115 independent metadynamics simulations, reassure us that likely there are no other kinetically relevant stable states lying on the dominant ligand unbinding pathways. We now describe the nature of the stabilizing interactions and mean lifetimes for the various states (Figs. 1 and 2). See SI Appendix for detailed statistical analyses of all lifetimes including associated errors.
State A.
This is the bound state reported by funnel metadynamics and various other previous studies, in good agreement with X-ray structures (see figure 1 of ref. 27 and SI Appendix for the details of relevant interactions in A). We find a mean lifetime of 42 ms for this state, reflecting its stability.
State B.
This state has an average lifetime of 5 and is slightly rotated with respect to the X-ray pose. However, the overall interactions established in pose A are conserved (SI Appendix). It is relevant to note that precisely the same pose was found in our previous metadynamics simulations where a different CV setting was used. Because the choice of CVs might bias the results, this finding lends confidence that the presence of state B is not an artifact of the CV choice. The basin B is around 4–5 kcal/mol higher in free energy than basin A, as estimated by taking the ratio of the respective mean lifetimes. This is higher than the value of 1–2 kcal/mol previously reported by funnel metadynamics. However, those calculations were performed with the rmsd fluctuations in the protein backbone constrained to preserve the shape of the ligand binding site. This constraint was functional for the accurate estimate of the binding energy of the A pose, which was the main objective of that calculation (27), whereas it slightly affected the free-energy estimate of state B.
State P.
The third stable state P has the much shorter mean lifetime of 49 ns. The most important feature of this state is that the ligand is rotated pointing the diamino group toward the solvent (Fig. 1). However, in this pose the ligand cannot yet leave the binding pocket. Given its important position on the unbinding pathways, we describe it in detail here. The phenyl ring of benzamidine is sandwiched between the Cα atoms of Cys191 and Trp211 where it can engage in hydrophobic contacts, whereas the polar diamino group is close to the triad formed by polar residues such as Gly212–Ser213–Gly214. As described in the next section, States S1 and S2, this triad plays an important role in deciding which unbinding pathway is adopted. In state P, the ligand engages in hydrogen bonds, water bridges, or both, with one or more of the residues from this triad. At variance with the poses A and B, here the ligand can rotate, changing the interacting partners within the triad residues. We have also calculated the mean lifetime of pose P with 25 independent unbiased MD runs starting in P, which gave an average lifetime of 29 ns, in excellent agreement with the metadynamics value.
States S1 and S2.
When the ligand is in the unbound and fully solvated state, the protein is found in two substates, which we call S1 and S2 (Figs. 2 and 3). In the first, the loop L is similar to its X-ray structure, whereas in the other it is conformationally distorted. The motion from S1 to S2 renders the protein temporarily inactive. To the best of our knowledge the distorted pose S2 has not been previously reported. It has a lifetime of around 0.7 ms. We checked that this state is not an artifact of the choice of CVs by doing metadynamics using different CVs, as well as two long unbiased MD runs (SI Appendix). Specifically, in each of these MD simulations the distorted pose lasted for at least 1.5 while still not showing any sign of recovery to the X-ray pose. Analyzing the protein motion, we note that S2 can form because in the presolvated state P, the triad 214–216 can undergo fast fluctuations involving switching of stabilizing hydrogen bond interactions. Often these interactions are temporarily stabilized by the formation of a small alpha loop (Fig. 2). This results in a partial collapse of the binding pocket that makes ligand reentry difficult, as we have explicitly observed in some funnel metadynamics runs where the system reached state S2 (SI Appendix). This happens because in the S2 state the ligand cannot form interactions with the triad 214–216. Interestingly, previous free-binding simulations with distributed computing resources have also found that during binding the system always goes through a state involving interaction with residues of this triad (38). Note that not taking this distortion into account has a very small effect on and given the small lifetime of .
Fig. 3.
State-to-state transition rates for trypsin–benzamidine unbinding. All rates are in s−1. The respective mean lifetimes for ligand binding states are also shown.
Dominant Unbinding Pathways and Rate-Limiting Steps.
To identify the dominant pathways during unbinding, we performed a flux analysis (56, 57) on the state-to-state transition matrix built from metadynamics simulations on the various stable states (Fig. 3). Solving for the dominant eigenvalue of this matrix gives a of 7.3 s−1, in excellent agreement with the full unbinding metadynamics simulations. Looking at Fig. 3 one can note that the state P is a mandatory stage during all of the unbinding pathways. As can be seen from the calculated flux (SI Appendix) (56, 57), we find that around 84% of the time the system prefers to unbind by going directly from A to B before going to P, whereas around 16% of the time it goes straight from A to P. Our matrix also shows that the two elementary steps, A to P and A to B, are the slowest steps in the whole unbinding process. We now describe the atomistic details of these rate-limiting steps along with the typical transition state structures (Fig. 4).
Fig. 4.
Typical mechanism of going from state A to P (Top) and A to B (Bottom). For each, typical TSE members as determined by committor analysis are also shown. Relevant residues and water molecules are also indicated. Note that the biological water in state A is removed in the pre-TS state for path 1 (Top) to highlight the role of water molecule coming from solvent. See main text for summary of key interactions, and SI Appendix for more details of the TSE and committor analysis.
A to P.
The exit from A to the presolvated pose P has a highly concerted and atomistically well-defined nature, assisted by solvent water molecules (Fig. 4). Specifically, one water molecule comes into the binding site from the outside, first breaking the direct hydrogen bonds between the carboxyl group of Asp189 and the benzamidine tail, and then screening the interaction between these two groups. This water bridge detaches the ligand from Asp189, allowing a higher number of contacts of the diamino group with the surrounding water molecules. Note how in the first intermediate subsequent to state A (Fig. 4), at the time of entry of the first water, the benzamidine tail can still engage in direct interaction with the triad 212–214. The ligand then rotates in the binding site in the direction of the triad, thus exposing its charged tail toward the solvent. Whereas Asp189 still forms hydrogen bonds with the water molecule, the ligand now forms a hydrogen bond or water-mediated interactions with one or more of the residues in the triad 212–214.
A to B.
This transition also involves a rotation of the ligand but is less pronounced and is in a direction opposite to that needed to go from A to P. A key role is played by not one but two water molecules entering from the solvent and by the orientation of the triad 212–214 (Fig. 4). Specifically, one water approaches the ligand in close proximity of Asp189 again mediating the interaction between the nitrogen atoms of the benzamidine diamino group and the carboxyl group of Asp189. In contrast with the A to P transition, here after the first water enters the binding pocket, the triad 212–214 is oriented such that it cannot engage in direct interaction with the benzamidine tail. This is the first intermediate subsequent to state A (Fig. 4). An additional water molecule now moves toward an inner position in the binding site where it can engage hydrogen bond interactions with the carbonyls of residues such as Val223 and Tyr224. The diamino group of the ligand follows this second water rotating its position in the binding pocket away from the triad and leading to the final B pose. In this state, the ligand still forms hydrogen bonds with Asp189 via a direct or water-mediated interaction and with residues including Val223 and Tyr224 through the second water molecule. The presence of water molecules at similar position in different X-ray structures of trypsin (PDB ID codes 1s0q and 3atl) had previously suggested a functional role of these waters that we are now able to explain.
Irrespective of how the state P is reached, the final unbinding involves breaking of the hydrogen bond between the ligand’s tail and the triad (Figs. 1 and 4).
TSEs for Rate-Limiting Steps.
To investigate further the nature of transition for the two rate-limiting steps, we analyzed the reactive trajectories from metadynamics corresponding to these steps (SI Appendix). We performed multiple short unbiased MD runs from different points along these trajectories, and through these we identified configurations which have a nearly 1/2 probability of going back into either A and P, or A and B, depending on the respective step being investigated. These configurations represent the true dynamical bottlenecks for the unbinding. Further details of these unbiased runs can be found in SI Appendix. We now describe the ligand and protein interactions formed in these transition states (Fig. 4).
The crucial and common feature is a partial solvation of the ligand’s tail or of specific residues in the protein, and partial breakage of shielded hydrogen bonds through water molecules coming from the bulk. For the A to P event, the typical TSE member as shown in Fig. 4 involves a water bridge formed between one of the nitrogen atoms of the benzamidine diamino group and oxygen atoms in Asp189. The same nitrogen atom is also interacting with one of triad 212–214 members. The other nitrogen atom of the diamino group is now partially solvent exposed, and the ligand is almost rotated outward of the binding pocket, but not yet fully. For the A to B event, the typical TSE member as shown in Fig. 4 involves a role played by two water molecules that previously were in the bulk solvent. Similar to the TSE for A to P, a water bridge is formed between one of the nitrogen atoms of the benzamidine diamino group and an oxygen atom in Asp189. However, the second nitrogen atom of the diamino group is now rotated inward and has started to engage in water bridge interaction with residues such as Val223 and Tyr224. In the TSE, the orientation of the ligand is closer to state B than to the docked pose (SI Appendix).
Discussion
In this work we have demonstrated the possibility of studying detailed unbinding kinetics of protein–ligand systems with all-atom molecular dynamics using a metadynamics-based strategy. We obtained multiple full unbinding trajectories for the trypsin–benzamidine complex starting in the X-ray pose and directly computed the unbinding rate . Our total simulated metadynamics time of 5, after taking into account the scaling factor of Eq. 1, corresponds to nearly 3 s of real-time evolution. We then enumerated the stable states found in the metadynamics runs and built a Markov model for transitions between these states. Through these we could describe the ensemble of unbinding pathways through the multiple intermediates and identify the rate-limiting steps. In combination with our previous work on funnel metadynamics that gave us accurate binding affinity (27), we could also calculate the binding rate . The validity of the rates at every step of the calculation was demonstrated using rigorous statistical analyses.
The calculated in this work is slower than the experimental measurement. This deviation is well within the error expected from the accuracy of current force fields. For instance, one reason for the being slower could be the lack of polarization in the force field (42). It is clear from our simulation that the rate-limiting step involves solvation of the ligand by external water molecules. Previous studies using polarizable force fields have suggested that polarization enhances the solvation of benzamidine in water and at the same time weakens the attraction between benzamidine and trypsin (42). Thus, our nonpolarizable force field (39, 40) which does not include these effects leads to a slower dissociation. Another shortcoming of the force field, which however could act in the direction of faster dissociation, is the diffusivity of three-site transferrable intermolecular potential water which is slightly faster than experiment (59). However, we expect that the effect of polarization in the protein–ligand interaction would dominate the effect due to diffusivity, especially because the binding pocket is fairly well exposed to the solvent. Our indirectly calculated is very close to reported experimental value but that close an agreement is most likely fortuitous due to cancellation of errors in and .
The unbinding kinetics of protein–ligand systems is a problem of immense practical interest, for calculating which no rigorous computational protocol has been available so far. Perhaps even more importantly than calculating the magnitude of , our protocol allows calculation of unbinding pathways, TSEs, and the residues that play a role in the dominant pathways. We hope this work will motivate the experimental community to investigate whether the unbinding is actually sensitive to mutations in these residues that we have identified or if it is just a force-field artifact. We expect that the metadynamics-based method proposed in this paper will open up new horizons in investigating mechanisms and computing rate constants for protein–ligand systems, having a great impact on drug design and lead optimization processes.
Supplementary Material
Acknowledgments
The computational time for this work was provided by the Swiss National Supercomputing Center and by the Eidgenössische Technische Hochschule Zürich Brutus cluster. We acknowledge European Union Grant ERC-2009-AdG-247075 for funding.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424461112/-/DCSupplemental.
References
- 1.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5(9):730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 2.Núñez S, Venhorst J, Kruse CG. Target-drug interactions: First principles and their application to drug discovery. Drug Discov Today. 2012;17(1-2):10–22. doi: 10.1016/j.drudis.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Pan AC, Borhani DW, Dror RO, Shaw DE. Molecular determinants of drug-receptor binding kinetics. Drug Discov Today. 2013;18(13-14):667–673. doi: 10.1016/j.drudis.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Woo H-J, Roux B. Calculation of absolute protein-ligand binding free energy from computer simulations. Proc Natl Acad Sci USA. 2005;102(19):6825–6830. doi: 10.1073/pnas.0409005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilson MK, Zhou H-X. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 6.Bolhuis PG, Dellago C, Chandler D. Reaction coordinates of biomolecular isomerization. Proc Natl Acad Sci USA. 2000;97(11):5877–5882. doi: 10.1073/pnas.100127697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geissler PL, Dellago C, Chandler D. Kinetic pathways of ion pair dissociation in water. J Phys Chem B. 1999;103(18):3706–3710. [Google Scholar]
- 8.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 9.Metzner P, Schütte C, Vanden-Eijnden E. Illustration of transition path theory on a collection of simple examples. J Chem Phys. 2006;125(8):084110–084119. doi: 10.1063/1.2335447. [DOI] [PubMed] [Google Scholar]
- 10.Poulsen TD, Garcia-Viloca M, Gao J, Truhlar DG. Free energy surface, reaction paths, and kinetic isotope effect of short-chain acyl-coa dehydrogenase. J Phys Chem B. 2003;107(35):9567–9578. [Google Scholar]
- 11.Bowman GR, Beauchamp KA, Boxer G, Pande VS. Progress and challenges in the automated construction of Markov state models for full protein systems. J Chem Phys. 2009;131(12):124101–124111. doi: 10.1063/1.3216567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman GR, Geissler PL. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc Natl Acad Sci USA. 2012;109(29):11681–11686. doi: 10.1073/pnas.1209309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002;99(20):12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barducci A, Bussi G, Parrinello M. Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Phys Rev Lett. 2008;100(2):020603–020606. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 15.Tiwary P, Parrinello M. From metadynamics to dynamics. Phys Rev Lett. 2013;111(23):230602–230606. doi: 10.1103/PhysRevLett.111.230602. [DOI] [PubMed] [Google Scholar]
- 16.Voter AF. Hyperdynamics: Accelerated molecular dynamics of infrequent events. Phys Rev Lett. 1997;78(20):3908–3911. [Google Scholar]
- 17.Grubmüller H. Predicting slow structural transitions in macromolecular systems: Conformational flooding. Phys Rev E. 1995;52(3):2893–2906. doi: 10.1103/physreve.52.2893. [DOI] [PubMed] [Google Scholar]
- 18.Huber T, Torda AE, van Gunsteren WF. Local elevation: A method for improving the searching properties of molecular dynamics simulation. J Comput Aided Mol Des. 1994;8(6):695–708. doi: 10.1007/BF00124016. [DOI] [PubMed] [Google Scholar]
- 19.Hansmann UH, Wille LT. Global optimization by energy landscape paving. Phys Rev Lett. 2002;88(6):068105–068108. doi: 10.1103/PhysRevLett.88.068105. [DOI] [PubMed] [Google Scholar]
- 20.Tiwary P, van de Walle A. Hybrid deterministic and stochastic approach for efficient atomistic simulations at long time scales. Phys Rev B. 2011;84(10):100301–100304. [Google Scholar]
- 21.Tiwary P, van de Walle A. Accelerated molecular dynamics through stochastic iterations and collective variable based basin identification. Phys Rev B. 2013;87(9):094304–094307. [Google Scholar]
- 22.Barducci A, Bonomi M, Parrinello M. Metadynamics. Wiley Interdiscip Rev: Comput Mol Sci. 2011;1(5):826–843. [Google Scholar]
- 23.Zheng L, Chen M, Yang W. Random walk in orthogonal space to achieve efficient free-energy simulation of complex systems. Proc Natl Acad Sci USA. 2008;105(51):20227–20232. doi: 10.1073/pnas.0810631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valsson O, Parrinello M. Variational approach to enhanced sampling and free energy calculations. Phys Rev Lett. 2014;113(9):090601–090605. doi: 10.1103/PhysRevLett.113.090601. [DOI] [PubMed] [Google Scholar]
- 25.Gervasio FL, Laio A, Parrinello M. Flexible docking in solution using metadynamics. J Am Chem Soc. 2005;127(8):2600–2607. doi: 10.1021/ja0445950. [DOI] [PubMed] [Google Scholar]
- 26.Limongelli V, et al. Molecular basis of cyclooxygenase enzymes (COXs) selective inhibition. Proc Natl Acad Sci USA. 2010;107(12):5411–5416. doi: 10.1073/pnas.0913377107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limongelli V, Bonomi M, Parrinello M. Funnel metadynamics as accurate binding free-energy method. Proc Natl Acad Sci USA. 2013;110(16):6358–6363. doi: 10.1073/pnas.1303186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Leva FS, Novellino E, Cavalli A, Parrinello M, Limongelli V. 2014. Mechanistic insight into ligand binding to g-quadruplex DNA. Nucl Acid Res, 10.1093/nar/gku247. [DOI] [PMC free article] [PubMed]
- 29.Limongelli V, et al. Sampling protein motion and solvent effect during ligand binding. Proc Natl Acad Sci USA. 2012;109(5):1467–1472. doi: 10.1073/pnas.1112181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grazioso G, et al. Investigating the mechanism of substrate uptake and release in the glutamate transporter homologue Glt(Ph) through metadynamics simulations. J Am Chem Soc. 2012;134(1):453–463. doi: 10.1021/ja208485w. [DOI] [PubMed] [Google Scholar]
- 31.Dama JF, Parrinello M, Voth GA. Well-tempered metadynamics converges asymptotically. Phys Rev Lett. 2014;112(24):240602–240605. doi: 10.1103/PhysRevLett.112.240602. [DOI] [PubMed] [Google Scholar]
- 32.Tiwary P, Parrinello M. 2014. A time-independent free energy estimator for metadynamics. J Phys Chem B, 10.1021/jp504920s.
- 33.Salvalaglio M, Tiwary P, Parrinello M. Assessing the reliability of the dynamics reconstructed from metadynamics. J Chem Theory Comput. 2014;10(4):1420–1425. doi: 10.1021/ct500040r. [DOI] [PubMed] [Google Scholar]
- 34.Bohner MU, Zeman J, Smiatek J, Arnold A, Kästner J. Nudged-elastic band used to find reaction coordinates based on the free energy. J Chem Phys. 2014;140(7):074109–074115. doi: 10.1063/1.4865220. [DOI] [PubMed] [Google Scholar]
- 35.Schneider J, Reuter K. 2014. Efficient calculation of microscopic dissolution rate constants: The aspirin–water interface. J Phys Chem Lett 5(21):3859–3862.
- 36.Sicard F, Destainville N, Manghi M. 2014. DNA denaturation bubbles: Free-energy landscape and nucleation/closure rates. J Chem Phy, 10.1063/1.4905668.
- 37.Guillain F, Thusius D. The use of proflavin as an indicator in temperature-jump studies of the binding of a competitive inhibitor to trypsin. J Am Chem Soc. 1970;92(18):5534–5536. doi: 10.1021/ja00721a051. [DOI] [PubMed] [Google Scholar]
- 38.Buch I, Giorgino T, De Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc Natl Acad Sci USA. 2011;108(25):10184–10189. doi: 10.1073/pnas.1103547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117(19):5179–5197. [Google Scholar]
- 40.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidtke P, Luque FJ, Murray JB, Barril X. Shielded hydrogen bonds as structural determinants of binding kinetics: Application in drug design. J Am Chem Soc. 2011;133(46):18903–18910. doi: 10.1021/ja207494u. [DOI] [PubMed] [Google Scholar]
- 42.Jiao D, Golubkov PA, Darden TA, Ren P. Calculation of protein-ligand binding free energy by using a polarizable potential. Proc Natl Acad Sci USA. 2008;105(17):6290–6295. doi: 10.1073/pnas.0711686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohlhoff KJ, et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat Chem. 2014;6(1):15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klippenstein SJ, Pande VS, Truhlar DG. Chemical kinetics and mechanisms of complex systems: A perspective on recent theoretical advances. J Am Chem Soc. 2014;136(2):528–546. doi: 10.1021/ja408723a. [DOI] [PubMed] [Google Scholar]
- 45.Bonomi M, Barducci A, Parrinello M. Reconstructing the equilibrium Boltzmann distribution from well-tempered metadynamics. J Comput Chem. 2009;30(11):1615–1621. doi: 10.1002/jcc.21305. [DOI] [PubMed] [Google Scholar]
- 46.Truhlar DG, Isaacson AD, Garrett BC. Generalized transition state theory. Theor Chem Reac Dyn. 1985;4:65–137. [Google Scholar]
- 47.Truhlar DG, Garrett BC. Variational transition state theory. Annu Rev Phys Chem. 1984;35:159–189. [Google Scholar]
- 48.Lelièvre T. Two mathematical tools to analyze metastable stochastic processes. In: Cangiani A, et al., editors. Numerical Mathematics and Advanced Applications 2011. Springer; Leicester, UK: 2013. pp. 791–810. [Google Scholar]
- 49.Tribello GA, Ceriotti M, Parrinello M. Using sketch-map coordinates to analyze and bias molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109(14):5196–5201. doi: 10.1073/pnas.1201152109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohrdanz MA, Zheng W, Maggioni M, Clementi C. Determination of reaction coordinates via locally scaled diffusion map. J Chem Phys. 2011;134(12):124116–124126. doi: 10.1063/1.3569857. [DOI] [PubMed] [Google Scholar]
- 51.Branduardi D, Gervasio FL, Parrinello M. From A to B in free energy space. J Chem Phys. 2007;126(5):054103–054112. doi: 10.1063/1.2432340. [DOI] [PubMed] [Google Scholar]
- 52.Bonomi M, Branduardi D, Gervasio FL, Parrinello M. The unfolded ensemble and folding mechanism of the C-terminal GB1 beta-hairpin. J Am Chem Soc. 2008;130(42):13938–13944. doi: 10.1021/ja803652f. [DOI] [PubMed] [Google Scholar]
- 53.Dickson BM, Makarov DE, Henkelman G. Pitfalls of choosing an order parameter for rare event calculations. J Chem Phys. 2009;131(7):074108–074113. doi: 10.1063/1.3204008. [DOI] [PubMed] [Google Scholar]
- 54.Bonomi M, et al. Plumed: A portable plugin for free-energy calculations with molecular dynamics. Comput Phys Commun. 2009;180(10):1961–1972. [Google Scholar]
- 55.Du W-N, Marino KA, Bolhuis PG. Multiple state transition interface sampling of alanine dipeptide in explicit solvent. J Chem Phys. 2011;135(14):145102–145111. doi: 10.1063/1.3644344. [DOI] [PubMed] [Google Scholar]
- 56.Berezhkovskii A, Hummer G, Szabo A. Reactive flux and folding pathways in network models of coarse-grained protein dynamics. J Chem Phys. 2009;130(20):205102–205106. doi: 10.1063/1.3139063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schafer NP, et al. Discrete kinetic models from funneled energy landscape simulations. PLoS ONE. 2012;7(12):e50635–e50642. doi: 10.1371/journal.pone.0050635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101–101107. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 59.Shen My MY, Freed KF. Long time dynamics of Met-enkephalin: Comparison of explicit and implicit solvent models. Biophys J. 2002;82(4):1791–1808. doi: 10.1016/s0006-3495(02)75530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.