Significance
This article describes a new method for generating large numbers of invariant natural killer T (iNKT) cells in mice through genetic engineering of blood stem cells. iNKT cells are potent immune cells that regulate many human diseases, including cancer, infections, allergies, and autoimmunity. However, both the study of iNKT cell biology and the clinical application of iNKT cells have been greatly hindered by their small numbers (∼0.01–1% in mouse and human blood). The method reported here provides a powerful new tool to study iNKT cell biology in a mouse model. It can also be applied to humans, opening a new avenue for iNKT cell-based immunotherapy that has the potential to provide patients with therapeutic levels of iNKT cells throughout life.
Keywords: genetic engineering, hematopoietic stem cells, HSCs, invariant natural killer T cells, iNKT cells
Abstract
Invariant natural killer T (iNKT) cells comprise a small population of αβ T lymphocytes. They bridge the innate and adaptive immune systems and mediate strong and rapid responses to many diseases, including cancer, infections, allergies, and autoimmunity. However, the study of iNKT cell biology and the therapeutic applications of these cells are greatly limited by their small numbers in vivo (∼0.01–1% in mouse and human blood). Here, we report a new method to generate large numbers of iNKT cells in mice through T-cell receptor (TCR) gene engineering of hematopoietic stem cells (HSCs). We showed that iNKT TCR-engineered HSCs could generate a clonal population of iNKT cells. These HSC-engineered iNKT cells displayed the typical iNKT cell phenotype and functionality. They followed a two-stage developmental path, first in thymus and then in the periphery, resembling that of endogenous iNKT cells. When tested in a mouse melanoma lung metastasis model, the HSC-engineered iNKT cells effectively protected mice from tumor metastasis. This method provides a powerful and high-throughput tool to investigate the in vivo development and functionality of clonal iNKT cells in mice. More importantly, this method takes advantage of the self-renewal and longevity of HSCs to generate a long-term supply of engineered iNKT cells, thus opening up a new avenue for iNKT cell-based immunotherapy.
Invariant natural killer T (iNKT) cells are a small population of αβ T lymphocytes highly conserved from mice to humans. Like conventional αβ T cells, iNKT cells are derived from hematopoietic stem cells (HSCs) and develop in the thymus. However, they differ from conventional T cells in several important aspects, including their display of NK cell markers, their recognition of glycolipid antigens presented by the nonclassical monomorphic major histocompatibility complex (MHC) molecule CD1d, and their expression of semi-invariant T-cell receptors (identical α chains paired with a limited selection of β chains) (1, 2). Despite their small numbers in vivo (∼0.1–1% in mouse blood and ∼0.01–1% in human blood), iNKT cells have been suggested to play important roles in regulating many diseases, including cancer, infections, allergies, and autoimmunity (3). When stimulated, iNKT cells rapidly release a large amount of effector cytokines like IFN-γ and IL-4, both as a cell population and at the single-cell level. These cytokines then activate various immune effector cells, such as natural killer (NK) cells and dendritic cells (DCs) of the innate immune system, as well as CD4 helper and CD8 cytotoxic conventional αβ T cells of the adaptive immune system via activated DCs (3, 4). Because of their unique activation mechanism, iNKT cells can attack multiple diseases independent of antigen and MHC restrictions, making them attractive universal therapeutic agents (3, 4). Notably, because of the capacity of effector NK cells and conventional αβ T cells to specifically recognize diseased tissue cells, iNKT cell-induced immune reactions result in limited off-target side effects (3, 4).
Restricted by their extremely low numbers, both the study of iNKT cells and their clinical applications have been challenging. iNKT T-cell receptor (TCR) transgenic mice (5, 6) and iNKT induced pluripotent stem (iPS) cell-derived transgenic mice (7) provide valuable tools to study iNKT cell biology in mice, but these methods are both costly and time-consuming. In addition, approaches using transgenic mice have no direct clinical application. As an alternative, a TCR-engineered HSC adoptive transfer strategy could overcome these limitations and become clinically applicable. Since its demonstration in mice in the early 2000s, this HSC-engineered T-cell strategy has been widely used to successfully generate both mouse and human antigen-specific conventional αβ T cells in multiple mouse and humanized mouse models (8–13). Human clinical trials testing this strategy for treating melanoma are also ongoing (14). Based on these previous works and the scientific principle that iNKT cells follow a “TCR instruction” development path similar to that of conventional αβ T cells (15), we hypothesized that HSCs could be engineered to express iNKT TCR genes and be programmed to develop into clonal iNKT cells. In the present report, we demonstrated the feasibility of this new HSC-engineered iNKT cell approach in mice and provided evidence to support its therapeutic potential in a mouse melanoma lung metastasis model.
Results
Cloning of iNKT TCR Genes and Construction of Retroviral Delivery Vectors.
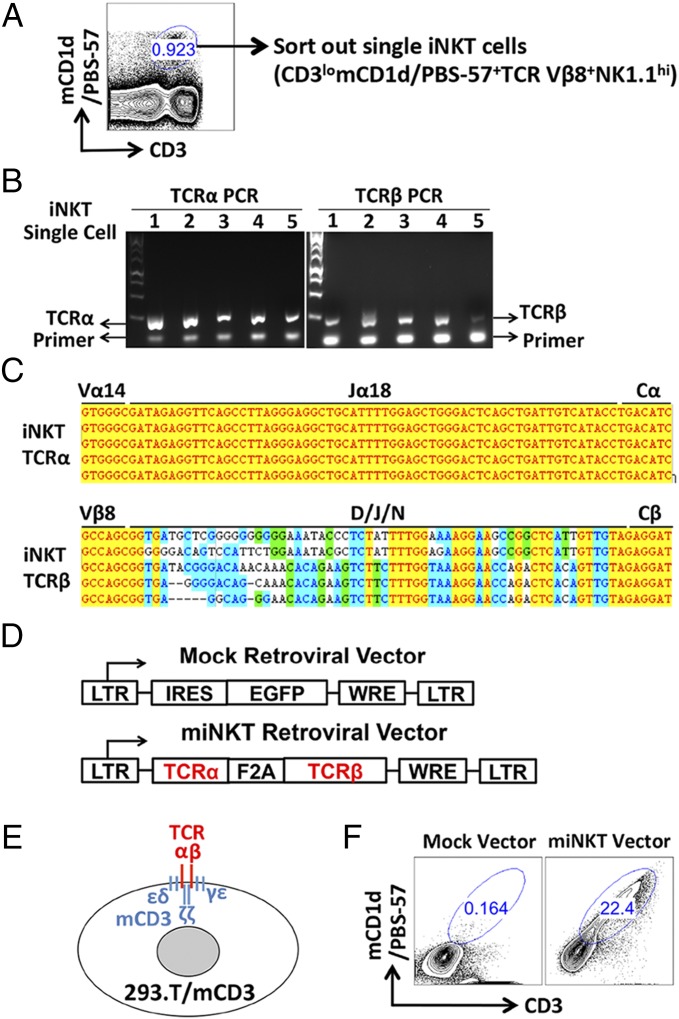
We used a robust and high-throughput single-cell TCR cloning technology recently established in our laboratories to obtain iNKT TCR genes (Materials and Methods). Single iNKT cells were sorted from mouse spleen cells using flow cytometry based on a stringent collection of surface markers gated as CD3lomCD1d/PBS-57+TCR Vβ8+NK1.1+ (Fig. 1A) (15). mCD1d/PBS-57 is the tetramer reagent that specifically identifies iNKT TCRs (16). We included TCR Vβ8 staining to focus on the dominant Vβ8+ population of mouse iNKT cells (1, 2). The sorted single iNKT cells were then subjected to TCR cloning (Fig. 1B). Several verified iNKT TCR α and β pairs were inserted into the murine stem cell virus (MSCV)-based retroviral vector to yield TCR gene delivery vectors (Fig. 1 C and D). Their vector-mediated expressions were then tested in 293.T/mCD3, a stable cell line that has been engineered to express mouse CD3 molecules that are required to support the surface display of mouse TCRs (Fig. 1E). One vector that mediated high expression of a high-affinity iNKT TCR was selected for the follow-up studies and was denoted as the miNKT vector (Fig. 1 D and F). The control MIG vector that encodes an EGFP reporter gene was denoted as the Mock vector (Fig. 1 D and F) (10).
Fig. 1.
Cloning of invariant natural killer T-cell receptor (iNKT TCR) genes and construction of retroviral delivery vectors. (A) Single iNKT cells were sorted out from mouse spleen cells using flow cytometry based on a stringent collection of surface markers (gated as CD3lomCD1d/PBS-57+TCR Vβ8+NK1.1hi). A representative FACS plot is shown. mCD1d/PBS-57 indicates the tetramer reagent that specifically stains mouse iNKT TCRs. (B) Sorted single iNKT cells were subjected to TCR cloning using a single-cell RT-PCR approach. Representative DNA gel pictures are presented showing the TCR α and β chain gene PCR products from five iNKT cells. (C) Representative sequencing results confirming the cloned single-cell iNKT TCR α and β chain genes. (D) Schematic representation of the retroviral vectors encoding either a control EGFP reporter gene (denoted as the Mock vector), or a pair of iNKT TCR α and β chain genes (denoted as the miNKT vector). LTR indicates long-term repeats; IRES, internal ribosome entry sites; EGFP, enhanced green fluorescence protein; F2A, foot-and-mouth disease virus 2A sequence; and WRE, woodchuck responsive element. (E) Schematic representation of the 293.T cell line that has been engineered to stably express mouse CD3 genes and so as to support the surface display of mouse TCRs (denoted as 293.T/mCD3). (F) Representative FACS plots showing the expression of clonal iNKT TCRs in 293.T/mCD3 cells transduced with the chosen miNKT vector.
Generation of Clonal iNKT Cells Through Genetic Engineering of HSCs.
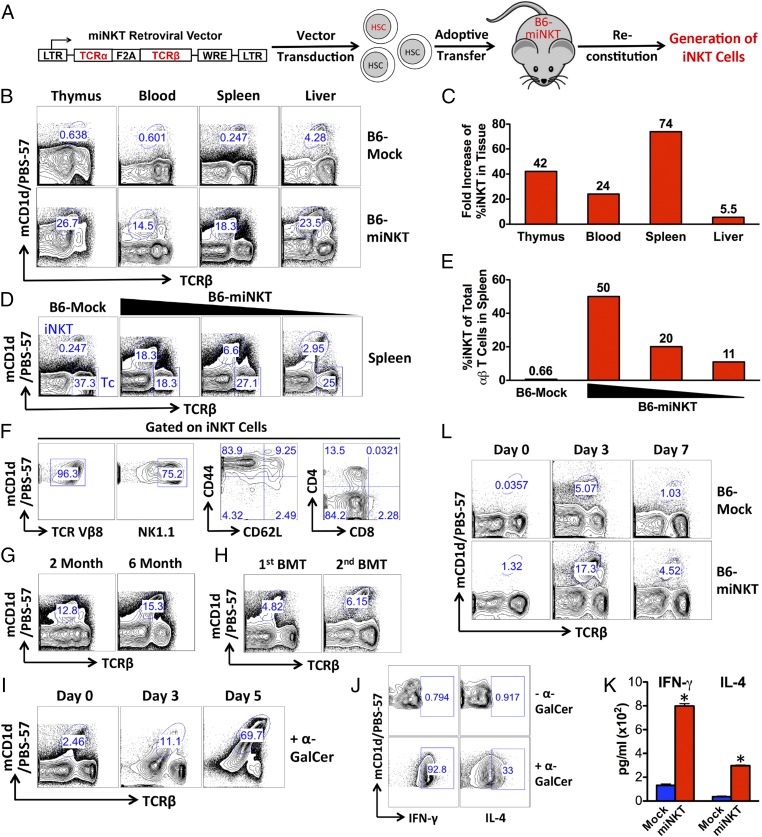
Following an established protocol (10), we performed miNKT-transduced bone marrow transfer in B6 mice to generate the recipient mice denoted as B6-miNKT (Fig. 2A). In brief, HSC-enriched bone marrow cells harvested from donor B6 mice were cultured in vitro, transduced with either Mock or miNKT retroviral vectors, then separately transferred into irradiated recipient B6 mice. The recipient mice were allowed to reconstitute their immune system over the course of 6–8 wk, followed by analysis to determine the presence of iNKT cells. Similar to the previously reported conventional αβ TCR engineering approach (10), we obtained desirable titers of the newly constructed miNKT retroviral vector [∼0.5–1 × 106 infectious units (IFU)/mL; Fig. S1] and achieved high efficiencies of HSC transduction (routinely over 50% of the cultured bone marrow cells). Compared with the Mock-engineered recipient mice, denoted as B6-Mock, we observed a significant increase of iNKT cells in the B6-miNKT mice from thymus to peripheral tissues, suggesting the successful generation of HSC-engineered iNKT cells (Fig. 2 B and C). Through titrating the miNKT vector-transduced HSCs used for bone marrow transfer, we were able to control the increase of the iNKT cells from as high as 50% of the total αβ T cells, down to a desired level in the B6-miNKT mice (Fig. 2 D and E). The ability to regulate the number of engineered iNKT cells can be valuable for clinical applications of this HSC-engineered iNKT cell strategy. Study of the iNKT cells from the B6-miNKT mice revealed that these iNKT cells displayed a typical phenotype of mouse iNKT cells in that they exhibited high expression of the NK1.1 marker, as well as a memory T-cell signature (CD62LloCD44hi) and a CD4+CD8− or CD4−CD8− coreceptor expression pattern (Fig. 2F) (17). Almost all of these iNKT cells showed positive staining for TCR Vβ8, indicating that they expressed the transgenic clonal iNKT TCR and suggesting that they were derived from the miNKT-engineered HSCs (Fig. 2F). The production of high levels of iNKT cells in the B6-miNKT mice persisted for up to 6 mo following the initial bone marrow transfer and also post secondary bone marrow transfer, highlighting the long-term effectiveness of this HSC-engineered iNKT cell strategy (Fig. 2 G and H).
Fig. 2.
Generation of functional iNKT cells through TCR gene engineering of hematopoietic stem cells (HSCs). B6 mice receiving adoptive transfer of HSCs transduced with either the Mock retroviral vector (denoted as B6-Mock mice) or miNKT retroviral vector (denoted as B6-miNKT mice) were allowed to reconstitute their immune system in a duration of 6–8 wk, followed by analysis. The experiments were repeated at least three times, and representative results are presented. iNKT cells were detected as TCRβlomCD1d/PBS-57+ using flow cytometry. (A) Schematic representation of the experimental design to generate HSC-engineered iNKT cells in mice. (B and C) Increase of iNKT cells in B6-miNKT mice compared with that in the control B6-Mock mice. (B) FACS plots showing the detection of iNKT cells in various tissues. (C) Bar graphs showing the fold increase of percent iNKT cells in the indicated tissues. (D and E) Control of iNKT cell numbers in B6-miNKT mice through titrating the miNKT vector-transduced HSCs used for adoptive transfer. (D) FACS plots showing the detection of iNKT cells in the spleens of various B6-miNKT recipient mice. Tc indicates the conventional αβ T cells (gated as TCRβ+mCD1d/PBS-57−). (E) Bar graphs showing the percent iNKT of total αβ T cells in spleen. (F) Phenotype of the HSC-engineered iNKT cells. FACS plots are presented showing the surface markers of iNKT cells detected in the liver of B6-miNKT mice. (G and H) Long-term production of HSC-engineered iNKT cells. FACS plots are presented showing the detection of iNKT cells in the spleen of B6-miNKT mice for up to 6 mo after initial HSC adoptive transfer (G) and at 2 mo after secondary bone marrow transfer (BMT) (H). (I–K) Functionality of the HSC-engineered iNKT cells tested in vitro. Spleen cells of B6-miNKT mice were cultured in vitro in the presence of α-GalCer (100 ng/mL). (I) FACS plots showing the time-course proliferation of iNKT cells. (J) FACS plots showing the cytokine production in iNKT cells on day 3, as measured by intracellular cytokine staining. (K) ELISA analysis of cytokine production in the cell culture medium at day 3. Data are presented as mean of duplicate cultures ± SEM, *P < 0.01 (B6-miNKT samples compared with the corresponding B6-Mock controls). (L) Functionality of the HSC-engineered iNKT cells tested in vivo. B6-Mock or B6-miNKT mice were given i.v. injection of 1 × 106 bone marrow-derived dendritic cells (BMDCs) loaded with α-GalCer (denoted as BMDC/α-GalCer) and then periodically bled to monitor iNKT cell responses. FACS plots are presented showing the change of iNKT cell frequencies in blood.
Functionality of the HSC-Engineered iNKT Cells.
We then analyzed the functionality of the HSC-engineered iNKT cells. When stimulated with the agonist glycolipid α-Galactosylceramide (α-GalCer) in vitro, the engineered iNKT cells proliferated vigorously by over 20-fold in 5 d and produced large amounts of the effector cytokines IFN-γ and IL-4 (Fig. 2 I–K). When B6-miNKT mice were immunized with bone marrow-derived dendritic cells (BMDCs) loaded with α-GalCer, the engineered iNKT cells mounted a strong and rapid response in vivo, expanding close to 20-fold in 3 d (Fig. 2L). Notably, the in vivo expansion of these cells peaked at day 3 postimmunization, compared with 7 d postimmunization for conventional αβ T cells (18). This speedy in vivo response is a signature of iNKT cells (17). These results indicate that the HSC-engineered iNKT cells are fully functional.
Development of the HSC-Engineered iNKT Cells.
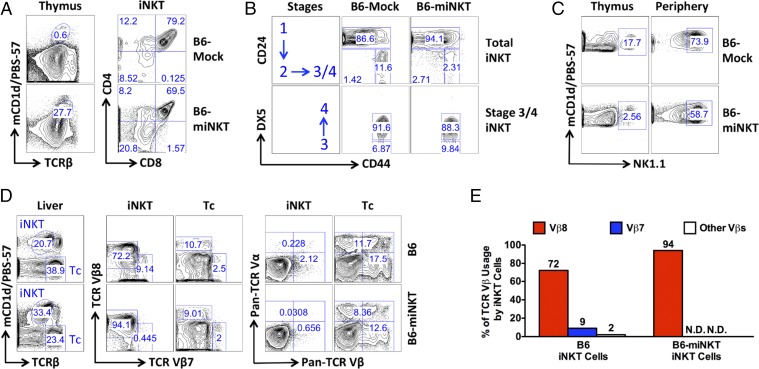
Next, we analyzed the development of the HSC-engineered iNKT cells. iNKT cell progenitors gated as TCRβlomCD1d/PBS-57+ were detected in the thymus of the B6-miNKT mice and were found to follow a classic developmental path similar to that observed for endogenous iNKT progenitor cells in the control B6-Mock mice (Fig. 3) (15). These progenitor cells appeared as CD4−CD8− (DN), CD4+CD8+ (DP), and CD4+CD8− (CD4 SP), corresponding with an iNKT development from DN to DP, then to CD4 SP or back to DN cells (Fig. 3A). The expression of CD24, CD44, and DX5 markers on iNKT progenitor cells further defined their development in thymus into four stages: Stage 1 (CD24+CD44−DX5−), Stage 2 (CD24−CD44−DX5−), Stage 3 (CD24−CD44+DX5−), and Stage 4 (CD24−CD44+DX5+) (15). Similar to their endogenous counterparts, HSC-engineered iNKT cell progenitors detected in the thymus of B6-miNKT mice followed a developmental path from Stages 1–4 (Fig. 3B). In addition to their development in thymus to gain TCR expression (Control Point 1), iNKT cells also differ from conventional αβ T cells in that they need to undergo an additional maturation step in the periphery to acquire the expression of NK1.1 (Control Point 2) (15). In B6-miNKT mice, iNKT cells detected in the periphery did up-regulate NK1.1 expression compared with iNKT cells detected in the thymus, similar to that observed for endogenous iNKT cells in the control B6-Mock mice (Fig. 3C).
Fig. 3.
Development of the HSC-engineered iNKT cells. B6-miNKT and control B6-Mock mice were analyzed for iNKT cell development at 6–8 wk post HSC transfer. The experiments were repeated at least three times, and representative results are presented. iNKT cells were detected as TCRβlomCD1d/PBS-57+ using flow cytometry. (A and B) FACS plots showing the characteristic development of iNKT cells in thymus. (C) FACS plots showing the maturation of iNKT cells in the periphery measured by the up-regulation of the NK1.1 marker. Comparisons of iNKT cells from thymus and periphery (liver) are shown. (D and E) FACS plots and bar graphs showing the exclusion of nontransgenic TCR expression on the HSC-engineered iNKT cells. Comparisons of iNKT and conventional αβ T (Tc) cells from the liver of B6-Mock or B6-miNKT mice are shown. Pan-TCR Vα panel includes Vα2, Vα3.2, and Vα8.3, whereas pan-TCR Vβ panel includes Vβ3, Vβ4, Vβ5, Vβ6, Vβ11, and Vβ13. N.D., not detected.
Overexpression of prerearranged αβ TCR genes in HSCs has been shown to induce allelic exclusion and block the rearrangements of endogenous TCR genes in the resulting conventional αβ T cells (13). Study of the iNKT cells generated in the B6-miNKT mice revealed that these cells expressed the transgenic TCR (Vβ8+), but not the other TCR Vβ chains analyzed in our experiment (Fig. 3 D and E). In particular, these HSC-engineered iNKT cells did not express the TCR Vβ7 used by ∼10% of endogenous iNKT cells (Fig. 3 D and E). Analysis of TCR α chain expression also showed an exclusion of other TCR Vα expression on the engineered iNKT cells (Fig. 3D). These results suggest that the iNKT TCR-engineered HSCs give rise to clonal iNKT cells that only express the transgenic iNKT TCRs, likely through an allelic exclusion mechanism during iNKT cell development in thymus.
We also studied the lineage differentiation of iNKT TCR-engineered HSCs. By detecting intracellular expression of transgenic iNKT TCRs (gated as Vβ8intra+), TCR-engineered HSCs and their progeny cells could be tracked (Fig. S2). Notably, because only T cells express the CD3 molecules that are required to support the surface display of TCRs and their signaling, the other cells that lack CD3 molecules can only express the transgenic iNKT TCRs intracellularly, and these TCRs are not functional. In addition to generating iNKT cells, our results show that TCR-engineered HSCs can also differentiate into all other blood cell lineages analyzed, including B cells (gated as CD19+), macrophages (gated as CD3−CD19−F4/80+), myeloid cells (gated as CD3−CD19−CD11b+), and granulocytes (gated as CD3−CD19−Gr-1+) (Fig. S2).
Antitumor Capacity of the HSC-Engineered iNKT Cells.
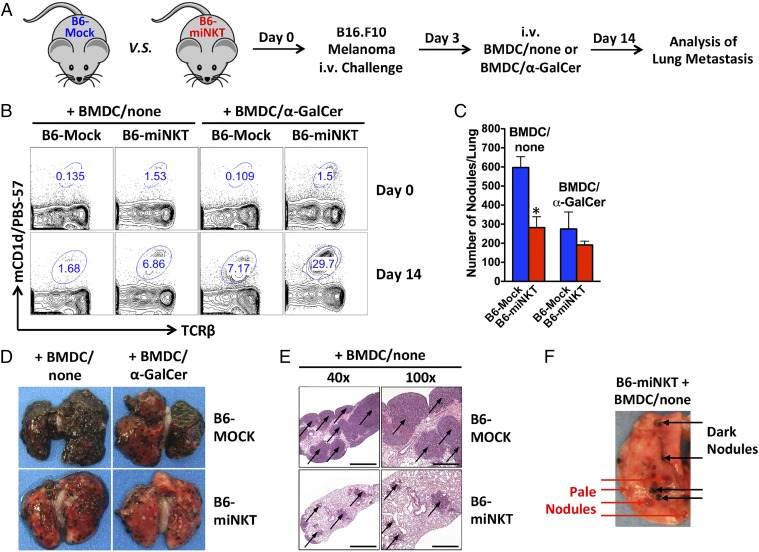
Finally, we studied the cancer therapy potential of the HSC-engineered iNKT cells. B6-miNKT mice and control B6-Mock mice were challenged with B16.F10 melanoma cells through i.v. injections and analyzed for lung metastasis 2 wk later (Fig. 4A). Experimental mice received immunization with either unloaded or α-GalCer–loaded BMDCs (denoted as BMDC/none or BMDC/α-GalCer, respectively) on day 3 post tumor challenge to boost iNKT cell activities and to mimic a therapeutic vaccination treatment (Fig. 4A). Monitoring of the HSC-engineered iNKT cells in the B6-miNKT mice showed that these cells actively responded to tumor challenge, evidenced by their expansion from ∼1.5% to ∼7% in blood (Fig. 4B). In comparison, endogenous iNKT cells in the control B6-Mock mice also responded to tumor challenge, but their limiting starting number (<0.2%) only allowed them to reach ∼1.7% in blood (Fig. 4B). We observed a significant protection from lung metastasis in the B6-miNKT mice compared with that in the control B6-Mock mice, as evidenced by the reduction of both the number and size of tumor nodules (Fig. 4 C–E). Inclusion of a BMDC/α-GalCer immunization further expanded the HSC-engineered iNKT cells (up to ∼30% in blood; Fig. 4B). However, no significant further reduction of lung tumor nodules was observed (Fig. 4C). We speculate that this may be due to a “saturation” of the antitumor capacity of iNKT cell-induced effector cells like NK cells and tumor-specific conventional αβ T cells that were limiting in mice. Total clearance of tumor metastasis likely requires combination therapy such as combining with adoptive transfer of additional effector cells. Notably, depigmentation of tumor nodules was observed in high numbers in the B6-iNKT mice (Fig. 4F). Key molecules in the pigment synthesis pathway are a major class of tumor antigens for melanoma, and mutating or down-regulating these molecules are common strategies by which melanoma tumor cells escape immune attack, often leading to depigmentation (19). The presence of a large fraction of depigmented tumor nodules in the B6-miNKT mice therefore suggests a strong immune response against these tumors, presumably induced by the HSC-engineered iNKT cells through activation of antitumor NK and conventional αβ T cells (Fig. 4F) (3, 4).
Fig. 4.
Protection from melanoma lung metastasis by the HSC-engineered iNKT cells. B6-miNKT and control B6-Mock mice were given i.v. injection of 0.5–1 × 106 B16.F10 melanoma cells on day 0 and analyzed for melanoma lung metastasis on day 14. On day 3, experimental mice received i.v. injection of 1 × 106 BMDCs either unloaded or loaded with α-GalCer (denoted as BMDC/none or BMDC/α-GalCer, respectively) to mimic a therapeutic vaccine treatment. The experiments were repeated twice (5–7 mice per group), and representative results are presented. (A) Schematic representation of the experimental design to study the cancer therapy potential of the HSC-engineered iNKT cells in the B16 melanoma lung metastasis mouse model. (B) FACS plots showing the expansion of iNKT cells in the blood of experimental mice in response to tumor challenge and BMDC/α-GalCer vaccination. (C–F) Analysis of melanoma lung metastasis in the experimental mice on day 14. (C) Enumeration of lung tumor nodules. Data are presented as mean ± SEM, *P < 0.01 (B6-miNKT samples compared with corresponding B6-Mock controls). (D) Photos of lung showing melanoma metastasis. (E) Immunohistology analysis showing the H-E staining of lung sections. Metastatic tumor nodules are indicated by arrows. Bars: 1,000 μm (40× magnification); 500 μm (100× magnification). (F) One image of lung from a representative B6-miNKT mouse showing the detection of pale, depigmented tumor nodules.
Discussion
In this report, we describe a new method of generating large numbers of iNKT cells in mice through iNKT TCR gene engineering of HSCs. Compared with existing iNKT TCR transgenic mouse technology and iNKT iPS cell-derived transgenic mouse technology, this new method is cost-effective and high-throughput. It is easy to implement through a standard retrovirus-transduced bone marrow transfer and has a fast turnover to generate iNKT cells within as few as 6 wk (Fig. 2). Most importantly, unlike transgenic mouse technologies, this method can be applied to humans through gene-modified CD34+ cell transfer and therefore has direct translational potential (20).
In our study, we showed that the HSC-engineered mouse iNKT cells followed a classical iNKT cell development path, Check Point 1 in the thymus to gain iNKT TCR expression and Check Point 2 in the periphery to gain NK1.1 expression (Fig. 3). They also displayed a typical iNKT cell phenotype (TCRβlomCD1d/PBS-57hiNK1.1hiCD62LloCD44hiCD4+/−CD8−) and exhibited full iNKT cell functionality with potent and fast response to antigen stimulation, both in vitro and in vivo (Fig. 2). These findings confirm the new HSC-engineered iNKT cell method as a powerful tool to study mouse iNKT cell biology. By increasing the iNKT cells to a suitable level, their development and function in health and disease conditions can be easily monitored. In particular, this method allows us to generate large numbers of clonal iNKT cells, thus enabling the investigation of similarities and differences between individual iNKT cell clones. For example, by studying the antigen recognition and functional differentiation of single iNKT cell clones, critical clues might be revealed to increase understanding of the origins of various iNKT cell subsets with distinct functions, such as those iNKT cell subsets biased to produce Th1, Th2, or Th17 effector cytokines (3). The flexibility of this method also allows the convenient generation of iNKT cells of different genomic backgrounds at a fast pace and an affordable cost, allowing examination of the functions of designated genes for regulating iNKT cell biology (8).
The therapeutic potential of this HSC-engineered iNKT cell approach is also promising. A broad range of applications, fast and strong responses, and the clinical availability of a potent stimulatory reagent α-GalCer make iNKT cells attractive therapeutic targets (4). In the past 2 decades, a series of iNKT cell-based clinical trials have been conducted, mainly targeting cancer (4, 21). A recent trial reported encouraging antitumor immunity in patients with head and neck squamous cell carcinoma, attesting to the potential of iNKT cell-based immunotherapies (22). However, most trials yielded unsatisfactory results (4, 21). Overall, these trials all worked through the direct stimulation or ex vivo expansion of patients’ endogenous iNKT cells, thus yielding only short-term, limited clinical benefits to a small number of patients. The low frequency and high variability of iNKT cells in humans (∼0.01–1% in blood), as well as the rapid depletion of these cells poststimulation, are considered to be the major stumbling blocks limiting the success of these trials. However, if successfully applied to humans, the reported new HSC-engineered iNKT cell approach has the potential to provide patients with a lifelong supply of therapeutic levels of iNKT cells, taking advantage of the longevity and self-renewal of HSCs (23), thus eliminating a key barrier against current iNKT cell-based immunotherapies. It is worthy to note that simply engineering conventional αβ T cells with iNKT TCR genes will not convert these cells into iNKT cells. The unique functions of iNKT cells can only be acquired during iNKT cell development, leaving HSC engineering the sole approach to produce functional engineered iNKT cells.
Materials and Methods
The full description of materials and methods is provided in SI Materials and Methods.
Single-Cell iNKT TCR Cloning.
The cloning method was performed based on an established protocol (24), with several modifications. Details are provided in SI Materials and Methods.
HSC Isolation, Transduction, Adoptive Transfer, and Secondary Bone Marrow Transfer.
The procedures were reported previously (10) and are provided in SI Materials and Methods.
Statistical Analysis.
Student’s two-tailed t test was used for paired comparisons. Data are presented as mean ± SEM, unless otherwise indicated. P < 0.01 was considered significant.
Supplementary Material
Acknowledgments
We thank the University of California, Los Angeles (UCLA), animal facility for providing animal support, the UCLA Broad Stem Cell Research Center (BSCRC) FACS Core for providing flow cytometry support, the UCLA Translational Pathology Core Laboratory (TPCL) for providing immunohistology support, the Laboratory of Donald Kohn, in particular, Eric Gschweng and Michael Kaufman, for providing technical help, the National Institutes of Health Tetramer Core Facility for providing the mCD1d/PBS-57 tetramer reagents, Donald Kohn and Gay Crooks for insightful discussion, and Pin Wang for critical reading of this manuscript. This work was supported by a UCLA BSCRC Innovation Award (to L.Y.), a UCLA SPORE in Prostate Cancer Career Development Award (NIH P50 CA092131, to L.Y.), a Concern Foundation Stem Cell Research Award (to L.Y.), a California Institute for Regenerative Medicine (CIRM) Basic Biology V Exploratory Concepts Award (RB5-07089, to L.Y.), and a National Institutes of Health (NIH) Director’s New Innovator Award (DP2 CA196335, to L.Y.). O.N.W. is an investigator of the Howard Hughes Medical Institute and is partially supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424877112/-/DCSupplemental.
References
- 1.Taniguchi M, Harada M, Kojo S, Nakayama T, Wakao H. The regulatory role of Valpha14 NKT cells in innate and acquired immune response. Annu Rev Immunol. 2003;21:483–513. doi: 10.1146/annurev.immunol.21.120601.141057. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 3.Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: Natural killer T cell defects and human disease. Nat Rev Immunol. 2011;11(2):131–142. doi: 10.1038/nri2904. [DOI] [PubMed] [Google Scholar]
- 4.Vivier E, Ugolini S, Blaise D, Chabannon C, Brossay L. Targeting natural killer cells and natural killer T cells in cancer. Nat Rev Immunol. 2012;12(4):239–252. doi: 10.1038/nri3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendelac A, Hunziker RD, Lantz O. Increased interleukin 4 and immunoglobulin E production in transgenic mice overexpressing NK1 T cells. J Exp Med. 1996;184(4):1285–1293. doi: 10.1084/jem.184.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi M, et al. Essential requirement of an invariant V alpha 14 T cell antigen receptor expression in the development of natural killer T cells. Proc Natl Acad Sci USA. 1996;93(20):11025–11028. doi: 10.1073/pnas.93.20.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren Y, et al. Generation of induced pluripotent stem cell-derived mice by reprogramming of a mature NKT cell. Int Immunol. 2014;26(10):551–561. doi: 10.1093/intimm/dxu057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Qin XF, Baltimore D, Van Parijs L. Generation of functional antigen-specific T cells in defined genetic backgrounds by retrovirus-mediated expression of TCR cDNAs in hematopoietic precursor cells. Proc Natl Acad Sci USA. 2002;99(9):6204–6209. doi: 10.1073/pnas.092154599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold PY, Burton AR, Vignali DA. Diabetes incidence is unaltered in glutamate decarboxylase 65-specific TCR retrogenic nonobese diabetic mice: Generation by retroviral-mediated stem cell gene transfer. J Immunol. 2004;173(5):3103–3111. doi: 10.4049/jimmunol.173.5.3103. [DOI] [PubMed] [Google Scholar]
- 10.Yang L, Baltimore D. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc Natl Acad Sci USA. 2005;102(12):4518–4523. doi: 10.1073/pnas.0500600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettini ML, Bettini M, Vignali DA. T-cell receptor retrogenic mice: A rapid, flexible alternative to T-cell receptor transgenic mice. Immunology. 2012;136(3):265–272. doi: 10.1111/j.1365-2567.2012.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vatakis DN, et al. Antitumor activity from antigen-specific CD8 T cells generated in vivo from genetically engineered human hematopoietic stem cells. Proc Natl Acad Sci USA. 2011;108(51):E1408–E1416. doi: 10.1073/pnas.1115050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giannoni F, et al. Allelic exclusion and peripheral reconstitution by TCR transgenic T cells arising from transduced human hematopoietic stem/progenitor cells. Mol Ther. 2013;21(5):1044–1054. doi: 10.1038/mt.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baltimore D, Witte ON, Yang L, Economou J, Ribas A. Overcoming barriers to programming a therapeutic cellular immune response to fight melanoma. Pigment Cell Melanoma Res. 2010;23(2):288–289. doi: 10.1111/j.1755-148X.2010.00666.x. [DOI] [PubMed] [Google Scholar]
- 15.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7(7):505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, et al. A modified alpha-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods. 2006;312(1-2):34–39. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Watarai H, Nakagawa R, Omori-Miyake M, Dashtsoodol N, Taniguchi M. Methods for detection, isolation and culture of mouse and human invariant NKT cells. Nat Protoc. 2008;3(1):70–78. doi: 10.1038/nprot.2007.515. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209(9):1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez-Montagut T, Turk MJ, Wolchok JD, Guevara-Patino JA, Houghton AN. Immunity to melanoma: Unraveling the relation of tumor immunity and autoimmunity. Oncogene. 2003;22(20):3180–3187. doi: 10.1038/sj.onc.1206462. [DOI] [PubMed] [Google Scholar]
- 20.Kohn DB, Pai SY, Sadelain M. Gene therapy through autologous transplantation of gene-modified hematopoietic stem cells. Biol Blood Marrow Transplant. 2013;19(1) Suppl:S64–S69. doi: 10.1016/j.bbmt.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 21.Pilones KA, Aryankalayil J, Demaria S. Invariant NKT cells as novel targets for immunotherapy in solid tumors. Clin Dev Immunol. 2012;2012:720803. doi: 10.1155/2012/720803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamasaki K, et al. Induction of NKT cell-specific immune responses in cancer tissues after NKT cell-targeted adoptive immunotherapy. Clin Immunol. 2011;138(3):255–265. doi: 10.1016/j.clim.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 24.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4(3):372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.