Significance
Hydrogenases are a source of environmentally benign bioenergy, catalyzing the reversible reduction of protons to form hydrogen. The most active subclass, the [FeFe]-hydrogenases, is dependent on a metallocofactor, the H cluster, which contains iron-bound CO and CN− ligands. Although the HydG maturase is known to catalytically form a CO- and CN−-bound iron precursor to the H cluster, mechanistic insight into this complex process has been hampered by the lack of structural information about HydG. We now describe the high-resolution crystal structure and EPR analysis of HydG. These results reveal a previously unreported [5Fe-5S] cluster that features a labile iron center proposed to provide the site of formation for a labile Fe(CO)2CN synthon, the precursor of the diiron subcluster hydrogenase H cluster.
Keywords: radical SAM enzyme, tyrosine lyase, H-cluster biosynthesis
Abstract
Hydrogenases use complex metal cofactors to catalyze the reversible formation of hydrogen. In [FeFe]-hydrogenases, the H-cluster cofactor includes a diiron subcluster containing azadithiolate, three CO, and two CN− ligands. During the assembly of the H cluster, the radical S-adenosyl methionine (SAM) enzyme HydG lyses the substrate tyrosine to yield the diatomic ligands. These diatomic products form an enzyme-bound Fe(CO)x(CN)y synthon that serves as a precursor for eventual H-cluster assembly. To further elucidate the mechanism of this complex reaction, we report the crystal structure and EPR analysis of HydG. At one end of the HydG (βα)8 triosephosphate isomerase (TIM) barrel, a canonical [4Fe-4S] cluster binds SAM in close proximity to the proposed tyrosine binding site. At the opposite end of the active-site cavity, the structure reveals the auxiliary Fe-S cluster in two states: one monomer contains a [4Fe-5S] cluster, and the other monomer contains a [5Fe-5S] cluster consisting of a [4Fe-4S] cubane bridged by a μ2-sulfide ion to a mononuclear Fe2+ center. This fifth iron is held in place by a single highly conserved protein-derived ligand: histidine 265. EPR analysis confirms the presence of the [5Fe-5S] cluster, which on incubation with cyanide, undergoes loss of the labile iron to yield a [4Fe-4S] cluster. We hypothesize that the labile iron of the [5Fe-5S] cluster is the site of Fe(CO)x(CN)y synthon formation and that the limited bonding between this iron and HydG may facilitate transfer of the intact synthon to its cognate acceptor for subsequent H-cluster assembly.
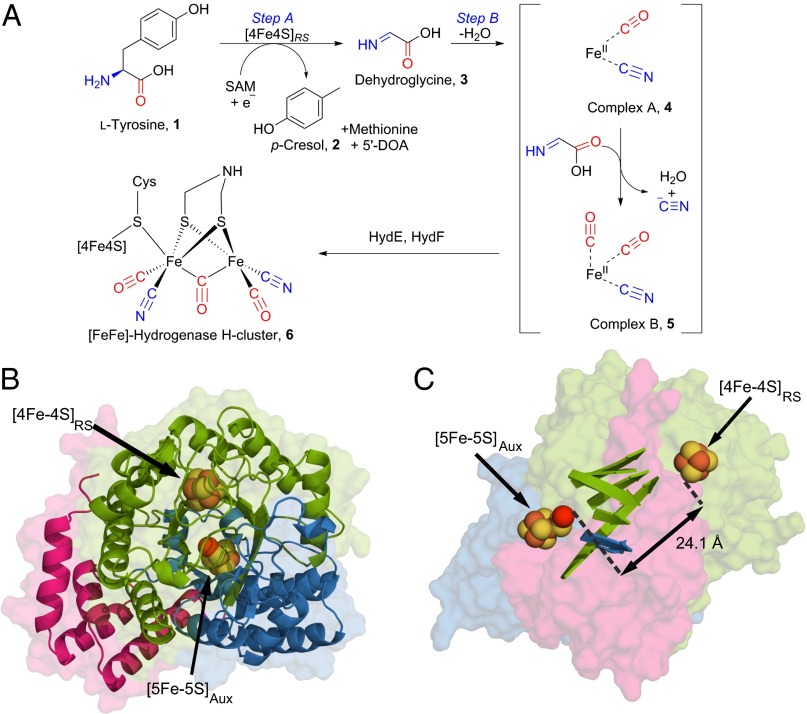
The assembly of the [FeFe]-hydrogenase diiron subcluster (1, 2) requires three maturase proteins, HydE, HydF, and HydG (3), and in vitro, they can assemble an active hydrogenase (4). The sequence and structure of the maturase HydE (5) indicates that it is a member of the radical S-adenosyl methionine (SAM) superfamily, although the biochemical function of HydE has not been experimentally determined. The GTPase HydF (6, 7) has been shown to transfer synthetic (8) or biologically derived (7, 9) diiron subclusters into apo-hydrogenase, suggesting that HydF functions as a template for diiron subcluster assembly. The tyrosine lyase HydG is also a member of the radical SAM superfamily and uses SAM and a reductant (such as dithionite) to cleave the Cα–Cβ bond of tyrosine, yielding p-cresol as the side chain-derived byproduct (10) and fragmenting the amino acid moiety into cyanide (CN−) (11) and carbon monoxide (CO) (12), which are ultimately incorporated as ligands in the H cluster of the [FeFe]-hydrogenase HydA (4). Two site-differentiated [4Fe-4S] clusters in HydG have been identified using a combination of spectroscopy and site-directed mutagenesis (12–16). The cluster bound close to the N terminus ([4Fe-4S]RS) by the CX3CX2C cysteine triad motif (SI Appendix, Fig. S1) is typical of the radical SAM superfamily (17, 18) and has been shown to catalyze the reductive cleavage of SAM (11, 13). The resultant highly reactive 5′-deoxyadenosyl radical is thought to abstract a hydrogen atom from tyrosine, thereby inducing Cα–Cβ-bond homolysis with release of dehydroglycine (DHG) and the spectroscopically characterized 4-oxidobenzyl radical anion (16), which is quenched to yield p-cresol (Fig. 1A, step A). The second (auxiliary) Fe-S cluster is proposed to promote the conversion of DHG into CO and CN− (Fig. 1A, step B) (13, 16). Two intermediates have been observed by stopped-flow IR spectroscopic analysis (19): an enzyme-bound organometallic species (complex A) (Fig. 1A, 4) that converts to a species that features an Fe(CO)2(CN) moiety (complex B) (Fig. 1A, 5). These results, combined with 57Fe electron-nuclear double resonance (ENDOR) studies that showed that iron from HydG is incorporated into mature hydrogenase, led to the proposal that an organometallic synthon with a minimum stoichiometry of [Fe(CO)2CN] is synthesized at the auxiliary cluster of HydG and eventually transferred to apo-hydrogenase (19).
Fig. 1.
Overall [FeFe]-hydrogenase H-cluster assembly and structure of TiHydG. (A) Formation of the Fe(CO)2CN synthon is proposed to occur at the auxiliary cluster of HydG (square brackets). (B) Overall fold of HydG with an end-on view of the TIM barrel showing the radical SAM core (green), the N-terminal extension (pink), and the C-terminal extension (blue). Monomer A is shown and contains a [4Fe-4S] cluster to catalyze the formation of the 5′-deoxyadenosyl radical from SAM and a [5Fe-5S] auxiliary cluster proposed to promote the conversion of DHG into cyanide and carbon monoxide. (C) The position of the two Fe-S clusters in TiHydG. The strands of the TIM barrel are shown. The orientation is rotated 90° from B.
Herein, we report the crystal structure of Thermoanaerobacter italicus HydG (TiHydG) complexed with SAM (the Protein Data Bank ID code for the structure of HydG is 4WCX). The structure, which contains two HydG monomers per asymmetric unit, reveals the auxiliary Fe-S cluster in two states: one monomer contains a [4Fe-5S] cluster, and the other monomer contains a structurally unprecedented [5Fe-5S] cluster consisting of a [4Fe-4S] cubane bridged by a μ2-sulfide to a mononuclear Fe(II) center (which we term the labile iron). To supplement the crystallographic studies of TiHydG, we also report EPR spectroscopic studies of Shewanella oneidensis HydG (SoHydG) that provide solution-state characterization of the [5Fe-5S] cluster and show its conversion to a [4Fe-4S] cluster in the presences of exogenous cyanide. Taken together, these results support a proposed mechanism for [FeFe]-hydrogenase maturation in which the labile iron of the [5Fe-5S] cluster is the site for Fe(CO)x(CN)y synthon assembly.
Results
TiHydG was heterologously expressed in Escherichia coli, purified, chemically reconstituted, and crystallized under strictly anaerobic conditions. Red-brown crystals of TiHydG diffracted to 1.59-Å resolution at the Diamond Light Source Beamline I03 (SI Appendix, Table S1), and the structure was solved by the single-wavelength anomalous diffraction method using data collected at the iron K edge (1.73891 Å) to reveal an asymmetric unit containing two HydG monomers related by a 90° rotation.
Structure of TiHydG.
The structure of TiHydG consists of a complete (βα)8 triosephosphate isomerase (TIM) barrel fold (Fig. 1B and SI Appendix, Fig. S2) flanked by helical N- and C-terminal extensions. In monomer A, residues 6–466 were modeled, and in monomer B, residues 4–466 were modeled, with the exception of a disordered region in the C-terminal domain (residues 345–351 in chain A and 348–360 in chain B) (SI Appendix, Fig. S2). A search for structural similarity (20) to HydG identified four radical SAM enzymes that are structurally most alike: the tryptophan lyase NosL (21), the [FeFe]-hydrogenase maturase HydE (5), methylornithine synthase (22), and biotin synthase (23); all five share a full (βα)8 TIM barrel and the two adjacent α-helices from the N-terminal extension. The HydG structure is exceptional in having a substantial (∼80 aa) C-terminal extension, which forms an additional helical domain bearing the auxiliary cluster.
In both TiHydG monomers, the separation of the [4Fe-4S]RS cluster and the auxiliary cluster is over 24 Å (Fig. 1C), a significantly greater separation than in other radical SAM enzymes bearing two cubane clusters [for example, RimO, 7.3 Å (24); LipA, 12.3 Å (25); BtrN, 15.8 Å (26); anSME and MoaA, 16.4 Å (27, 28)]. The Fe-S clusters and ligands in monomers A and B differ substantially, and the anomalous difference Fourier map calculated from data collected at the iron Κ edge was used to locate the iron atoms (Fig. 2 and SI Appendix, Fig. S1 and Table S2). In monomer A, the [4Fe-4S]RS cluster is bound to a methionine ligand by the α-amino and α-carboxy groups, which is likely formed by turnover of SAM in either the crystallization solution or the crystal itself. In monomer B, SAM is bound by the α-amino and α-carboxy groups, with the sulfonium sulfur atom poised 3.4 Å from the proximal iron. The interactions of SAM with the [4Fe-4S]RS cluster as well as several canonical sequence motifs, including the CX3CXϕC, the glycine-rich region GGE and the adenine structural binding motifs GxIGxxE and strand β-6 (SI Appendix, Fig. S1), are shared with other members of the radical SAM superfamily (17, 29).
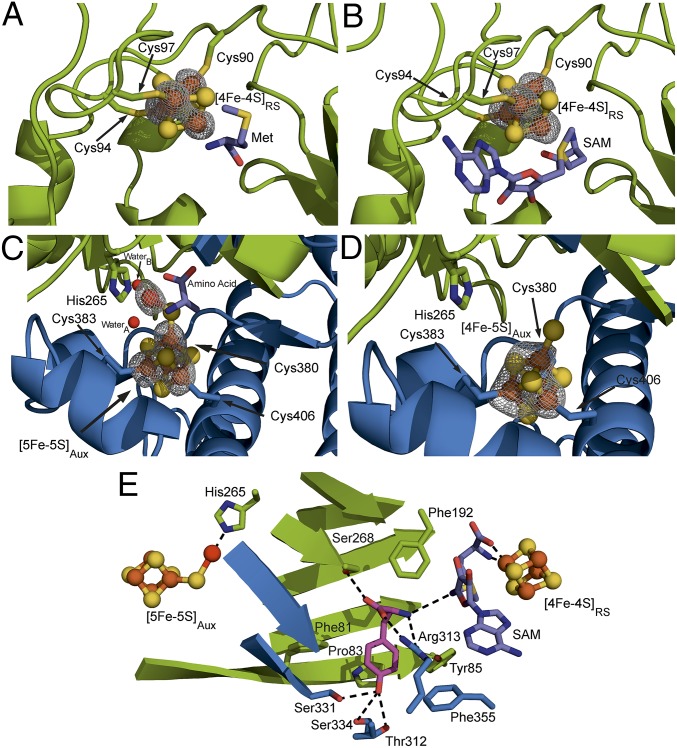
Fig. 2.
TiHydG Fe-S clusters and ligands. Residues from the radical SAM core are shown in green, and residues from the C-terminal domain are shown in blue. The labile iron is shown in bright red, tyrosine is in purple, and SAM is in lilac. Iron atoms were located using the anomalous difference Fourier map shown in gray and contoured in A–D at 5.0σ. (A) The [4Fe-4S]RS site of monomer A with methionine bound. (B) The [4Fe-4S]RS site of monomer B with SAM bound (same orientation as monomer A). (C) The [5Fe-5S]Aux cluster from monomer A showing the coordinating water molecules, amino acid ligand, and His265. (D) The [4Fe-5S]Aux cluster of monomer B. (E) A composite model of tyrosine and SAM binding to HydG.
For the auxiliary cluster, located at the far end of the TIM barrel but enclosed by the C-terminal helical domain, the positions of five iron atoms were identified in monomer A, whereas only four iron atoms were identified in monomer B (Fig. 2 C and D). Additional refinement of the structures confirmed the monomer A auxiliary cluster as a [5Fe-5S] cluster that can be regarded as a conventional [4Fe-4S] cubane ligated by three cysteine residues (380, 383, and 406) and connected to the fifth iron atom by a μ2 sulfide ion. There is significant asymmetry in the Fe-S bond lengths to the bridging sulfide ion, with the cluster to μ2 sulfide bond length being 2.2(5) Å and the labile iron to μ2 sulfide bond length being 2.5 Å (a long but not unprecedented interatomic distance) (30). The fifth iron has an estimated occupancy of 0.73 and adopts an approximately octahedral geometry, with a single protein-derived ligand, His265, positioned trans to the sulfide ion. This residue is highly conserved in HydG (SI Appendix, Fig. S1) but not present in the related radical SAM tyrosine lyases ThiH (31, 32) and CofH (33). The four remaining ligand sites around this iron are occupied by two water molecules and a nonproteinaceous amino acid bound in a bidentate manner through the α-amino and α-carboxy groups (ligand interaction distances are summarized in SI Appendix, Table S2). The side chain of the amino acid ligand is not sufficiently well-ordered to be identified and was modeled as alanine. The amino acid is unlikely to be tyrosine, because it was not included in the crystallization condition, but it may be methionine derived from SAM turnover or degradation during crystallization. Monomer B contains a [4Fe-5S] auxiliary cluster bound by the same three cysteine residues. Refinement with different ligands attached to the fourth iron atom gave a best fit for hydrosulfide with an occupancy of 0.79 and a typical distance for an iron-sulfur bond (∼2.3 Å). In contrast to monomer A, monomer B has no well-resolved density between the hydrosulfide and His265 (Fig. 2D and SI Appendix, Fig. S3D). The fifth iron’s partial occupancy in monomer A and absence in monomer B underscore its lability and suggest its viability as the site for Fe(CO)x(CN)y synthon formation and release.
EPR Spectroscopy.
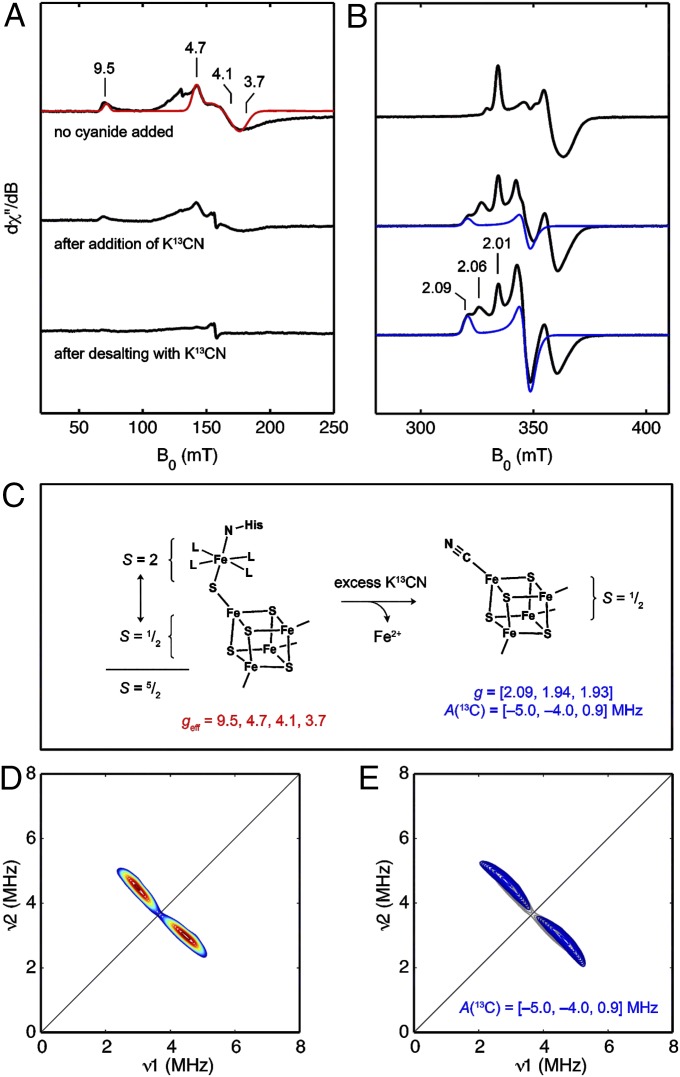
Because the auxiliary [5Fe-5S] cluster in HydG is structurally unprecedented and likely plays a central role in [FeFe]-hydrogenase maturation, we sought to supplement our crystallographic studies with solution-state EPR studies to characterize the electronic structure and reaction chemistry of this unique cluster. The EPR spectrum of reduced SoHydG in the presence of SAM displays two distinct signals (Fig. 3 A and B, Top): an S = 1/2 signal with g = [2.01, 1.88, 1.85] and an S = 5/2 signal with geff values of 9.5, 4.7, 4.1, and 3.7. The g = [2.01, 1.88, 1.85] signal has been previously identified as the SAM-bound form of the N-terminal [4Fe-4S] cluster (13, 16), and the S = 5/2 signal may be ascribed to the auxiliary [5Fe-5S] cluster on the basis of the experiments described below.
Fig. 3.
EPR spectroscopic studies of SoHydG. (A and B) X-band EPR spectra of SoHydG recorded under the following conditions: 9.38 GHz, 10 K, and (A) 5.00 or (B) 0.126 mW. Top represents dithionite-reduced HydG in the presence of 3 mM SAM. Middle represents dithionite-reduced HydG in the presence of 3 mM SAM and 20 mM K13CN. Bottom represents dithionite-reduced HydG after desalting three times with buffer containing 3 mM SAM, 15 mM K13CN, and 10 mM dithionite as described in the text. Red trace is the simulation of the S = 5/2 species assigned to the [5Fe-5S] cluster with D = +4.5 cm−1 and E/D = 0.255. Blue traces are the simulations of the species assigned to a 13CN-bound [4Fe-4S] cluster with g = [2.09, 1.94, 1.93] and σ = (0.03, 0.02, 0.04). The g1 components of the species described in the text are indicated above Bottom. (C) Exchange coupling scheme for an S = 5/2 [5Fe-5S] cluster and cyanide-induced [5Fe-5S] to [4Fe-4S] cluster conversion. (D) X-band HYSCORE spectrum of HydG after three successive desalting steps with buffer containing 10 mM dithionite, 3 mM SAM, and 15 mM K13CN recorded at g = 2.09. Data were recorded at 10 K and 9.73 GHz with π/2 = π = 16 ns and τ = 132 ns. (E) Simulation of the spectrum with A(13C) = [−5.0, −4.0, 0.9] MHz and Euler angles of [−90°, −40°, 0°] (blue); experimental data (gray contours) are reproduced for clarity.
To explain how the crystallographically observed [5Fe-5S] cluster could give rise to an S = 5/2 spin system, a simple exchange–coupling model (Fig. 3C) may be invoked, in which ferromagnetic coupling between an S = 1/2 [4Fe-4S]+ cluster and an S = 2 Fe2+ center gives rise to the observed spin system. (Antiferromagnetic coupling to give an S = 3/2 spin system may also be possible depending on the [4Fe-4S]-S-Fe geometry and other factors. Some of the intensity around geff = 5.5 could be ascribed to such a spin system.) Such an arrangement has parallels with a proposal for the geometric and electronic structure of the oxygen-evolving complex of photosystem II, in which a dangling manganese is coupled by a μ2 oxoligand to a strongly coupled Mn3Ca cubane (34). The zero-field splitting parameters for the S = 5/2 signal in SoHydG were determined by spectral simulation (35) of the EPR spectrum recorded at multiple temperatures (SI Appendix, Fig. S4). The rhombicity parameter, E/D = 0.255, can be ascertained from the geff values observed in the EPR spectrum (36) and was confirmed by spectral simulation. The geff value of the lowest field position (geff = 9.5) is less than 9.8, which means that this feature arises from the mS = ±1/2 doublet and that the zero-field splitting parameter D is, therefore, positive (36). Examination of the temperature dependence of the EPR spectrum allows for D to be estimated, because the populations of the mS = ±1/2 doublet (with signal intensity at geff = 9.5) and mS = ±3/2 doublet (with signal intensity at geff = 4.7, 4.1, 3.8) are dictated by the energy difference between the two manifolds, 2D, and follow the Boltzmann distribution. Simulations of these temperature-dependent data (SI Appendix, Fig. S4) give an approximate value of D = +4.5 cm−1. It is important to note that the S = 5/2 signal in SoHydG is not consistent with the highly rhombic signal for junk Fe3+ that is often observed by EPR spectroscopy. Moreover, because this S = 5/2 signal becomes more intense on reduction, it cannot arise from any reasonable mononuclear Fe3+ species and therefore, likely corresponds to a form of an iron-sulfur cluster.
To test the hypothesis that the S = 5/2 signal arises from the auxiliary cluster, we prepared a mutant (SoHydGXN), in which the three Cys residues that bind the N-terminal cluster are mutated to Ser residues (SI Appendix). This mutant, which binds only the auxiliary cluster, shows an S = 5/2 signal, with similar geff values to that observed in WT SoHydG (SI Appendix, Fig. S5). As expected, the S = 5/2 signal is absent from spectra of the previously studied SoHydG mutant (SoHydGSxxS) that cannot bind the auxiliary cluster, because two of the Cys residues that coordinate the auxiliary cluster are mutated to Ser residues (16).
Because iron lability is proposed to be a key feature of the HydG reaction (19), chemical conversion of the S = 5/2 [5Fe-5S] cluster to an S = 1/2 [4Fe-4S] cluster with concomitant loss of the labile iron was pursued. Treatment of HydG with dithionite, SAM, and K13CN results in a new set of S = 1/2 EPR signals (with g values of [2.09, 1.94, 1.93] and [2.06, 1.95, 1.93]) (SI Appendix, Fig. S6 shows a full simulation) and a decrease in intensity of the S = 5/2 signal assigned to the [5Fe-5S] C-terminal cluster (Fig. 3A, Middle). This speciation is indicative of a multicomponent equilibrium, wherein a fraction of the S = 5/2 species is converted to the two new S = 1/2 species with EPR properties that resemble [4Fe-4S] clusters (vide infra). Such an interpretation suggests that this equilibrium could be further driven toward the S = 1/2 species if SoHydG is diluted with buffer containing dithionite, SAM, and K13CN, then concentrated, and desalted to remove the labilized iron. Indeed, EPR spectra of solutions of SoHydG desalted three times in the manner described above show negligible intensity of the S = 5/2 signal intensity corresponding to the [5Fe-5S] cluster; only the S = 1/2 SAM-bound, N-terminal [4Fe-4S] cluster signal and the two new S = 1/2 signals are present (Fig. 3A, Bottom).
The g values of the new S = 1/2 signals are indicative of [4Fe-4S] clusters, and we pursued pulse EPR studies to further support this structural assignment. X-band HYSCORE spectra acquired at multiple field positions—including at g = 2.09, where only the g = [2.09, 1.94, 1.93] EPR signal has appreciable intensity—show intense cross-peaks centered at the 13C Larmor frequency for K13CN-treated samples (Fig. 3D and SI Appendix, Fig. S7). These cross-peaks are well-simulated (Fig. 3E and SI Appendix, Fig. S7) by a pseudoaxial hyperfine tensor, A(13C) = [−5.0, −4.0, 0.9], which corresponds to aiso = −2.7 MHz and T = 1.8 MHz, where A = aiso + [T1, T2, T3] and 2T ≡ T3 = −T1 − T2 (neglecting the slight rhombicity of the tensor). This hyperfine tensor is notably similar to that of the 13CN− ligand bound to the [4Fe-4S] cluster in Pyrococcus furiosus (Pf) ferredoxin: [−4.5, −4.5, 0.1] MHz corresponding to aiso = −2.9 MHz and T = 1.5 MHz (37). Moreover, the g tensor for the new CN-bound species in SoHydG (g = [2.09, 1.94, 1.93]) is nearly identical to that reported for the CN-bound form of Pf ferredoxin (g = [2.09, 1.95, 1.92]) (37). The spectral similarities between 13CN-treated WT HydG and 13CN-treated Pf ferredoxin support the structural assignment of the new species in WT SoHydG as a 13CN-bound [4Fe-4S] cluster. Importantly, the same EPR signal and 13C HYSCORE cross-peaks are observed in samples of SoHydGXN treated with K13CN (SI Appendix, Fig. S8), suggesting that this new 13CN-bound [4Fe-4S] cluster occupies the C-terminal cluster binding site. We have not yet unambiguously assigned the identity of the other new S = 1/2 signal with g = [2.06, 1.95, 1.93], although it could correspond to the crystallographically observed [4Fe-5S] cluster form of the auxiliary cluster or a CN-bound form of the [4Fe-4S]RS cluster.
Modeling Tyrosine Binding.
Cocrystallization of tyrosine bound at the active site of HydG was not achieved. It has been proposed that tyrosine binds to the auxiliary cluster of HydG (16). However, modeling of tyrosine bound to the auxiliary cluster in this structure leaves a gap of at least 12 Å between the tyrosine substrate and the 5′ carbon of SAM, the site of radical generation. Furthermore, we could not identify apposite residues to bridge this gap through a radical transfer pathway (38). We, therefore, considered an alternative binding site for tyrosine close to the SAM binding site and permitting direct hydrogen atom abstraction. Pocket analysis indicated an extended active-site cavity stretching between the iron-sulfur clusters of HydG and running approximately down the axis of the TIM barrel (Figs. 1C and 2E). The tyrosine ligand was modeled into the active-site pocket by superimposing the HydG crystal structure with that of NosL (21) and verifying that there was a comparable amino acid binding pocket capable of encapsulating tyrosine in the HydG structure. After tyrosine had been manually built into the pocket in the same pose as tryptophan in NosL, the ligand and neighboring residue side chains were minimized using the Merck molecular force field MMFF94 (39) to a gradient of 0.1 kcal/mol per angstrom. Crystallographic waters that overlapped with the tyrosine position were removed.
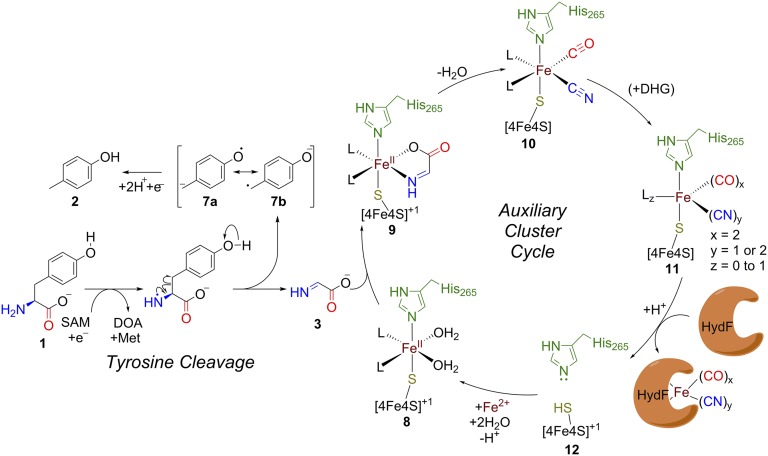
Analysis of the resulting structure identified a network of hydrogen bonds from Arg327 (SI Appendix, Fig. S1) (conserved in ThiH and HydG) to the tyrosine amino and carboxyl groups and potential hydrogen bonds to the side chains of Ser282, Ser345, and Thr326. The phenolic side chain occupies a pocket between the side chains of Phe95 and Phe369. This model positions the tyrosine near to SAM, with a distance between the 5′ carbon of SAM and the amino group of tyrosine of 5.1 Å, only slightly longer than that observed (3.7–4.1 Å) in substrate complex structures of other radical SAM enzymes (23, 26, 40, 41). This arrangement would enable a tyrosine cleavage reaction, in which the SAM-derived 5′-deoxyadenosyl radical abstracts an α-amino hydrogen atom (by analogy to a recent mechanistic proposal for NosL) (21) followed by homolysis of the tyrosine Cα–Cβ bond (Fig. 4) to yield the spectroscopically characterized (16) 4-oxidobenzyl radical anion 7a↔7b and DHG 3. This model includes an unobstructed pathway from the proposed site of DHG formation to the auxiliary iron sulfur cluster. Such a mechanism is consistent with the observations that both Clostridium acetobutylicum HydGΔCTD (a truncated HydG lacking the C-terminal domain and associated auxiliary cluster) (15) and ThiH are able to cleave tyrosine to p-cresol and DHG (step A in Fig. 1A) (13, 32), suggesting that the auxiliary cluster is not required for the initial tyrosine cleavage step.
Fig. 4.
Working mechanistic model of HydG catalysis. The 5′-deoxyadenosyl radical initiates tyrosine cleavage. The resultant DHG may then be captured by the labile iron of the auxiliary cluster and rearranged to Fe-bound CO and CN−. Turnover of a second equivalent of DHG has been observed by stopped-flow IR (19) to yield an organometallic synthon 11, which can be transferred to the cognate acceptor. For the intermediates 8–11, the ligands (L) to the labile iron cannot be unequivocally assigned from our results but may include water or amino acid side chains that line the active-site cavity. DOA, deoxyadenosine.
Discussion
The X-ray structure of HydG reveals that the auxiliary cluster binding site can accommodate either a [5Fe-5S] or a [4Fe-5S] cluster. EPR studies support the presence of the unprecedented [5Fe-5S] cluster in solution and show the lability of the fifth iron on addition of exogenous CN−. Previous work has shown that the auxiliary cluster of reconstituted HydG can bind an S = 1/2 [4Fe-4S] cluster (13, 16) rather than an S = 5/2 [5Fe-5S] cluster as suggested by the crystallographic and EPR data presented here. These seemingly conflicting observations may be reconciled by ascribing the previously reported S = 1/2 EPR signal for the auxiliary cluster to a [4Fe-4S] or [4Fe-5S] form, in which the labile iron is not present.
The discovery that HydG can accommodate [4Fe-5S] or [5Fe-5S] as the auxiliary cluster sheds light on how Fe(CO)x(CN)y moieties may be formed en route to the mature H cluster. Whereas previous studies (19) pointed to formation of Fe(CO)x(CN)y intermediates by decoration of the site-differentiated iron of a [4Fe-4S] auxiliary cluster with CO and CN−, the structural and spectroscopic data presented here suggest a working hypothesis that involves elaboration of the labile iron of a [5Fe-5S] cluster (Fig. 4). The labile iron of the auxiliary cluster nearly occludes the channel between the iron sulfur clusters of HydG (Fig. 2E) and is, therefore, well-positioned to capture the intermediate DHG (Fig. 4, 3→9), potentially binding it in a bidentate manner resembling the amino acid ligand observed in monomer A (Fig. 2C and SI Appendix, Fig. S3C). With only one sulfide and one histidine ligand, the labile iron can, in principle, retain approximately octahedral geometry while accommodating up to four diatomic ligands, leading to complexes 10 and 11 (complexes A and B as observed previously by stopped-flow IR) (19).
With only one proteinacious ligand to the labile iron—the highly conserved His265—it is likewise poised for release to its accepter. Two mechanisms can be envisaged by which an acceptor protein can access the newly synthesized Fe(CO)xCNy moiety. In one model, a local rearrangement of the helical region enclosing and supporting the auxiliary cluster (approximately residues 379–421) may reveal the labile iron. Alternatively, binding of the requisite diatomic ligands to the labile iron may result in the release of either the μ2-sulfide or His265 ligands, leaving only the peptide chain linking the TIM barrel to the C-terminal domain. Pivoting on a putative hinge region (approximately residues 373–376) between the radical SAM core and C-terminal domain would expose the interdomain interface, including the labile iron.
The structure also gives clues as to how tyrosine cleavage might proceed. Although we did not obtain cocrystals of HydG and tyrosine, computational analysis of the active-site cavity within the TIM barrel identified a putative tyrosine binding site with the α-amino H atom oriented toward the [4Fe-4S]RS cluster and the amino acid group poised at the start of the channel leading to the labile iron, the proposed binding site for the DHG intermediate. The (βα)8 TIM barrel fold permits the isolation of small molecule substrates and radical intermediates from the external medium [in BioB (23) and PylB (22), for example]. In HydG, the modeled tyrosine is fully enclosed, and the hydrophobic nature of this region of the cavity may help to exclude water, thereby protecting DHG from hydrolysis. In a conceptually related manner, ThiH forms a noncovalent complex with the next enzyme in the thiazole biosynthetic pathway, ThiG (42), an arrangement that is proposed to sequester the DHG away from the aqueous medium and permit direct transfer of DHG from ThiH to ThiG (31).
Finally, it is interesting to note the relationship between the maturation of [FeFe]-hydrogenases and that of [NiFe]-hydrogenases (43). For the latter, it is thought that two CN− ligands and one CO ligand are delivered in a stepwise fashion to a mononuclear iron site on a complex of HypC and HypD before insertion into the large subunit of apo–[NiFe]-hydrogenase (44, 45). The labile iron in the [5Fe-5S] cluster of HydG is proposed to be the analogous site for [FeFe]-hydrogenase maturation, with one significant difference being that the iron scaffold in HydG is bound to a [4Fe-4S] cluster by a bridging sulfide. One possible role for this electronically coupled [4Fe-4S] cluster is to act as a redox partner to the labile iron, which may participate directly in the conversion of DHG to CO, CN−, and water. In this manner, two-electron reactions at the labile iron (e.g., oxidative addition)—which otherwise may be challenging for a mononuclear iron site—could be facilitated, whereby one electron is supplied by the labile iron and one is supplied by the [4Fe-4S] subcluster. Additional experimental and computational work is underway to investigate this possibility.
We anticipate that the results described in this report will stimulate additional research to understand the precise mechanism of CO and CN− formation and the downstream processing of the organometallic product of HydG to yield the H cluster. Particular challenges include understanding how HydG interfaces with other H-cluster biosynthetic proteins to permit organometallic synthon transfer as well as the exact nature of the transferred synthon, the unequal numbers of equivalents of CO and CN− observed in the mature H cluster (despite HydG apparently yielding CO and CN− in a 1:1 ratio), and the origin of the azadithiolate ligand (10).
Materials and Methods
Recombinant TiHydG was expressed in E. coli BL21(DE3) cells. All steps involving HydG solutions were maintained under anaerobic conditions. The protein was purified using nickel affinity agarose and S-75 Superdex chromatography. The protein was reconstituted (23) and crystallized by the sitting-drop vapor diffusion method. Recombinant SoHydG samples (WT and mutants) were prepared as described previously (46). The structure was determined by single-wavelength anomalous diffraction at the iron Κ edge. The modeled tyrosine was manually inserted based on pocket analysis performed on the crystal structure with SAM present. Continuous wave and pulse EPR experiments were performed at low temperature using Bruker Biospin EleXys E500 and E580 spectrometers, respectively. More detailed experimental protocols can be found in SI Appendix, Methods.
Note Added in Proof.
While this paper was under revision, Nicolet et al. (47) reported the crystal structure of Carboxydothermus hydrogenoformans HydG containing the [4Fe4S]RS.
Supplementary Material
Acknowledgments
We thank the staff at the Diamond Light Source Synchrotron for assistance and access to Beamline I03 under Proposal 8889. The research was supported by the University of Southampton, National Institutes of Health Grants F32GM111025 (to D.L.M.S.) and GM104543 (to R.D.B.), Division of Material Sciences and Engineering Grant DE-FG02-09ER46632 (to J.R.S.), and Biotechnology and Biological Sciences Research Council Grant BB/J017302/1 (to J.W.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4WCX).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417252112/-/DCSupplemental.
References
- 1.Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC. Desulfovibrio desulfuricans iron hydrogenase: The structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7(1):13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 2.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282(5395):1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 3.Posewitz MC, et al. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem. 2004;279(24):25711–25720. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 4.Kuchenreuther JM, George SJ, Grady-Smith CS, Cramer SP, Swartz JR. Cell-free H-cluster synthesis and [FeFe] hydrogenase activation: All five CO and CN⁻ ligands derive from tyrosine. PLoS ONE. 2011;6(5):e20346. doi: 10.1371/journal.pone.0020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolet Y, et al. X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima. J Biol Chem. 2008;283(27):18861–18872. doi: 10.1074/jbc.M801161200. [DOI] [PubMed] [Google Scholar]
- 6.Brazzolotto X, et al. The [Fe-Fe]-hydrogenase maturation protein HydF from Thermotoga maritima is a GTPase with an iron-sulfur cluster. J Biol Chem. 2006;281(2):769–774. doi: 10.1074/jbc.M510310200. [DOI] [PubMed] [Google Scholar]
- 7.Shepard EM, et al. Synthesis of the 2Fe subcluster of the [FeFe]-hydrogenase H cluster on the HydF scaffold. Proc Natl Acad Sci USA. 2010;107(23):10448–10453. doi: 10.1073/pnas.1001937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berggren G, et al. Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature. 2013;499(7456):66–69. doi: 10.1038/nature12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGlynn SE, et al. HydF as a scaffold protein in [FeFe] hydrogenase H-cluster biosynthesis. FEBS Lett. 2008;582(15):2183–2187. doi: 10.1016/j.febslet.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 10.Pilet E, et al. The role of the maturase HydG in [FeFe]-hydrogenase active site synthesis and assembly. FEBS Lett. 2009;583(3):506–511. doi: 10.1016/j.febslet.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Driesener RC, et al. [FeFe]-hydrogenase cyanide ligands derived from S-adenosylmethionine-dependent cleavage of tyrosine. Angew Chem Int Ed Engl. 2010;49(9):1687–1690. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 12.Shepard EM, et al. [FeFe]-hydrogenase maturation: HydG-catalyzed synthesis of carbon monoxide. J Am Chem Soc. 2010;132(27):9247–9249. doi: 10.1021/ja1012273. [DOI] [PubMed] [Google Scholar]
- 13.Driesener RC, et al. Biochemical and kinetic characterization of radical S-adenosyl-L-methionine enzyme HydG. Biochemistry. 2013;52(48):8696–8707. doi: 10.1021/bi401143s. [DOI] [PubMed] [Google Scholar]
- 14.Rubach JK, Brazzolotto X, Gaillard J, Fontecave M. Biochemical characterization of the HydE and HydG iron-only hydrogenase maturation enzymes from Thermatoga maritima. FEBS Lett. 2005;579(22):5055–5060. doi: 10.1016/j.febslet.2005.07.092. [DOI] [PubMed] [Google Scholar]
- 15.Nicolet Y, Martin L, Tron C, Fontecilla-Camps JC. A glycyl free radical as the precursor in the synthesis of carbon monoxide and cyanide by the [FeFe]-hydrogenase maturase HydG. FEBS Lett. 2010;584(19):4197–4202. doi: 10.1016/j.febslet.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Kuchenreuther JM, et al. A radical intermediate in tyrosine scission to the CO and CN- ligands of FeFe hydrogenase. Science. 2013;342(6157):472–475. doi: 10.1126/science.1241859. [DOI] [PubMed] [Google Scholar]
- 17.Vey JL, Drennan CL. Structural insights into radical generation by the radical SAM superfamily. Chem Rev. 2011;111(4):2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29(5):1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchenreuther JM, et al. The HydG enzyme generates an Fe(CO)2(CN) synthon in assembly of the FeFe hydrogenase H-cluster. Science. 2014;343(6169):424–427. doi: 10.1126/science.1246572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolet Y, Zeppieri L, Amara P, Fontecilla-Camps JC. Crystal structure of tryptophan lyase (NosL): Evidence for radical formation at the amino group of tryptophan. Angew Chem Int Ed Engl. 2014;53(44):11840–11844. doi: 10.1002/anie.201407320. [DOI] [PubMed] [Google Scholar]
- 22.Quitterer F, List A, Eisenreich W, Bacher A, Groll M. Crystal structure of methylornithine synthase (PylB): Insights into the pyrrolysine biosynthesis. Angew Chem Int Ed Engl. 2012;51(6):1339–1342. doi: 10.1002/anie.201106765. [DOI] [PubMed] [Google Scholar]
- 23.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303(5654):76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forouhar F, et al. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat Chem Biol. 2013;9(5):333–338. doi: 10.1038/nchembio.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmer JE, et al. Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem J. 2014;464(1):123–133. doi: 10.1042/BJ20140895. [DOI] [PubMed] [Google Scholar]
- 26.Goldman PJ, Grove TL, Booker SJ, Drennan CL. X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc Natl Acad Sci USA. 2013;110(40):15949–15954. doi: 10.1073/pnas.1312228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman PJ, et al. X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc Natl Acad Sci USA. 2013;110(21):8519–8524. doi: 10.1073/pnas.1302417110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann P, Schindelin H. Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci USA. 2006;103(18):6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowling DP, Vey JL, Croft AK, Drennan CL. Structural diversity in the AdoMet radical enzyme superfamily. Biochim Biophys Acta. 2012;1824(11):1178–1195. doi: 10.1016/j.bbapap.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macedo S, et al. Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation. J Biol Inorg Chem. 2002;7(4-5):514–525. doi: 10.1007/s00775-001-0326-y. [DOI] [PubMed] [Google Scholar]
- 31.Challand MR, Martins FT, Roach PL. Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli. J Biol Chem. 2010;285(8):5240–5248. doi: 10.1074/jbc.M109.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriek M, Martins F, Challand MR, Croft A, Roach PL. Thiamine biosynthesis in Escherichia coli: Identification of the intermediate and by-product derived from tyrosine. Angew Chem Int Ed Engl. 2007;46(48):9223–9226. doi: 10.1002/anie.200702554. [DOI] [PubMed] [Google Scholar]
- 33.Decamps L, et al. Biosynthesis of F0, precursor of the F420 cofactor, requires a unique two radical-SAM domain enzyme and tyrosine as substrate. J Am Chem Soc. 2012;134(44):18173–18176. doi: 10.1021/ja307762b. [DOI] [PubMed] [Google Scholar]
- 34.Peloquin JM, et al. 55Mn ENDOR of the S2-state multiline EPR signal of photosystem II: Implications on the structure of the tetranuclear Mn cluster. J Am Chem Soc. 2000;122(44):10926–10942. [Google Scholar]
- 35.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178(1):42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Hagen WR. EPR spectroscopy of iron—sulfur proteins. In: Cammack R, editor. Advances in Inorganic Chemistry. Vol 38. Elsevier; Amsterdam: 1992. pp. 165–222. [Google Scholar]
- 37.Telser J, et al. Cyanide binding to the novel 4Fe ferredoxin from Pyrococcus furiosus - investigation by EPR and ENDOR spectroscopy. J Am Chem Soc. 1995;117(18):5133–5140. [Google Scholar]
- 38.Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc Chem Res. 2013;46(11):2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17(5-6):490–519. [Google Scholar]
- 40.Vey JL, et al. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc Natl Acad Sci USA. 2008;105(42):16137–16141. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. The x-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc Natl Acad Sci USA. 2005;102(39):13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonardi R, Fairhurst SA, Kriek M, Lowe DJ, Roach PL. Thiamine biosynthesis in Escherichia coli: Isolation and initial characterisation of the ThiGH complex. FEBS Lett. 2003;539(1-3):95–99. doi: 10.1016/s0014-5793(03)00204-7. [DOI] [PubMed] [Google Scholar]
- 43.Lubitz W, Ogata H, Rüdiger O, Reijerse E. Hydrogenases. Chem Rev. 2014;114(8):4081–4148. doi: 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- 44.Bürstel I, et al. A universal scaffold for synthesis of the Fe(CN)2(CO) moiety of [NiFe] hydrogenase. J Biol Chem. 2012;287(46):38845–38853. doi: 10.1074/jbc.M112.376947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stripp ST, et al. HypD is the scaffold protein for Fe-(CN)2CO cofactor assembly in [NiFe]-hydrogenase maturation. Biochemistry. 2013;52(19):3289–3296. doi: 10.1021/bi400302v. [DOI] [PubMed] [Google Scholar]
- 46.Kuchenreuther JM, et al. High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli. PLoS ONE. 2010;5(11):e15491. doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolet Y, et al. 2014 Crystal structure of HydG from Carboxydothermus hydrogenoformans: A trifunctional [FeFe]-hydrogenase maturase. Chembiochem, 10.1002/cbic.201402661. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.