Significance
MHC class I molecules select and present a limited set of peptides from a broad repertoire provided by TAP. How MHC class I makes this selection is unclear. We show that MHC class I H-2Kb molecules initially bind many peptides because of highly flexible binding pockets. Peptide binding is followed by a selection step wherein a large fraction of these peptides is released, leaving the canonical peptides for presentation. The peptide presentation has a remarkable temperature dependency and explains the low-affinity peptides found associated to MHC class I molecules in cells cultured at low temperature. Our data suggest that MHC class I goes through rounds of considering and rejecting peptides until peptides with high affinity are acquired for presentation.
Keywords: antigen presentation, peptide binding, anchor residues, dynamics, entropy
Abstract
MHC class I molecules present a variable but limited repertoire of antigenic peptides for T-cell recognition. Understanding how peptide selection is achieved requires mechanistic insights into the interactions between the MHC I and candidate peptides. We find that, at first encounter, MHC I H-2Kb considers a wide range of peptides, including those with expanded N termini and unfitting anchor residues. Discrimination occurs in the second step, when noncanonical peptides dissociate with faster exchange rates. This second step exhibits remarkable temperature sensitivity, as illustrated by numerous noncanonical peptides presented by H-2Kb in cells cultured at 26 °C relative to 37 °C. Crystallographic analyses of H-2Kb–peptide complexes suggest that a conformational adaptation of H-2Kb drives the decisive step in peptide selection. We propose that MHC class I molecules consider initially a large peptide pool, subsequently refined by a temperature-sensitive induced-fit mechanism to retain the canonical peptide repertoire.
MHC class I molecules present a wide array of peptides to cytotoxic T lymphocytes, allowing the immune system to scan for intracellular pathogens and mutated proteins. These peptides are not chosen at random, but rather are selected for their ability to bind to the polymorphic MHC class I peptide-binding groove. Antigenic peptide precursors are produced by the proteasome and further trimmed by cytosolic aminopeptidases. They are translocated into the endoplasmic reticulum (ER) lumen by the peptide transporter TAP that has broad peptide specificity. Peptides can be further trimmed in the ER lumen by ER aminopeptidases before selection by a defined MHC class I allele (reviewed in ref. 1). Only few peptides from a broad peptidome are presented by a given MHC class I allele. How a defined MHC I allele selects the correct peptides for presentation out of a large and diverse peptide pool is unclear.
In principle, MHC class I molecules could consider only “optimal” peptides and ignore the remainder of the TAP-translocated peptidome. Alternatively, MHC class I could bind all available peptides followed by a selection step for the optimal candidates for presentation. To discriminate between these scenarios, we studied peptide association (on-rates) and dissociation (off-rates) from the mouse MHC class I molecule H-2Kb. To characterize the biophysical basis of discrimination between candidate peptides we complemented the kinetic data with five crystal structures of H-2Kb with peptide variants. A comprehensive analysis of our in vitro data indicated that discrimination against suboptimal peptides by MHC class I may exhibit a strong temperature dependency, as illustrated by the H-2Kb peptidome from cells cultured at 26 °C versus 37 °C. We thus arrive at a two-step model for peptide selection by MHC class I molecules, which explains how not so “empty MHC class I molecules come out in the cold” (2).
Results
Expanding the H-2Kb–Associated Peptidome Under Experimental Conditions.
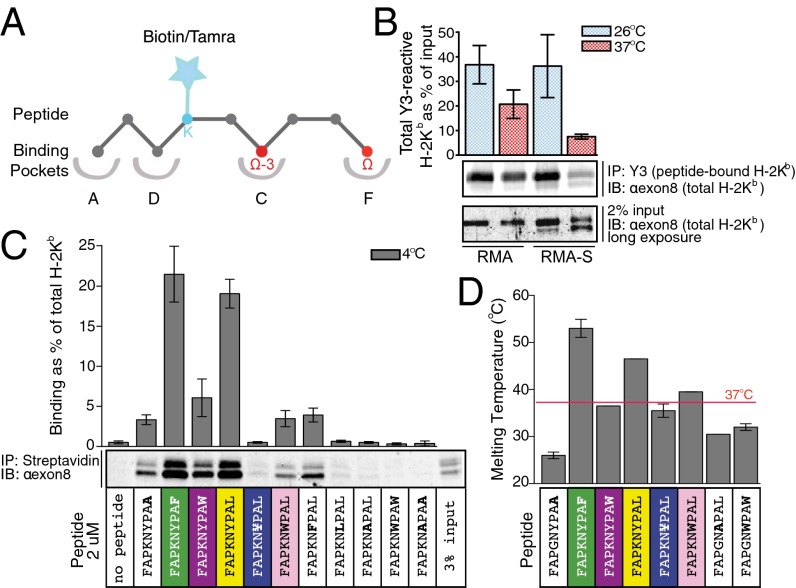
Peptide loading is a limiting factor for MHC class I complex assembly in cells (3). To study the initial steps of this process, we aimed to alleviate such limitation experimentally. First, the effect of temperature on the production of H-2Kb–peptide complexes in vivo was assessed, using TAP-deficient (RMA-S) and control (RMA) mouse leukemia cells. The levels of peptide-bound H-2Kb increased significantly in RMA-S and RMA cells when cultured at 26 °C compared with 37 °C. (Fig. 1B), as observed previously (2, 4, 5).
Fig. 1.
Binding and stability of H-2Kb in association with suboptimal peptides. (A) Schematic representation of H-2Kb binding groove with 9-mer peptide. Anchor residues at position P(Ω–3) and P(Ω) (red) bind into the C and F pockets, respectively. Residue P4 was exchanged to Lys (cyan) to attach biotin/TAMRA for isolation/detection purposes. (B) RMA and RMA-S cells were cultured at 26 °C or 37 °C and lysed and peptide-bound forms of H-2Kb were isolated using Y3 antibody and examined by immunoblotting (IB) with anti-exon8 antibody (recognizing all forms of H-2Kb). Efficiency of peptide binding is expressed as Y3-reactive H-2Kb as percentage of total H-2Kb. A representative experiment is shown; quantification represents the average ± SD of two independent experiments. (C) Peptides with nonclassical anchor residues bind to H-2Kb in lysates. RMA-S cells cultured at 37 °C were lysed and incubated with biotinylated peptide. Peptides used were FAPKNYPAL or its derivatives: FAPKNXPAL [where X is 3,5-diiodo–Tyr (¥), Trp, Phe, Leu, or Ala]; FAPKNYPAX (where X is Trp, Phe, or Ala); or FAPKNXPAX (where X is Trp or Ala) (Table S1). H-2Kb–peptide complexes were isolated with streptavidin beads and examined by IB with anti-exon8 antibody. Binding is represented as the amount of isolated H-2Kb relative to total H-2Kb. A representative experiment is shown; quantification represents the average ± SD of four independent experiments. (D) Thermal denaturation of various H-2Kb–peptide complexes was determined using the barycentric mean fluorescence (BCM) (Fig. S1). Fit of BCM to the first derivative yields melting temperature Tm (Fig. S1D). The average Tm ± SD of four independent experiments for all H-2Kb–peptide complexes is shown.
To explore whether the increase in peptide-bound H-2Kb is attributable to an altered peptide repertoire we evaluated binding of suboptimal peptides to H-2Kb. Optimal MHC class I peptides are typically nine residues in length with defined residues at particular positions that dock into specialized pockets within the MHC I peptide-binding groove (6, 7) (Fig. 1A). As a model peptide, we used FAPGNYPAL (Sendai virus nucleoprotein peptide 324–332) (5) and designed variants with anchor residues Tyr at position P(Ω–3) (third amino acid from the C terminus) and Leu at the C-terminal position P(Ω) substituted by either smaller or larger residues (Table S1). In addition, we replaced Gly with Lys (yielding FAPKNYPAL) to attach a biotin for the isolation of peptide complexes (Fig. 1A). Biotinylated peptide variants were incubated with lysates of RMA-S cells at 4 °C and isolated with streptavidin beads, and bound H-2Kb was examined by immunoblotting with exon8 (Fig. 1C). Exchanging the Tyr anchor at P(Ω–3) for a similar or larger amino acid reduced (Tyr→Phe/Trp) or prevented [Tyr→3,5-diiodo-l-Tyr (¥)] peptide binding to H-2Kb. Replacement of Tyr by smaller amino acids (Leu/Ala) did not yield detectable H-2Kb complexes. Substitution of Leu at P(Ω) with Phe was allowed (Fig. 1C). Other replacements (Leu→Trp/Ala) significantly decreased binding to H-2Kb. This suggests that under steady-state conditions binding of peptides to H-2Kb is not strictly limited to the presence of canonical anchor residues, as would be expected from the binding rules extracted from the H-2Kb–associated peptidome (6).
Because steady-state association of modified peptides with H-2Kb in cell lysates was assayed at 4 °C, no information was obtained regarding stability of the resulting complexes. Therefore, H-2Kb complexes with altered peptides were produced (Table S2) for thermal stability experiments, where intrinsic Trp fluorescence and light scattering at 473 nm were monitored over a temperature range to measure protein unfolding and aggregation, respectively (Fig. S1A). H-2Kb–FAPGNYPAL complex showed two transitions, the first at ∼25–26 °C and the second at ∼46–52 °C, which corresponds to its reported melting temperature (Tm) (8). Aggregation was not observed before reaching the latter temperature range (Fig. S1 A–C). Two distinct folding events were also observed for H-2Kb in complex with suboptimal peptides, with the first conformational change at ∼26 °C, and Tm ranged between 30 °C and 40 °C, depending on the peptide (Fig. 1D and Fig. S1D). There was one exception, H-2Kb–FAPGNYPAF, with a Tm of ∼53 °C (similar to the original peptide FAPGNYPAL), consistent with the efficient binding to H-2Kb in lysates (Fig. 1C).
These biophysical experiments suggest that H-2Kb molecules can associate with a broader set of peptides than defined strictly on the basis of canonical anchor residues. Most of these peptides do not stabilize H-2Kb sufficiently to survive at physiological temperatures.
Association and Disassociation Kinetics of 9-Mer Peptide Variants Binding to H-2Kb.
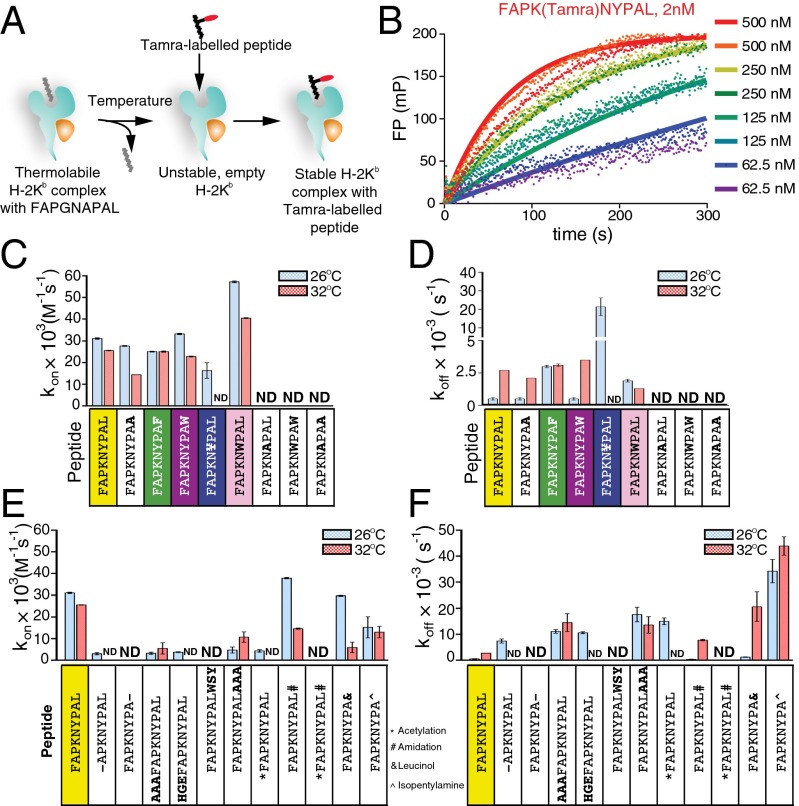
To better understand the peptide selection process by MHC I we explored the kinetic association (kon) and dissociation (koff) rates of our noncanonical peptides. Because empty MHC class I molecules are unstable (9), thermolabile H-2Kb–FAPGNAPAL complex (Tm ∼30.5 °C, Fig. 1D) was used as a source of peptide-receptive MHC I (Fig. 2A). We monitored the binding in time of TAMRA-labeled peptides to H-2Kb at 26 °C or 32 °C using fluorescence polarization (FP) (Fig. 2B) and calculated kon, koff, and the equilibrium dissociation constant (Kd = koff/kon) (Table S3). FAPKNYPAL is characterized by a high on-rate and a low off-rate, resulting in stable binding to H-2Kb (Kd ∼16 nM, Table S3) (10).
Fig. 2.
Kinetics of modified peptide binding to H-2Kb as a function of temperature. (A) Schematic representation of peptide exchange by H-2Kb. FAPGNAPAL from thermolabile H-2Kb–FAPGNAPAL complex is exchanged for TAMRA-labeled peptide. (B) Primary data of FP measurements of FAPGNAPAL in H-2Kb complex exchanged for FAPK(TAMRA)NYPAL. H-2Kb–FAPGNAPAL was used at concentrations indicated and peptide FAPK(TAMRA)NYPAL at 2 nM. From the binding curves, association rates (kon), dissociation rates (koff), and Kd were calculated (see SI Experimental Procedures). (C–F) kon and koff values of peptides with variable anchor residues (C and D) or altered or extended N or C termini (E and F; for sequence details see Fig. S2C). FP measurements were performed at 26 °C and 32 °C, as indicated (also see Table S3). Average kon, and koff values ± SD from at least two independent experiments are shown. ND, no binding resolved at the highest concentration of H-2Kb (4 μM).
We first considered the effect of anchor residue size at P(Ω–3) and P(Ω) (Fig. 2 C and D and Table S3). As in the cell lysate experiments (Fig. 1C), replacing Tyr for a small Ala residue abolished association with H-2Kb. Expanding the anchor residue with Trp or 3,5-diiodo-l-Tyr (¥) allowed efficient association, whereas dissociation increased with size of the anchor, resulting in low affinity. Importantly, exchange of the C-terminal Leu to a smaller (Ala) or larger (Phe/Trp) residue incurred minor effects on binding kinetics to H-2Kb, suggesting that the peptide-binding groove is receptive to more peptides than it ultimately presents. However, its flexibility is not unlimited, because H-2Kb ignored the FAPKNWPAW peptide with two expanded anchors. These data indicate that H-2Kb molecules can sample a variety of noncanonical peptides, as long as those alterations are compatible with productive association.
Comparison of peptide on- and off-rates (Fig. 2 C and D, respectively) revealed that although most peptides bind with similar on-rates both at 26 °C and 32 °C off-rates can differ significantly between these temperatures. This implies that despite some peptide selection by MHC class I at the level of initial binding (on-rate) the most significant filtering step is dictated by peptide dissociation (off-rate).
We next varied anchor residue charge (Tyr→Arg/Asp, Leu→Arg/Asp, Fig. S2 A and B). The F pocket (with net neutral charge) responsible for accommodation of the C-terminal anchor residue (11) did not allow any charged amino acid at the P(Ω). The net negatively charged C pocket (12) accommodating residue at P(Ω–3) accepted positively but not negatively charged residues. Interestingly, although FAPKNRPAL has an on-rate comparable to that of FAPKNYPAL it displays a considerably higher off-rate, reducing overall binding affinity (Fig. S2 A and B and Table S3), which may explain the underrepresentation of such peptides in the H-2Kb peptidome under physiological conditions (6).
Finally, we considered the role of secondary anchors by replacing most residues in FAPKNYPAL for Ala. Such peptides still associated with H-2Kb, given the presence of canonical anchors, but showed higher off-rates. Rather surprisingly, although the single P(Ω–3) Tyr→Ala mutant peptide FAPKNAPAL was ignored by H-2Kb (Fig. 2 C and D), poly-Ala peptides (FAPKAAAAL and AAPKAAAAL) had association rates similar to that of FAPKNYPAL. Their dissociation rates, however, were markedly elevated, resulting in compromised binding to H-2Kb (Fig. S2 A and B and Table S3). This suggests that peptides with a small anchor residue at P(Ω–3) are considered by H-2Kb if placed within an otherwise favorable sequence context. Furthermore, selection of peptides for presentation is determined mostly by higher dissociation rates of suboptimal peptides and consequent retention of those that manage to stay bound.
Peptide Size Selection During the First Binding Steps to H-2Kb.
MHC class I typically harbors peptides of nine amino acids in length, restricted by the hydrogen bonds formed between MHC class I and the αNH2- and αCOOH-terminal ends of the peptide. Longer peptides then bulge outward to favorably accommodate their N and C termini within the binding groove (13–16). To establish how modification of peptide length impinges on the individual binding parameters to H-2Kb we reduced or increased peptide length (Fig. 2 E and F and Table S3). Removal of N-terminal Phe or N-terminal extension (AAA/HGE) resulted in low but detectable on-rates and high off-rates. Removal of C-terminal Leu abolished binding to H-2Kb, whereas C-terminal extensions affected kon and koff values in a sequence-dependent manner (compare extensions AAA and WSY). This suggests a role for C-terminal anchor docking for the first step of peptide binding to MHC class I, although both termini need to be properly positioned to produce a stable complex with H-2Kb. This could be due to the peptide NH2- and COOH-terminal groups that interact with the peptide-binding groove. We modified these groups to reduce the number of potential hydrogen donors or acceptors (Fig. S2C). Acetylation of the N terminus (*FAPKNYPAL) reduced on- and increased off-rates similarly to removal of the N-terminal Phe residue. Amidation of the C-terminal COOH group (FAPKNYPAL#) or its conversion into an alcohol (FAPKNYPA&) did not significantly affect peptide on- and off-rates. Replacement of Leu by isopentylamine (FAPKNYPA^) to eliminate all C-terminal hydrogen donors reduced peptide on-rates twofold and enhanced off-rates 70-fold. Only when both N- and C-terminal ends were blocked (*FAPKNYPAL#) was the peptide was completely ignored (Fig. 2 E and F and Table S3). Our data suggest that the C-terminal amino acid (for all noncharged amino acids tested) has to fill the F pocket in order for the peptide to be considered by H-2Kb. The free N and C termini of the peptide are of lesser importance for initial binding unless both groups are prevented from making hydrogen bonds. This then suggests that both termini have to dock into the MHC class I peptide-binding groove for detection by our FP assays.
Collectively, these data suggest that MHC class I considers peptides of different sequences and sizes during the first binding step, as evidenced by measurable association constants. This, however, is followed by a second step, wherein peptides of canonical sequence and size remain due to low off-rates.
Structural Flexibility in the Peptide-Binding Groove of H-2Kb.
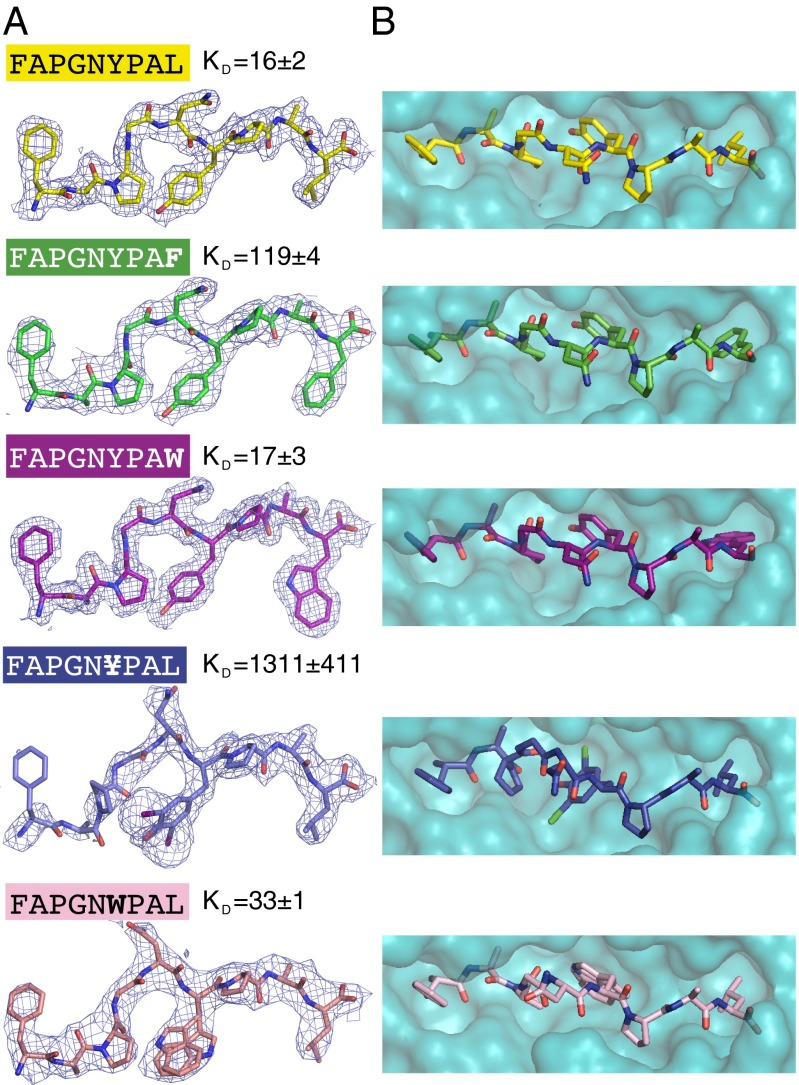
The peptide-binding groove should be flexible to accommodate expanded anchors (17–22). To visualize this, we crystallized H-2Kb molecules in complex with the original FAPGNYPAL peptide and its variants (FAPGNWPAL, FAPGN¥PAL, FAPGNYPAF, and FAPGNYPAW). We failed to obtain crystals of H-2Kb with FAPGNAPAL or FAPGNYPAA. The crystal structures were determined to resolutions from 2.0 to 2.8 Å, all with excellent Rfree values in the 23.4–28.1 range, no Ramachandran outliers, and all belonging to the 99th or 100th percentile of the MolProbity score (Table S4). In all structures the peptides could be resolved and fit into the electron density (Fig. 3A). No major changes in the H-2Kb structures were observed as a function of different peptides, indicating that the pockets in the peptide-binding groove of H-2Kb can accommodate amino acid side chains that are considerably larger than the canonical ones (Fig. S3). A closer examination of the H-2Kb peptide structures reveals differences that may explain the nonselective first peptide binding step and a second more selective step required for the selection of stable peptides.
Fig. 3.
Binding of peptides with expanded anchor residues to H-2Kb. (A) The electron density maps of various peptides crystallized in complex with H-2Kb. The peptides are represented as sticks and the electron density maps are contoured at 1σ level, with a carve of 1.5 Å. The N-terminal region of FAPGN¥PAL is disordered as observed by poor electron density. Trp in the FAPGNWPAL peptide was modeled with two discrete conformers. Kd values are shown for peptide binding to H-2Kb as determined by FP (Table S3). (B) Surface representation of the space-filling model of H-2Kb with the peptide represented as sticks in top view. Expanded anchor residues exert only minimal effects on the peptide conformation in H-2Kb.
An expanded anchor at P(Ω–3) hampers H-2Kb interactions with the peptide.
Significant structural changes are observed within the H-2Kb–FAPGN¥PAL complex, containing 3,5-diiodo-l-Tyr (¥). The Pro ring at P3 rotates ∼90° in comparison with the other structures, and three N-terminal residues (Phe, Ala, and Pro) become significantly disordered, as evidenced by lack of clear electron density. Additionally, ¥ anchor slightly changes the width of the peptide-binding groove, especially around the N terminus and the middle part of the peptide (Fig. S3 C and D). Fewer hydrogen bonds and hydrophobic interactions between FAPGN¥PAL and the H-2Kb are observed than for other complexes (Table S5). The diiodo-Tyr substitution has a relatively small twofold effect on the on-rate, indicating that the expanded anchor residue can easily access H-2Kb during the initial binding step. The off-rate, however, is 40-fold higher (Fig. 2 C and D and Table S3), implying that the peptide cannot adapt well to the H-2Kb pocket.
An expanded anchor at P(Ω) alters the F pocket.
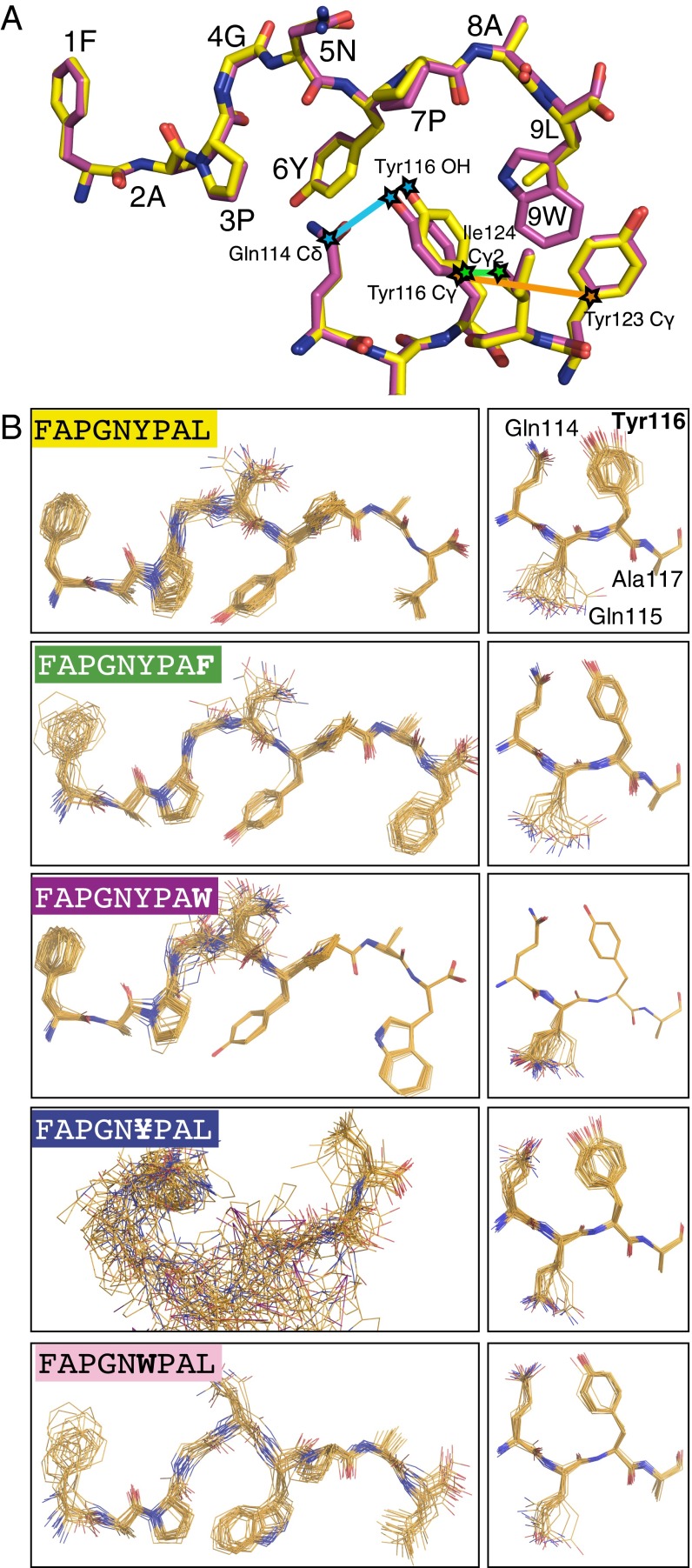
Additional structural changes are observed within the H-2Kb–FAPGNYPAW complex, where the large side chain of Trp in position P(Ω) “pushes away” Tyr116 of the H-2Kb peptide-binding groove to create space in the F pocket (Fig. 4A). In the H-2Kb–FAPGNYPAL complex, Tyr116 interacts with Gln114 via a water molecule and thus shapes the C pocket, occupied by the Tyr anchor at P(Ω–3) of the peptide. In addition, Tyr116 with Ile124 and Tyr123 comprise the F pocket (Fig. 4A), occupied by the Leu anchor at P(Ω). Binding of FAPGNYPAW induces interactions between Tyr116 and Gln114. This plasticity in the H-2Kb peptide-binding groove may allow binding of unusual anchor residues without a significant decrease in the association rate.
Fig. 4.
Dynamics and the gatekeeper’s role of Tyr116 in peptide binding. (A) Zoom-in into the C-terminal end of the peptide-binding groove of H-2Kb in complex with FAPGNYPAL (in yellow) and FAPGNYPAW (in magenta). Model shows the peptide and H-2Kb residues Gln114 as well as Tyr116, Tyr123, and Ile124 that form part of the C and F pockets, respectively. Trp of FAPGNYPAW repositions Tyr116 of H-2Kb. (B) Representative ensemble of peptides (Left) and H-2Kb amino acids 114–117 (Right). Note the selective restrained effect of expanded anchor residues at P(Ω–3) and P(Ω) on flexibility of H-2Kb amino acids Gln114 and Tyr116. The 3,5-diiodo-Tyr–containing peptide is largely disordered (quantification in Fig. S4).
Dynamics and entropy are factors controlling stable interactions.
Although FAPGNYPAW and other suboptimal peptides associate to H-2Kb they are also swiftly released from H-2Kb (Fig. 2 and Table S3). We considered the dynamics within the H-2Kb peptide structures by exploring the X-ray data and generated a representative structural ensemble for each of the H-2Kb peptide structures by time-averaged refinement, in which local molecular vibrations are sampled with molecular dynamics (23). Here, the dynamics are calculated from the X-ray data rather than being based entirely on force field calculations. Zooming into the H-2Kb peptide-binding groove with bound FAPGNYPAL revealed considerable flexibility of Tyr116. FAPGNYPAX peptide, where X is Phe or Trp, restricted the dynamics of Tyr-116 by increased filling of the F pocket (Fig. 4B and Fig. S4B). This implies that the F pocket can accommodate larger anchor residues owing to intrinsic flexibility of Tyr116—a conserved residue found in many MHC class I alleles (11). Tyr116 dynamics contributes to accommodation of the larger than canonical anchors also in C pocket as observed for FAPGNWPAL peptide. Beyond a certain size of the anchor residue—as shown here for diiodo-Tyr (¥)—long-range effects on the H-2Kb peptide-binding groove and increased peptide flexibility are observed (Fig. 4B and Fig. S4B). This may drive the release of FAPGN¥PAL from H-2Kb (Fig. 2D).
Peptides with Noncanonical Motifs Join the H-2Kb Peptidome at 26 °C.
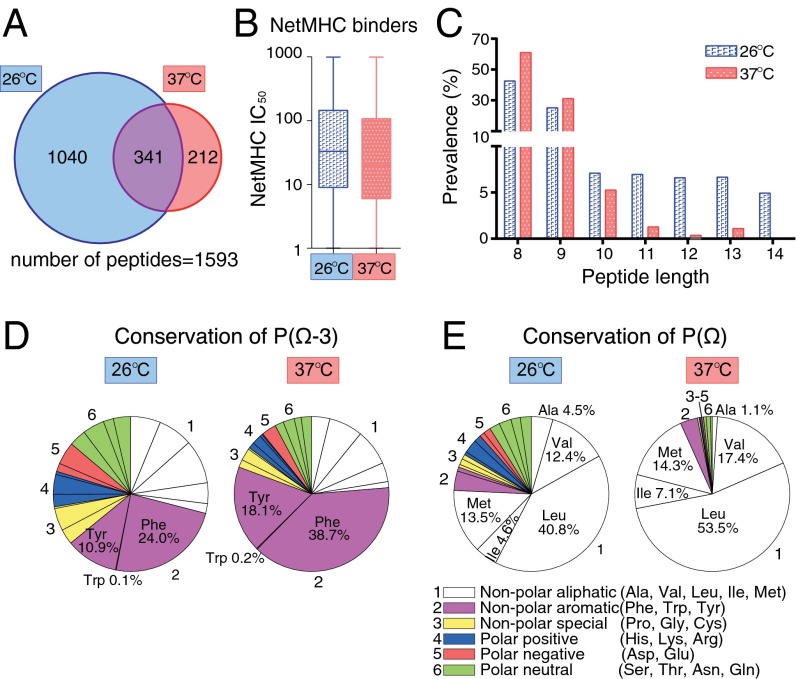
Because suboptimal peptides bind to H-2Kb at 26 °C (Table S3), and because H-2Kb–peptide complexes are more abundant at 26 °C than 37 °C, even in the absence of TAP (Fig. 1B) (2), H-2Kb could present a significant fraction of suboptimal peptides at 26 °C. RMA cells were cultured for 3 days at either 26 °C or 37 °C, and H-2Kb–associated peptidomes were analyzed by mass spectrometry, as described (24). We identified 1,381 and 553 peptides bound to H-2Kb molecules isolated from cells cultured at 26 °C and 37 °C, respectively, with 341 peptides in common (Fig. 5A and Dataset S1). NetMHC algorithm (25–27) predicted peptides presented by H-2Kb at 26 °C to be worse binders (higher IC50) relative to those presented at 37 °C (Fig. 5B), harboring differences in sequence and length. As expected, 8-mer, 9-mer, and 10-mer peptides constituted the largest pool at both temperatures. However, a substantial fraction of longer peptides (25% of total) was isolated from H-2Kb molecules from cells cultured at 26 °C, compared with only 3% at 37 °C (Fig. 5C). Moreover, the presence of canonical anchor residues Phe or Tyr at P(Ω–3) was reduced from 57% (at 37 °C) to 35% (at 26 °C) (Fig. 5D). The presence of canonical aliphatic amino acids at P(Ω) was reduced from 93% (at 37 °C) to 76% (at 26 °C) (Fig. 5E).
Fig. 5.
Characterization of H-2Kb-associated peptidome in cells as a function of temperature. Peptides associated with H-2Kb were isolated from RMA cells cultured at 26 °C or 37 °C and identified by mass spectrometry. (A) Venn diagrams shows the number of unique and shared peptides between H-2Kb peptidomes of RMA cells cultured at 26 °C and 37 °C (for peptide sequences see Dataset S1). (B) Distribution and median IC50 values of peptides in the peptidome of H-2Kb from cells cultured at 26 °C or 37 °C predicted using the NetMHC algorithm (median IC50 = 33 for 26 °C and 20 for 37 °C). Distribution of predicted IC50 values for the peptides identified at 26 °C and 37 °C were significantly different (P = 0.0009) according to the nonparametric Mann–-Whitney test. (C) Distribution according to size of the H-2Kb–associated peptidome of RMA cells cultured at 26 °C or 37 °C. The total pool at given temperature is set at 100%. (D and E) Sequence conservation of anchor residue at P(Ω–3) (D) and P(Ω) (E) in the H-2Kb–associated peptidome of RMA cells cultured at 26 °C or 37 °C. Pie charts illustrate the proportion of nonpolar aliphatic (Ala, Val, Leu, Ile, and Met, white), nonpolar aromatic (Phe, Trp, and Tyr, magenta), nonpolar special (Pro, Gly, and Cys, yellow), polar positive (His, Lys, and Arg, blue), polar negative (Asp and Glu, red), and polar neutral (Ser, Thr, Asn, and Gln, green) amino acids at P(Ω–3). Amino acids are represented in the following order: Ala, Val, Leu, Ile, Met, Phe, Trp, Tyr, Pro, Gly, Cys, His, Lys, Arg, Asp, Glu, Ser, Thr, Asn, and Gln starting from the 12:00 position on the pie chart.
Thus, the H-2Kb peptidome at 26 °C has many aspects in common with the suboptimal peptides that bound efficiently to H-2Kb but were also swiftly released in our biophysical experiments (Fig. 2 and Table S3). These observations also indicate that at lower temperature the peptide repertoire of MHC class I can be expanded beyond the consensus sequences and lengths, resulting in presentation of a broader peptide pool.
Discussion
It has long been appreciated that one peptide transporter, TAP, feeds a wide variety of MHC class I alleles. Because these peptides may greatly differ in sequence, a central question therefore arises as to how a defined MHC class I allele (H-2Kb in our study) is able to select the correct peptides for presentation. One possible option could be that binding of only those peptides that are ultimately presented is considered. This should then result from an intrinsic quality of the MHC class I molecule. Alternatively, H-2Kb may in fact first consider a range of available peptides, followed by a selection step for presentation
In an effort to advance our understanding of the events occurring during the initial confrontation of MHC class I molecules with candidate peptides we engineered systematic variations within a model peptide, FAPGNYPAL, to study the effects of altered sequence, length, and termini accessibility on peptide selection by H-2Kb. Evaluation of association and dissociation parameters of a battery of peptides, containing targeted departures from the model, revealed that peptide acquisition by H-2Kb takes place in two distinct phases. During the first phase, the peptide-binding groove of H-2Kb accepts a wide range of candidates, including even amino acid side chains much larger than their optimal size, which implies significant flexibility in the anchor residue-accepting pockets (17, 19–22). Following the initial association event, ill-suited peptides exhibit greater propensity for dissociation and are thus selected against.
There are several reasons for selecting peptides for presentation. The pockets for anchor residues show considerable flexibility. This flexibility is in turn associated with higher entropic costs (Fig. 4), which may yield elevated dissociation rates and a compromised overall stability of the complex. This point is particularly apparent for Tyr116, an amino acid conserved in many MHC class I alleles, essential for tapasin-independent peptide loading (28, 29) and influencing MHC I–T-cell receptor interactions (30). Ensemble refinements calculated on the basis of X-ray diffraction data, where local low resolution essentially corresponds to higher local dynamics, indicate that mobility of Tyr116 is markedly restricted by larger than typical peptide anchors (Fig. 4B). These observations indicate how MHC class I molecules can accommodate a great variety of peptides in the first step of binding and subsequently release suboptimal candidates.
It stands to reason that if the entropic constraints within the peptide-binding groove contribute to higher dissociation rates of peptides harboring expanded anchors, then alleviating the necessity to maintain such flexibility by lowering the temperature during complex assembly could shift the peptidome equilibrium away from the cannon. Indeed, we observed a considerable reduction in dissociation rates for peptides acquired at 26 °C, relative to those at 32 °C, with on-rates remaining only marginally affected in this temperature range (Fig. 2). Moreover, comparing the peptidomes of H-2Kb molecules from cells cultured at 26 °C and 37 °C also showed more elongated peptides and the loss of conservation of anchor residues (Fig. 5). This pattern is similar to that observed in cells where peptide loading is obstructed by tapasin deficiency or where peptide trimming is reduced by inactivation of endoplasmic reticulum aminopeptidase associated with antigen processing (ERAAP) (31, 32).
These observations hint at temperature-dependent conformational adaptations in the H-2Kb peptide-binding groove, following the initial binding event. In fact, we detect a conformational change in H-2Kb at around 26 °C by Trp fluorescence (Fig. S1). This may constitute a step wherein H-2Kb molecules are able to stably acquire noncanonical peptides at 26 °C that would otherwise be entropically disfavored in the second phase of peptide consideration. Importantly, a shift of these MHC class I complexes to higher temperatures results in a swift release of suboptimal peptides, producing a high yield of peptide-receptive MHC class I molecules (4), an observation advantageous in vaccination studies (33, 34).
On the way to understanding the nature of peptide selectivity by MHC class I our work defines a number of simple rules: (i) noncanonical peptides with expanded anchor residues associate with rates similar to those of their canonical counterparts, owing to accommodation by the flexible pockets within the peptide-binding groove of MHC class I; (ii) peptides harboring incompatible charges are ignored, likely by simple repulsion; and (iii) stable complex formation between MHC class I and peptide requires each peptide terminus to contribute at least two hydrogen bonds, as well as the F pocket to be properly occupied by an amino acid side chain. The composition of the peptide–MHC class I repertoire is determined primarily by peptide-binding properties of MHC class I itself, but tapasin and ERAAP further shape the peptide repertoire presented (Fig. 5) (31, 32). Of note, Duan et al. (35) showed that cancer neopeptides that elicit CD8+ T-cell immunity may reflect the peptide pool detected at 26 °C and then expand the MHC class I-associated peptidome considered by the immune system. Collectively, this suggests that, at physiological temperature, MHC class I undergoes rounds of consideration and rejection until high-affinity peptides are acquired. This mechanism simultaneously allows for a wide range of sampling while ensuring that only the best available options are chosen for presentation.
Materials and Methods
Purification, Characterization, and Crystallization of H-2Kb Peptide Complexes in Vitro.
H-2Kb was produced in Escherichia coli, refolded, and purified. For kinetic measurements, H-2Kb–FAPGNAPAL complexes were incubated at 26 °C or 32 °C with various TAMRA-labeled peptides for FP experiments. Proteins were crystallized and diffraction data solved by molecular replacement to 2.0–2.8 Å resolution. Protein dynamics were done by ensemble refinement modeling (23).
H-2Kb–Associated Peptidome.
H-2Kb molecules were isolated from 1010 RMA cells, cultured at 26 °C or 37 °C for 72 h. Peptides were analyzed by a conventional Agilent 1100 gradient HPLC system coupled to a Q Exactive mass spectrometer and matched against the International Protein Index database using the Mascot search engine.
Supplementary Material
Acknowledgments
We thank J. de Widt for antibody production; H. Hilkmann and D. El Atmioui for peptides; M. Maletta and M. Zacharias for scientific discussions; B. Rodenko for help with images; beamline scientists at European Synchrotron Radiation Facility and Swiss Light Source for support; and P. Gros for support with molecular dynamics. This work was supported by EMBO Long-Term Fellowship Grant ALTF_230-2010 (to M.A.G.), Netherlands Organisation for Scientific Research (NWO) Veni Grant 722.011.011 (to R.P.J.), and NWO-TOP (91213078) and the Gravity Program Institute for Chemical Immunology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4PG9, 4PGB, 4PGC, 4PGD, and 4PGE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416543112/-/DCSupplemental.
References
- 1.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.Ljunggren HG, et al. Empty MHC class I molecules come out in the cold. Nature. 1990;346(6283):476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 3.Neefjes JJ, Hämmerling GJ, Momburg F. Folding and assembly of major histocompatibility complex class I heterodimers in the endoplasmic reticulum of intact cells precedes the binding of peptide. J Exp Med. 1993;178(6):1971–1980. doi: 10.1084/jem.178.6.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Silva AD, et al. Thermolabile H-2Kb molecules expressed by transporter associated with antigen processing-deficient RMA-S cells are occupied by low-affinity peptides. J Immunol. 1999;163(8):4413–4420. [PubMed] [Google Scholar]
- 5.Schumacher TN, et al. Direct binding of peptide to empty MHC class I molecules on intact cells and in vitro. Cell. 1990;62(3):563–567. doi: 10.1016/0092-8674(90)90020-f. [DOI] [PubMed] [Google Scholar]
- 6.Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 7.Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 8.Saini SK, et al. Not all empty MHC class I molecules are molten globules: tryptophan fluorescence reveals a two-step mechanism of thermal denaturation. Mol Immunol. 2013;54(3–4):386–396. doi: 10.1016/j.molimm.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Bouvier M, Wiley DC. Structural characterization of a soluble and partially folded class I major histocompatibility heavy chain/beta 2m heterodimer. Nat Struct Biol. 1998;5(5):377–384. doi: 10.1038/nsb0598-377. [DOI] [PubMed] [Google Scholar]
- 10.Matsumura M, Saito Y, Jackson MR, Song ES, Peterson PA. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules isolated from transfected Drosophila melanogaster cells. J Biol Chem. 1992;267(33):23589–23595. [PubMed] [Google Scholar]
- 11.Zhang C, Anderson A, DeLisi C. Structural principles that govern the peptide-binding motifs of class I MHC molecules. J Mol Biol. 1998;281(5):929–947. doi: 10.1006/jmbi.1998.1982. [DOI] [PubMed] [Google Scholar]
- 12.Young AC, Zhang W, Sacchettini JC, Nathenson SG. The three-dimensional structure of H-2Db at 2.4 A resolution: Implications for antigen-determinant selection. Cell. 1994;76(1):39–50. doi: 10.1016/0092-8674(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 13.Bulek AM, et al. Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes. Nat Immunol. 2012;13(3):283–289. doi: 10.1038/ni.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo HC, et al. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature. 1992;360(6402):364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- 15.Liu YC, et al. The energetic basis underpinning T-cell receptor recognition of a super-bulged peptide bound to a major histocompatibility complex class I molecule. J Biol Chem. 2012;287(15):12267–12276. doi: 10.1074/jbc.M112.344689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6(11):1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 17.Garstka MA, et al. Tapasin dependence of major histocompatibility complex class I molecules correlates with their conformational flexibility. FASEB J. 2011;25(11):3989–3998. doi: 10.1096/fj.11-190249. [DOI] [PubMed] [Google Scholar]
- 18.Mage MG, et al. The peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics. J Immunol. 2012;189(3):1391–1399. doi: 10.4049/jimmunol.1200831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieker F, Springer S, Zacharias M. Comparative molecular dynamics analysis of tapasin-dependent and -independent MHC class I alleles. Protein Sci. 2007;16(2):299–308. doi: 10.1110/ps.062568407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieker F, Straatsma TP, Springer S, Zacharias M. Differential tapasin dependence of MHC class I molecules correlates with conformational changes upon peptide dissociation: A molecular dynamics simulation study. Mol Immunol. 2008;45(14):3714–3722. doi: 10.1016/j.molimm.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Zacharias M, Springer S. Conformational flexibility of the MHC class I alpha1-alpha2 domain in peptide bound and free states: A molecular dynamics simulation study. Biophys J. 2004;87(4):2203–2214. doi: 10.1529/biophysj.104.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hein Z, et al. Peptide-independent stabilization of MHC class I molecules breaches cellular quality control. J Cell Sci. 2014;127(Pt 13):2885–2897. doi: 10.1242/jcs.145334. [DOI] [PubMed] [Google Scholar]
- 23.Burnley BT, Afonine PV, Adams PD, Gros P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife. 2012;1:e00311. doi: 10.7554/eLife.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassan C, et al. The human leukocyte antigen-presented ligandome of B lymphocytes. Mol Cell Proteomics. 2013;12(7):1829–1843. doi: 10.1074/mcp.M112.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundegaard C, et al. NetMHC-3.0: Accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36(web server issue):W509-12. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24(11):1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16(4):509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 29.Zernich D, et al. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med. 2004;200(1):13–24. doi: 10.1084/jem.20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart-Jones GB, et al. Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms. Proc Natl Acad Sci USA. 2012;109(50):E3483–E3492. doi: 10.1073/pnas.1207896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchard N, et al. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J Immunol. 2010;184(6):3033–3042. doi: 10.4049/jimmunol.0903712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanaseki T, et al. ERAAP and tapasin independently edit the amino and carboxyl termini of MHC class I peptides. J Immunol. 2013;191(4):1547–1555. doi: 10.4049/jimmunol.1301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. Mamu-A01/K(b) transgenic and MHC Class I knockout mice as a tool for HIV vaccine development. Virology. 2009;387(1):16–28. doi: 10.1016/j.virol.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vierboom MP, et al. Tumor eradication by wild-type p53-specific cytotoxic T lymphocytes. J Exp Med. 1997;186(5):695–704. doi: 10.1084/jem.186.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan F, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211(11):2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.