Significance
Gene and whole-genome duplications are important evolutionary forces promoting organismal diversification. Teleost fishes, for example, possess many gene duplicates responsible for photoreception (opsins), which emerged through gene duplication and allow fishes to adapt to the various light conditions of the aquatic environment. Here, we reevaluate the evolutionary history of the violet-blue–sensitive opsins [short wavelength-sensitive 2 (SWS2)] in modern teleosts using next generation genome sequencing. We uncover a gene duplication event specific to the most diverse lineage of vertebrates (the percomorphs) and show that SWS2 evolution was highly dynamic and involved gene loss, pseudogenization, and gene conversion. We, thus, clarify previous discrepancies regarding opsin annotations. Our study highlights the importance of integrative approaches to help us understand how species adapt and diversify.
Keywords: gene duplication, gene conversion, gene resurrection, Percomorpha, SWS2
Abstract
Single-gene and whole-genome duplications are important evolutionary mechanisms that contribute to biological diversification by launching new genetic raw material. For example, the evolution of animal vision is tightly linked to the expansion of the opsin gene family encoding light-absorbing visual pigments. In teleost fishes, the most species-rich vertebrate group, opsins are particularly diverse and key to the successful colonization of habitats ranging from the bioluminescence-biased but basically dark deep sea to clear mountain streams. In this study, we report a previously unnoticed duplication of the violet-blue short wavelength-sensitive 2 (SWS2) opsin, which coincides with the radiation of highly diverse percomorph fishes, permitting us to reinterpret the evolution of this gene family. The inspection of close to 100 fish genomes revealed that, triggered by frequent gene conversion between duplicates, the evolutionary history of SWS2 is rather complex and difficult to predict. Coincidentally, we also report potential cases of gene resurrection in vertebrate opsins, whereby pseudogenized genes were found to convert with their functional paralogs. We then identify multiple novel amino acid substitutions that are likely to have contributed to the adaptive differentiation between SWS2 copies. Finally, using the dusky dottyback Pseudochromis fuscus, we show that the newly discovered SWS2A duplicates can contribute to visual adaptation in two ways: by gaining sensitivities to different wavelengths of light and by being differentially expressed between ontogenetic stages. Thus, our study highlights the importance of comparative approaches in gaining a comprehensive view of the dynamics underlying gene family evolution and ultimately, animal diversification.
Gene and whole-genome duplications facilitate the acquisition of novel biological functions (1, 2) and are, hence, considered important forces to achieve major evolutionary transitions (3). For example, whole-genome duplications in the respective ancestors of yeast (4), vertebrates (5), and teleost fishes (6) are thought to have laid the genomic foundation for many key characteristics crucial to the evolutionary success of these lineages. More common, however, are single-gene duplications, which often act as a springboard for adaptive diversification of entire gene families as exemplified by the immune-regulatory MHC genes in hominids (7), hemoglobins in tetrapods (8) and bony fishes (9), or opsins in mantis shrimps (10), fishes (11), and primates (12).
Opsins are at the core of animal vision, an important sensory system involved in, for example, food gathering, communication, predator avoidance, mate selection, and navigation. In vertebrates, opsins are expressed primarily in ciliary photoreceptor cells (c-opsins) and encode for G protein-coupled receptors that bind to a light-absorbing, vitamin A-derived nonprotein retinal chromophore (13). The evolution of opsin genes is a prime textbook example of how changes at a molecular level—in the form of duplications (11, 12), mutations (14), and changes in gene expression (11)—drive adaptation to divergent photic environments (15), which may ultimately lead to speciation (16). In addition, because of the possibility to directly link opsin genotypes to functional visual phenotypes (i.e., spectral sensitivities), opsins are among the best studied and functionally best characterized gene families in vertebrates (15, 17).
Other than rhodopsin (RH1), the rod-based visual pigment often used for scotopic vision, vertebrates possess four basic types of cone opsin genes, which mediate color vision: two short wavelength (UV-blue)-sensitive (SWS) genes (SWS1 and SWS2), a midwavelength (green)-sensitive gene (RH2), and a long wavelength (yellow-red)-sensitive gene (LWS) (17). Unlike in tetrapods, where this basic opsin setup remained relatively constant, teleost opsins have duplicated extensively, leading to an astonishing richness of opsin genes (18). Opsins are particularly diverse in spiny-rayed fishes [Acanthomorpha (18)]—with >18,000 species, it is the most species-rich taxon of vertebrates that also includes the highly diverse percomorphs (19).
Opsin duplications in teleosts occur at all taxonomic levels (18), affecting the visual systems of entire families (20), genera (21), or individual species (22). In addition, opsin diversity increases because of differences in the evolutionary fate of duplicates (18). In many fishes, novel opsins become pseudogenes (i.e., still detectable functionally disrupted genes) or are lost shortly after emerging through duplication (i.e., nonfunctionalization). However, novel opsins can persist if they acquire new functions (i.e., neofunctionalization). Neofunctionalization is primarily achieved through changes in amino acids at key tuning sites (typically of the retinal binding pocket), leading to shifts in the peak absorbance (λmax) of opsin proteins and consequently, sensitivities to different wavelengths of light (17, 18). However, neofunctionalization can also include differential expression of genes throughout ontogeny (20). Finally, opsin duplicates might be subject to gene conversion (18, 21), a common form of reticulate evolution that serves as an important homogenizing force or a repair mechanism between paralogous genes (23). Gene conversion typically occurs between functional paralogs, but it may also involve pseudogenized genes, thus leading to their resurrection (24).
The majority of known opsin gene duplications affecting a large number of fish species involve the midwavelength and long wavelength-sensitive genes (RH1, RH2, and LWS), whereas only one major duplication event of an SWS gene, that of the blue opsin SWS2 (SWS2A and SWS2B) at the base of the spiny-rayed fishes, has been described (18). However, phylogenetic and functional comparisons between different opsin gene families suggest that the evolutionary history of SWS2 might be more complex than previously thought. To begin with, based on a predicted duplication rate of approximately one duplication event every 100 My (25) and the estimated age of the clade [teleosts started to diversify in the Carboniferous to Permian 330–260 Mya (26, 27)], a larger number of SWS2 duplicates is to be expected. Furthermore, teleost SWS2 genes show surprisingly high rates of amino acid substitutions (28) but comparatively low rates of diversification postduplication (18), indicating major discrepancies in current SWS2 gene annotations.
Against this background, we reevaluate the evolutionary history of SWS2 in teleosts using next generation sequencing and open access data mining. We explored transcriptomic and genomic information on SWS2 from a phylogenetically representative set of close to 100 fish species and examined in detail the different evolutionary scenarios (gene duplication, loss, and conversion) that have shaped the SWS2 diversity in teleosts, with a particular focus on acanthomorphs. In doing so, we uncover a major SWS2A duplication, which coincides with the radiation of percomorph fishes. Using a combination of microspectrophotometric (MSP) and quantitative real-time RT-PCR (qRT-PCR) experiments in a species that retained both paralogs, the dusky dottyback Pseudochromis fuscus, we provide evidence for neofunctionalization in SWS2A. We finally show that the SWS2A duplication was followed by a complex pattern of gene loss and gene conversion in the different lineages of this highly diverse group of fish, offering an explanation for why this duplication event remained undetected so far.
Results and Discussion
SWS2 Duplication, Gene Synteny, and Phylogenetic Reconstruction.
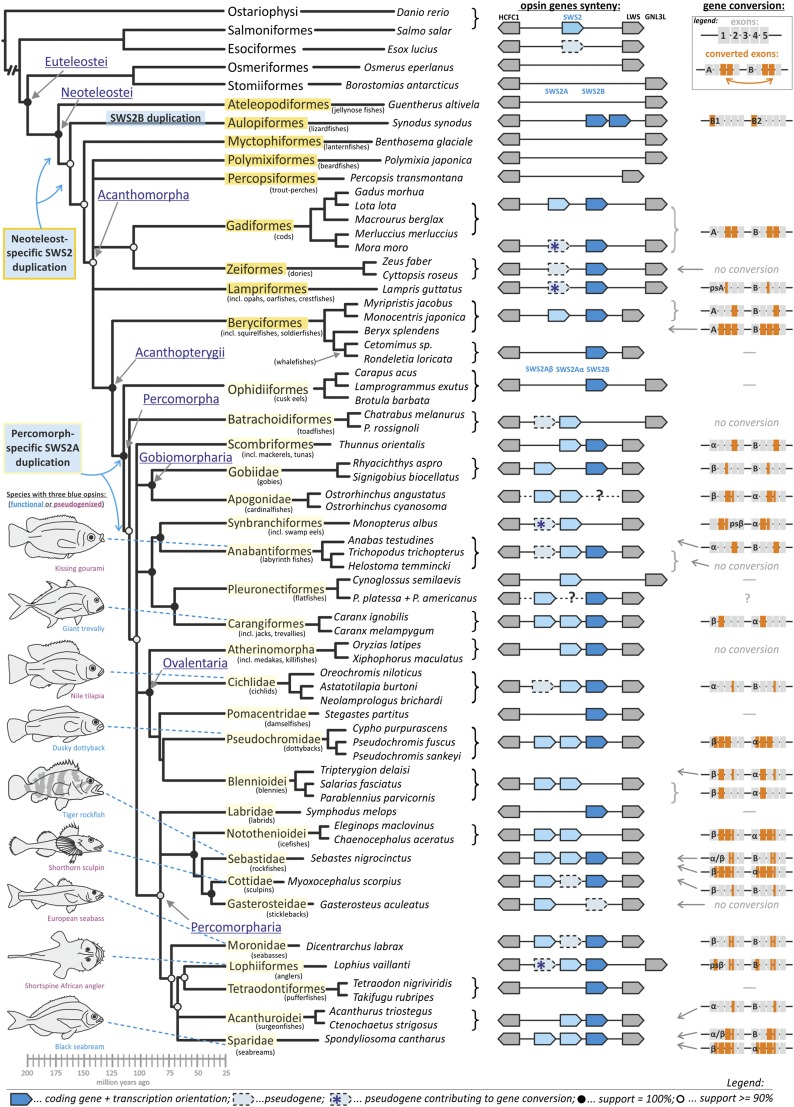
Using a phylogenetic representative sample of 97 fish species (Table S1) covering most of the currently recognized neoteleost lineages [Neoteleostei (19)], we first show that a duplication of SWS2 into SWS2A and SWS2B occurred around the appearance of the first neoteleosts 190–170 Mya (26, 27), thereby shifting the previously described acanthomorph-specific origin of this duplication deeper into the teleost phylogeny (18, 21) (Fig. 1). A closer inspection of the genomic region (∼30 kb) between the highly conserved HCFC1 gene upstream and LWS or GNL3L downstream of SWS2 revealed two additional duplication events and the retention of up to three SWS2 genes in some fish lineages (Fig. 1).
Fig. 1.
Evolutionary history of SWS2 in teleost fishes. A first ancestral duplication of SWS2 into SWS2A and SWS2B happened at the base of the Neoteleostei (orange), which was followed by a second percomorph-specific duplication of SWS2A into SWS2Aα and SWS2Aβ (yellow). A lineage-specific SWS2B duplication was further discerned in lizardfishes (Aulopiformes). SWS2 gene synteny is schematically shown by blue polygons pointing out the direction of transcription, and the highly conserved HCFC1 upstream and LWS or GNL3L (in case of LWS loss) downstream genes are shown in gray; missing polygons equal gene loss. A dotted line with a question mark indicates a lineage for which genomic data of the target region could not be obtained. Gene conversion is depicted on an exon by exon basis in orange. Phylogenetic reconstruction, including age estimation, is based on the consensus of the most recent global fish phylogenies (19, 27). Fig. S1 shows the SWS2 gene phylogeny.
More ancestral fish only possess one SWS2 gene [Anguilliformes, Ostariophysi, and Salmoniformes (18)], whereas most of the basal neoteleosts have lost SWS2 entirely (Osmeriformes, Stomiiformes, Ateleopodiformes, and Myctophiformes) (Fig. 1). However, we discovered two SWS2 genes in lizardfishes (Aulopiformes), which cluster together with SWS2B from more derived taxa and therefore, mark the earliest appearance of SWS2B in the phylogeny (Fig. S1). Because the two SWS2B paralogs were only recovered in lizardfish and not in other neoteleosts, this duplication is likely lineage-specific (Fig. 2 and Fig. S1). Most interestingly, we discovered a duplication of SWS2A that is associated with the emergence of the first percomorph fishes 110–130 Mya (19, 27), the most species-rich clade of teleosts (Figs. 1 and 2 and Fig. S1). Several percomorph groups, including jacks (Carangiformes), dottybacks (Pseudochromidae), rockfishes (Sebastidae), and seabreams (Sparidae), retained three complete copies of SWS2 (SWS2Aα, SWS2Aβ, and SWS2B), whereas others, including tunas (Scombriformes; SWS2Aα and SWS2B), pufferfishes (Tetradodontiformes; SWS2B), stickleback (Gasterosteiformes; SWS2Aβ and SWS2B pseudogene), and cichlids (Cichlidae; SWS2Aα, SWS2B, and SWS2Aβ pseudogene), have secondarily lost one or two SWS2 copies and/or feature pseudogenized SWS2 paralogs (Figs. 1 and 2). The earliest indication for an SWS2A-specific duplication was found in toadfishes (Batrachoidiformes), which have a complete SWS2Aα copy and an SWS2Aβ pseudogene (Fig. 1).
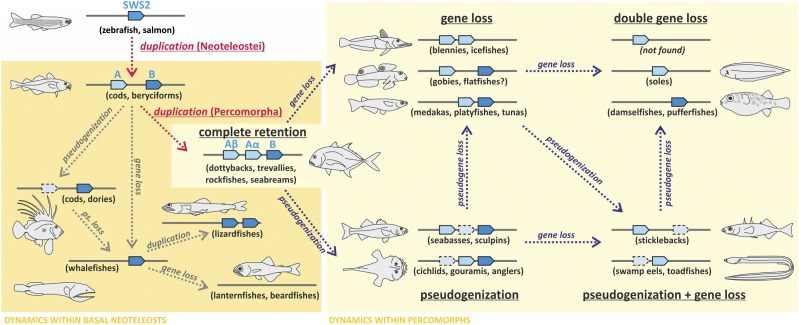
Fig. 2.
Schematic of the evolutionary dynamics affecting SWS2 in teleosts. The orange box indicates lineages affected by the initial Neoteleostei-specific duplication of SWS2 (SWS2A and SWS2B); the yellow box shows the lineages additionally affected by the Percomorpha-specific duplication of SWS2A (SWS2Aα and SWS2Aβ). Note that gene loss and pseudogenization happened repeatedly and independently between fish lineages (examples shown in parentheses), causing various stages of SWS2 retention in extant taxa. The missing genomic target region for flatfishes is marked with a question mark. Interestingly, no complete gene loss of SWS2 has been found within percomorphs.
Evolutionary History of SWS2.
In general, teleosts vary substantially in the retention of SWS2 opsins between but also within lineages (e.g., Beryciformes, Gobiomorpharia, and Pleuronectiformes) (Figs. 1 and 2). Furthermore, high rates of gene conversion seem to promote the evolutionary dynamics in this gene family (Fig. 1). Using single-exon phylogenies (Fig. 3A and Fig. S2) and a sliding window analysis to measure the neutral divergence along SWS2 [rate of synonymous substitutions per synonymous sites (dS)] (Fig. 3B and Fig. S3), we found that gene conversion affects SWS2 copies in almost all fish lineages. However, the extent of gene conversion differed between SWS2 paralogs. When two SWS2A paralogs were involved, conversion affected larger sections of genes (mostly of exons 2 and 3) compared with SWS2B (mostly of exon 4) (Fig. 1 and Figs. S2 and S3). These differences could be explained by a higher similarity of SWS2A copies because of the additional duplication event in percomorphs, which is likely to increase the chances and extent of gene conversion (23).
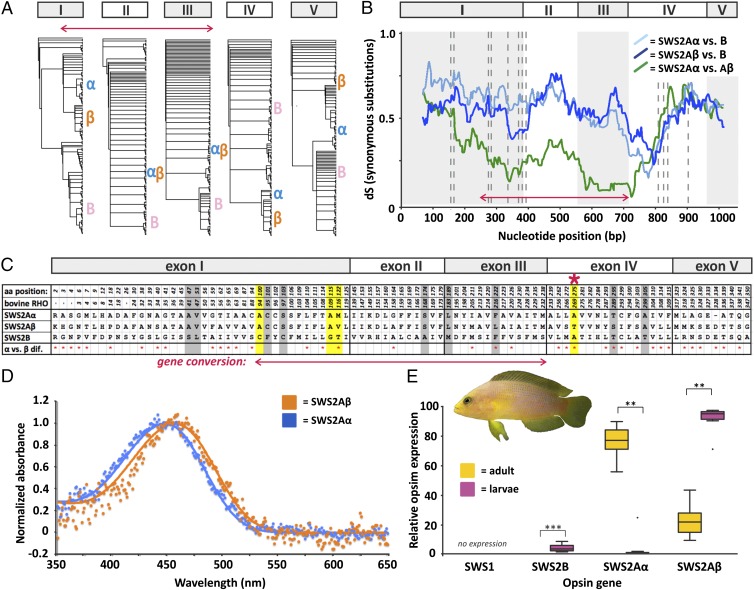
Fig. 3.
Integrative approach to study opsin gene evolution exemplified in the dusky dottyback P. fuscus. (A–C) Gene conversion approach. (A) Single-exon phylogenies show distinct phylogenetic placements of SWS2 copies when exon 1, 4, or 5 is used, whereas SWS2A copies are resolved as sister groups when exons 2 and 3 are analyzed. Letters α, β, and B mark the position of the corresponding dottyback gene in the trees. (B) Sliding window analysis. Pairwise dS rate between SWS2 copies calculated with a window of 30 and a step size of 1. The red arrow depicts low dS rates between SWS2A copies in exons 2 and 3 and part of exon 1, corresponding to gene conversion. (C) Amino acid alignment of known key tuning (yellow) (17) and retinal binding pocket sites, showing all variable positions across dottyback SWS2s. Additional putative key substitutions that were identified across fish families are highlighted in gray. The red asterisk marks the substitution A269T in SWS2Aβ, which is known to cause a positive shift in visual sensitivity of 6 nm (17). (D) MSP of adult and larval dottybacks. Orange shows spectral absorbance curves for adult and larval single cones at 457 nm λmax (n = 4), and blue shows spectral absorbance curves for adult-specific single cones at 448 nm λmax (n = 11). (E) Relative SWS gene expression measured by qRT-PCR in adult (n = 12) and larval (n = 10) dottybacks. Note that larvae almost exclusively express SWS2Aβ, whereas adults predominantly express SWS2Aα. **P < 0.01; ***P < 0.001.
Surprisingly, in the common mora (Mora mora) and the roughhead grenadier (Macrourus berglax; both Gadiformes), SWS2A is pseudogenized, but some exons do not contain stop codons or frame shifts; compared with the functional SWS2B, these parts produce highly congruent nucleotide alignments (>90% identical in both cases). A similar pattern was found for the SWS2A pseudogene in the opah (Lampris guttatus; Lampriformes; 86% identical to SWS2B) and the SWS2Aβ pseudogene in the Asian swamp eel (Monopterus albus; Synbranchiformes), and the shortspine African angler (Lophius vaillanti; Lophiiformes; >94% identical to SWS2Aα in both cases). Sliding window analyses revealed that, in these species, gene conversion occurred between pseudogenized and complete paralogs (Fig. S3). Moreover, using phylogenetic approaches, we could show that, at least for the grenadier and the swamp eel, the conversion occurred in the direction from the pseudogene to the potentially functional gene, providing what may be the first evidence, to our knowledge, for gene resurrection in vertebrate opsins (Fig. S4). However, a broader taxonomic sampling and functional approaches using expression analyses are needed to fully sustain our findings. Notably, in beryciforms, the dS values between SWS2A and SWS2B are very low, indicating that an almost complete conversion between those genes has recently occurred (Fig. S3). This observation is supported by the SWS2 phylogeny, where the beryciform SWS2A clusters close to the SWS2B clade and outside of the remaining SWS2As, thus suggesting that, in this case, a conversion occurred from SWS2B to SWS2A (Fig. S1).
Overall, the postduplication dynamics of SWS2 do not follow a phylogenetic pattern (Fig. 1) and are much more complex than previously reported for other opsin genes in fishes (18). The high rates of gene conversion were unexpected, because gene conversion usually affects larger gene families [more than five members (23)]. Our findings have strong implications for the interpretation of SWS2 gene evolution. Initially, we reconstructed gene phylogenies based on full coding regions; however, the SWS2Aα clade in particular was poorly resolved, showing low or a lack of support for many of the nodes (Fig. S5). In contrast, when the converted regions were removed (based on the sliding window analysis) (Fig. S3), clades became well-resolved and supported (Fig. S1). Most importantly, high and variable rates of conversion even within lineages (e.g., Anabantiformes) (Fig. S3) constantly homogenize gene copies, making it impossible to reconstruct their evolutionary history on the basis of traditional phylogenetic methods.
If high rates of gene conversion are, in fact, a much more common phenomenon affecting not only opsin evolution but the evolution of many other gene families alike, then our results could have even farther reaching implications in that previous analyses based on common methods of gene evolution should potentially be reassessed.
Neofunctionalization of SWS2 Genes.
The ancestral SWS2 was predicted to have had a λmax between 400 and 440 nm (17). However, SWS2As and SWS2B diversified and became maximally sensitive within the blue light (440–480 nm) and the violet light (400–440 nm) spectra, respectively (11). Comparing known (17) and potential key tuning sites (i.e., retinal binding pocket sites) between SWS2 copies (Fig. 3C and Fig. S6) combined with ancestral state reconstruction, we identified 11 amino acid sites with clade specificity (Fig. S1). Five of these sites also differed in physical properties between one another, making them prime candidates for sites under adaptive divergence by spectral tuning (29) (Fig. S1). In agreement with the older age of the initial neoteleost-specific SWS2 duplication, we found that most of these amino acid substitutions occurred between SWS2B and both SWS2A paralogs (n = 9 of 11 sites) (Fig. S1). The remaining two substitutions were found to be SWS2Aβ-specific, whereas no SWS2Aα-specific amino acid could be identified, thus suggesting that functional divergence between SWS2A paralogs occurred through a shift in spectral sensitivity in SWS2Aβ (Fig. S1).
Interestingly, only three of the newly identified sites coincide with the eleven previously known key tuning sites of SWS2 (17): 94, 109, and 116 (amino acid positions standardized to bovine rhodopsin). Therefore, our approach highlights the importance of comparative approaches across a large number of species to identify amino acid substitutions that might have a more general impact on opsins.
Neofunctionalization in the Percomorph-Specific SWS2A Duplicates.
MSP measurements of the dusky dottyback retina (P. fuscus; Pseudochromidae) revealed that dottybacks possess single cone cells with two distinct visual sensitivities, which fall within the expected range of SWS2A. Although both adult and larval dottybacks were found to have single cone cells sensitive to 457 nm λmax (prebleach λmax mean ± SE: 456.78 ± 1.53 nm; n = 4 cells), adult dottybacks were additionally found to have single cone cells sensitive to 448 nm λmax (447.51 ± 0.91 nm; n = 11 cells) (Fig. 3D). Therefore, SWS2A paralogs may be differentially expressed between ontogenetic stages in dottybacks, a pattern that has previously been described from the cichlid-specific green opsin duplicates (RH2Aα and RH2Aβ), which feature a similar difference in λmax (∼11 nm) to the one found here (20).
To elaborate on the possibility of ontogenetic neofunctionalization in dottybacks and because single cones mostly express SWS1 and SWS2 (30), we compared the relative levels of gene expression across all SWS genes between adult and larval dottybacks. We found that the UV-sensitive SWS1 gene was not expressed (Fig. 3E), which is supported with transmission measurements that show UV-impermeable lenses in adult dottyback eyes (31). Likewise, although SWS2B expression was found to differ between ontogenetic stages (percentage of total SWS expression mean ± SE: adults, 0.04% ± 0.01%; larvae, 4.45% ± 0.73%; two-tailed t test, t9 = −9.43, P < 0.001), it is probably not relevant for dottyback vision, because it is expressed in very low levels overall (Fig. 3E). Consequently, dottyback single cones mostly express SWS2As. Importantly, the SWS2A paralogs differed substantially in their relative levels of gene expression between ontogenetic stages: adults primarily expressed SWS2Aα (adults, 75.91% ± 2.96%; larvae, 2.66% ± 2.46%; Wilcox test, Z = −2.803, P = 0.005), whereas larvae mostly expressed SWS2Aβ (adults, 24.05% ± 2.95%; larvae, 92.87% ± 2.53%; Z = −2.803, P = 0.005) (Fig. 3E). These results together with the MSP measurements (see above) suggest that SWS2Aβ is the longer wavelength-tuned paralog, which is consistent with the occurrence of an amino acid substitution at site A269T that is known to induce a positive shift in spectral sensitivity of 6 nm (17). Coincidentally, A269T is the only amino acid substitution within our dataset for which the resulting shift in spectral sensitivity has been experimentally confirmed by in vitro mutagenesis (17). Moreover, the A269T amino acid substitution was never found in the putatively more conserved SWS2Aα copy, but it arose multiple times independently in other SWS2 copies of fishes, including in SWS2Aβ of jacks and seabreams (two other families that retained a full set of SWS2 copies) and one of the SWS2B duplicates of lizardfishes (Fig. S1).
Although at this point, we can only speculate about the biological significance of the ∼10-nm shift in spectral sensitivity between the dottyback SWS2A copies, small spectral shifts in sensitivity of other opsin genes in fishes (4–15 nm) have previously been implicated to drive ecological adaptations to various light environments (32) or in some cases, even lead to speciation (16, 33). Similarly, the biological significance of the ontogenetic neofunctionalization of SWS2A copies remains to be investigated but could be tied to major life history changes when larval dottybacks transition in light environment and/or food source from a pelagic life in the open water to a benthic adult life on shallow coral reefs.
Summary and Significance of Findings.
Despite the importance of opsin genes as key components of the animal visual system, little is known about the evolutionary history of this gene family within a larger phylogenetic context. Here, we examine the molecular evolution of SWS2 opsins across teleost fishes. We report multiple gene duplication events in SWS2, including a newly discovered duplication of SWS2A that is specific to the most species-rich lineage of vertebrates (percomorph fish), and provide a novel classification of teleost SWS2 genes, calling for the reinterpretation of previous results. Furthermore, we uncover a complex pattern of gene loss, pseudogenization, and gene conversion (in some cases, possibly leading to the resurrection of pseudogenized gene copies) after SWS2 duplications in fishes. Finally, we provide evidence for functional (adaptive) divergence through neofunctionalization between the percomorph-specific SWS2A paralogs. Our study highlights, once more, the importance of comparative approaches in gaining a comprehensive understanding of the dynamics underlying gene family evolution and ultimately, speciation.
Materials and Methods
Detailed methods are described in SI Materials and Methods.
Data Collection, Gene Synteny, and Phylogenetic Analysis.
Our analyses focused on the genomic region containing SWS2 genes between the upstream HCFC1 and downstream LWS or GNL3L genes (∼30 kbp) in 97 fish species. Genomes, transcriptomes, or single SWS2 genes of 44 species were accessed from public databases at GenBank (www.ncbi.nlm.nih.gov/genbank/) and Ensembl (www.ensembl.org/index.html) (Table S1); the sequences for 53 taxa are new to this study. Raw reads of 38 teleost genomes were used to BLAST search and assemble the target genomic region; for nine species, we sequenced the region using PGM IonTorrent (www.lifetechnologies.com) (Table S2). PGM IonTorrent was also used to generate a reference transcriptome for the dusky dottyback, and an Illumina HiSeq 2000 DNA sequencer (www.illumina.com) was used to generate retina-specific transcriptomes for five additional species (Table S1). Coding regions of the SWS2 genes were individually retrieved from the genomic region containing SWS2. To test for gene conversion, we used single-exon gene phylogenies and combined them with a sliding window approach on one member of each fish family to compare the dS ratio of gene copies (Figs. S2 and S3). Bayesian phylogenetic analyses were performed on coding regions of SWS2 genes (exons one to five; excluding converted parts) (Fig. S1). Results from synteny, gene phylogeny, and conversion approaches were subsequently mapped onto a consensus of the latest fish phylogenies (19, 27) (Fig. 1).
Functional Analysis.
Potentially functional amino acid substitutions were searched for by comparing known key tuning (17) and retinal binding pocket sites of genes from one fish species per family (based on alignments in ref. 34) and extracting those sites that differed in the clade consensus (applying a majority rule consensus after removal of the converted parts) between paralogs (Fig. S6). Mesquite v.3.0 (35) was used to reconstruct the ancestral state of 15 identified sites, which confirmed that 11 of them were clade-specific. Key amino acids were then mapped onto the SWS2 gene phylogeny (standardized to bovine rhodopsin) (Fig. S1). Additionally, we also marked those species with a substitution of A269T, which is known to cause a positive shift in visual sensitivity of 6 nm (17) (Fig. S1). A functional analysis of the percomorph-specific SWS2A duplicates was conducted in the dusky dottyback using a combination of MSP (36) and qRT-PCR approaches (29) (Fig. 3 D and E and Table S2).
Supplementary Material
Acknowledgments
We thank Nicolas Boileau for support with IonTorrent sequencing, Michael Matschiner for help during data mining, the staff at the Lizard Island Research Station for logistical help, and two anonymous referees for valuable suggestions. F.C. was supported by an Australian Endeavour Research Fellowship, Swiss National Science Foundation (SNSF) Doc.Mobility Fellowship P1BSP3_148460, and a Doctoral Fellowship from the Lizard Island Research Station, a facility of the Australian Museum. Z.M. was supported by a Novartis–University of Basel Excellence Scholarship for Life Sciences. N.S.H. was supported by Australian Research Council (ARC) Queen Elizabeth II Research Fellowship DP0558681. M.M. was supported by a Doctoral Fellowship from the Molecular Life Science Foundation. The Teleost Genome Project lead by S.J. is supported by Research Council of Norway Grant 222378. K. L. Cheney and N.J.M. were supported by the ARC. W.S. was supported by the SNSF and European Research Council Grants StG “INTERGENADAPT” and CoG “CICHLID∼X.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: New SWS2 sequences, SWS2 genomic regions, and the transcriptomic raw reads of the P. fuscus reference transcriptome have been deposited in the GenBank database (accession nos. KM978043–KM978047, KP004247–KP004345, and SRX736911).
See Commentary on page 1252.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417803112/-/DCSupplemental.
References
- 1.Taylor JS, Raes J. Duplication and divergence: The evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by Gene Duplication. Springer; Heidelberg: 1970. [Google Scholar]
- 3.Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007;8(12):921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387(6634):708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 5.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3(10):e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer A, Van de Peer Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27(9):937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 7.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13(2):133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grispo MT, et al. Gene duplication and the evolution of hemoglobin isoform differentiation in birds. J Biol Chem. 2012;287(45):37647–37658. doi: 10.1074/jbc.M112.375600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Souza PC, Bonilla-Rodriguez GO. Fish hemoglobins. Braz J Med Biol Res. 2007;40(6):769–778. doi: 10.1590/s0100-879x2007000600004. [DOI] [PubMed] [Google Scholar]
- 10.Porter ML, Zhang Y, Desai S, Caldwell RL, Cronin TW. Evolution of anatomical and physiological specialization in the compound eyes of stomatopod crustaceans. J Exp Biol. 2010;213(Pt 20):3473–3486. doi: 10.1242/jeb.046508. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann CM, Carleton KL. Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr Comp Biol. 2009;49(6):630–643. doi: 10.1093/icb/icp079. [DOI] [PubMed] [Google Scholar]
- 12.Dulai KS, von Dornum M, Mollon JD, Hunt DM. The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Res. 1999;9(7):629–638. [PubMed] [Google Scholar]
- 13.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 14.Sugawara T, et al. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proc Natl Acad Sci USA. 2005;102(15):5448–5453. doi: 10.1073/pnas.0405302102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowmaker JK, Hunt DM. Evolution of vertebrate visual pigments. Curr Biol. 2006;16(13):R484–R489. doi: 10.1016/j.cub.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Seehausen O, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455(7213):620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama S. Evolution of dim-light and color vision pigments. Annu Rev Genomics Hum Genet. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- 18.Rennison DJ, Owens GL, Taylor JS. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 2012;62(3):986–1008. doi: 10.1016/j.ympev.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 19.Near TJ, et al. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci USA. 2013;110(31):12738–12743. doi: 10.1073/pnas.1304661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spady TC, et al. Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Mol Biol Evol. 2006;23(8):1538–1547. doi: 10.1093/molbev/msl014. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura Y, et al. Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. Proc Natl Acad Sci USA. 2013;110(27):11061–11066. doi: 10.1073/pnas.1302051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minamoto T, Shimizu I. Molecular cloning of cone opsin genes and their expression in the retina of a smelt, Ayu (Plecoglossus altivelis, Teleostei) Comp Biochem Physiol B Biochem Mol Biol. 2005;140(2):197–205. doi: 10.1016/j.cbpc.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 23.Katju V, Bergthorsson U. Genomic and population-level effects of gene conversion in caenorhabditis paralogs. Genes (Basel) 2010;1(3):452–468. doi: 10.3390/genes1030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468(2-3):109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- 25.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 26.Near TJ, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA. 2012;109(34):13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betancur-R R, et al. The tree of life and a new classification of bony fishes. PLoS Curr. 2013;5:5. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gojobori J, Innan H. Potential of fish opsin gene duplications to evolve new adaptive functions. Trends Genet. 2009;25(5):198–202. doi: 10.1016/j.tig.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann CM, et al. The eyes have it: Regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biol. 2009;7(12):e1000266. doi: 10.1371/journal.pbio.1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng CL, Flamarique IN. Chromatic organization of cone photoreceptors in the retina of rainbow trout: Single cones irreversibly switch from UV (SWS1) to blue (SWS2) light sensitive opsin during natural development. J Exp Biol. 2007;210(Pt 23):4123–4135. doi: 10.1242/jeb.009217. [DOI] [PubMed] [Google Scholar]
- 31.Siebeck UE, Marshall NJ. Ocular media transmission of coral reef fish—can coral reef fish see ultraviolet light? Vision Res. 2001;41(2):133–149. doi: 10.1016/s0042-6989(00)00240-6. [DOI] [PubMed] [Google Scholar]
- 32.Jokela M, Vartio A, Paulin L, Fyhrquist-Vanni N, Donner K. Polymorphism of the rod visual pigment between allopatric populations of the sand goby (Pomatoschistus minutus): A microspectrophotometric study. J Exp Biol. 2003;206(Pt 15):2611–2617. doi: 10.1242/jeb.00472. [DOI] [PubMed] [Google Scholar]
- 33.Carleton KL, Parry JWL, Bowmaker JK, Hunt DM, Seehausen O. Colour vision and speciation in Lake Victoria cichlids of the genus Pundamilia. Mol Ecol. 2005;14(14):4341–4353. doi: 10.1111/j.1365-294X.2005.02735.x. [DOI] [PubMed] [Google Scholar]
- 34.Carleton KL, Spady TC, Cote RH. Rod and cone opsin families differ in spectral tuning domains but not signal transducing domains as judged by saturated evolutionary trace analysis. J Mol Evol. 2005;61(1):75–89. doi: 10.1007/s00239-004-0289-z. [DOI] [PubMed] [Google Scholar]
- 35.Maddison WP, Maddison DR. 2014 Mesquite: A Modular System for Evolutionary Analysis, Version 3.0. Available at mesquiteproject.org. Accessed September 7, 2014.
- 36.Hart NS, Theiss SM, Harahush BK, Collin SP. Microspectrophotometric evidence for cone monochromacy in sharks. Naturwissenschaften. 2011;98(3):193–201. doi: 10.1007/s00114-010-0758-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.