Significance
We show that the assembly/disassembly of the purinosome is cell cycle-dependent and correlates with cellular demands for purine biosynthesis encountered during the cell cycle. The number of purinosome-containing cells fluctuates: it peaks in G1 phase in HeLa cells cultured under purine-depleted conditions, but remains high across G1, S, and G2/M phases in fibroblasts incapable of purine salvage owing to a deficiency in hypoxanthine-guanine phosphoribosyltransferase. Thus, purinosome function is a cellular biomarker for increased metabolic flux through the de novo purine biosynthetic pathway in response to cellular purine requirements.
Keywords: purinosome, de novo purine biosynthesis, cell cycle, metabolism, fluorescence microscopy
Abstract
The de novo purine biosynthetic pathway relies on six enzymes to catalyze the conversion of phosphoribosylpyrophosphate to inosine 5′-monophosphate. Under purine-depleted conditions, these enzymes form a multienzyme complex known as the purinosome. Previous studies have revealed the spatial organization and importance of the purinosome within mammalian cancer cells. In this study, time-lapse fluorescence microscopy was used to investigate the cell cycle dependency on purinosome formation in two cell models. Results in HeLa cells under purine-depleted conditions demonstrated a significantly higher number of cells with purinosomes in the G1 phase, which was further confirmed by cell synchronization. HGPRT-deficient fibroblast cells also exhibited the greatest purinosome formation in the G1 phase; however, elevated levels of purinosomes were also observed in the S and G2/M phases. The observed variation in cell cycle-dependent purinosome formation between the two cell models tested can be attributed to differences in purine biosynthetic mechanisms. Our results demonstrate that purinosome formation is closely related to the cell cycle.
Enzymes have been shown to form clusters in a cell to regulate metabolic processes (1–3). More recently, the concept of multienzyme complexes has been expanded to include mesoscale protein assemblies that appear to be substantially larger than a single protein (4). Depending on the metabolic or developmental state of the cells, these types of protein clusters range from transiently associated to very rigid and well defined (4). Examples of such enzyme clusters include the EF-Tu cytoskeleton, RNA degradosome, CTP synthase, carboxysomes, and nucleolus (5–9).
In humans, purine nucleotides are synthesized by two different mechanisms. The first mechanism, de novo purine biosynthesis, converts phosphoribosylpyrophosphate (PRPP) to inosine 5′-phosphate (IMP) in 10 highly conserved steps catalyzed by six enzymes. These six enzymes include one trifunctional enzyme (TrifGART: GARS, GART, and AIRS domains), two bifunctional enzymes (PAICS: CAIRS and SAICARS domains; ATIC: AICART and IMPCH domains), and three monofunctional enzymes (PPAT, FGAMS, and ASL). ASL is also necessary for the conversion of IMP to AMP and may be classified as bifunctional (10). The second mechanism uses nucleotide salvage pathways to either phosphorylate a nucleoside (e.g., thymidine kinase) or add a purine base to ribose 5′-phosphate to regenerate the respective monophosphate. For example, hypoxanthine/guanine phosphoribosyl transferase (HGPRT) catalyzes the conversion of hypoxanthine to IMP and the conversion of guanine to guanosine 5′-phosphate (GMP). The de novo pathway is more energy-intensive, with the synthesis of 1 mole of IMP requiring 5 moles of ATP. Therefore, the salvage pathway is the preferred pathway for purine biosynthesis. Cells deficient in HGPRT, such as those that exhibit a Lesch–Nyhan disease (LND) phenotype, rely primarily on the de novo purine biosynthetic pathway to generate purine nucleotides (11–15).
Recently, enzymes in the de novo purine biosynthetic pathway were shown to organize and reversibly assemble into punctate cellular bodies known as “purinosomes” under purine-depleted conditions (16). Further investigations into the organization of the purinosome showed that several of the enzymes form a core structure (PPAT, TrifGART, and FGAMS), whereas others appear to interact peripherally (PAICS, ASL, and ATIC) (17). The presence of this protein assembly and its putative function(s) clearly suggest that the spatial organization of pathway enzymes into purinosomes within a cell play an important role in meeting the cellular demand for purines (18). Although many studies have implied up-regulation of the de novo purine biosynthetic pathway during cell cycle progression (14, 19–21), here we used time-lapse microscopy to determine whether a correlation exists between the cell cycle stage and the number of cells with purinosomes or the cells’ purinosome content. Two different cell types, HeLa and LND cells, were used, with the latter deficient in purine salvage, to assess the effect of increased demand on the de novo pathway for purine biosynthesis and the attendant consequences for purinosome assembly.
Results
Purinosome Formation in HeLa Cells in Different Phases of the Cell Cycle.
In previous studies, we demonstrated reversibility, characterized the approximate spatial organization of the proteins, and identified putative signaling pathways for the purinosome at steady state (17, 18, 22–24). Many cellular processes, including metabolism, undergo significant changes as the cells progress through the cell cycle (15, 20, 25, 26). Cells’ nutrient and energy needs vary as they prepare for growth (G1), duplicate their genetic materials (S), get ready for division (G2), and divide (M). Given that HeLa cells have an approximate doubling time of 24 h, we anticipated that these cells would be in different phases of the cell cycle over the time course studied.
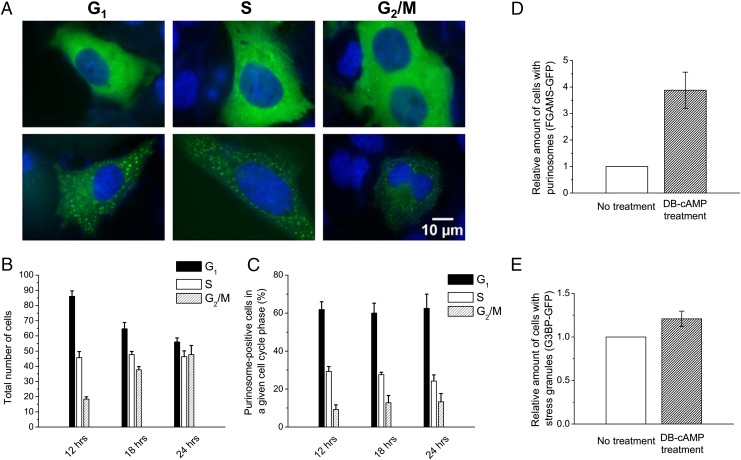
To establish how purinosome formation might vary in different phases of the cell cycle, we captured representative images of purinosome-negative and -positive HeLa cells in different phases of the cell cycle (Fig. 1A). By quantifying the fluorescence intensity of Vybrant DyeCycle staining, we determined the phase of the cell cycle for each set of cells imaged at 12, 18, or 24 h posttransfection (Figs. S1 and S2). At each time point, 150 cells were selected at random, measured, and classified based on their cell cycle status (Fig. 1B). During the time frame studied, the number of cells in G1 phase decreased. The purinosome-positive cells in G1, S, and G2/M phases of the cell cycle were ∼60%, 25%, and 15%, respectively, of the total number of purinosome-positive cells independent of the time points studied (Fig. 1C). This observation is consistent with the changing metabolic requirements of the cell as it prepares to grow and duplicate its genetic material (20), and provides the first direct evidence of a strong correlation between cell cycle stage and the number of purinosome-containing cells.
Fig. 1.
Cell cycle dependence of the relative proportion of HeLa cells with purinosomes. (A) Fluorescence microscope images of HeLa cells without purinosomes (Upper) or with purinosomes (Lower) in different phases of the cell cycle (green, FGAMS-GFP; blue, Vybrant DyeCycle). (B) Number of HeLa cells (both purinosome-negative and -positive) as a function of the cell cycle at at 12, 18, and 24 h posttransfection. A total of 150 cells were analyzed at each time point (n = 3). (C) Percentage of purinosome-positive HeLa cells in a given phase of the cell cycle at 12, 18, and 24 h (n = 3). (D) Comparison of the relative amount of cells with purinosomes when synchronized with DB-cAMP (n = 3). A total of 50 transfected cells were analyzed in each independent experiment. (E) Comparison of the relative amount of cells with stress granules using G3BP-GFP as a marker when synchronized with DB-cAMP (n = 3).
Synchronization of HeLa Cells in G1 Phase.
To confirm that purinosomes form in the cells predominately at G1 phase, HeLa cells were synchronized during G1 phase with dibutyryl-cAMP (DB-cAMP), and the number of purinosome-containing cells was determined. Under these conditions, a 3.8-fold increase in purinosome-positive cells was observed (Fig. 1D). To address whether the increased purinosome content was related to an increase in cellular stress introduced by DB-cAMP, we monitored the formation of stress granules in synchronized cells at G1 phase under the same conditions. RasGAP-associated endoribonuclease G3BP-GFP was used as the stress granule marker in the control experiment (27, 28). We observed only a 1.2-fold increase in stress granule formation in cells treated with DB-cAMP (Fig. 1E). Images of HeLa cells transfected with FGAMS-GFP and G3BP-GFP also revealed the morphological differences between the two types of markers (Fig. S3). Thus, the increase in number of purinosome-positive cells at the G1 phase is not the result of extra stress on the cells.
Temporal Analysis of Relative Purinosome Formation in HeLa Cells.
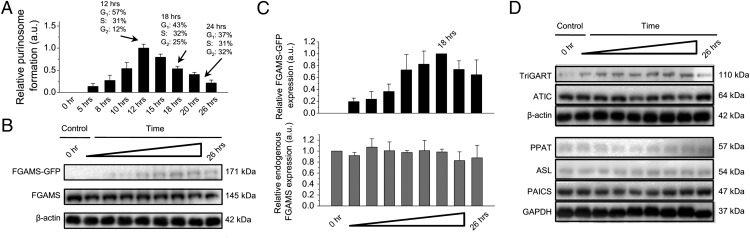
To gain additional insight into the relationship between purinosome formation and the cell cycle, we investigated the temporal aspects of relative purinosome formation in HeLa cells from 5 h to 26 h posttransfection. Cell cycle analysis demonstrated that on this time scale, HeLa cells should be progressing through various stages of the cell cycle (Fig. 1B). Relative purinosome formation is defined as the number of purinosome-positive cells over the total number of transfected cells at a given time point. The number of purinosome-positive cells was determined based on a random sampling of 100 cells. Purinosome formation increased steadily, reaching a maximum at 12 h, and then diminished to ∼20% of the maximum at 26 h (Fig. 2A). The percentage of the cells in different cell cycle phases was also evaluated at 12, 18, and 24 h. The drop in the relative amount of purinosome-positive cells was correlated with progression of the HeLa cells into the S and G2/M phases.
Fig. 2.
Temporal dynamics of purinosome formation in HeLa cells. (A) Relative proportion of HeLa cells with purinosomes in purine-depleted medium observed by fluorescence microscopy at different time points posttransfection with FGAMS-GFP. The relative proportion of purinosome-positive cells was based on 100 transfected cells. The percentages of the cells in different cell cycle phases for 12, 18, and 24 h were calculated from Fig. 1B. (B) Protein expression levels of FGAMS-GFP and endogenous FGAMS posttransfection as a function of time. β-actin served as a loading control. (C) Statistical analyses of protein expression levels of FGAMS-GFP and endogenous FGAMS as a function of time. (D) Protein expression levels of endogenous TrifGART, ATIC, PPAT, ASL, and PAICS as a function of time posttransfection of FGAMS-GFP. GAPDH and β-actin served as loading controls.
Purinosome Formation Is Not a Function of Protein Expression.
We performed Western blot analysis to identify any correlation between FGAMS-GFP expression and the relative amount of purinosome-positive cells. Statistical analyses of the protein expression levels of FGAMS-GFP and endogenous FGAMS showed that the FGAMS-GFP expression level reached a maximum at 18 h posttransfection and remained constant thereafter (Fig. 2 B and C). Statistical analyses of endogenous FGAMS expression also remained constant throughout the time course, indicating that overexpression of FGAMS-GFP did not alter the endogenous level of the protein (n = 3).
To confirm that the expression of the other enzymes in the de novo purine biosynthetic pathway was not altered posttransfection, we also monitored the endogenous protein expression level of the five remaining enzymes. A shown in Fig. 2D and Fig. S4, the protein expression levels of the members enzymes remained constant over the time course studied.
We performed single-cell analysis to evaluate the total fluorescence intensity in cells with and without purinosomes under a purine-depleted condition. No difference in the average fluorescence intensity per cell was observed between cells classified as purinosome-negative and those classified as purinosome-positive (Fig. S5). This result suggests that highly fluorescent cells (correlated with high protein expression of FGAMS-GFP) do not show a higher propensity to form purinosomes. Therefore, the formation of purinosome in the cells is not governed by protein expression level.
Purinosome Characterization in Cell Models.
We used an LND fibroblast model to evaluate the influence of the parallel salvage pathway in HeLa cells on purinosome appearance and levels in the phase of the cell cycle. These LND cells are HGPRT-deficient and rely primarily on the de novo purine biosynthetic pathway to meet purine demand.
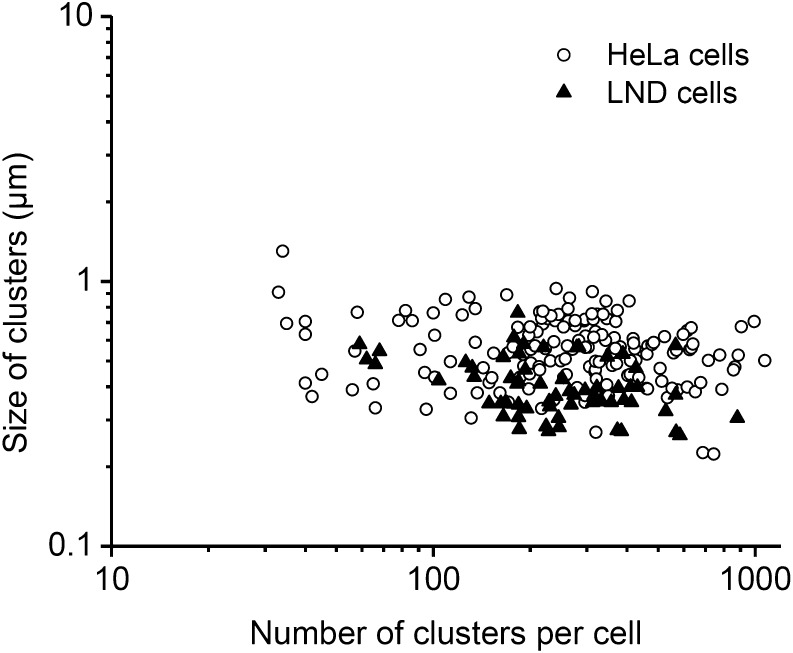
To properly classify purinosome-containing cells in the two cell models used for this study, we performed the basic morphological characterization of purinosomes in both HeLa and LND cells. We chose the average size and number of purinosomes in a given cell as the physical criteria to distinguish purinosomes from other cellular bodies. Purinosome diameter varied between 0.2 and 0.9 μm, with an average of 0.56 ± 0.16 μm in HeLa cells (Fig. 3). The median number of purinosomes inside purinosome-positive HeLa cells was 278 (Fig. 3). We found no correlations between fluorescence intensity in a purinosome-positive cells and the average size and number of purinosomes in that cell (Fig. S6). For added measure, we evaluated the spatial organization of purinosomes in cells using superresolution stochastic optical reconstruction microscopy (STORM) (29). The size distribution in HeLa cells detected using STORM was consistent with previous observations (Fig. 3 and Fig. S7).
Fig. 3.
Purinosome characterization in cell models. Shown are the general size and number distribution of purinosomes in HeLa cells and LND cells after single-cell analysis (n = 200 for HeLa cells; n = 50 for LND cells).
Finally, we subjected nontransfected fixed LND cells to immunofluorescence imaging of the enzymes ASL and FGAMS, which demonstrated their clustering into purinosome punctates (Fig. S8). In LND cells, the average diameter of purinosomes was 0.41 ± 0.11 μm, and the median number of purinosomes inside LND purinosome-positive cells was 235. The results show that purinosomes formed in LND cells are of similar size and number distribution as those formed in HeLa cells (Fig. 3). Therefore, the results are in accordance with the observation of the same cellular body, the purinosomes, in both cell types.
Cell Cycle Dependency of HGPRT-Deficient Cells.
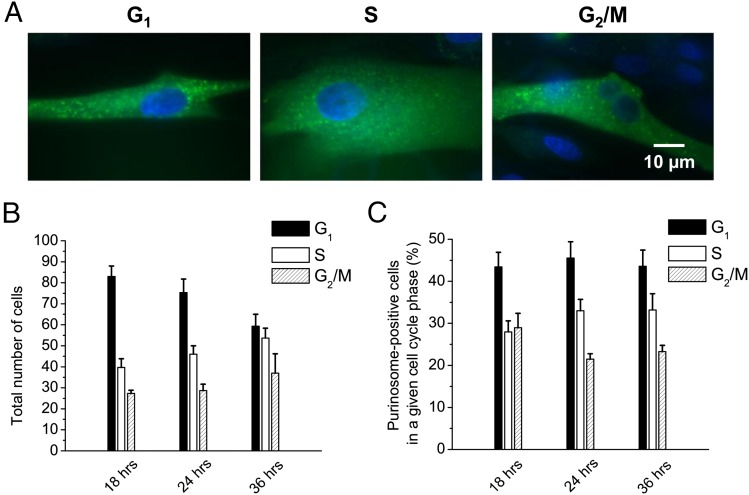
LND fibroblast cells were transfected with FGAMS-GFP, and representative images of purinosome-positive cells in different phases of the cell cycle were acquired (Fig. 4A). The phases of the cell cycle were determined for each set of cells imaged at 18, 24, and 36 h posttransfection. We evaluated the cell population in different phases of cell cycle shift after 24 h posttransfection (Fig. 4B). Similar to HeLa cells, in LND cells the majority of purinosome formation was observed in the G1 phase of the cell cycle (45% for LND vs. 60% for HeLa); however, higher proportions of purinosome-positive LND cells were observed in the S and G2/M phases (30% and 25%, respectively, for LND vs. 25% and 15% for HeLa) (Figs. 1C and 4C).
Fig. 4.
Cell cycle dependency of HGPRT-deficient LND fibroblast cells on purinosome formation. (A) Fluorescence microscope images of LND cells with purinosomes in different phases of the cell cycle (green, FGAMS-GFP; blue, Vybrant DyeCycle). (B) Number of LND cells (both purinosome-negative and -positive) in different phases of the cell cycle at 18, 24, and 36 h. A total of 150 cells were analyzed at each time point (n = 3). (C) Purinosome-positive LND cells in different phases of the cell cycle at 18, 24, and 36 h (n = 3).
In addition, we investigated the average size and number of purinosomes per cell in different cell cycle phases in both HeLa and LND fibroblast cells. Fig. S9 A and B illustrates the distribution of the average size of purinosomes in the three phases of the cell cycle, and Fig. S9 C and D shows the number of purinosomes per cell. No correlation between the average size and number of purinosomes per cell was observed across the different phases of the cell cycle (Fig. S9).
Discussion
Previous findings have demonstrated that de novo purine biosynthesis is closely related to the cell cycle (19, 20, 25, 30–33). Studies of other enzyme complexes have suggested that the assembly or disassembly of an enzyme cluster may be correlated with cellular events, such as developmental cues or metabolic states of the cell (33); for example, the replitase, a six-enzyme complex involved in DNA replication, has been shown to exist only during S phase (34). In the present study, we aimed to understand purinosome formation as a function of the cell cycle phases. Through the use of time-lapse fluorescence microscopy, we observed that HeLa and fibroblast cells had the highest number of purinosome-positive cells in the G1 phase of the cell cycle (Fig. 1 C and D). The de novo purine biosynthetic pathway is the predominant pathway in G1 phase when purine demand for the cells is highest (19). This observation further supports our hypothesis that purinosomes are assembled to meet the cellular demand for purines via the de novo biosynthetic pathway.
The proportion of purinosome-positive cells in HeLa cells dropped after 12 h posttransfection (Fig. 2A). Our Western blot data suggest that this decrease is not related to the degradation of FGAMS-GFP or to changes in endogenous protein expression levels (Fig. 2). Furthermore, we observed a decrease in the percentage of purinosome-positive cells in the S and G2/M phases (Fig. 1C). We attribute the decline shown in Fig. 2A to a shift in the population of cells out of the G1 phase. This shift underscores the reversibility of purinosome assembly.
In this study, HeLa cells were grown and maintained in a purine-depleted condition. Extracellular purine depletion exacerbates the dependency on the de novo purine biosynthetic pathway, owing to a lack of free nucleosides that normally would promote salvage pathway use (14, 15, 17, 25, 31). As a result, more purinosome-positive cells are observed in the G1 phase when the purine demand is highest. However, once purines are produced by the cells using the de novo pathway, HeLa cells can leverage the salvage pathway to recycle the purines thus generated. As the purine demand decreases throughout the cell cycle, recycled purines are sufficient for cell growth. This would account for the decrease in purinosome-positive cells in S and G2/M phases of the cell cycle.
Compared with HeLa cells, higher percentages of purinosome-positive LND cells were maintained in the S and G2/M phases (Fig. 4C). This difference can be attributed to the fact that LND cells rely primarily on the de novo pathway to meet intracellular purine demand (13, 35). Thus, LND cells provide support to justify our hypothesis that purinosome formation correlates with enhanced reliance on the de novo purine biosynthetic pathway.
Future studies will focus on understanding the variability in the size of purinosome punctates as a consequence of changes in their enzyme composition. Although to date at least six enzymes as well as Hsp70/90 have been identified in the purinosomes, the size of the punctate suggests that they are present in multiples. Other, as-yet unknown proteins may be involved as well. One advantage of enzyme clustering is to accelerate the processing of pathway intermediates and differs from direct channeling (36). Another advantage is that the clusters act as delivery or receptor vehicles. The results of the work detailed herein will provide valuable tools to facilitate these future investigations.
Materials and Methods
Cell Culture.
HeLa cells were obtained from the American Type Culture Collection, maintained in MEM, and transitioned for 1 wk into purine-depleted medium [RPMI 1640 with 5% (vol/vol) dialyzed FBS] as described previously (16). LND fibroblasts were provided by Dr. H. A. Jinnah (Emory University, Atlanta, GA) and maintained in high-glucose DMEM (Life Technologies) supplemented with 2 mM glutamine and 15% (vol/vol) FBS (Invitrogen).
Plasmids for Transient Transfection.
Plasmids of FGAMS-GFP and FGAMS-mEos2 were constructed as described previously (16). G3BP-GFP was a generous gift from Dr. Jamal Tazi (Institut de Genetique Moleculaire de Montpellier, Montpellier, France).
Transient Transfection and Live Cell Fluorescence Imaging of Mammalian Cells.
HeLa cells grown in purine-depleted media were transiently transfected with plasmid DNA (concentration ≥1 μg/μL) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. LND fibroblasts grown in normal media were electroporated using the Neon Transfection System MPK5000 (Life Technologies) following the manufacturer’s optimized protocol. For each 10-µL reaction, 2.5‒3 × 105 cells were used. Reactions were performed under the conditions of 1400 V/20 ms/1 pulse. Cells were then transferred to Minimum Essential Medium (Corning) supplemented with 10% (vol/vol) FBS (Atlanta Biological). For live cell fluorescence imaging, cells were imaged under physiological conditions (∼37 °C, 5% CO2) in a cell incubation chamber (Tokai Hit) with a 60× Nikon Apo TIRF oil immersion objective using a S484/15x excitation filter and a S517/30m emission filter for GFP (Chroma Technology).
Cell Synchronization.
To synchronize the cells at the G1 phase, 0.5 mM of DB-cAMP (Sigma-Aldrich) was added to the cell culture medium at 5 h after transfection of FGAMS-GFP or G3BP-GFP and incubated for an additional 18 h. Medium containing DB-cAMP was exchanged with purine-depleted medium without phenol red before imaging. A total of 50 cells were selected at random, and the number of cells containing purinosomes or stress granules was determined with and without synchronization.
Western Blot Analysis of Purinosome Enzymes.
After the transfection of FGAMS-GFP, cell lysates were prepared using M-PER mammalian protein extraction reagent (Thermo Scientific) at different time points. SDS/PAGE was performed on the extracted protein (20 μg/lane), followed by transferrence to a PVDF membrane, probing, and detection by chemiluminescence (Millipore). Primary antibodies for the six purinosome enzymes were purchased from either Abcam or LifeSpan Biosciences. HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology. Quantification of protein expression levels from Western blot analyses was performed with the Bio-Rad ChemiDoc XRS+ system.
Single-Cell Analysis.
Cell cycle status was determined using Vybrant DyeCycle (Life Technologies) according to the manufacturer’s protocols. Fluorescence microscopy images were captured using the DAPI filter channel. The intensity in the cell with two nuclei served as a reference for the G2/M phase of the cell cycle. A cell in the G1 phase of the cell cycle was characterized as having one-half of the reference fluorescence intensity. Any cell with a fluorescence intensity falling between the G1 and G2/M phases was classified as being in S phase. Purinosome density was defined as the number of purinosomes per cell, and purinosome size was defined as the average size within a single cell. More details are provided in SI Materials and Methods.
Immunofluorescence Imaging of Fixed Cells.
A total of 3.4 × 104 LND fibroblast cells were plated on a 35-mm2 glass-bottom plate (MatTek) and incubated at 37 °C (5% CO2) for 16 h. Adherent cells were fixed with freshly prepared 4% (vol/vol) formaldehyde (Electron Microscopy Sciences) prepared in 1× Dulbecco’s PBS (Corning) for 10 min in the dark at room temperature, and then permeabilized with 0.2% (vol/vol) Triton X-100 prepared in 1× Dulbecco’s PBS for 10 min at room temperature. Cells were blocked with 5% (vol/vol) normal goat serum (Jackson ImmunoResearch) for 30 min at room temperature. Primary antibodies for ASL (Santa Cruz Biotechnology; catalog no. sc-365623) and FGAMS (Bethyl Laboratories; catalog no. A304-218A) were directly labeled using Lightning-Link antibody conjugation kits (Novus Biologics) with Atto488 and Cy3, respectively. Cells were stained with a 1:100 (vol/vol) dilution of Atto488-labeled PPAT antibody or Cy3-labeled FGAMS antibody for 2 h at room temperature in the dark. All antibody dilutions were performed in 1× Dulbecco’s PBS. Cells were washed 3 × 2 mL with 1× Dulbecco’s PBS and placed into HBSS before imaging. Imaging was performed with a 60× Nikon Apo TIRF oil immersion objective (Nikon) using a S484/15x excitation filter and a S517/30m emission filter for Atto488 detection and a S555/25x excitation filter and a S605/40m emission filter for Cy3 detection (Chroma Technology). Images were analyzed in ImageJ.
STORM.
HeLa cells were transiently transfected with FGAMS-mEos2 (FGAMS linked to photoactivatable fluorescent proteins; mEos2) under the aforementioned conditions. STORM was performed on fixed cells at 18 h posttransfection. Purinosome size was determined by analysis algorithms as described previously (29).
Supplementary Material
Acknowledgments
We thank Dr. Yanhui Zhao, Dr. Peng Li, and Feng Guo (The Pennsylvania State University) for helpful discussions. Funding for this study was provided by National Institutes of Health Grant GM024129 (to S.J.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423009112/-/DCSupplemental.
References
- 1.Wilson MZ, Gitai Z. Beyond the cytoskeleton: Mesoscale assemblies and their function in spatial organization. Curr Opin Microbiol. 2013;16(2):177–183. doi: 10.1016/j.mib.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norris V, Turnock G, Sigee D. The Escherichia coli enzoskeleton. Mol Microbiol. 1996;19(2):197–204. doi: 10.1046/j.1365-2958.1996.373899.x. [DOI] [PubMed] [Google Scholar]
- 3.Mathews CK, Moen LK, Wang Y, Sargent RG. Intracellular organization of DNA precursor biosynthetic enzymes. Trends Biochem Sci. 1988;13(10):394–397. doi: 10.1016/0968-0004(88)90182-x. [DOI] [PubMed] [Google Scholar]
- 4.Wombacher H. Molecular compartmentation by enzyme cluster formation: A view over current investigations. Mol Cell Biochem. 1983;56(2):155–164. doi: 10.1007/BF00227216. [DOI] [PubMed] [Google Scholar]
- 5.Mayer F. Cytoskeletons in prokaryotes. Cell Biol Int. 2003;27(5):429–438. doi: 10.1016/s1065-6995(03)00035-0. [DOI] [PubMed] [Google Scholar]
- 6.Taghbalout A, Rothfield L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci USA. 2007;104(5):1667–1672. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12(8):739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: Their properties and paradoxes. BioEssays. 2008;30(11-12):1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendes P, Kell DB, Welch GR. Metabolic channeling in organized enzyme systems: Experiments and models. Adv Mol Cell Biol. 1995;11:1–19. [Google Scholar]
- 10.Smith GK, Mueller WT, Wasserman GF, Taylor WD, Benkovic SJ. Characterization of the enzyme complex involving the folate-requiring enzymes of de novo purine biosynthesis. Biochemistry. 1980;19(18):4313–4321. doi: 10.1021/bi00559a026. [DOI] [PubMed] [Google Scholar]
- 11.McKeran RO, Watts RWE. Use of phytohaemagglutinin-stimulated lymphocytes to study effects of hypoxanthine-guanine phosphoribosyltransferase (HGPRT) deficiency on polynucleotide and protein synthesis in the Lesch-Nyhan syndrome. J Med Genet. 1976;13(2):91–95. doi: 10.1136/jmg.13.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinnah HA, De Gregorio L, Harris JC, Nyhan WL, O’Neill JP. The spectrum of inherited mutations causing HPRT deficiency: 75 new cases and a review of 196 previously reported cases. Mutat Res. 2000;463(3):309–326. doi: 10.1016/s1383-5742(00)00052-1. [DOI] [PubMed] [Google Scholar]
- 13.Fu R, et al. Lesch-Nyhan Disease International Study Group Genotype-phenotype correlations in neurogenetics: Lesch-Nyhan disease as a model disorder. Brain. 2014;137(Pt 5):1282–1303. doi: 10.1093/brain/awt202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker MA, Kim M. Regulation of purine synthesis de novo in human fibroblasts by purine nucleotides and phosphoribosylpyrophosphate. J Biol Chem. 1987;262(30):14531–14537. [PubMed] [Google Scholar]
- 15.Becker MA, Losman MJ, Kim M. Mechanisms of accelerated purine nucleotide synthesis in human fibroblasts with superactive phosphoribosylpyrophosphate synthetases. J Biol Chem. 1987;262(12):5596–5602. [PubMed] [Google Scholar]
- 16.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 17.Deng Y, et al. Mapping protein-protein proximity in the purinosome. J Biol Chem. 2012;287(43):36201–36207. doi: 10.1074/jbc.M112.407056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verrier F, et al. GPCRs regulate the assembly of a multienzyme complex for purine biosynthesis. Nat Chem Biol. 2011;7(12):909–915. doi: 10.1038/nchembio.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo M, et al. The rate of cell growth is regulated by purine biosynthesis via ATP production and G(1) to S phase transition. J Biochem. 2000;128(1):57–64. doi: 10.1093/oxfordjournals.jbchem.a022730. [DOI] [PubMed] [Google Scholar]
- 20.Fridman A, et al. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biochem J. 2013;454(1):91–99. doi: 10.1042/BJ20130153. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, French JB, Fang Y, Benkovic SJ. The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem Commun (Camb) 2013;49(40):4444–4452. doi: 10.1039/c3cc41437j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci USA. 2010;107(29):12872–12876. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J Biol Chem. 2010;285(15):11093–11099. doi: 10.1074/jbc.M110.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.French JB, et al. Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc Natl Acad Sci USA. 2013;110(7):2528–2533. doi: 10.1073/pnas.1300173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaoka T, et al. Feedback inhibition of amidophosphoribosyltransferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses. J Biol Chem. 2001;276(24):21285–21291. doi: 10.1074/jbc.M011103200. [DOI] [PubMed] [Google Scholar]
- 26.Quéméneur L, et al. Differential control of cell cycle, proliferation, and survival of primary T lymphocytes by purine and pyrimidine nucleotides. J Immunol. 2003;170(10):4986–4995. doi: 10.4049/jimmunol.170.10.4986. [DOI] [PubMed] [Google Scholar]
- 27.Tourrière H, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160(6):823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Tourrière H, et al. RasGAP-associated endoribonuclease G3Bp: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21(22):7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143(7):1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwahana H, et al. Rat genomic structure of amidophosphoribosyltransferase, cDNA sequence of aminoimidazole ribonucleotide carboxylase, and cell cycle-dependent expression of these two physically linked genes. Biochim Biophys Acta. 1995;1261(3):369–380. doi: 10.1016/0167-4781(95)00036-g. [DOI] [PubMed] [Google Scholar]
- 31.Yamaoka T, et al. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J Biol Chem. 1997;272(28):17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- 32.Shedden K, Cooper S. Analysis of cell cycle-specific gene expression in human cells as determined by microarrays and double-thymidine block synchronization. Proc Natl Acad Sci USA. 2002;99(7):4379–4384. doi: 10.1073/pnas.062569899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Field MS, Anderson DD, Stover PJ. Mthfs is an essential gene in mice and a component of the purinosome. Front Genet. 2011;2:36. doi: 10.3389/fgene.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA. 1980;77(6):3312–3316. doi: 10.1073/pnas.77.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dauphinot L, et al. Transcriptomic approach to Lesch-Nyhan disease. Nucleosides Nucleotides Nucleic Acids. 2014;33(4-6):208–217. doi: 10.1080/15257770.2014.880477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castellana M, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014;32(10):1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.