Abstract
This study was designed to assess the influence of three soil DNA extraction procedures, namely the International Organization for Standardization (ISO-11063, GnS-GII and modified ISO procedure (ISOm), on the taxonomic diversity and composition of soil bacterial and fungal communities. The efficacy of each soil DNA extraction method was assessed on five soils, differing in their physico-chemical characteristics and land use. A meta-barcoded pyrosequencing approach targeting 16S and 18S rRNA genes was applied to characterize soil microbial communities. We first observed that the GnS-GII introduced some heterogeneity in bacterial composition between replicates. Then, although no major difference was observed between extraction procedures for soil bacterial diversity, we saw that the number of fungal genera could be underestimated by the ISO-11063. In particular, this procedure underestimated the detection in several soils of the genera Cryptococcus, Pseudallescheria, Hypocrea and Plectosphaerella, which are of ecological interest. Based on these results, we recommend using the ISOm method for studies focusing on both the bacterial and fungal communities. Indeed, the ISOm procedure provides a better evaluation of bacterial and fungal communities and is limited to the modification of the mechanical lysis step of the existing ISO-11063 standard.
Introduction
During the last three decades, the challenge to better characterize soil microbial communities has led to the development of culture-independent techniques that are well suited to deciphering the huge diversity of soil microbes as they provide access to previously hidden genetic resources (Martin-Laurent et al., 2001). These methods are based essentially on the direct extraction and characterization of soil DNA. In this context, most efforts have been devoted to optimizing the soil DNA extraction procedure in order to obtain suitable representative extracts for quantitative and qualitative characterization of the microbial communities (Roesch et al., 2007; Rajendhran and Gunasekaran, 2008; Terrat et al., 2012). These efforts led to the development of various homemade DNA extraction protocols and even commercial kits (Zhou et al., 1996; Martin-Laurent et al., 2001; Delmont et al., 2011a; Terrat et al., 2012). However, each method had its own advantages and potential biases, leading to variations in DNA representativeness and consequently to effects on soil microbial assessments, making comparisons between studies impossible (Zhou et al., 1996; Martin-Laurent et al., 2001; Terrat et al., 2012). To deal with this issue, Delmont and colleagues (2011b) suggested that several soil sampling and DNA extraction strategies should be combined to access the whole soil microbial metagenome in terms of species richness. However, this approach is clearly not applicable or relevant to wide-scale studies, where time and cost constraints make the need to use a standardized single DNA extraction procedure obvious (Dequiedt et al., 2011).
In this context, a standardized ‘ISO-11063: Soil quality – Method to directly extract DNA from soil’ was developed and validated by independent laboratories to efficiently recover bacterial DNA from various soil samples (Philippot et al., 2010; Petric et al., 2011). However, archaeal and fungal groups also constitute a significant proportion of the soil microbial biodiversity and are key organisms for soil processes. In a previous study, we tested the sensitivity of the ISO-11063 method for the detection of these groups (Plassart et al., 2012). Briefly, three different procedures were compared on five soils with contrasting land-use and physico-chemical properties: (i) the ISO-11063 standard; (ii) a modified ISO procedure (ISOm) that includes a particular mechanical lysis step (a FastPrep®-24 lysis step instead of the recommended bead beating using a mini bead-beater cell disruptor); and (iii) a custom procedure called GnS-GII, which also includes the FastPrep®-24 mechanical lysis step. This evaluation revealed that the ISO-11063 procedure yielded significantly less overall microbial DNA, (corroborated by measurement of the bacterial, archaeal and fungal densities by real-time PCR), whatever the soil is (Plassart et al., 2012). Furthermore, the analysis of fungal communities' structure with terminal restriction fragment length polymorphism (T-RFLP) patterns showed that the two non-ISO methods clearly outperformed the ISO-11063 method, leading to more significant variations because of soil type and management. Finally, one major conclusion of this study was that the non-ISO methods provided a better representativeness of soil DNA mainly due to use of the FastPrep®-24 bead-beating system, achieving lysis of the majority of cells with tough walls and particularly fungal cells, more efficiently than the usual bead beating (Ranjard et al., 2010; Rousk et al., 2010; Yarwood et al., 2010; Dequiedt et al., 2011; Plassart et al., 2012). Nevertheless, this comparative study was carried out using classical molecular approaches, i.e., quantitative PCR and community DNA fingerprinting through T-RFLP. Nowadays, high throughput sequencing technologies (e.g. 454 or Illumina) are readily available to assess microbial diversity with greater precision by obtaining hundreds of thousands of ribosomal rRNA gene sequences from a single metagenomic DNA (Roesch et al., 2007; Will et al., 2010; Maron et al., 2011). Nonetheless, the DNA extraction techniques previously described has never been evaluated with these new technologies, in terms of efficiency and representativeness, despite their widespread use in soil microbial diversity studies.
In the present study, the same three DNA extraction procedures, coupled with high throughput sequencing technology, were evaluated to identify a technique suitable to characterize the diversity and composition of bacterial and fungal communities simultaneously. The guideline standard ISO-11063, the custom GnS-GII and a custom DNA extraction procedure derived from the ISO-11063 standard (ISOm), were used to extract template DNA from five different soils with contrasting land-use and physico-chemical properties (Plassart et al., 2012). A meta-barcoded pyrosequencing technique, targeting the 16S and 18S rRNA genes, was used to characterize bacterial and fungal communities' richness [based on the number of operational taxonomic units (OTUs) and genera detected], diversity (using Shannon and Evenness indices) and composition (taxonomic affiliation of OTUs). We also measured the phylogenetic distance between sets of OTUs in a phylogenetic tree using the unifrac method to determine whether bacterial and fungal community compositions were influenced by the DNA extraction procedures.
Results and discussion
Since the development of molecular tools to study soil microbial communities, it has been largely demonstrated that the characterization of these communities might be influenced by the method used to recover soil metagenomic DNA (Delmont et al., 2011b; Terrat et al., 2012). It is consequently essential to test the representativeness of soil DNA extraction methods in terms of bacterial and fungal organisms, which constitute a major part of the soil microbial community. Here, the efficacy of three soil DNA extraction methods (ISO-11063, ISOm and GnS-GII) was assessed on five soils with different physico-chemical characteristics and land use (Table 1) using a meta-barcoded pyrosequencing technique targeting bacterial and fungal communities. This approach was chosen because it is a recently developed powerful technique widely used for detailed phylogenetic and taxonomic surveys of microbial communities (Roesch et al., 2007; Rousk et al., 2010; Will et al., 2010; Lienhard et al., 2013a).
Table 1.
Origin, physical and chemical parameters of the five French soils used
| Soil | Collection site | Origin | Clay | Fine loam | Coarse loam | Fine sand | Coarse sand | Organic carbon | Total N | C/N | CaCO3 | pH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | Agricultural Site (Champdotre, Burgundy) | Crop soil | 504 | 180 | 145 | 73 | 98 | 24.9 | 2.8 | 9 | 102 | 7.75 |
| E | INRA Experimental Site (Epoisses, Burgundy) | Crop soil | 392 | 320 | 228 | 34 | 26 | 16.5 | 1.65 | 10 | 2 | 7 |
| F | Forest Observatory Plot (La Mailleraye-sur-Seine, Normandy) | Forest soil | 101 | 167 | 205 | 217 | 310 | 103.3 | 3.1 | 34 | < 1 | 3.8 |
| L | INRA Experimental Site SOERE-ACBB (Lusignan, Poitou) | Grassland | 175 | 369 | 304 | 73 | 79 | 13.2 | 1.33 | 9.92 | < 1 | 6.6 |
| R | INRA Experimental Site (Pierrelaye, Ile-de-France) | Crop soil | 79 | 66 | 44 | 315 | 496 | 50.2 | 2.16 | 23.3 | 22 | 7.5 |
Clay, fine loam, coarse loam, fine sand and coarse sand, organic carbon, total N and calcium carbonate are given in mg g−1. Originally published and extracted from (Plassart et al., 2012).
Influence of soil DNA extraction procedure on bacterial richness and diversity
Bacterial rRNA gene sequences were successfully amplified by PCR and sequenced from all soils using each of the three DNA extraction procedures (Table 2). After bioinformatic filters, 2322 high-quality reads per sample were kept, analyzed and taxonomically identified using a curated database derived from SILVA (Quast et al., 2013) (Table 3). Rarefaction curves of bacterial richness demonstrated that our sequencing depth allowed accurate description of the bacterial community diversity in each soil sample studied (Supporting Information Fig. S1).
Table 2.
Bacterial richness and diversity indices of the five soils used
| Number of genera | OTUs (95% of similarity) | Shannon | Evenness | ||
|---|---|---|---|---|---|
| C | GnS | 207.67 (± 5.73) 2 | 485.67 (± 49.13) 1,2 | 5.17 (± 0.13) 1,2 | 0.84 (± 0.01) 1,2 |
| ISO | 207.33 (± 4.19) 1,2 | 524.33 (± 15.11) 1,2 | 5.15 (± 0.01) 1,2 | 0.82 (± 0.00) 1,2 | |
| ISOm | 205.50 (± 0.5) 1,2 | 521.5 (± 2.5) 1,2 | 5.24 (± 0.03) 1,2 | 0.84 (± 0.01) 1,2 | |
| E | GnS | 205.67 (± 7.59) 2 | 522.67 (± 50.37) 1,2 | 5.16 (± 0.12) 1,2 | 0.82 (± 0.01) 1,2 |
| ISO | 194.00 (± 5.89) 1,2 | 545.33 (± 31.48) 1,2 | 5.22 (± 0.06) 1,2 | 0.83 (± 0.00) 1,2 | |
| ISOm | 200.00 (± 7.48) 1,2 | 498.67 (± 21.64) 1,2 | 5.16 (± 0.02) 1,2 | 0.80 (± 0.00) 1,2 | |
| F | GnS | 102.67 (± 11.14) 1 | 329.33 (± 50.31) 1 | 4.12 (± 0.26) 1 | 0.71 (± 0.03) 1 |
| ISO | 111.00 (± 1.63) 1 | 358.33 (± 31.56) 1 | 4.33 (± 0.21) 1 | 0.74 (± 0.03) 1 | |
| ISOm | 97.33 (± 2.87) 1 | 281.33 (± 12.5) 1 | 3.99 (± 0.13) 1 | 0.71 (± 0.02) 1 | |
| L | GnS | 234.33 (± 14.27) 1,2 | 658.3 (± 22.48) 2 | 5.58 (± 0.04) 2 | 0.86 (± 0.00) 2 |
| ISO | 232.67 (± 2.87) 2 | 668.67 (± 38.69) 2 | 5.68 (± 0.06) 2 | 0.87 (± 0.00) 2 | |
| ISOm | 231.67 (± 11.09) 1,2 | 692 (± 50.34) 2 | 5.62 (± 0.09) 2 | 0.86 (± 0.01) 2 | |
| R | GnS | 219.00 (± 9.80) 2 | 561.67 (± 72.67) 1,2 | 5.31 (± 0.15) 1,2 | 0.84 (± 0.01) 1,2 |
| ISO | 223.33 (± 6.13) 2 | 653.33 (± 39.35) 1,2 | 5.63 (± 0.07) 1,2 | 0.87 (± 0.00) 1,2 | |
| ISOm | 231.00 (± 6.98) 2 | 653.67 (± 33.89) 1,2 | 5.5 (± 0.04) 1,2 | 0.85 (± 0.00) 1,2 | |
The means were calculated with three replicates per soil (C, E, F, L and R) and procedure (ISO, ISOm and GnS-GII), and the standard errors of the means are indicated in parentheses. Significant differences between soils for the same procedure are indicated with numbers (1 – 1,2 – 2).
Table 3.
Bioinformatic parameters and databases used in the analysis of bar-coded pyrosequencing results
| Step | Parameter | Targeted rDNA Gene | |
|---|---|---|---|
| 16S | 18S | ||
| Preprocessing | Length threshold | 370 | 300 |
| Number of ambiguities tolerated | 0 | 0 | |
| Detection of proximal primer sequence | Complete and perfect | Complete and perfect | |
| Detection of distal primer sequence | No | Perfect, but potentially incomplete | |
| Clustering | Chosen level of similarity (%) | 95 | 95 |
| Ignoring differences in homopolymer lengths | Yes | Yes | |
| Filtering | Chosen clustering similarity threshold | 95 | 95 |
| Used taxonomic database | SILVA (r114) | SILVA (r111) | |
| Chosen taxonomic level | Phylum | Phylum | |
| Similarity or confidence threshold (%) | 90 | 85 | |
| Homogenization | High-quality reads kept for each sample | 2322 | 4378 |
| Taxonomy | Used taxonomic database | SILVA (r114) | SILVA (r111) |
| Method or tool of comparison | usearch | megablast | |
| Similarity or confidence threshold (%) | 80 | 80 | |
| Analysis | Chosen level of similarity (%) | 95 | 95 |
| Ignoring differences in homopolymer lengths | Yes | Yes | |
| Computation of a unifrac distance matrix | Yes | Yes | |
No significant differences were found between the three DNA extraction methods for the number of bacterial genera detected, the number of bacterial OTUs or for the Shannon and Evenness indices in any of the soils (Table 2). This means that neither the mechanical lysis step (using a mini bead-beater cell disruptor or the FastPrep®-24) nor the complete DNA extraction procedures had a significant effect on the evaluation of bacterial diversity parameters by meta-barcoding for the wide range of soil types and land uses tested (Table 1).
On the other hand, soil type did have an impact on bacterial richness and diversity indices, as significant differences were highlighted between soils, whatever the DNA extraction procedure (Table 2). Indeed, F and L soils (respectively the sandy acidic forest soil and the loamy grassland soil) were significantly different (P < 0.001) based on the number of OTUs, Shannon and Evenness indices. More precisely, the F soil had the lowest richness (number of OTUs and genera) and diversity, with for example a Shannon index of about 4.1 against 5.6 for the L soil (Table 2). This observation can be linked to particular physico-chemical characteristics, because the F soil had a pH of 3.8 and a C/N ratio of 34 (Dequiedt et al., 2011; Lienhard et al., 2013b). Several studies have highlighted that bacterial richness had a positive correlation with soil pH (Fierer and Jackson, 2006; Lauber et al., 2008; Terrat et al., 2012) and a negative correlation with C/N ratio (Kuramae et al., 2012). Indeed, a high C/N ratio is generally typical of a large recalcitrant organic matter content that is unfavourable for bacterial growth (Boer et al., 2005). However, the sandy crop soil R, also harbouring a C/N ratio of the same magnitude (23.3), holds a greater richness of OTUs and genera than the forest soil (Table 2). This might partly be due to either the high sand content (Table 1), which increases soil microscale heterogeneity and stimulates the bacterial richness (Chau et al., 2011) or an alkaline pH (7.5) favouring bacterial richness (Fierer and Jackson, 2006; Lauber et al., 2008; Terrat et al., 2012).
Altogether, our results confirmed that bacterial diversity and richness can be strongly linked to soil characteristics and especially soil pH, organic matter and texture (Fierer and Jackson, 2006; Lauber et al., 2008; Kuramae et al., 2012; Terrat et al., 2012; Lienhard et al., 2013b). All DNA extraction procedures tested gave enough and similar sensitivity to detect changes between indigenous bacterial communities of soils differing by their characteristics and management. These data also support the idea that a study limited to these diversity indices could not be sufficient to determine whether a DNA extraction procedure is more powerful than another to describe soil bacterial communities and that it might be completed by a more detailed bacterial community composition analysis.
Influence of soil DNA extraction procedure on fungal richness and diversity
Using the same DNA extracts as for the bacterial analysis (three DNA extraction procedures applied to five soils), 18S rRNA gene sequences were successfully amplified and sequenced from all samples (Table 4). Homogenized high-quality reads (4378 per sample) were then analyzed using taxonomically dependent and independent analyses to determine fungal richness and diversity (Table 3). As for bacteria, the rarefaction curves of fungal richness confirmed that the number of high-quality reads allowed accurate description of the fungal community diversity in each soil sample studied (Supporting Information Fig. S1).
Table 4.
Fungal richness and diversity indices of the five French soils used
| Number of genera | OTUs (95% of similarity) | Shannon | Evenness | ||
|---|---|---|---|---|---|
| C | GnS | 116.00 (± 11.43) 1 | 350.33 (± 42.32) 1,2 | 3.72 (± 0.05) 1,2 | 0.64 (± 0.01) 1,2 |
| ISO | 92.33 (± 15.69) 1 | 273.67 (± 63.67) 1 | 3.31 (± 0.33) 1 | 0.59 (± 0.04) 1,2 | |
| ISOm | 118.67 (± 9.67) 1 | 340.67 (± 68.23) 1,2 | 3.73 (± 0.15) 1,2 | 0.64 (± 0.01) 1,2 | |
| E | GnS | 128.00 (± 13.74) 1 | 287.33 (± 30.58) 1 | 3.39 (± 0.08) 1,2 | 0.6 (± 0.01) 1,2 |
| ISO | 108.33 (± 9.81) 1 | 239.33 (± 31.54) 1 | 3.34 (± 0.22) 1 | 0.61 (± 0.03) 1,2 | |
| ISOm | 125.67 (± 6.55) 1 | 289 (± 33.66) 1 | 3.54 (± 0.18) 1,2 | 0.63 (± 0.02) 1,2 | |
| F | GnS | 129.67 (± 8.34) 1 | 249.67 (± 11.15) 1 | 2.98 (± 0.05) 1 | 0.54 (± 0.01) 1 |
| ISO | 136.00 (± 7.79) 2 | 312 (± 35.36) 1 | 3.19 (± 0.12) 1 | 0.56 (± 0.01) 1 | |
| ISOm | 140.33 (± 10.14) 1 | 267 (± 24.91) 1 | 3.27 (± 0.18) 1 | 0.59 (± 0.03) 1 | |
| L | GnS | 127.33 (± 9.29) a.1 | 416.33 (± 89.46) 2 | 4.05 (± 0.21) 2 | 0.67 (± 0.01) 2 |
| ISO | 89.67 (± 5.79) b.1 | 353.33 (± 51.45) 1 | 3.89 (± 0.09) 1 | 0.66 (± 0.03) 1,2 | |
| ISOm | 129.00 (± 6.48) a.1 | 382.33 (± 71.82) 2 | 3.74 (± 0.49) 2 | 0.63 (± 0.06) 2 | |
| R | GnS | 141.33 (± 13.82) 1 | 407.33 (± 84.94) 2 | 3.9 (± 0.2) 1,2 | 0.65 (± 0.01) 1,2 |
| ISO | 111.00 (± 11.00) 1,2 | 399.00 (± 30.00) 2 | 4.14 (± 0.01) 1 | 0.69 (± 0.01) 2 | |
| ISOm | 135.00 (± 12.68) 1 | 407 (± 67.38) 2 | 3.94 (± 0.12) 1,2 | 0.66 (± 0.01) 1,2 | |
The means were calculated with three replicates per soil (C, E, F, L and R) and procedure (ISO, ISOm and GnS-GII), and the standard errors of the means are indicated in parentheses. Significant differences between procedures for the same soil are indicated by letters (a, b), and significant differences between soils for the same procedure are indicated with numbers (1 – 1,2 – 2).
With regard to the number of detected genera, the numbers of OTUs and the computed indices (Shannon and Evenness), significant differences among the three DNA extraction procedures were recorded only for the L soil (Table 4), in which a lower number of fungal genera was significantly detected using the ISO procedure (P = 0.003). Moreover, in all the other soils but F, the number of genera recovered followed the same trend, with lower values detected for the ISO, than what was observed for the two other procedures. These genera missed by the ISO demonstrate that fungal diversity can be skewed using this procedure. As the main difference among the ISO and the two other procedures is the soil-grinding step; we can hypothesize that the traditional bead-beating system is not sufficient to lyze some fungal cells. Indeed, many fungi have cell walls that impede lysis and the recovery of nucleic acids (Fredricks et al., 2005). The mechanical lysis step of the ISOm and GnS-GII procedures was strongly optimized in terms of type and size of the glass beads as well as in terms of the strength and duration of grinding using the FastPrep®-24 (Terrat et al., 2012).
When fungal diversity was compared between soils, significant differences in the Shannon and Evenness indices (P < 0.05) and in the number of OTUs (P < 0.1) were observed whatever the DNA extraction procedure (Table 4). More precisely, the acidic forest soil F harboured the lowest richness and diversity, and the alkaline sandy crop soil R the highest (Table 4). These differences could be explained by several soil physico-chemical parameters, namely their contrasting pH (3.8 against 7.75), but also their C/N ratio (34 against 23.3) (Table 1). Although extreme environments like acidic soils may provide suitable biotopes for fungi (Baker and Banfield, 2003; Butinar et al., 2005), the lowest richness and diversity was detected in the acidic forest soil F, indicating that other physico-chemical parameters can limit fungal communities. Thus, a high C/N ratio is typical of soil systems with a low rate of organic matter degradation because of the presence of a high proportion of recalcitrant organic matter (Kuramae et al., 2012). Strickland and Rousk (2010) demonstrated in a previous study that the optimal C/N for fungi is expected to range from 5 to 15; i.e. closer to the C/N of the sandy crop soil R than to the ratio of the forest soil F, which has a higher carbon content. Focusing on the number of fungal genera recovered by the three DNA extraction procedures, only the ISO allowed the detection of significant differences between soils. This finding has to be seriously questioned because we demonstrated in the previous paragraph that the ISO underestimates the number of fungal genera.
Influence of soil DNA extraction procedure on bacterial community composition
The bacterial community composition in the five soils was compared by computing the unifrac distances on a phylogenetic tree (Lozupone and Knight, 2005). In addition to analyzing the phylogenetic distances, we also compared the bacterial communities' compositions based on the relative abundance of the bacterial genera detected in the samples (Fig. 1). Due to the size and variability of the genus table, only the most highly represented bacterial genera in the samples (i.e. only those for which the sum of the relative abundances of the genus in all samples was higher than 5%) were identified and mapped.
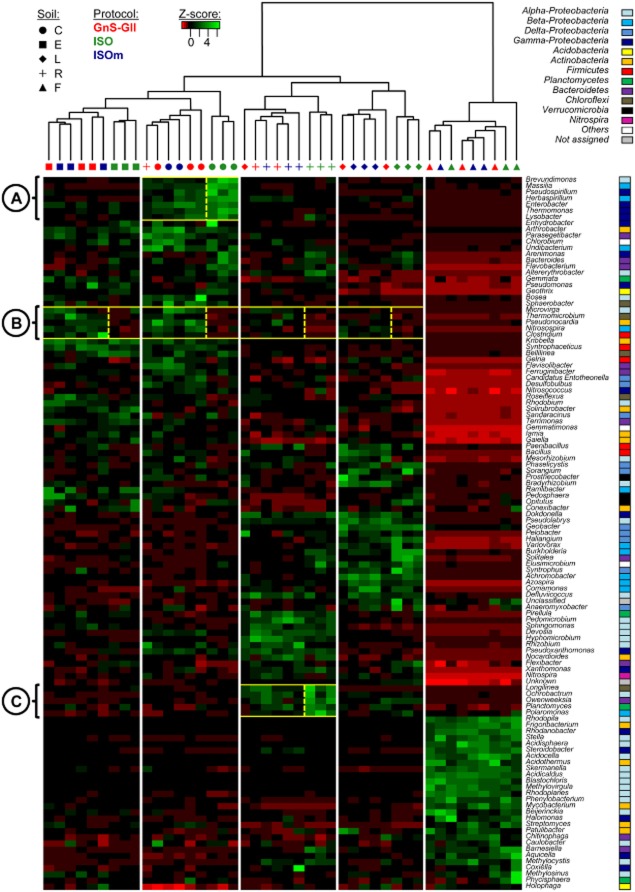
Fig 1.
Heat map comparison of the dominant bacterial genera detected in soils according to extraction procedures. The five different soils (C, E, F, L, R) were organized based on the UPGMA dendrogram of unifrac distances (weighted and normalized) between soil samples according to the three DNA extraction procedures (ISO-11063, GnS-GII and ISOm). The legend shows the Z-scores (relative abundances are expressed as median centred Z-scores between all samples, and the colours scaled to standard deviations). Subcells A, B and C in the heat map have been highlighted by yellow squares and numbered to identify significant differences in the relative abundance of particular bacterial genera according to DNA extraction procedure.
The clustering of soil bacterial communities indicated that replicates from the same soil were more similar to each other than replicates from other soils, whatever the DNA extraction procedure (Fig. 1). This observation demonstrated the good reproducibility between replicates for each type of soil DNA extraction procedure even if, surprisingly, two GnS-GII replicates from soils L and R seemed to be erroneously clustered. More precisely, two main clusters were identified, sorting the samples from the acidic forest soil F (which hosted a very different bacterial composition) apart from the four other soils (Fig. 1). Four sub-clusters could also be defined, each one grouping samples from each of the four soils and confirming that the studied soils hosted distinct bacterial communities, as already demonstrated by DNA fingerprinting approach (Plassart et al., 2012). This observation demonstrated the good reproducibility between replicates for ISO and ISOm procedures. However, even if clustering revealed that soil type had a more important effect on bacterial composition than the DNA extraction procedure, it is interesting to note that this latter could induce significant variations (Fig. 1). For all soils (except the forest soil F), the bacterial diversity profiles resulting from the ISOm and GnS-GII DNA extraction procedures grouped together (i.e. were not discriminated by the unifrac analysis), but were different from those obtained with the ISO-11063 procedure (Fig. 1). These observations confirm the influence of soil DNA extraction procedure on soil bacterial composition and especially the clear distinction between ISO-11063 and the two other procedures, potentially explained by differences in the soil-grinding methods (as discussed above for fungal richness and diversity). These differences were also confirmed by a more detailed analysis of bacterial composition (Fig. 1, subcells A–C). For example, the genus Brevundimonas was more detected (P < 0.05) with the ISO-11063 procedure than with the two others in the clayey crop soil C (Fig. 1, subcell A), as were the genera Massilia, Pseudospirillum, Herbaspirillum, Enterobacter, Thermomonas and Lysobacter. Similarly, the genus Polaromonas was more detected in the sandy crop soil R (Fig. 1, subcell C), but not in the other soils. On the contrary, the genera Clostridium, Nitrosospira, Microvirga and Pseudonocardia were respectively less detected (P < 0.05) with ISO-11063 than with the ISOm and GnS-GII procedures in soils C, E, L and R (Fig. 1, subcell B). Because the genera Clostridium or Pseudonocardia are known to be potentially recalcitrant to mechanical lysis, because of their spore-forming ability (Kaewkla and Franco, 2011; Yang and Ponce, 2011), their lower detection with the ISO-11063 procedure may be explained by the less efficient mechanical lysis (bead beating) of this procedure, compared with the two others, which are based on FastPrep®-24 grinding (Plassart et al., 2012; Terrat et al., 2012).
Influence of soil DNA extraction procedure on fungal community composition
As with the bacterial communities, the fungal communities' composition in all soils was compared by using the unifrac distances and determining the most highly represented fungal genera in the samples (Fig. 2). The unifrac dendrogram revealed a better discrimination of fungal composition between soils than between DNA extraction methods, demonstrating a good reproducibility between replicates for all procedures (Fig. 2). Moreover, as for the bacterial communities, the same clustering organization was obtained for fungal communities, revealing a significant distinction between the forest soil and the other soils. This observation corroborates other studies in which soil characteristics (e.g. pH, texture, C/N) were shown to impact fungal community diversity and composition (Rousk et al., 2010; Strickland and Rousk, 2010; McGuire et al., 2013). The fungal populations in this acidic soil clearly differed from those of the other soils, with a dominance of the Basidiomycota phylum (e.g. genera Sebacina, Boletus, Pleurotus or Hericium), which is common in forest soils (Buée et al., 2009).
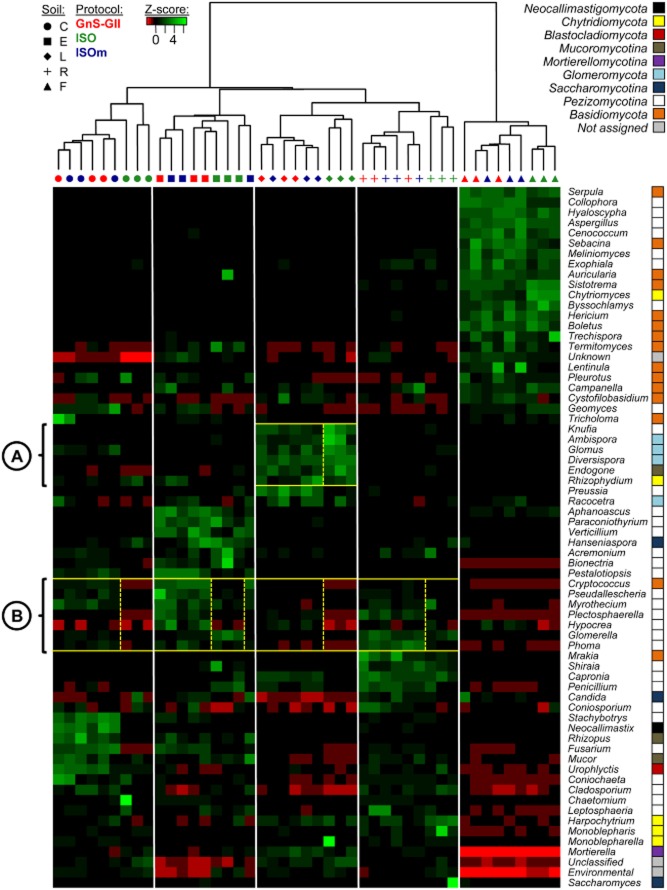
Fig 2.
Heat map comparison of the dominant fungal genera detected in soils according to extraction procedures. The five different soils (C, E, F, L, R) were organized based on the UPGMA dendrogram of unifrac distances (weighted and normalized) between soil samples according to the three DNA extraction procedures (ISO-11063, GnS-GII and ISOm). The legend shows the Z-scores (relative abundances are expressed as median centred Z-scores between all samples, and the colours scaled to standard deviations). Subcells A and B in the heat map have been highlighted by yellow squares and numbered to identify significant differences in the relative abundance of particular fungal genera according to DNA extraction procedure.
For soils F, L and R, the patterns of the fungal communities resulting from the ISO-11063 procedure were discriminated from those obtained with the two non-ISO protocols (Fig. 2). This observation evidenced that in these soils, the fungal community compositions detected with the ISO-11063 differed from those detected with the non-ISO procedures. More precisely, in the loamy grassland soil L, several genera (e.g. Knufia and Diversispora) were more detected (P < 0.1) with the ISO-11063 protocol (Fig. 2, subcell A). However, this positive impact of the ISO-11063 procedure was only visible for this particular soil. On the contrary, the genera Myrothecium, Cryptococcus, Glomerella and Plectosphaerella were respectively less detected (P < 0.05) with the ISO-11063 protocol than with the other methods in soils C, E, L and R (Fig. 2, subcell B), as Pseudallescheria in soils C, E and R, and Hypocrea in soils E and L (P < 0.1). This would be of great importance in ecological studies as some of these genera (e.g. Cryptococcus, Pseudallescheria, Hypocrea and Plectosphaerella) are saprotrophic fungi known to play key roles in organic matter turnover (Martínez et al., 2003; Jaklitsch et al., 2005; Buée et al., 2009; McGuire et al., 2013). Moreover, the genus Pseudallescheria, which has been found in compost-amended or heavily hydrocarbon-polluted soils, can be used as an indicator of soil disturbance (April et al., 1998). Therefore, the ISO-induced underrepresentation of these genera could lead to a misinterpretation of the functioning of an ecosystem.
This difference in community composition is, together with the lower number of fungal genera recovered with the ISO described earlier, a clue indicating that the ISO procedure may not be the most appropriate to investigate soil fungal communities. Besides, these differences are thought to be due to the less efficient mechanical lysis of soil with the ISO-11063 procedure; the classical system seems not to break open as many cells as the FastPrep®-24 bead-beating system, particularly in the case of fungal cells with tough walls. This is why the ISOm and GnS-GII methods are thought to be more efficient at extracting fungal DNA from different types of soils. This conclusion strengthens the idea that the physical lysis step is of crucial importance in a soil DNA extraction procedure (Feinstein et al., 2009; İnceoğlu et al., 2010; Delmont et al., 2011b). This finding is in agreement with previous comparisons of these DNA extraction procedures based on quantitative PCR and community DNA fingerprinting (Plassart et al., 2012).
Conclusion
In the context of modern microbial ecology, where investigations to describe the whole soil microbiota in numerous samples are carried out on a very large scale, the importance of using a single, standardized soil DNA extraction procedure is paramount. Among the three DNA extraction procedures evaluated in this study, the GnS-GII introduced some heterogeneity in bacterial composition between replicates, and the ISO-11063 DNA caused an underrepresentation of several fungal groups of ecological interest. Therefore, the ISOm procedure provides a better snapshot of bacterial and fungal communities.
Experimental procedures
Soil samples
Five soils were chosen for their contrasting land-use and physico-chemical characteristics (Table 1) (Plassart et al., 2012). All necessary permits were obtained from the respective land owners (INRA, ADEME and private owners). For each soil, three independent replicates were collected at a depth of 20 cm [fully described in (Plassart et al., 2012)]. Physico-chemical characteristics (pH, texture, organic carbon, total N and CaCO3) were analyzed, using international standard procedures, by the Soil Analysis Laboratory at INRA (Arras, France, http://www.lille.inra.fr/las).
Soil DNA extraction, purification and quantification
Three different procedures were tested: the GnS-GII protocol, the ISO-11063 standard and the ISOm. All three procedures are adapted to extract DNA from 1 g of soil (dry weight) and have already been described by Plassart and colleagues (2012).
ISO-11063 procedure
This protocol is a version of the ISO-11063 standard (Martin-Laurent et al., 2001; Petric et al., 2011). Soil was added to a bead-beating tube containing 2 g of glass beads of 106 μm diameter and eight glass beads of 2 mm diameter. Each soil sample was mixed with a solution of 100 mM Tris-HCl (pH 8), 100 mM EDTA (pH 8), 100 mM NaCl, 2% (w/v) polyvinylpyrrolidone (40 g mol−1) and 2% (w/v) sodium dodecyl sulfate. The tubes were then shaken for 30 s at 1600 r.p.m. in a mini bead-beater cell disruptor (Mikro-Dismembrator, Braun Biotech International), then incubated for 10 min at 70°C and centrifuged at 14,000g for 1 min. After removing the supernatant, proteins were precipitated, with 1/10 volume of 3 M sodium acetate prior to centrifugation (14,000g for 5 min at 4°C). Finally, nucleic acids were precipitated by adding 1 volume of ice-cold isopropanol. The DNA pellets obtained after centrifugation (14,000g for 5 min at 4°C) were washed with 70% ethanol (full details are described in (Martin-Laurent et al., 2001; Philippot et al., 2010; Petric et al., 2011).
ISOm procedure
This protocol is a modified version of ISO-11063 standard as it includes a different mechanical lysis step (FastPrep® bead-beating instead of the recommended bead beating). Soil was added to 15 ml of Falcon tube containing 2.5 g of 1.4 mm diameter ceramic beads, 2 g of 106 μm diameter silica beads and four glass beads of 4 mm diameter. Each soil sample was mixed with a solution of 100 mM Tris-HCl (pH 8), 100 mM EDTA (pH 8), 100 mM NaCl, 2% (w/v) polyvinylpyrrolidone (40 g mol−1) and 2% (w/v) sodium dodecyl sulfate. The tubes were then shaken for 3 × 30 s at 4 m sec−1 in a FastPrep®-24 (MP-Biomedicals, NY, USA), before incubation for 10 min at 70°C and centrifugation at 14,000g for 1 min. After removing the supernatant, proteins were precipitated with 1/10 volume of 3 M sodium acetate prior to centrifugation (14,000g for 5 min at 4°C). Finally, nucleic acids were precipitated by adding 1 volume of ice-cold isopropanol. The DNA pellets obtained after centrifugation (14,000g for 5 min at 4°C) were washed with 70% ethanol.
GnS-GII procedure
This DNA extraction procedure was initially developed and optimized by the GenoSol platform (Terrat et al., 2012). Soil was added to 15 ml of Falcon tube containing 2.5 g of 1.4 mm diameter ceramic beads, 2 g of 106 μm diameter silica beads and four glass beads of 4 mm diameter. Each soil sample was mixed with a solution of 100 mM Tris-HCl (pH 8), 100 mM EDTA (pH 8), 100 mM NaCl, 2% (w/v) and 2% (w/v) sodium dodecyl sulfate. The tubes were then shaken for 3 × 30 s at 4 m sec−1 in a FastPrep®-24 (MP-Biomedicals, NY, USA), before incubation for 30 min at 70°C and centrifugation at 7,000g for 5 min at 20°C. After removing the supernatant, proteins were precipitated with 1/10 volume of 3 M sodium acetate prior to centrifugation (14,000g for 5 min at 4°C). Finally, nucleic acids were precipitated by adding 1 volume of ice-cold isopropanol. The DNA pellets obtained after centrifugation (14,000g for 5 min at 4°C) were washed with 70% ethanol.
Purification and quantification procedure
As the DNA purification step is not part of the evaluated protocols to avoid additional biases among the three procedures and only compare the extraction step, all crude soil DNA extracts were purified and quantified using the same procedure (Ranjard et al., 2003; Plassart et al., 2012). Briefly, 100 μl aliquots of crude DNA extracts were loaded onto PVPP (polyvinylpolypyrrolidone) Microbiospin minicolumns (Bio-Rad) and centrifuged for 4 min at 1000g and 10°C. Eluates were then collected and purified for residual impurities using the Geneclean Turbo kit (MP-Biomedicals, NY, USA). Purified DNA extracts were quantified using the PicoGreen staining Kit (Molecular Probes, Paris, France).
Pyrosequencing of 16S and 18S rRNA gene sequences
Microbial diversity was determined for each biological replicate and for each soil (C, F, E, L and R) by 454 pyrosequencing of ribosomal genes. A 16S rRNA gene fragment with sequence variability and appropriate size (about 450 bases) for 454 pyrosequencing was amplified using the primers F479 (5′-CAGCMGCYGCNGTAANAC-3′) and R888 (5′-CCGYCAATTCMTTTRAGT-3′) (Supporting Information Table S1 for in silico match analysis, Terrat et al. 2014). For each sample, 5 ng of DNA were used for a 25 μl of PCR conducted under the following conditions: 94°C for 2 min, 35 cycles of 30 s at 94°C, 52°C for 30 s and 72°C for 1 min, followed by 7 min at 72°C. The PCR products were purified using a MinElute gel extraction kit (Qiagen, Courtaboeuf, France) and quantified using the PicoGreen staining Kit (Molecular Probes, Paris, France). Similarly, an 18S rRNA gene fragment of about 350 bases was amplified using the primers FR1 (5′-ANCCATTCAATCGGTANT-3′) and FF390 (5′-CGATAACGAACGAGACCT-3′) (Prevost-Boure et al., 2011) under the following PCR conditions: 94°C for 3 min, 35 cycles of 1 min at 94°C, 52°C for 1 min and 72°C for 1 min, followed by 5 min at 72°C. A second PCR of nine cycles was then conducted twice for each sample under similar PCR conditions with purified PCR products and 10 base pair multiplex identifiers added to the primers at 5′ position to specifically identify each sample and avoid PCR bias. Finally, the duplicate PCR products were pooled, purified and quantified as previously described. Pyrosequencing was then carried out on a GS FLX Titanium (Roche 454 Sequencing System) by Genoscreen (Lille, France).
Bioinformatic analysis of 16S and 18S rRNA gene sequences
Bioinformatic analyses were done using the GnS-PIPE initially developed by the Genosol platform (INRA, Dijon, France) (Terrat et al., 2012) and recently optimized. The parameters chosen for each bioinformatic step can be found in Table 3. First, all the 16S and 18S raw reads were sorted according to the multiplex identifier sequences. The raw reads were then filtered and deleted based on (i) their length, (ii) their number of ambiguities (Ns) and (iii) their primer(s) sequence(s). A perl program was then applied for rigorous dereplication (i.e. clustering of strictly identical sequences). The dereplicated reads were then aligned using infernalalignment (Cole et al., 2009), and clustered into OTU using a perl program that groups rare reads to abundant ones, and does not count differences in homopolymer lengths. A filtering step was then carried out to check all single singletons (reads detected only once and not clustered, which might be artefacts, such as PCR chimeras) based on the quality of their taxonomic assignments. Finally, in order to compare the data sets efficiently and avoid biased community comparisons, the reads retained were homogenized by random selection closed to the lowest dataset.
The retained high-quality reads were used for (i) taxonomy-independent analyses, determining several diversity and richness indices using the defined OTU composition at the genus level and (ii) taxonomy-based analysis using similarity approaches against dedicated reference databases from SILVA (Quast et al., 2013) (see Table 3). The raw data sets are available on the European Bioinformatics Institute database system under project accession number PRJEB4825.
Statistical analyses
The effects of the DNA extraction procedure on bacterial and fungal diversities were tested by analysis of variance (multiple paired comparisons). The effects of the DNA extraction procedure on bacterial and fungal community compositions were assessed by Kruskal–Wallis tests. All statistical analyses were performed under xlstat software (Addinsoft®). The bacterial and fungal communities from all samples were also compared by using unifrac (Lozupone and Knight, 2005), based on the 16S and 18S phylogenetic trees computed with fasttree(Price et al., 2010).
Acknowledgments
Thanks to the National Research Infrastructure ‘Agro-écosystèmes, Cycles Biogéochimique et Biodiversité (SOERE-ACBB http://www.soere-acbb.com/fr/) for providing support during the sampling campaign and making available the physico-chemical soil properties data. The funders [National Agency for Research (through ‘Investments for the Future’ program), Regional Council of Burgundy and European Commission) had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
None declared.
Supporting Information
Fig. S1. Rarefaction curves of bacterial and fungal OTUs detected in soils according to extraction procedures.
Table S1. Detailed hit frequencies (%) of the in silico analysis of the F479/R888 primer set for Bacteria, Archaea and Eukaryota.
References
- April TM, Abbott SP, Foght JM. Currah RS. Degradation of hydrocarbons in crude oil by the ascomycete Pseudallescheria boydiiMicroascaceae. Can J Microbiol. 1998;44:270–278. doi: 10.1139/w97-152. [DOI] [PubMed] [Google Scholar]
- Baker BJ. Banfield JF. Microbial communities in acid mine drainage. FEMS Microbiol Ecol. 2003;44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- Boer Wd, Folman LB, Summerbell RC. Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S. Martin F. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009;184:449–456. doi: 10.1111/j.1469-8137.2009.03003.x. [DOI] [PubMed] [Google Scholar]
- Butinar L, Santos S, Spencer-Martins I, Oren A. Gunde-Cimerman N. Yeast diversity in hypersaline habitats. FEMS Microbiol Lett. 2005;244:229–234. doi: 10.1016/j.femsle.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Chau JF, Bagtzoglou AC. Willig MR. The effect of soil texture on richness and diversity of bacterial communities. Environ Forensics. 2011;12:333–341. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, et al. Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol. 2011a;77:1315–1324. doi: 10.1128/AEM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Robe P, Clark I, Simonet P. Vogel TM. Metagenomic comparison of direct and indirect soil DNA extraction approaches. J Microbiol Methods. 2011b;86:397–400. doi: 10.1016/j.mimet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Dequiedt S, Saby NPA, Lelievre M, Jolivet C, Thioulouse J, Toutain B, et al. Biogeographical patterns of soil molecular microbial biomass as influenced by soil characteristics and management. Glob Ecol Biogeogr. 2011;20:641–652. [Google Scholar]
- Feinstein LM, Sul WJ. Blackwood CB. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl Environ Microbiol. 2009;75:5428–5433. doi: 10.1128/AEM.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N. Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN, Smith C. Meier A. Comparison of six DNA extraction methods for recovery of fungal DNA as assessed by quantitative PCR comparison of six DNA extraction methods for recovery of Fungal DNA as assessed by quantitative PCR. J Clin Microbiol. 2005;43:5122–5128. doi: 10.1128/JCM.43.10.5122-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- İnceoğlu Ö, Hoogwout EF, Hill P. van Elsas JD. Effect of DNA extraction method on the apparent microbial diversity of soil. Appl Environ Microbiol. 2010;76:3378–3382. doi: 10.1128/AEM.02715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP. Druzhinina IS. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycol. 2005;97:1365–1378. doi: 10.3852/mycologia.97.6.1365. [DOI] [PubMed] [Google Scholar]
- Kaewkla O. Franco CMM. Pseudonocardia eucalypti sp. nov, an endophytic actinobacterium with a unique knobby spore surface, isolated from roots of a native Australian eucalyptus tree. Int J Syst Evol Microbiol. 2011;61:742–746. doi: 10.1099/ijs.0.022327-0. [DOI] [PubMed] [Google Scholar]
- Kuramae EE, Yergeau E, Wong LC, Pijl AS, van Veen JA. Kowalchuk GA. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol Ecol. 2012;79:12–24. doi: 10.1111/j.1574-6941.2011.01192.x. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Strickland MS, Bradford MA. Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem. 2008;40:2407–2415. [Google Scholar]
- Lienhard P, Terrat S, Mathieu O, Levêque J, Prévost-Bouré NC, Nowak V, et al. Soil microbial diversity and C turnover modified by tillage and cropping in Laos tropical grassland. Environ Chem Lett. 2013a;11:1–8. [Google Scholar]
- Lienhard P, Tivet F, Chabanne A, Dequiedt S, Lelievre M, Sayphoummie S, et al. No-till and cover crops shift soil microbial abundance and diversity in Laos tropical grasslands. Agron Sustain Dev. 2013b;33:375–384. [Google Scholar]
- Lozupone C. Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ, et al. Digging the New York City skyline: soil fungal communities in green roofs and city parks. PLoS ONE. 2013;8:e58020. doi: 10.1371/journal.pone.0058020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron P-A, Mougel C. Ranjard L. Soil microbial diversity: methodological strategy, spatial overview and functional interest. C R Biol. 2011;334:403–411. doi: 10.1016/j.crvi.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Martínez M, López-Solanilla E, Rodríguez-Palenzuela P, Carbonero P. Díaz I. Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (Gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol Plant Microbe Interact. 2003;16:876–883. doi: 10.1094/MPMI.2003.16.10.876. [DOI] [PubMed] [Google Scholar]
- Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G. Catroux G. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol. 2001;67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petric I, Philippot L, Abbate C, Bispo A, Chesnot T, Hallin S, et al. Inter-laboratory evaluation of the ISO standard 11063 ‘Soil quality – Method to directly extract DNA from soil samples’. J Microbiol Methods. 2011;84:454–460. doi: 10.1016/j.mimet.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Philippot L, Abbate C, Bispo A, Chesnot T, Hallin S, Lemanceau P, et al. Soil microbial diversity: an ISO standard for soil DNA extraction. J Soils Sediments. 2010;10:1344–1345. [Google Scholar]
- Plassart P, Terrat S, Thomson B, Griffiths R, Dequiedt S, Lelievre M, et al. Evaluation of the ISO standard 11063 DNA extraction procedure for assessing soil microbial abundance and community structure. PLoS ONE. 2012;7:e44279. doi: 10.1371/journal.pone.0044279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost-Boure NC, Christen R, Dequiedt S, Mougel C, Lelievre M, Jolivet C, et al. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS ONE. 2011;6:e24166. doi: 10.1371/journal.pone.0024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS. Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendhran J. Gunasekaran P. Strategies for accessing soil metagenome for desired applications. Biotechnol Adv. 2008;26:576–590. doi: 10.1016/j.biotechadv.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Ranjard L, Lejon DPH, Mougel C, Schehrer L, Merdinoglu D. Chaussod R. Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environ Microbiol. 2003;5:1111–1120. doi: 10.1046/j.1462-2920.2003.00521.x. [DOI] [PubMed] [Google Scholar]
- Ranjard L, Dequiedt S, Jolivet C, Saby NPA, Thioulouse J, Harmand J, et al. Biogeography of soil microbial communities: a review and a description of the ongoing French national initiative. Agron Sustain Dev. 2010;30:359–365. [Google Scholar]
- Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- Strickland MS. Rousk J. Considering fungal: bacterial dominance in soils – methods, controls, and ecosystem implications. Soil Biol Biochem. 2010;42:1385–1395. [Google Scholar]
- Terrat S, Christen R, Dequiedt S, Lelievre M, Nowak V, Regnier T, et al. Molecular biomass and MetaTaxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure. Microb Biotechnol. 2012;5:135–141. doi: 10.1111/j.1751-7915.2011.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrat S, Dequiedt S, Horrigue W, Lelievre M, Cruaud C, Saby N, et al. Improving soil bacterial taxa-area relationships assessment using DNA meta-barcoding. Heredity. 2014 doi: 10.1038/hdy.2014.91. . In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C, Thürmer A, Wollherr A, Nacke H, Herold N, Schrumpf M, et al. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl Environ Microbiol. 2010;76:6751–6759. doi: 10.1128/AEM.01063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-W. Ponce A. Validation of a Clostridium endospore viability assay and analysis of Greenland ices and Atacama desert soils. Appl Environ Microbiol. 2011;77:2352–2358. doi: 10.1128/AEM.01966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood S, Bottomley P. Myrold D. Soil microbial communities associated with Douglas-fir and red alder stands at high- and low-productivity forest sites in Oregon, USA. Microb Ecol. 2010;60:606–617. doi: 10.1007/s00248-010-9675-9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA. Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Rarefaction curves of bacterial and fungal OTUs detected in soils according to extraction procedures.
Table S1. Detailed hit frequencies (%) of the in silico analysis of the F479/R888 primer set for Bacteria, Archaea and Eukaryota.