Abstract
Polyglutamine diseases, including spinocerebellar ataxia type 3 (SCA3), are caused by CAG repeat expansions that encode abnormally long glutamine repeats in the respective disease proteins. While the mechanisms underlying neurodegeneration remain uncertain, evidence supports a proteotoxic role for the mutant protein dictated in part by the specific genetic and protein context. To further define pathogenic mechanisms in SCA3, we generated a mouse model in which a CAG expansion of 82 repeats was inserted into the murine locus by homologous recombination. SCA3 knockin mice exhibit region-specific aggregate pathology marked by intranuclear accumulation of the mutant Atxn3 protein, abundant nuclear inclusions and, in select brain regions, extranuclear aggregates localized to neuritic processes. Knockin mice also display altered splicing of the disease gene, promoting expression of an alternative isoform in which the intron immediately downstream of the CAG repeat is retained. In an independent mouse model expressing the full human ATXN3 disease gene, expression of this alternatively spliced transcript is also enhanced. These results, together with recent findings in other polyglutamine diseases, suggest that CAG repeat expansions can promote aberrant splicing to produce potentially more aggregate-prone isoforms of the disease proteins. This report of a SCA3 knockin mouse expands the repertoire of existing models of SCA3, and underscores the potential contribution of alternative splicing to disease pathogenesis in SCA3 and other polyglutamine disorders.
Introduction
Spinocerebellar ataxia type 3 (SCA3), also known as Machado-Joseph disease, is the most common dominantly inherited ataxia and one of at least nine neurodegenerative diseases caused by polyglutamine-encoding CAG repeat expansions (1). In SCA3, this expansion occurs in the ATXN3 gene which encodes the deubiquitinase ataxin-3 (ATXN3) (2). Like other polyglutamine diseases, SCA3 is a disabling and ultimately fatal disorder characterized by selective degeneration in specific brain regions and age-dependent intraneuronal accumulation and aggregation of the mutant protein (3–5). While neuronal inclusions formed by the disease protein may not be directly toxic in polyglutamine disease (6,7), evidence supports the view that inclusions are a marker for accumulated, misfolded polyglutamine protein that is proteotoxic and contributes to neuronal dysfunction and cell loss in disease (8–12). Unfortunately, despite recent advances in understanding polyglutamine diseases, no preventive treatments are available for any of them.
Animal models have been instrumental in providing insight into polyglutamine disease pathogenesis and suggesting routes to potential therapy. Existing animal models of SCA3 have advanced the field in many ways, but all of them overexpress mutant ATXN3 above physiological concentrations (13). While overexpression models are particularly good at recapitulating robust aggregation pathology and behavioral abnormalities, they may mask early molecular changes important to pathogenesis. Moreover, models overexpressing a single isoform of ATXN3 from cDNA do not permit investigations of the potential disease contribution of splicing changes in the mutant transcript. In contrast, ‘knockin’ models in which the CAG repeat expansion is inserted precisely into the endogenous murine locus have proved useful in understanding various aspects of polyglutamine disease (14–23) including altered splicing of mutant transcripts (20,23). For example, Sathasivam et al. (23) recently discovered that the CAG expansion in Huntington's disease knockin mice promotes aberrant splicing of the Htt transcript and the production of a truncated amino-terminal fragment of the disease protein, Htt. Genetically precise knockin mouse models express the mutant protein from the endogenous promoter in the proper genomic context, including all regulatory elements that influence mutant gene expression.
Knockin mouse models have been generated for most polyglutamine diseases, but none has existed for SCA3. Here, we report a SCA3 knockin mouse model, generated by replacing the endogenous murine CAG repeat with an 82 repeat CAG expansion. Expressing physiological levels of mutant Atxn3, the SCA3 knockin mice exhibit robust Atxn3 accumulation both in regions known to be affected in human disease (e.g. brainstem and cerebellum) and in regions not previously described (e.g. hippocampus). Intriguingly, SCA3 knockin mice also display altered splicing of the mutant Atxn3 transcript that results in the formation of a previously described alternative ATXN3 transcript in human disease (24). We further show that CAG expansion results in similar altered splicing in another mouse model expressing the full-length human ATXN3 disease gene. In summary, the SCA3 knockin mouse model reported here recapitulates several important molecular features of disease and should facilitate the study of early pathogenic events in this polyglutamine disease.
Results
Generation of the SCA3 knockin mouse model and mutant Atxn3 expression
SCA3 is caused by CAG expansions ranging from ∼60 to 87 repeats in the ATXN3 gene (25). We inserted a CAG repeat expansion at the upper end of this disease range into the endogenous murine Atxn3 locus on a C57BL/6 background (Fig. 1A; Supplemental Material, Fig. S1). By homologous recombination, we replaced the endogenous murine sequence CAA(CAG)5 with (CAG)2(CAAAAG)(CAG)82 which encodes 85 glutamines interrupted by a lysine at the fourth residue (i.e. Q3KQ82), replicating the polyglutamine stretch found in human mutant ATXN3 (24). Polymerase chain reaction (PCR) across the endogenous CAG repeat confirmed successful insertion of the pathogenic expansion in heterozygous (Atxn3Q82/Q6) and homozygous (Atxn3Q82/Q82) SCA3 knockin mice (Fig. 1B). Sequencing of the intronic region upstream of the repeat expansion confirmed the removal of the neomycin cassette and the presence of the remaining FRT sequence (Supplemental Material, Fig. S2), and sequencing of exon 10, all of intron 10 and exon 11 did not reveal any differences compared with wild-type mice other than the expansion (data not shown). CAG repeat length sizing in offspring of heterozygous Atxn3Q82/Q6 mice revealed a modest tendency toward CAG repeat contraction upon maternal transmission of the mutant allele and stabilization or mild expansion upon paternal transmission (Fig. 1C). Similar intergenerational repeat instability has been reported in other polyglutamine disease mouse models, including a transgenic mouse model of SCA3 (14,18,26,27).
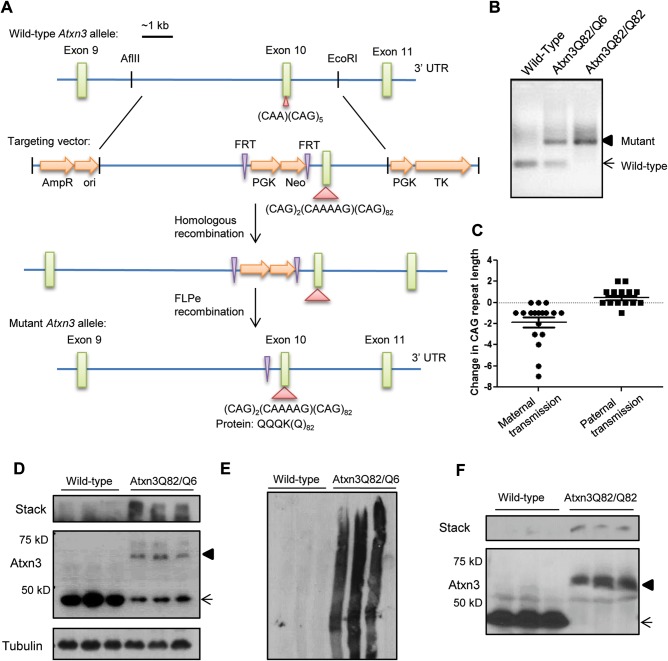
Figure 1.
Generation of a SCA3 knockin mouse expressing mutant Atxn3 (Q82). (A) Schematic of the generation of the SCA3 knockin mouse in which the endogenous murine (CAA)(CAG)5 was replaced with a human CAG-expanded sequence, (CAG)2(CAAAAG)(CAG)82, by homologous recombination. The neomycin (neo) selection cassette, flanked by FRT sites, was removed by FLPe recombination. (B) PCR across the CAG repeat shows the expanded repeat in heterozygous (Atxn3Q82/Q6) and homozygous (Atxn3Q82/Q82) SCA3 knockin mice. (C) SCA3 knockin mice show modest intergenerational repeat length instability with a tendency for CAG repeat contraction upon maternal transmission. (D) Western blotting shows expression of mutant Atxn3 accompanied by increased aggregates in the stacking gel in 1-year-old Atxn3Q82/Q6 hindbrain lysates. (E) Electrophoresis of lysates from (D) on 3% SDS–PAGE further illustrates high-molecular-weight aggregates. (F) About 30-week-old homozygous Atxn3Q82/Q82 mice express only mutant Atxn3 with aggregates in the stack. (D and F) Arrow indicates wild-type (WT) Atxn3 and arrowhead indicates mutant Atxn3.
Mutant Atxn3 is expressed widely throughout the SCA3 knockin mouse, including in brain, heart, liver, muscle and spleen (Supplemental Materials, Fig. S3). Western blot analysis of hindbrain lysates from 1-year-old Atxn3Q82/Q6 mice confirmed mutant Atxn3 expression in the brain, as well as aggregated Atxn3 protein in the stacking gel (Fig. 1D). Electrophoresis of brain lysates from knockin mice on low percentage (3%) SDS–PAGE further illustrated a range of high-molecular-weight aggregate species (Fig. 1E). Analysis of hindbrain lysates from homozygous knockin mice revealed exclusively mutant (expanded) Atxn3 expression with a corresponding loss of wild-type Atxn3, confirming that mutant Atxn3 is expressed from the endogenous allele (Fig. 1F). The increased high-molecular-weight aggregate species are consistent with detergent-resistant aggregates previously reported in an SCA3 transgenic mouse model (8,28). In heterozygous mice, the decreased intensity of mutant Atxn3 monomer compared with wild-type Atxn3 monomer is also consistent with the aggregation propensity of expanded polyglutamine proteins (29).
Neuropathological and behavioral characterization of the SCA3 knockin mouse
To begin defining neuropathological changes in SCA3 knockin mice, we immunostained for Atxn3 in SCA3 knockin brain. Compared with other polyglutamine disease knockin mouse models in which CAG expansions within the human disease range often elicit relatively modest neuropathological findings (30–33), SCA3 knockin mice display comparatively robust aggregate pathology. In heterozygous mice, enhanced intranuclear staining for Atxn3 is present by 10 weeks in several neuronal populations, including the deep cerebellar nuclei (DCN) (Fig. 2A), and is often accompanied by small intranuclear puncta. We do not reliably detect immunohistochemical changes in Atxn3Q82/Q6 mice that are less than ∼6 weeks of age. One-year-old heterozygous knockin mice display more prominent intranuclear puncta, as well as more frequent solitary inclusions that are characteristic of the human disease (Fig. 2B). Neuronal inclusions in the SCA3 knockin mice often co-localize with p62, an ubiquitin-binding protein implicated in autophagy and previously reported to localize to intranuclear inclusions in SCA3 disease brains (5,34,35) (Fig. 2C; Supplemental Materials, Fig. S4).
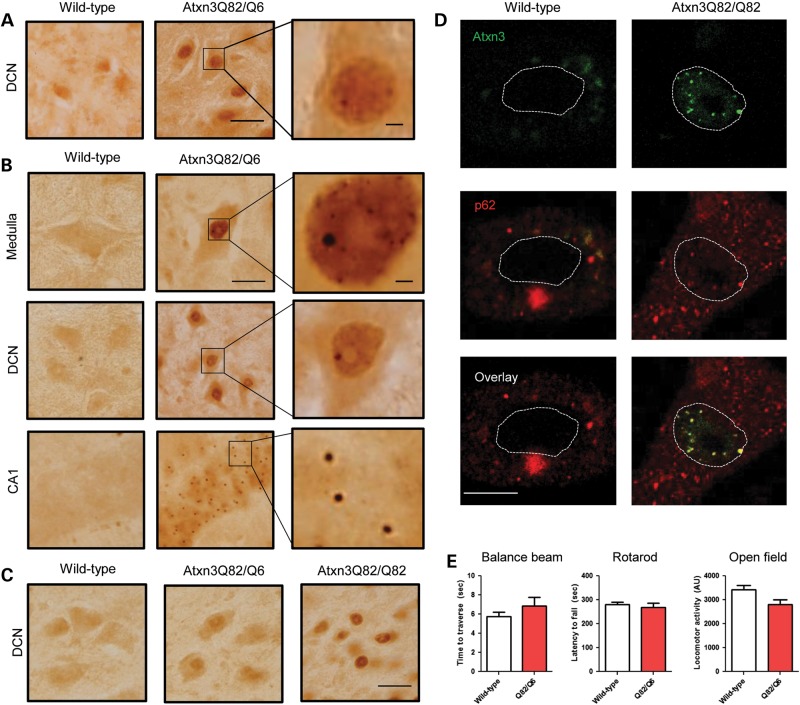
Figure 2.
Mutant Atxn3 accumulation in the SCA3 knockin mouse brain. (A) Immunohistochemical staining (IHC) for Atxn3 shows nuclear accumulation in the DCN of a 10-week-old Atxn3Q82/Q6 mouse. Scale bar = 20 μm, inset scale bar = 2 μm. (B) IHC of 1-year-old Atxn3Q82/Q6 mice shows increased diffuse nuclear staining and intranuclear puncta/inclusions in several brain regions, including the brain stem (medulla shown), DCN and the hippocampus (CA1 region is shown). Higher magnification insets show puncta and inclusions in nuclei. (C) Intranuclear accumulation of Atxn3 (DCN shown) is accelerated in an ∼7-month-old homozygous Atxn3Q82/Q82 mouse compared with a heterozygous littermate. Scale bar = 20 μm (D) Intranuclear puncta and inclusions in brainstem neurons from SCA3 knockin mice frequently co-stain with p62, whereas p62 is predominantly cytoplasmic in wild-type neurons. The nuclear border is outlined by a dashed line. Scale bar = 10 μm. (E) Performance of 1-year-old Atxn3Q82/Q6 (Q82/Q6) mice (n = 9) did not differ from age-matched WT mice (n = 10) on motor behavior tasks, including from left to right, 5 mm balance beam, accelerating rotarod and open field exploration. Graphs represent mean + SE.
SCA3 knockin mice display striking regional differences in mutant Atxn3 deposition. Table 1 qualitatively reports the intensity of diffuse nuclear staining and frequency of intranuclear inclusions in various brain regions in 1-year-old heterozygous mice (n = 4). Neurons of the hindbrain, including the DCN and several brain stem nuclei, which are known to be vulnerable targets in the human disease, show strong diffuse nuclear staining that is often accompanied by multiple intranuclear puncta and less frequently by distinct solitary large inclusions. In contrast, neurons of the forebrain, including the hippocampus, cortex and striatum, have moderately increased diffuse nuclear staining with frequent solitary intranuclear inclusions. Despite widespread aberrant accumulation of mutant Atxn3, including in the hindbrain, 1-year-old Atxn3Q82/Q6 mice (n = 9) performed equally well as age-matched wild-type mice (n = 10) on various motor tasks (Fig. 2D). Examination of cresyl violet-stained brains of 1-year-old Atxn3Q82/Q6 mice (n = 3) did not reveal obvious degenerative changes in any brain regions. For example, a close examination of the DCN, a region known to be consistently affected in SCA3, did not suggest neuronal loss at 1 year of age (Supplemental Material, Fig. S5).
Table 1.
Regional differences in Atxn3 accumulation in SCA3 knockin mouse brain
| Region | Diffuse nuclear staining | Intranuclear inclusions | Extranuclear inclusions |
|---|---|---|---|
| Purkinje cells | – | + | – |
| Granular cells | + | – | – |
| DCN | ++ | ++ | – |
| Medulla | ++ | ++ | + |
| Pons | ++ | ++ | + |
| Substantia nigra | + | + | – |
| Red nucleus | ++ | ++ | – |
| Spinal cord | ++ | ++ | + |
| Striatum | + | +++ | + |
| Hippocampus | + | +++ | +++ |
| Amygdala | + | – | ++ |
| Cortex | + | +++ | ++ |
| Thalamus | + | – | – |
One-year-old Atxn3Q82/Q6 mice (n = 3) were immunohistochemically stained for Atxn3. For diffuse nuclear staining, increasing + numbers indicate neurons with denser nuclear staining of Atxn3. For intranuclear and extranuclear inclusions, increasing + numbers indicate higher frequency of inclusions. Regions in knockin mice that are indistinguishable from wild-type mice are indicated by –.
We also observed striking extranuclear neuronal aggregates in select brain regions, summarized in Table 1. In 1-year-old knockin mice, for example, ubiquitin-positive extranuclear inclusions are especially abundant in the stratum radiatum of the hippocampus, which also shows frequent ubiquitin-positive intranuclear inclusions in pyramidal neurons (Fig. 3A and B). Extranuclear inclusions are also present in the subiculum, central amygdala and bed nucleus of the stria terminalis. Extranuclear inclusions in the hippocampus often co-localize with anti-MAP2 and SMI32 antibodies, suggesting that they reside in dendrites (Fig. 3C). At 3 months of age, they are detected as small puncta in the hippocampal neuropil and grow over time into large, often irregular structures by 1 year of age. In a 2-year-old Atxn3Q82/Q6 mouse, many of these large inclusions co-localized with reticulon-3 (RTN3), a potential marker for dystrophic neurites (36,37) (Fig. 3D). We also examined soluble lysates of SCA3 knockin mouse hindbrain and hippocampus by western blot, but did not detect regional differences in levels of soluble, monomeric mutant Atxn3 or of high-molecular-weight, aggregated Atxn3 species (Supplemental materials, Fig. S6).
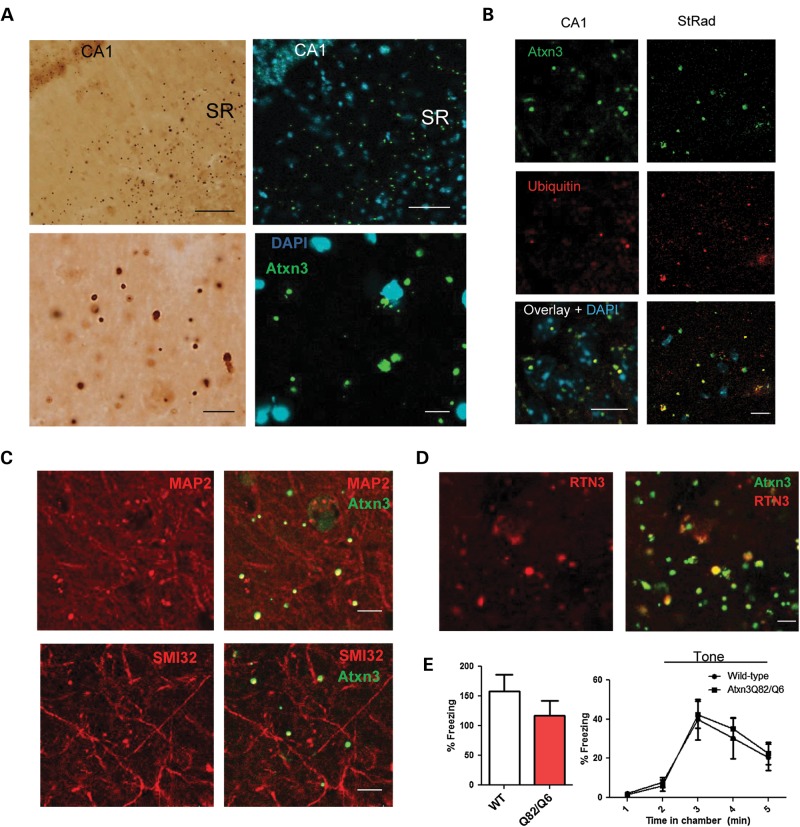
Figure 3.
Extranuclear inclusions in the hippocampus of SCA3 knockin mice. (A) Large extranuclear inclusions are concentrated in the stratum radiatum (SR) of the hippocampus 1-year-old Atxn3Q82/Q82 mice. Right, immunofluorescence of a different Atxn3Q82/Q82 mouse showing that inclusions do not colocalize with nuclear DAPI. Bottom panel shows magnified view of inclusions. (B) Hippocampal aggregates often stain ubiquitin positive, including both intranuclear inclusions in CA1 pyramidal neurons (left) and extranuclear inclusions in the stratum radiatum (right). (C) Extranuclear inclusions show overlap with dendritic markers MAP2 and SMI32. (D) Large Atxn3 extranuclear inclusions in the stratum radiatum of a 2-year-old Atxn3Q82/Q6 mouse stain for RTN3, a marker for dystrophic neurites. (E) 1-year-old Atxn3Q82/Q6 mice (n = 9) did not differ from wild-type mice (n = 9) in tests of fear conditioning, including freezing to context (left) and tone (right). Scale bars in (A) top and bottom panels are 50 and 10 μm, respectively. Scale bars in (B)–(D) are 10 μm.
The presence of robust hippocampal pathology prompted us to perform a test of fear conditioning to shock, which broadly assesses hippocampal- and amygdala-dependent memory consolidation (38). We did not observe significant differences in freezing to context or tone in 1-year-old Atxn3Q82/Q6 mice (n = 9) compared with wild-type littermates (n = 9) (Fig. 3E). All SCA3 knockin mice tested in this experiment were later confirmed to contain extensive extranuclear aggregates in the hippocampus and central amygdala nucleus (Supplementary material, Fig. S7). In summary, SCA3 knockin mice exhibit robust aggregate pathology throughout the brain upon physiological expression of polyglutamine-expanded Atxn3 and thus are well suited to explore the early molecular changes contributing to disease pathogenesis.
Alternative processing of mutant ataxin-3 transcript in SCA3
Many polyglutamine diseases and mouse models of disease are associated with transcriptional changes that may contribute to disease pathogenesis (39,40). To identify early transcriptional and splicing changes in SCA3 knockin mice, including potential alterations in the Atxn3 transcript itself, we performed RNA-sequencing on a vulnerable brain region in SCA3, the pons, from 6-month-old wild-type (n = 7) and homozygous SCA3 knockin mice (n = 7). A full analysis of transcriptional changes will be reported later. In this study, we focus on intriguing differences noted in expression of the Atxn3 transcript itself.
The human ATXN3 and murine Atxn3 genes both contain 11 exons, with the CAG repeat residing in exon 10. At least two major ATXN3 transcript variants are expressed in humans: a full-length, 11 exon ATXN3 (ATXN3-11e) transcript that contains the entire ATXN3 protein coding sequence, and a 10 exon (ATXN3-10e) transcript that retains intron 10 and lacks exon 11 (24,41–43) (Fig. 4A). The ATXN3-11e transcript (RefSeq NM_004993) encodes the ATXN3 isoform MJD1c (41,44), whereas the ATXN3-10e transcript (RefSeq S75313), initially identified by Kawaguchi et al. when they reported the disease mutation in SCA3 (24), encodes isoform MJD1a or MJD1b depending on a polymorphism that alters the location of the stop codon. In this study, we refer to these transcripts and corresponding protein isoforms by the number of exons (10e and 11e). We previously reported that ATXN3-11e is normally the most abundantly expressed isoform in the brain and that the less abundant ATXN3-10e isoform is more unstable and aggregation-prone in cell models (45). The unstable, aggregation-prone nature of ATXN3-10e may reflect the presence of a C-terminal hydrophobic domain that is encoded by read-through into the intron downstream of exon 10 (Fig. 2A). This domain is absent from ATXN3-11e, which instead ends with a domain containing an ubiquitin-interacting motif.
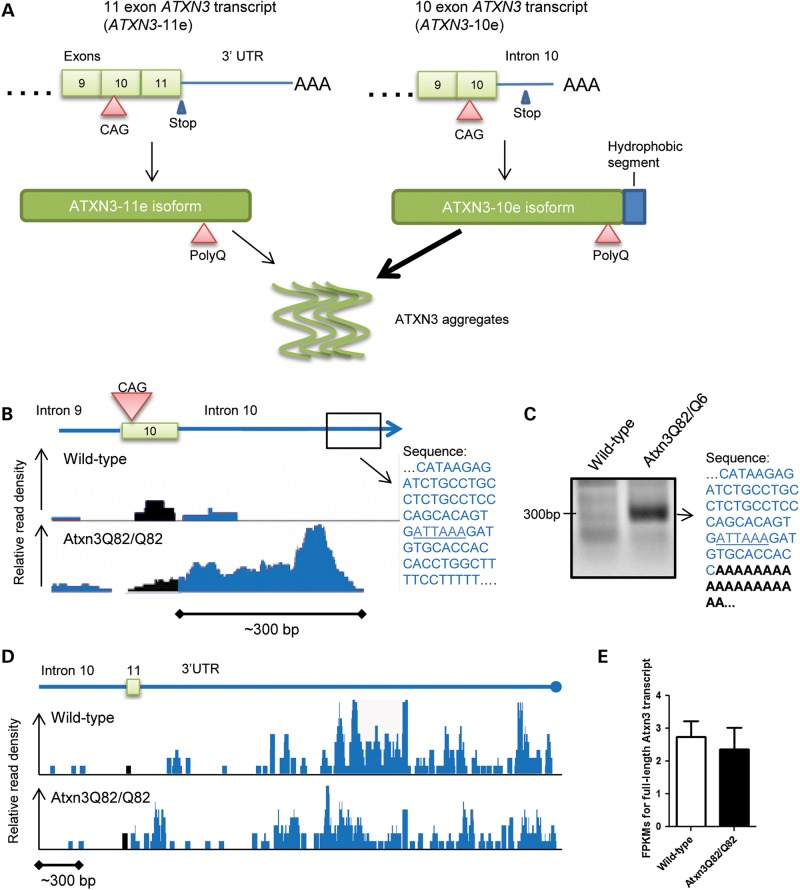
Figure 4.
Alternative splicing of the mutant Atxn3 transcript is enhanced in SCA3 knockin mice. (A) Diagram of 3′ alternative splicing of the human ATXN3 transcript, showing the 10 exon-containing ATXN3 transcript (right) generated from retention of intron 10, which encodes a hydrophobic segment that may accelerate mutant ATXN3 aggregation. (B) RNA-sequencing on the pons of Atxn3Q82/Q82 mice shows elevated reads in intron 10 following the CAG repeat-containing exon 10. Genomic sequence near the end of the reads in intron 10 (boxed) contains a putative polyadenylation (polyA) site ATTAAA (underlined). (C) 3′ RACE in an Atxn3Q82/Q6 mouse amplified a 300 bp band containing this putative polyA site (underlined) followed by a polyA tail (black, bold). (D) Wild-type and homozygous Atxn3Q82/Q82 mice showed similar frequency of sequencing reads in exon 11 and the 3′ UTR of the SCA3 knockin mice. (E) Levels of predicted full-length Atxn3 transcript from the RNA-sequencing (FPKMs) do not significantly differ between wild-type (n = 7) and Atxn3Q82/Q82 mice (n = 7). Graph represents mean + SD.
Unexpectedly, comparison of aligned RNA sequencing reads (46,47) revealed markedly elevated reads into intron 10 of Atxn3 in SCA3 knockin mice, whereas few intronic reads were present in wild-type mice (Fig. 4B). These increased reads continue ∼300 bp into intron 10, ending near a putative polyadenylation site (ATTAAA) and suggesting that the transcript could terminate and become polyadenylated at this site. Indeed, 3′ RACE on Atxn3Q82/Q6 mouse RNA to the beginning of intron 10 revealed an ∼300 bp band, which sequencing confirmed is a putative Atxn3-10e transcript polyadenylated at this predicted polyA site in intron 10 (Fig. 4C). Importantly, despite the increased expression of the Atxn3-10e transcript, SCA3 knockin mice still express the Atxn3-11e variant. RNA-sequencing reads containing exon 11 and the established Atxn3 3′ UTR appear to be equally present in wild-type and SCA3 knockin mice (Fig. 4D): normalized levels (fragments per kilobase of exon per million fragments mapped, FPKMs) of predicted full-length Atxn3-11e transcript (accession NM_029705.3) did not differ significantly between wild-type and mutant mice (Fig. 4E). Except for the differences noted at the exon 10/intron 10 junction, we did not observe striking or consistent changes in reads across the entire Atxn3 gene (Supplemental Material, Fig. S8).
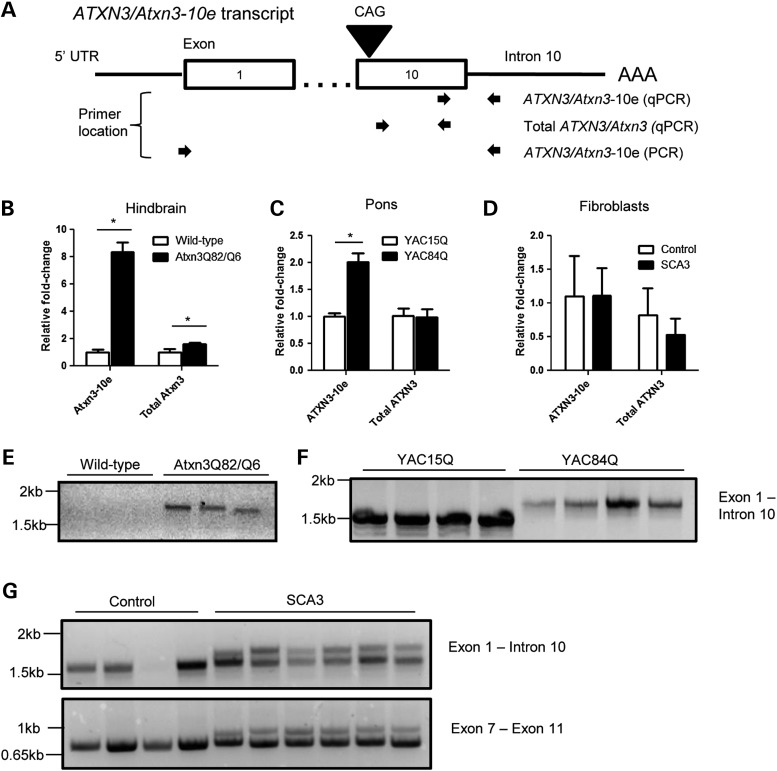
To further quantify Atxn3-10e transcript levels in SCA3 knockin mice, we performed quantitative PCR (qPCR) on reverse-transcribed RNA from brain using primers against the exon 10/intron 10 junction. We also separately performed qPCR using primers that amplify only exon 10 to estimate total (exon 10-containing) Atxn3 transcript levels (as diagrammed in Fig. 5A). Atxn3-10e transcript levels proved to be markedly increased in 3-month-old Atxn3Q82/Q6 mice compared with age-matched wild-type mice, whereas total exon 10-containing transcripts (i.e. 10e and 11e variants combined) showed a more modest upregulation in knockin mice (Fig. 5B). In addition, we found that Atxn3-10e transcript levels were still further elevated in a homozygous Atxn3Q82/Q82 mouse, which also shows enhanced nuclear accumulation of Atxn3 (Supplemental Material, Fig. S9). The basis for the upregulation of total Atxn3 transcript in SCA3 knockin mice is not clear but may reflect differences in the regulation of Atxn3-10e and Atxn3-11e transcripts since they utilize different 3′UTRs. We also cannot exclude the possibility that the manipulation of the endogenous murine Atxn3 gene, including the presence of the FRT site, alters its expression.
Figure 5.
Expression of 10 exon Atxn3/ATXN3 transcript in SCA3 mouse models and SCA3 human fibroblasts. (A) Diagram of 10 exon ATXN3/Atxn3 (ATXN3/Atxn3-10e) transcript and arrows indicating location of primer pairs used for qPCR and non-qPCR on reversed-transcribed RNA. (B) Atxn3-10e transcript is highly upregulated (∼8-fold) while total Atxn3 transcript is only modestly upregulated (∼1.5-fold) in heterozygous SCA3 knockin mice (n = 3). (C) ATXN3-10e transcript is also upregulated (∼2-fold) in YAC mice expressing the full human ATXN3 gene with 84 CAG repeats (YAC84Q, n = 4) compared with 15 CAG repeats (YAC15Q, n = 4). (D) ATXN3-10e transcript levels in SCA3 fibroblasts (n = 6) did not significantly differ from non-disease control fibroblasts (n = 4). qPCR in (B)–(D) was normalized to Gapdh, TRIP11 and ACTB, respectively. (E) PCR reveals the presence of mutant Atxn3-10e cDNA in SCA3 knockin mice only. (F and G) PCR shows expanded and non-expanded ATXN3-10e in both YAC mouse lines, all SCA3 fibroblast lines, and three of four non-disease control fibroblast lines. Amplification of ATXN3 exon 7 to exon 11 indicates the presence of ATXN3-11e in all fibroblast lines. Graphs represent the mean + SD. *P < 0.02 by an unpaired t-test.
To determine whether the increased levels of this alternative isoform reflect the presence of a CAG expansion, we investigated an independent SCA3 mouse model: YAC transgenic mice that express the full human ATXN3 gene with either a normal repeat (ATXN3Q15) or an expanded repeat (ATXN3Q84) (48). Employing qPCR with primers against ATXN3 (Fig. 5A), we observed significantly elevated ATXN3-10e transcript levels in the pons of ∼5–month-old ATXN3Q84-YAC mice compared with age-matched ATXN3Q15-YAC mice (Fig. 5C). These results suggest that CAG repeat expansions are associated with increased production of the ATXN3-10e transcript. To further explore this possibility, we analyzed ATXN3 transcripts in fibroblasts derived from SCA3 patients (n = 6) or non-disease controls (n = 4) (Fig. 5D). By qPCR, however, ATXN3-10e transcript expression levels did not differ significantly between disease and control fibroblasts.
To verify that the ATXN3/Atxn3-10e transcripts observed above contain the rest of the N-terminal protein coding sequence, we performed non-qPCR using primers to amplify from exon 1 to intron 10 (Fig. 5A) on the same reverse-transcribed RNA preparations described above. We note that this PCR is non-quantitative because large GC-rich repeats (i.e. expanded CAG repeats) are poorly amplified by the polymerase. PCR amplified a single band from Atxn3Q82/Q6 mice but none from wild-type mice (Fig. 5E). Sequencing confirmed this band to be Atxn3-10e cDNA containing the first 10 exons followed by read through into intron 10. Thus, SCA3 knockin mice express the full Atxn3-10e transcript with a CAG repeat expansion, analogous to the ATXN3-10e transcript in humans. Importantly, non-qPCR showed that the complete ATXN3-10e transcript is also expressed in YAC mice expressing normal or expanded ATXN3, in three of four control fibroblast lines, and in all SCA3 disease fibroblast lines, including from the non-expanded allele (Fig. 5F and G). The one control line that did not produce a detectable PCR signal (lane 3) showed reduced levels of total RNA by qPCR, which may prevent detection of the less abundant ATXN3-10e transcript by PCR. This sample demonstrated proportionally lower levels of ATXN3-10e, total ATXN3 and ACTB transcripts. PCR amplification from ATXN3 exon 7–11 demonstrated that ATXN3-11e is present in this and all other fibroblast lines (Fig. 5G).
The expression of the ATXN3-10e transcript even in normal human fibroblasts suggests that the relatively larger CAG repeat length of the normal human ATXN3 gene (20–42 repeats) may support the production of the ATXN3-10e transcript. In contrast, wild-type mice, which have a CAG repeat of only five repeats, show nearly undetectable levels of Atxn3-10e transcript, leading to a dramatically higher fold change in the SCA3 knockin mice. Tissue-specific differences could further mask CAG repeat-dependent splicing changes in SCA3 fibroblasts.
Discussion
The SCA3 knockin mouse reported here expresses mutant Atxn3 with an expansion in the human disease range and exhibits robust accumulation and aggregation of mutant Atxn3 in the brain, detected biochemically and immunohistochemically. In addition to replicating the intranuclear accumulation and inclusions described in human disease tissue, SCA3 knockin mice also develop extensive neuritic aggregates in select brain regions, most notably the hippocampus. SCA3 knockin mice harboring a CAG expansion also display marked retention of intron 10 in Atxn3 transcripts, a finding we confirmed in an independent mouse model of SCA3 expressing the full human disease gene. Based on these results, the SCA3 knockin mouse model reported here represents an important addition to existing models of disease that should prove particularly useful for the study of early molecular changes including mutant protein aggregation and alternative splicing.
Despite only modest expression of the mutant protein from the endogenous locus, SCA3 knockin mice manifest relatively early signs of Atxn3 accumulation in the brain, including increased concentration in neuronal nuclei and the formation of intranuclear puncta and larger inclusions. Atxn3 accumulation is noted especially in neurons of the hindbrain known to be affected in SCA3, such as DCN and brainstem neurons (49). Nuclear localization of the mutant protein is believed to contribute to disease pathogenesis in various polyglutamine diseases including SCA3 (50–52), and this new model could facilitate the study of factors acting early to regulate nuclear trafficking and handling of mutant Atxn3. The knockin mouse model also should assist the dissection of posttranslational modifications occurring to mutant Atxn3 in vivo including proteolytic cleavage, phosphorylation and ubiquitination, as well as their impact on disease protein behavior.
SCA3 knockin mice also develop striking neuritic inclusions, particularly in the synapse-rich stratum radiatum of the hippocampus. The mechanisms driving the formation of these aggregates remain unclear. Deficits in trafficking and/or handling of misfolding proteins in the distal reaches of pyramidal neurons may precipitate the formation of inclusions in subcellular compartments far removed from clearance mechanisms concentrated in the cell soma. Alternatively, neuritic aggregates could arise as a consequence of neuronal activity, which has been proposed to promote cleavage and aggregation of mutant ATXN3 at synapses (53). While the hippocampus is not considered a major disease target in SCA3, the study of the anatomically layered and easily accessible hippocampus could provide unique opportunities to study misfolded protein handling in axons and dendrites. Furthermore, an increasing number of studies report cognitive dysfunction in SCA3, which may reflect forebrain pathology (54–56). Axonal and cytoplasmic inclusions have been reported in SCA3 (57), but the extent to which they affect neuronal function is unknown. Future studies on the formation of extranuclear aggregates in SCA3 knockin hippocampus may provide insight into the handling of misfolded protein in neurites. Our findings also underscore the need for further pathological characterization of SCA3 disease tissue, including of the cortex and hippocampus.
The absence of behavioral deficits and overt neuropathological changes in heterozygous SCA3 knockin mice despite prominent disease protein aggregation is perhaps not surprising given the low-level, physiological expression of the Atxn3 disease gene. Knockin mice with CAG repeat lengths in the human disease range, as in this study, often display mild or no behavioral deficits (18,30,31,58). For example, SCA1 78Q knockin mice manifested mild abnormalities only when homozygous for the mutant allele (18). It will be important to look for motor deficits and molecular markers of degenerative change in aged homozygous SCA3 knockin mice, which we are in the process of generating. The absence of an overt behavioral phenotype in this and many other age-related neurodegenerative disease models highlights the point that such models are best at providing insight into pathogenic mechanisms that precede neuronal loss.
Intriguingly, SCA3 knockin mice exhibit altered splicing of the mutant Atxn3 transcript, mirroring the formation of a known alternative ATXN3 transcript in humans (24). Increased retention of intron 10 in mutant Atxn3 transcripts parallels the findings of Sathasivam et al., who observed the retention of intron 1 in mutant Htt transcripts following the CAG repeat-containing exon 1 in a knockin mouse model of HD and suggested that differential binding of splicing factors mediates aberrant splicing of the mutant transcript (23,59). Alternatively, differences in the kinetics of transcription through an expanded repeat may shift polyadenylation site usage (60–62). Shared mechanisms across polyglutamine diseases may dictate aberrant splicing of CAG repeat-containing transcripts, potentially producing more toxic isoforms of disease proteins in several polyglutamine disorders. We previously reported that even a non-expanded form of the ATXN3-10e variant is more aggregate-prone and less stable than the full-length isoform, suggesting that the higher hydrophobicity of the C-terminus encoded by translational read-through into intron 10 increases overall aggregation propensity (45). If identified, factors that shift splicing to favor production of the full-length (i.e. 11e) ATXN3 isoform might represent targets through which to lessen mutant protein aggregation. The SCA3 knockin mouse should facilitate studies to identify such factors.
Unfortunately, the high degree of similarity between the 10e and 11e ATXN3 isoforms raises challenges in distinguishing the two encoded proteins. We previously showed that these two isoforms have slightly different electrophoretic properties, but were unable to distinguish the ATXN3-10e isoform from ATXN3-11e isoform in ATXN3Q84-YAC mice (45). In addition, decreased solubility of the ATXN3-10e isoform may impede its detection by conventional biochemical methods. While both the ATXN3-11e and ATXN3-10e transcripts are presumably made in the human brain, their relative abundance in the SCA3 disease brain is not known. Future studies will be needed to quantify the expression of alternative ATXN3 transcripts and ATXN3 protein isoforms in human brain in order to determine their relative contribution to disease pathogenesis.
Materials and Methods
Generation of SCA3 knockin mice
Genomic murine Atxn3 DNA from a C57BL/6-tyr(c-2J) albino embryonic stem (ES) cell line (Millipore, Cat SCR011) was used to amplify a 4 kb upstream flanking arm derived from intron 9 to 10 and a 2.6 kb downstream flanking arm that spans exon 10. NotI and SalI restriction sites were engineered to flank the upstream flanking arm and a novel KpnI restriction site was engineered onto the 5′ end of the downstream flanking arm (Supplemental Material, Fig. S1). The 5′ flanking arm was then subcloned into pBY49a upstream of an FRT-PGK-Neo-FRT-positive selection cassette. The 3′ flanking arm was subcloned into pBluescript SK(−) and the CAG repeat was expanded using a modified QuickChange approach (63). Briefly, a human-expanded CAG template (At3-Q129-GFP and At3-Q166-GFP) was amplified with partially complementary primers to generate an expanded (CAG)n ‘megaprimer’ flanked by murine genomic sequence. This double-stranded megaprimer was used to insert an expanded (CAG)n repeat into the Atxn3 gene using the QuickChange Mutagenesis method (Stratagene). One clone generated was chosen for additional repeat expansion through splicing by overlap extension (64). We inserted the 3′ flanking arm from one clone of the megaprimer expansion series, which contained a Q3KQ82-encoding expansion, into the targeting vector between the FRT-PGK-Neo-FRT-positive selection cassette and the PGK-TK-negative selection cassette, using KpnI and EcoRI restriction sites (Supplemental Materials, Fig. S1).
The complete targeting vector was purified with NdeI and electroporated into Bruce4.G9 ES cells (65). G418 and ganciclovir selection were used to enrich for ES cell colonies with the positive selection cassette and loss of the negative selection cassette (Fig. 1A). Each colony was screened for homologous recombination and CAG repeat expansion by PCR- and Southern blot-based strategies. ES cell clones containing the expansion and confirmed to be sufficiently euploid were microinjected into homozygous albino B6(Cg)-Tyr<c-2J>/J blastocysts. Microinjected blastocysts were introduced then into the uterine horns of pseudopregnant female mice. Chimeras were crossed to albino B6(Cg)-Tyr<c-2J>/J mice, and all black pups were assayed for germline transmission of the knockin allele by PCR.
For in vivo excision of the FRT-site flanked-positive selection cassette, F1 heterozygous knockin animals were crossed with homozygous FLP recombinase transgenic mice driving FLPe expression under the human β-actin promoter (Jackson Laboratories, strain B6.Cg-g(ACTFLPe)9205Dym/J). F2 mice positive for the Q82 knockin expansion were crossed with C57BL/6 mice (Jackson Laboratories) to remove the FLPe transgene and outbred for at least four generations. SCA3 knockin mice were submitted to Jackson Laboratories (strain B6.Cg-Atxn3(tm1hlp)/J).
Genotyping and CAG repeat sizing
DNA was extracted from clipped mouse tails using a DNeasy Tissue kit (Qiagen) and used for genotyping by PCR. Primers flanked the endogenous CAG repeat of exon 10 of Atxn3 (forward primer: 5′-TTCACGTTTGAATGTTTCAGG-3′, reverse primer: 5′-ATATGAAAGGGGTCCAGGTCG-3′). The PCR conditions are as follows: 95°C/2 min, 30 cycles of 95°/30 s, 51°C/30 s, 72°C/1 m, 1 cycle 72°C/10 min. Products were run on a 1.5% agarose gel and visualized with ethidium bromide. Purified tail DNA was submitted for CAG repeat sizing to Laragen, Inc. (Culver City, CA, USA).
Tissue lysate preparation, SDS–PAGE, SDS–agarose and western blot analyses
Tissue lysates were prepared as previously described (8) with minor modifications. Briefly, frozen tissue was homogenized in 10 vol RIPA + protease inhibitor (PI) cocktail containing RIPA buffer (50 mm Tris–HCl, pH 7.4, 150 mm sodium chloride, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40) and PIs (Complete Mini Tablets, Roche). Homogenates were centrifuged at 1500g for 15 min at 4°C, and supernatants (soluble fraction) was transferred to a new tube and assayed for protein concentration using bicinchoninic acid (Pierce). The soluble fractions were diluted to a final concentration of 4 μg/μl in RIPA + PI + 6× loading buffer (ingredients), and boiled for 5 min. Samples were separated by 10 or 3% of SDS–PAGE, transferred onto polyvinylidene fluoride membrane (PVDF) and blotted as previously described (8). The 3% gel was transferred onto PVDF using the Bio-Rad TransBlot SD semi-dry apparatus on 10 V for 30 min. Antibodies: 1H9 1 : 2000, anti-MJD 1 : 10 000, β-tubulin 1 : 5000 (Cell signaling).
Immunohistochemistry, immunofluorescence and scoring Atxn3 accumulation
Mice were transcardially perfused with chilled PBS and the brains removed. One hemisphere was fixed in 4% paraformaldehyde at 4°C for 48 h and the other hemisphere was frozen on dry ice and stored in −80°C for biochemical experiments. Fixed brains were transferred to 30% sucrose in 0.1 m phosphate buffer for at least 48 h at 4°C. Brains were then serially sectioned and stained as previously described (8). Antibodies include anti-ataxin-3 clone 1H9 1 : 1000 (MAB5360, Millipore), anti-MJD 1 : 5000 (66), anti-RTN3 R458 1 : 2000 (kindly provided by Riqiang Yan, Cleveland Clinic, Cleveland, OH, USA), anti-MAP2 1 : 2000 (M4403, Sigma), SMI32 1 : 2000 (NE1023, Calbiochem), anti-p62 1 : 500 (P62-C DS-160211, Progen) and NeuN 1 : 500 (ABN78, Millipore). Immunofluorescent images were taken by Zeiss LSM 510-META Laser Scanning Confocal Microscope at the University of Michigan Microscopy & Image Analysis Core.
Three heterozygous SCA3 knockin brains immunohistochemically stained for Atxn3 were systematically scored for diffuse nuclear staining, intranuclear inclusions and extranuclear inclusions. Staining patterns indistinguishable from wild-type mice were scored as −. The presence of diffuse nuclear staining was represented by + or ++. Frequency of solitary intranuclear inclusions or extranuclear inclusions were scored by +, ++ or +++. Intranuclear versus extranuclear inclusions were further confirmed by co-immunofluorescent staining with nuclear DAPI. Examples of scoring diffuse nuclear staining and intranuclear inclusions are shown in Supplemental Material, Figure S4.
Motor behavior tasks and fear conditioning
The examiner was blinded to the genotype in all behavioral experiments. Motor function was assessed on SCA3 knockin mice using tests as previously reported (67), including performances on accelerating rotarod, balance beam and locomotor activity in an open field chamber for 30 min. We tested female Atxn3Q82/Q6 (n = 9) and wild-type (n = 10) mice ranging from 47 to 57 weeks of age. The average weight ± ***SEM of the Atxn3Q82/Q6 mice and wild-type mice were 24.9 ± 0.34 and 22.8 g ± 0.82, respectively. Atxn3Q82/Q6 mice were slightly, but significantly, heavier than wild-type mice. CAG repeat sizes for the tested mice ranged from 79 to 87, but found no correlation between repeat length and performance on these tasks.
Fear conditioning was performed on Atxn3Q82/Q6 mice (n = 7 males and 2 females) and wild-type littermates (n = 7 males and 2 females) ranging from 52 to 60 weeks of age using the protocol previously described for single-day experiments (68). These mice were then sacrificed and examined for aggregates in the hippocampus and amygdala.
RNA extraction, RNA-sequencing, qPCR and 3′ RACE
RNA was extracted from pons or half of the hindbrain of wild-type and SCA3 knockin mice using TRIzol® (Life Technologies) and further purified using the RNeasy kit with on-column DNase I digestion (Qiagen). Purified RNA was submitted to the University of Michigan Bioinformatics Core for library generation and Illumina-Solexa RNA-sequencing. The sequencing reads for each mouse were aligned using TopHat and the output was sorted and indexed using SAMtools to generate a BAM file. We visualized Atxn3 transcript reads in each BAM file using Integrative Genomics Viewer (46,47). FPKMs reported for the full-length Atxn3 transcript (accession NM_029705) were created by CuffLinks. We used a locally developed R script, in conjunction with CummeRbund, to output a table of all expression values (raw, externally normalized and FPKM).
DNase I-treated RNA samples from six SCA3 fibroblast lines and one non-disease control fibroblast line were kindly provided by Dr Guangbin Xia and Dr Tetsuo Ashizawa (University of Florida, Gainesville, FL, USA). RNA was also isolated from three additional non-disease fibroblast lines that were graciously provided by Dr Eva Feldman (University of Michigan, Ann Arbor, MI, USA).
For qPCR, 1 μg of RNA was reverse-transcribed using iScript™. 0.5 μl was used with the SYBR® Green Master Mix and each reaction was performed in duplicate. qPCR was performed on the Bio-Rad iCycler with MyIQ single color real-time PCR detection system module with the following parameters: 95°C at 3 min, (95°C 10 s, 55°C 30 s) × 40, 95°C 1 min, 55°C 1 min. The fold change in transcript levels was calculated using the ΔΔCt method (69). Gapdh and ACTB were used as controls for SCA3 knockin mice and patient fibroblasts, respectively. TRIP11 was used for the SCA3 YAC mice (48), since the ATXN3Q15-YAC and ATXN3Q84-YAC mice contain different numbers of the same integrated YAC construct, which also expresses the human TRIP11 gene. 3′RACE was performed on 2 μg of RNA from an Atxn3Q82/Q6 mouse hindbrain per the manufacturer's instructions (Life Technologies). 3′ RACE products were run on an agarose gel and the 300 bp band of interest was extracted, TA-cloned (Life Technologies) and sequenced. The primers used for RT-PCR and 3′RACE are listed below.
Primers
| Amplicon | Forward primer (5′–3′) | Reverse primer (5′–3′) | Experiment |
| Atxn3 exon 10—intron 10 | GGACGTAGGAGCGACCAAG | CGAGGATCTTGGGTATCGAGT | qPCR |
| Atxn3 exon 10 | TAGACCGACCTGGACCCCTT | CTTGGTCGCTCCTACGTCC | qPCR |
| ATXN3 exon 10—intron 10 | GAGCACTTGGGAGTGATCTAG | ATCACATGGAGCTCGTATGTCAG | qPCR |
| ATXN3 exon 10 | GACCTATCAGGACAGAGTTCAC | CTAGATCACTCCCAAGTGCTCC | qPCR |
| Atxn3 exon 1—intron 10 | GACAAATAAACATGGAGTCCATCTTC | CGAGGATCTTGGGTATCGAGT | PCR |
| ATXN3 exon 1—intron 10 | GACAAATAAACATGGAGTCCATCTTC | ATCACATGGAGCTCGTATGTCAG | PCR |
| ATXN3 exon 7—exon 11 | GAAGCTGACCAACTCCTGC | CGCATTGTTCCACTTTCCCATCA | PCR |
| Atxn3 intron 10 | TACTCGATACCCAAGATCCTCGTC | Provided by 3′ RACE kit (Invitrogen) | 3′RACE |
| Gapdh | CTTTGTCAAGCTCATTTCCTG | TCTTGCTCAGTGTCCTTG | qPCR |
| TRIP11 | GCCAGTCTCTGGGTCAAGTC | AATTCTGCTTCCACTTCCTCCG | qPCR |
| ACTB | CGTCCACACCCGCCG | CCACCATCACGCCCTGG | qPCR |
Statistical analyses
Statistics were performed with Prism® software (San Diego, CA, USA). For motor behavior results, statistical significance was analyzed between wild-type and SCA3 knockin mice using a Student's t-test with a Bonferroni post hoc correction considering the three different behavior tasks on the same group of mice. Freezing to context and all qPCR were analyzed by an unpaired t-test. Freezing to tone was analyzed using a one-way ANOVA.
Ethical use of animals
All mice are maintained by veterinarians and animal care staff from the University of Michigan Unit for Laboratory Animal Medicine. All manipulations and handling were performed in strict accordance with guidelines set forth by the University of Michigan Committee on Use and Care of Animals.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (RO1NS038712, RO3NS072967 to H.L.P.; F31NS083167 to B.R.; the University of Iowa Medical Scientist Training Program to G.M.H.) and the University of Michigan Rackham School of Graduate Studies to B.R.
Supplementary Material
Acknowledgements
We thank members of the Paulson laboratory and Ravi Chopra, as well as two anonymous reviewers, for thoughtful and helpful comments on the manuscript. We are grateful to Dr Tetsuo Ashizawa, Dr Eva Feldman and Dr Riqiang Yan for providing reagents. This work utilized the Sequencing and Bioinformatics Core Services provided by the Biomedical Research Core Facilities at the University of Michigan.
Conflict of Interest statement. None declared.
References
- 1.Todd T.W., Lim J. Aggregation formation in the polyglutamine diseases: protection at a cost? Mol. Cells. 2013;36:185–194. doi: 10.1007/s10059-013-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 3.Paulson H.L., Perez M.K., Trottier Y., Trojanowski J.Q., Subramony S.H., Das S.S., Vig P., Mandel J., Fischbeck K.H., Pittman R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 4.Seidel K., Siswanto S., Brunt E.R.P., den Dunnen W., Korf H.-W., Rüb U. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124:1–21. doi: 10.1007/s00401-012-1000-x. [DOI] [PubMed] [Google Scholar]
- 5.Seidel K., Meister M., Dugbartey G.J., Zijlstra M.P., Vinet J., Brunt E.R.P., van Leeuwen F.W., Rüb U., Kampinga H.H., den Dunnen W.F.A. Cellular protein quality control and the evolution of aggregates in spinocerebellar ataxia type 3 (SCA3) Neuropathol. Appl. Neurobiol. 2012;38:548–558. doi: 10.1111/j.1365-2990.2011.01220.x. [DOI] [PubMed] [Google Scholar]
- 6.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 7.Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 8.Williams A.J., Knutson T.M., Colomer Gould V.F., Paulson H.L. In vivo suppression of polyglutamine neurotoxicity by C-terminus of Hsp70-interacting protein (CHIP) supports an aggregation model of pathogenesis. Neurobiol. Dis. 2009;33:342–353. doi: 10.1016/j.nbd.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labbadia J., Novoselov S.S., Bett J.S., Weiss A., Paganetti P., Bates G.P., Cheetham M.E. Suppression of protein aggregation by chaperone modification of high molecular weight complexes. Brain. 2012;135:1180–1196. doi: 10.1093/brain/aws022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sontag E.M., Lotz G.P., Agrawal N., Tran A., Aron R., Yang G., Necula M., Lau A., Finkbeiner S., Glabe C., et al. Methylene blue modulates huntingtin aggregation intermediates and is protective in Huntington's disease models. J. Neurosci. 2012;32:11109–11119. doi: 10.1523/JNEUROSCI.0895-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 12.Malik B., Nirmalananthan N., Gray A.L., La Spada A.R., Hanna M.G., Greensmith L. Co-induction of the heat shock response ameliorates disease progression in a mouse model of human spinal and bulbar muscular atrophy: implications for therapy. Brain. 2013;136:926–943. doi: 10.1093/brain/aws343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colomer Gould V.F. Mouse models of spinocerebellar ataxia type 3 (Machado-Joseph disease) Neurotherapeutics. 2012;9:285–296. doi: 10.1007/s13311-012-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishiguro H., Yamada K., Sawada H., Nishii K., Ichino N., Sawada M., Kurosawa Y., Matsushita N., Kobayashi K., Goto J., et al. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington's disease gene. J. Neurosci. Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- 15.Heng M.Y., Duong D.K., Albin R.L., Tallaksen-Greene S.J., Hunter J.M., Lesort M.J., Osmand A., Paulson H.L., Detloff P.J. Early autophagic response in a novel knock-in model of Huntington disease. Hum. Mol. Genet. 2010;19:3702–3720. doi: 10.1093/hmg/ddq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z., Dadgar N., Albertelli M., Gruis K., Jordan C., Robins D.M., Lieberman A.P. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J. Clin. Invest. 2006;116:2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menalled L.B., Sison J.D., Dragatsis I., Zeitlin S., Chesselet M.-F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzetti D., Watase K., Xu B., Matzuk M.M., Orr H.T., Zoghbi H.Y. Repeat instability and motor incoordination in mice with a targeted expanded CAG repeat in the Sca1 locus. Hum. Mol. Genet. 2000;9:779–785. doi: 10.1093/hmg/9.5.779. [DOI] [PubMed] [Google Scholar]
- 19.Lam Y.C., Bowman A.B., Jafar-Nejad P., Lim J., Richman R., Fryer J.D., Hyun E.D., Duvick L.A., Orr H.T., Botas J., et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Watase K., Barrett C.F., Miyazaki T., Ishiguro T., Ishikawa K., Hu Y., Unno T., Sun Y., Kasai S., Watanabe M., et al. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc. Natl Acad. Sci. USA. 2008;105:11987–11992. doi: 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo S.Y., Pennesi M.E., Weeber E.J., Xu B., Atkinson R., Chen S., Armstrong D.L., Wu S.M., Sweatt J.D., Zoghbi H.Y. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron. 2003;37:383–401. doi: 10.1016/s0896-6273(02)01190-x. [DOI] [PubMed] [Google Scholar]
- 22.Yang S., Huang S., Gaertig M.A., Li X.-J., Li S. Age-dependent decrease in chaperone activity impairs MANF expression, leading to Purkinje cell degeneration in inducible SCA17 mice. Neuron. 2014;81:349–365. doi: 10.1016/j.neuron.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Housman D.E., Bates G.P. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. USA. 2012;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I., et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 25.Maciel P., Costa M.C., Ferro A., Rousseau M., Santos C.S., Gaspar C., Barros J., Rouleau G.A., Coutinho P., Sequeiros J. Improvement in the molecular diagnosis of Machado-Joseph disease. Arch. Neurol. 2001;58:1821–1827. doi: 10.1001/archneur.58.11.1821. [DOI] [PubMed] [Google Scholar]
- 26.Watase K., Venken K.J.T., Sun Y., Orr H.T., Zoghbi H.Y. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum. Mol. Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 27.Silva-Fernandes A., Costa M.D.C., Duarte-Silva S., Oliveira P., Botelho C.M., Martins L., Mariz J.A., Ferreira T., Ribeiro F., Correia-Neves M., et al. Motor uncoordination and neuropathology in a transgenic mouse model of Machado-Joseph disease lacking intranuclear inclusions and ataxin-3 cleavage products. Neurobiol. Dis. 2010;40:163–176. doi: 10.1016/j.nbd.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 28.Goti D., Katzen S.M., Mez J., Kurtis N., Kiluk J., Ben-Haïem L., Jenkins N.A., Copeland N.G., Kakizuka A., Sharp A.H., et al. A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J. Neurosci. 2004;24:10266–10279. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watase K., Weeber E.J., Xu B., Antalffy B., Yuva-Paylor L., Hashimoto K., Kano M., Atkinson R., Sun Y., Armstrong D.L., et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler V.C., White J.K., Gutekunst C., Vrbanac V., Weaver M., Li X., Li S., Yi H., Vonsattel J., Gusella J.F., et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in Hdh Q92 and Hdh Q111 knock-in mice. Hum. Mol. Genet. 2000;9:503–514. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 31.Damrath E., Heck M.V., Gispert S., Azizov M., Nowock J., Seifried C., Rüb U., Walter M., Auburger G. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 2012;8:e1002920. doi: 10.1371/journal.pgen.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menalled L.B., Sison J.D., Wu Y., Olivieri M., Li X., Li H., Zeitlin S. Early motor dysfunction and striosomal distribution of Huntingtin microaggregates in Huntington's disease knock-in mice. Hum. Mol. Genet. 2002;22:8266–8276. doi: 10.1523/JNEUROSCI.22-18-08266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelbourne P.F., Killeen N., Hevner R.F., Johnston H.M., Tecott L., Lewandoski M., Ennis M., Ramirez L., Li Z., Iannicola C., et al. Huntington's disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum. Mol. Genet. 1999;8:763–774. doi: 10.1093/hmg/8.5.763. [DOI] [PubMed] [Google Scholar]
- 34.Mori F., Tanji K., Odagiri S., Toyoshima Y., Yoshida M., Kakita A., Takahashi H., Wakabayashi K. Autophagy-related proteins (p62, NBR1 and LC3) in intranuclear inclusions in neurodegenerative diseases. Neurosci. Lett. 2012;522:134–138. doi: 10.1016/j.neulet.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 35.Nascimento-Ferreira I., Santos-Ferreira T., Sousa-Ferreira L., Auregan G., Onofre I., Alves S., Dufour N., Colomer Gould V.F., Koeppen A., Déglon N., et al. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain. 2011;134:1400–1415. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- 36.Hu X., Shi Q., Zhou X., He W., Yi H., Yin X., Gearing M., Levey A., Yan R. Transgenic mice overexpressing reticulon 3 develop neuritic abnormalities. EMBO J. 2007;26:2755–2767. doi: 10.1038/sj.emboj.7601707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Q., Hu X., Prior M., Yan R. The occurrence of aging-dependent reticulon 3 immunoreactive dystrophic neurites decreases cognitive function. J. Neurosci. 2009;29:5108–5115. doi: 10.1523/JNEUROSCI.5887-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maren S. Neurobiology of Pavlovian fear conditioning. Annu. Rev. Neurosci. 2001;24:897–931. doi: 10.1146/annurev.neuro.24.1.897. [DOI] [PubMed] [Google Scholar]
- 39.Seredenina T., Luthi-Carter R. What have we learned from gene expression profiles in Huntington's disease? Neurobiol. Dis. 2012;45:83–98. doi: 10.1016/j.nbd.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi T., Katada S., Onodera O. Polyglutamine diseases: where does toxicity come from? What is toxicity? Where are we going? J. Mol. Cell Biol. 2010;2:180–191. doi: 10.1093/jmcb/mjq005. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt T., Landwehrmeyer G.B., Schmitt I., Trottier Y., Auburger G., Laccone F., Klockgether T., Völpel M., Epplen J.T., Schöls L., et al. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 1998;8:669–679. doi: 10.1111/j.1750-3639.1998.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chai Y., Koppenhafer S.L., Shoesmith S.J., Perez M.K., Paulson H.L. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 43.Chai Y., Wu L., Griffin J.D., Paulson H.L. The role of protein composition in specifying nuclear inclusion formation in polyglutamine disease. J. Biol. Chem. 2001;276:44889–44897. doi: 10.1074/jbc.M106575200. [DOI] [PubMed] [Google Scholar]
- 44.Goto J., Watanabe M., Ichikawa Y., Yee S.B., Ihara N., Endo K., Igarashi S., Takiyama Y., Gaspar C., Maciel P., et al. Machado-Joseph disease gene products carrying different carboxyl termini. Neurosci. Res. 1997;28:373–377. doi: 10.1016/s0168-0102(97)00056-4. [DOI] [PubMed] [Google Scholar]
- 45.Harris G.M., Dodelzon K., Gong L., Gonzalez-Alegre P., Paulson H.L. Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS ONE. 2010;5:e13695. doi: 10.1371/journal.pone.0013695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cemal C.K., Carroll C.J., Lawrence L., Lowrie M.B., Ruddle P., Al-Mahdawi S., King R.H.M., Pook M.A., Huxley C., Chamberlain S. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum. Mol. Genet. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- 49.Rüb U., Schöls L., Paulson H., Auburger G., Kermer P., Jen J.C., Seidel K., Korf H.-W., Deller T. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog. Neurobiol. 2013;104:38–66. doi: 10.1016/j.pneurobio.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Ataxin-1 nuclear localization and aggregation : role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 51.Bichelmeier U., Schmidt T., Hübener J., Boy J., Rüttiger L., Häbig K., Poths S., Bonin M., Knipper M., Schmidt W.J., et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J. Neurosci. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters M.F., Nucifora F.C.N., Jr, Kushi J., Seaman H.C., Cooper J.K., Herring W.J., Dawson V.L., Dawson T.M., Ross C.A. Nuclear targeting of mutant Huntingtin increased toxicity. Mol. Cell. Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- 53.Koch P., Breuer P., Peitz M., Jungverdorben J., Kesavan J., Poppe D., Doerr J., Ladewig J., Mertens J., Tüting T., et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 54.Feng L., Chen D.B., Hou L., Huang L.H., Lu S.Y., Liang X.L., Li X.H. Cognitive impairment in native Chinese with spinocerebellar ataxia type 3. Eur. Neurol. 2014;71:262–270. doi: 10.1159/000357404. [DOI] [PubMed] [Google Scholar]
- 55.Lopes T.M., D'Abreu A., França M.C., Yasuda C.L., Betting L.E., Samara A.B., Castellano G., Somazz J.C., Balthazar M.L.F., Lopes-Cendes I., et al. Widespread neuronal damage and cognitive dysfunction in spinocerebellar ataxia type 3. J. Neurol. 2013;260:2370–2379. doi: 10.1007/s00415-013-6998-8. [DOI] [PubMed] [Google Scholar]
- 56.Roeske S., Filla I., Heim S., Amunts K., Helmstaedter C., Wüllner U., Wagner M., Klockgether T., Minnerop M. Progressive cognitive dysfunction in spinocerebellar ataxia type 3. Mov. Disord. 2013;28:1435–1438. doi: 10.1002/mds.25512. [DOI] [PubMed] [Google Scholar]
- 57.Seidel K., den Dunnen W.F.A., Schultz C., Paulson H., Frank S., de Vos R.A., Brunt E.R., Deller T., Kampinga H.H., Rüb U. Axonal inclusions in spinocerebellar ataxia type 3. Acta Neuropathol. 2010;120:449–460. doi: 10.1007/s00401-010-0717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin C., Tallaksen-Greene S., Chien W., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X., Albin R.L., Detloff P.J. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 59.Gipson T.A., Neueder A., Wexler N.S., Bates G.P., Housman D.E. Aberrantly spliced HTT, a new player in Huntington's disease pathogenesis. RNA Biol. 2013;10:1–6. doi: 10.4161/rna.26706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinto P.A.B., Henriques T., Freitas M.O., Martins T., Domingues R.G., Wyrzykowska P.S., Coelho P.A., Carmo A.M., Sunkel C.E., Proudfoot N.J., et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Mata M., Alonso C.R., Fededa J.P., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 62.Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 63.Geiser M., Cebe R., Drewello D., Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–90. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- 64.Laccone F., Maiwald R., Bingemann S. A fast polymerase chain reaction-mediated strategy for introducing repeat expansions into CAG-repeat containing genes. Hum. Mutat. 1999;13:497–502. doi: 10.1002/(SICI)1098-1004(1999)13:6<497::AID-HUMU10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 65.Hughes E.D., Qu Y.Y., Genik S.J., Lyons R.H., Pacheco C.D., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm. Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 66.Paulson H.L., Das S.S., Crino P.B., Perez M.K., Patel S.C., Gotsdiner D., Fischbeck K.H., Pittman R.N. Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann. Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
- 67.Heng M.Y., Tallaksen-Greene S.J., Detloff P.J., Albin R.L. Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington's disease. J. Neurosci. 2007;27:8989–8998. doi: 10.1523/JNEUROSCI.1830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perkowski J.J., Murphy G.G. Deletion of the mouse homolog of KCNAB2, a gene linked to monosomy 1p36, results in associative memory impairments and amygdala hyperexcitability. J. Neurosci. 2011;31:46–54. doi: 10.1523/JNEUROSCI.2634-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.