Abstract
Hutchinson–Gilford progeria syndrome (HGPS) is a segmental progeroid syndrome with multiple features suggestive of premature accelerated aging. Accumulation of progerin is thought to underlie the pathophysiology of HGPS. However, despite ubiquitous expression of lamin A in all differentiated cells, the HGPS mutation results in organ-specific defects. For example, bone and skin are strongly affected by HGPS, while the brain appears to be unaffected. There are no definite explanations as to the variable sensitivity to progeria disease among different organs. In addition, low levels of progerin have also been found in several tissues from normal individuals, but it is not clear if low levels of progerin contribute to the aging of the brain. In an attempt to clarify the origin of this phenomenon, we have developed an inducible transgenic mouse model with expression of the most common HGPS mutation in brain, skin, bone and heart to investigate how the mutation affects these organs. Ultrastructural analysis of neuronal nuclei after 70 weeks of expression of the LMNA c.1824C>T mutation showed severe distortion with multiple lobulations and irregular extensions. Despite severe distortions in the nuclei of hippocampal neurons of HGPS animals, there were only negligible changes in gene expression after 63 weeks of transgenic expression. Behavioral analysis and neurogenesis assays, following long-term expression of the HGPS mutation, did not reveal significant pathology. Our results suggest that certain tissues are protected from functional deleterious effects of progerin.
Introduction
Hutchinson–Gilford progeria syndrome (HGPS, progeria) is a very rare genetic disease characterized by multiple features of accelerated premature aging in children (1). The affected individuals display no signs of disease at birth, but within their first few years of life, they gradually develop an appearance often referred to as aged-like. HGPS affects diverse body systems including skeleton, body fat, skin, hair and the cardiovascular system. On average, death occurs at the age of 13, with at least 90% of HGPS subjects dying from progressive atherosclerosis of the coronary and cerebrovascular arteries (2).
HGPS was first described by Jonathan Hutchinson and then by Hastings Gilford in the late 19th century, but it was not until 2003 that the genetic basis of HGPS was discovered (3,4). The underlying genetic defect for 90% of HGPS patients was found to be a de novo heterozygous point mutation in exon 11 of the LMNA gene (c.1824C>T, p.G608G). Although this mutation does not result in any changes in the amino acid sequence, it causes aberrant mRNA splicing and generates a truncated and partially processed pre-lamin A protein named progerin. Unlike mature lamin A, progerin lacks 50 amino acids, which include the internal ZMPSTE24 cleavage site, and so remains permanently farnesylated and carboxymethylated. Due to this structural change, progerin remains attached to the nuclear membrane, consequently causing a disruption in the integrity of the nuclear lamina. Indeed, HGPS patient cells have a number of abnormalities in nuclear structure and function. Using indirect immunofluorescence labeling with antibodies directed against lamins A and C, fibroblasts from individuals with HGPS were characterized by the presence of dysmorphic nuclei with altered size and shape, lobules, wrinkles, herniations of the nuclear envelope, thickening of the nuclear lamina, loss of peripheral heterochromatin and clustering of nuclear pores (4,5). These features worsen with passages in cell culture and are correlated with an apparent intranuclear accumulation of progerin (5). Progerin has also been found to impair the cell cycle by interfering with the disassembly and reassembly of the nucleus during the M phase and after the G1 phase, respectively (6). For this reason, accumulation of progerin is thought to play a major role in the pathophysiology of HGPS.
Progerin has also been found to interfere with cellular functions of normally aging cells. Cells from HGPS patients and normally aged individuals share several common nuclear defects, and cells from unaffected individuals also express progerin (7). Additionally, a small amount of progerin protein was detected in protein extracts derived from elderly individuals but was absent in samples from the young (8). Rodriguez et al. quantified the levels of progerin transcripts using real-time quantitative reverse transcriptase polymerase chain reaction (RT-PCR) and showed that the progerin transcript is present in unaffected elderly individuals, although at a very low level compared with HGPS patients, and this level increased with in vitro aging, similar to HGPS cells (9). Recently, Olive and others reported that progerin is present in the coronary arteries of non-HGPS aging individuals and significantly increases with advancing age (10). On the whole, accumulation of progerin, which is formed sparsely over time as a result of the aging process, appears to be partially responsible for the cellular senescence and genomic instability observed in aging cells.
Despite the presence of multiple premature aging symptoms, not all aging processes are advanced in HGPS children. For example, progeria patients have no defects in their mental and intellectual abilities, which suggests that the brain may be protected from, and/or insensitive to, and/or unaffected by, the expression of the progeria mutation. However, there are, to our knowledge, no published studies that have investigated the neuronal accumulation of progerin in HGPS. In order to verify these hypotheses, this study presents an inducible transgenic mouse model that specifically expresses the most common LMNA c.1824C>T HGPS mutation in neurons as well as in cells of bone, skin and heart. The results from this study demonstrated that long-term expression of the common HGPS mutation did not result in any apparent neuropathological defects despite severe nuclear distortions including nuclear blebbing and invaginations of the hippocampal neurons. These intriguing results suggest that post-mitotic cells are less sensitive to induced changes in the nuclear lamina and that severe nuclear distortions do not necessarily induce changes in gene expression or functional changes in hippocampal activity.
Results
Correctly targeted and localized transgenic lamin A/progerin in neurons and cells of the skin, bone and heart
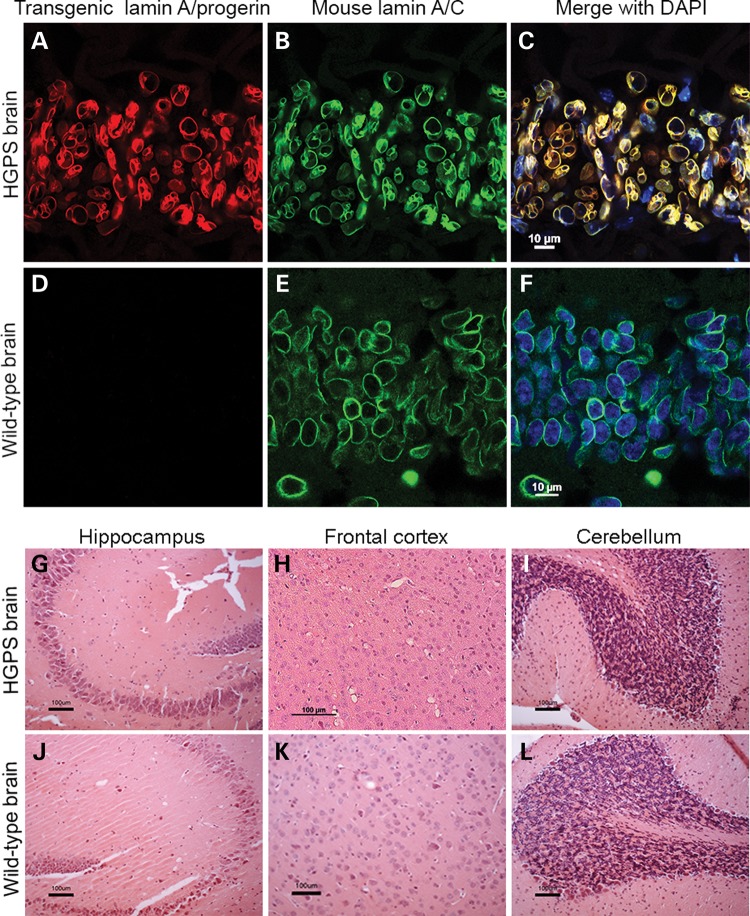
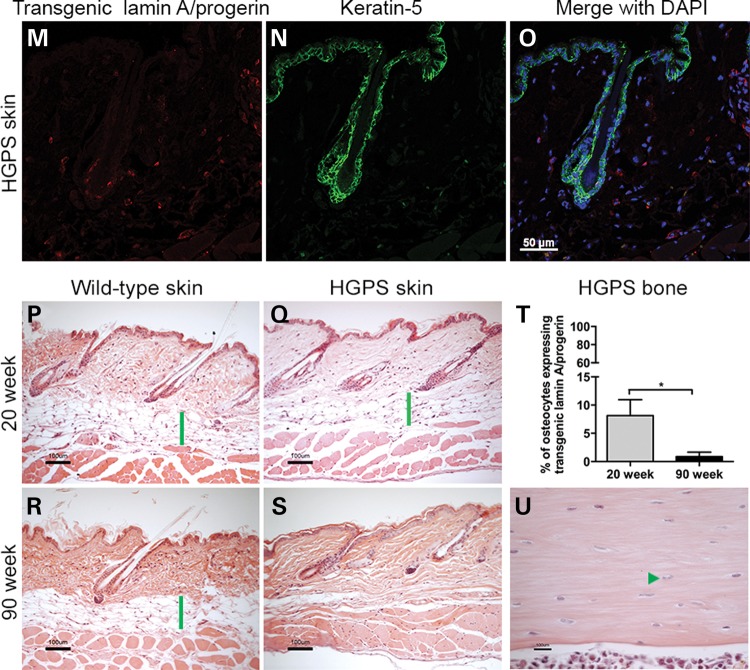
The transgenic expression was evaluated using immunofluorescence and immunohistochemistry in the brain, skin and bone (Fig. 1). The hippocampal region of the brain from 90-week HGPS animals was analyzed for the expression pattern of transgenic lamin A/progerin in combination with mouse lamin A/C (Fig. 1A–F). It was clear that the transgenic lamin A/progerin was properly targeted to its correct location in the nucleus, that is, the nuclear envelope, and formed a ‘ring’ structure around the 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclear DNA (Fig. 1A and C). Furthermore, transgenic lamin A/progerin expression substantially overlapped the mouse lamin A/C expression (Fig. 1B and C). Specific expression of the transgenic lamin A/progerin was also confirmed by its absence in the wild-type animals (Fig. 1D). The skin from 20-week HGPS animals was double-labeled for the transgenic lamin A/progerin and keratin-5 (Fig. 1M–O); keratin-5 was labeled only for structural guidance (Fig. 1N). The transgenic lamin A/progerin was observed in the keratinocytes of the bulb of the hair follicle and scattered in the dermal and hypodermal layers of the skin (Fig. 1M and O). Immunohistochemical labeling of the transgenic lamin A/progerin in the 20- and 90-week HGPS femur (Supplementary Material, Fig. S1) showed that only 8.1% of HGPS osteocytes expressed the transgene at 20 weeks, and this expression decreased to a negligible level by 90 weeks of age (P = 0.048; Fig. 1T), suggesting that the vast majority of cells in the bone tissue did not express the transgene. In addition, there were also a few osteoblasts that showed transgenic expression; however, the amount of cells was very low and therefore not quantified. To be able to compare results to our previously characterized HGPS model using a different transactivator, but with a severe bone phenotype (11), we counted femoral osteocytes in 3-week Tetop-LAG608G+; Sp7-tTA+, F1-line VF1-07 and wild-type littermates. Our results showed that the majority of osteocytes (92%) expressed the transgene when using the Sp7-tTA-transactivator.
Figure 1.
Expression pattern of the transgenic lamin A/progerin in brain, skin and bone and subsequent histopathological analyses. (A–F) Immunofluorescent labeling of 90-week CA1 hippocampal brain regions from HGPS and wild-type animals. (G–L) Hematoxylin and eosin staining showed no noticeable pathology in various regions of the brain in 90-week HGPS animals compared with wild-type animals. Representative images from hippocampus (CA3 region), frontal cortex and cerebellum. (M–O) Immunofluorescent labeling of 20-week skin showed that transgenic lamin A/progerin expression was observed in keratinocytes of the proximal part of the bulb of the hair follicle, scattered fibroblast-like cells in the dermis and cells of the hypodermis. (P–S) Skin tissues stained with hematoxylin and eosin showed that the hypodermal layer (green bars), which is used to store subcutaneous fat, was lost in the 90-week HGPS animals. (T) Percentage of osteocytes that expressed the transgenic lamin A/progerin in femur from 20- and 90-week-old HGPS animals. (U) Hematoxylin and eosin staining of the femur from 90-week-old HGPS animals with osteocytes in most lacunae (arrowhead) and a healthy bone marrow. Values represent mean ± SEM (*P < 0.05).
Expression of the LMNA c.1824C>T mutation in mice
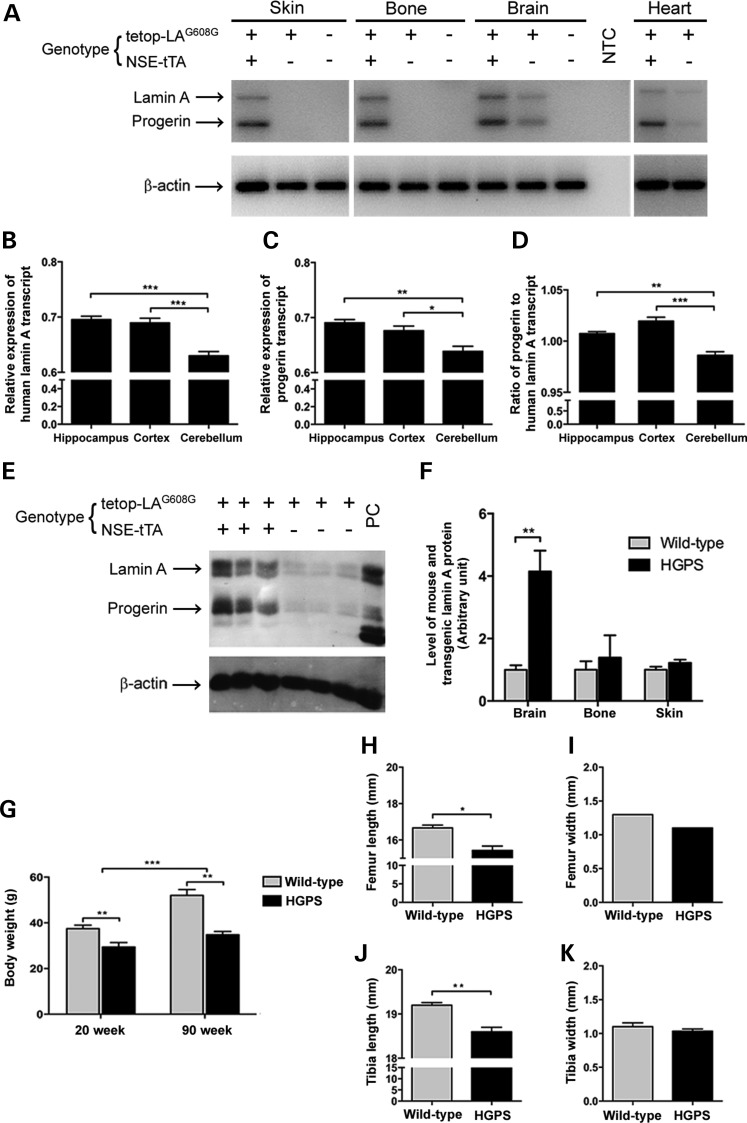
Human lamin A and progerin transcripts were expressed in skin, bone, brain and heart tissues of the HGPS animals (Tetop-LAG608G+; NSE-tTA+) but were absent in wild-type animals (Tetop-LAG608G−; NSE-tTA−) (Fig. 2A). There were also low levels of transgenic expression in the brain and heart tissues of the single-transgenic animals (Tetop-LAG608G+; NSE-tTA−) (Fig. 2A). In order to further examine the expression level of the transgene in different regions of the brain, we focused on three brain regions, hippocampus, cortex and cerebellum. The level of transgenic lamin A and progerin was measured by quantitative RT-PCR (qRT-PCR) using primers specific for human lamin A and progerin that do not cross-react with mouse lamin A (Fig. 2B–D). The levels of both human lamin A and progerin differed between the three regions, of which the cerebellum had the lowest expression compared with the hippocampus or the cortex, relative to β-actin (for lamin A: P = 0.0004 compared with hippocampus, P = 0.0009 compared with cortex; for progerin: P = 0.003 compared with hippocampus, P = 0.021 compared with cortex). However, the levels of human lamin A and progerin expression in the hippocampus and cortex were not significantly different (Fig. 2B–C). The ratio between the progerin and human lamin A also displayed a similar pattern; the cerebellum had the lowest level of progerin relative to human lamin A, while the hippocampus and cortex had higher, but similar, expression levels compared with cerebellum (P = 0.003 compared with hippocampus, P = 0.0001 compared with cortex; Fig. 2D).
Figure 2.
Transgenic lamin A/progerin expression in mouse. (A) RT-PCR of skin, bone, brain and heart tissues from HGPS, single transgenic and wild-type mice using a human lamin A and progerin-specific assay. Presence or absence of the transgene is indicated with + or –, respectively. (B–D) Relative transcript level of human lamin A and progerin in three different regions of the brain: hippocampus, cortex and cerebellum. (E) Western blot analysis of protein extracts from mouse brain tissue using a transgene-specific lamin antibody (anti-human lamin A/C, clone JoL2). Protein extracts from a HGPS patient were loaded as a positive control (PC). (F) Quantification of relative protein levels using densitometry. (G) Body weights of the HGPS and wild-type animals. (H–K) Bone length and width in HGPS compared with wild-type animals. NTC, no template control. Values represent mean ± SEM (*P < 0.05, **P < 0.01–0.001, ***P < 0.001).
The human lamin A and progerin proteins were detected in the brain, bone and skin of the HGPS animals (Fig. 2E and data not shown). The level of the transgenic overexpression of lamin A in the 20-week HGPS animals was quantified using an antibody that targets both mouse and human lamin A (Fig. 2F). It showed that in the brain, there was an increase of 3.15 over the level of lamin A in the HGPS animals compared with wild-type animals (P = 0.01), while there was only an increase of 0.39 and 0.22 in the bone and skin, respectively (Fig. 2F). There were also low levels of transgenic protein expression in brain tissue of the single transgenic animal (Fig. 2E). This leaky expression corresponded to 11% of the transgenic protein expression in the brain when quantified by densitometry.
Lower body weight and shorter bone length in HGPS mice
HGPS animals expressed the LMNA c.1824C>T mutation from embryonic day 12.5 until postnatal week 90, the maximum age analyzed. Animals were born at the expected Mendelian frequency, and there was no significant effect on life span compared with wild-type littermates (P = 0.42, animals were allowed to age until postnatal week 95). No abnormal behavior or obvious external phenotype was observed, except for the slightly smaller size of the HGPS animals. The body weights of the HGPS animals were significantly lower than those of wild-type animals at both 20 and 90 weeks of age (P = 0.004 for 20 weeks; P = 0.001 for 90 weeks; Fig. 2G). Furthermore, while body weights of the wild-type animals increased significantly with age (P = 0.0001), the HGPS animals did not significantly gain weight as they aged (Fig. 2G).
Bone length and width were measured at 20 weeks (Fig. 2H–K), and the results showed that the HGPS animals had significantly shorter femurs and tibias compared with those of wild-type animals (P = 0.011 and P = 0.007, respectively; Fig. 2H and J). However, there was no difference in femur and tibia width between the HGPS and wild-type animals (Fig. 2I and K).
Loss of subcutaneous fat in HGPS mice
Despite the presence of multiple premature aging symptoms in multiple organs of HGPS patients (for example, bone and skin), other organs, such as liver, kidney, lung, brain, gastrointestinal tract and bone marrow, appeared unaffected (1,2). In this study, samples from brain, skin, bone and heart tissue were stained with hematoxylin and eosin (H&E) for histopathological analysis (Fig. 1). In spite of long-term expression of the LMNA c.1824C>T mutation, 90-week HGPS animals did not exhibit any neuropathological changes compared with wild-type animals (Fig. 1G–L). To rule out the possibility of apoptosis, which was previously reported in HGPS, 90-week HGPS and wild-type brain tissues were labeled for cleaved caspase-3. There were no indications of apoptosis (Supplementary Material, Fig. S2A and B). Immunohistochemistry with an antibody for the glial fibrillary acidic protein (GFAP) did not reveal any significant differences between the HGPS and wild-type brain, with no indication of astrocyte activation in the hippocampus or frontal cortex in 20- or 90-week HGPS animals (Supplementary Material, Fig. S3). Interestingly, there was a considerable loss of the fat layer in the skin of 90-week HGPS animals (Fig. 1S) compared with wild-type animals (green bar, Fig. 1R), which may partly explain the lower body weight of HGPS animals compared with wild-type animals (Fig. 2G). Nevertheless, there was no apparent difference in the skin of 20-week animals (Fig. 1P and Q). Twenty or 90 weeks of LMNA c.1824C>T mutation expression did not cause any major pathological changes in bone (Fig. 1U and Supplementary Material, Fig. S1A and B). Most lacunae were filled with osteocytes (arrowheads) and the bone marrow appeared unaffected (Fig. 1U and Supplementary Material, Fig. S1A and B). Empty osteocyte lacunae were counted in femoral sections from HGPS and wild-type animals, but there were no significant differences. There was a slight increase in empty osteocyte lacunae in HGPS animals with increased age, 3.49% at 20 weeks compared with 6.06% empty osteocyte lacunae at 90 weeks. A similar increase was seen in the sections from femur from wild-type animals (3.32% at 20 weeks compared with 7.01% empty osteocyte lacunae at 90 weeks). Furthermore, animals were examined for the presence of callus formation in the rib cage because this was previously associated with HGPS (11), but neither HGPS nor wild-type animals had any calluses between 20 and 70 weeks of age (Supplementary Material, Fig. S1C–F). H&E and Masson's trichrome staining of the heart tissue demonstrated no obvious defects in the heart after 20 or 74 weeks of LMNA c.1824C>T mutation expression (Supplementary Material, Fig. S4). Furthermore, heart tissues from 74-week HGPS and wild-type animals showed no signs of apoptosis (Supplementary Material, Fig. S2C and D).
Progerin accumulates in the neuronal cells of the hippocampus and frontal cortex with aging
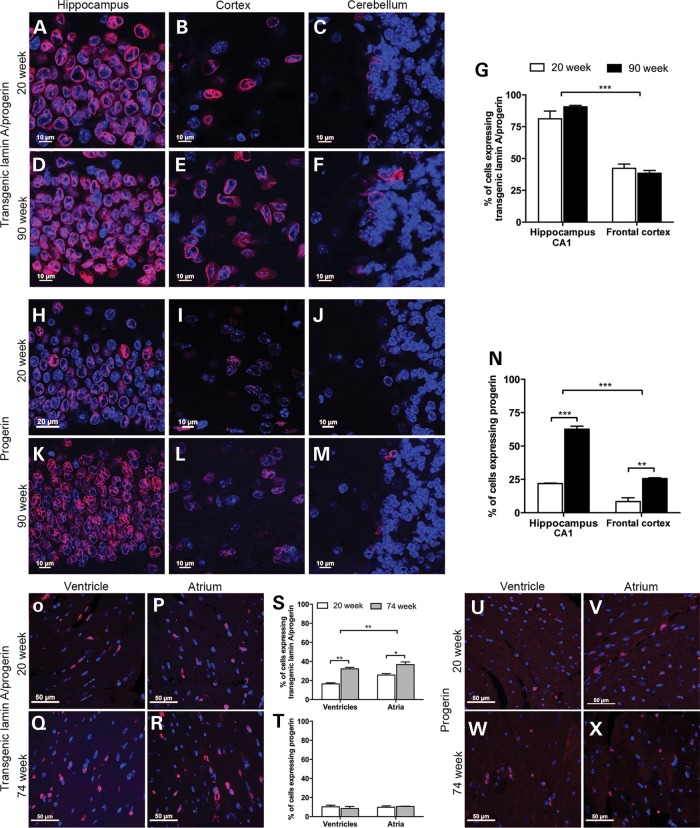
One feature identified in HGPS that is believed to contribute to disease progression is the accumulation of progerin during disease progression (5,10). To investigate whether transgenic lamin A and progerin proteins accumulate in different types of tissue, immunofluorescence was performed on the brain and heart using a transgenic lamin A/progerin or progerin antibody, and the number of positive cells was quantified (Fig. 3). The number of cells expressing transgenic lamin A/progerin and progerin was counted in the frontal cortex and the CA1 region of the hippocampus in 20- and 90-week-old HGPS animals. Because very few cells expressed the transgene in the cerebellum (Fig. 3C, F, J and M), the quantification was performed only on the CA1 region of the hippocampus and the frontal cortex. The labeling profile of both transgenic lamin A/progerin- and progerin-positive cells consisted of a nuclear rim (red) filled with DAPI nuclear staining (blue) (Fig. 3). The number of neurons expressing the transgenic lamin A/progerin in the CA1 region of the hippocampus was significantly higher than in the frontal cortex (P < 0.0001), but there was no difference between age groups (Fig. 3G). There was a significant increase in progerin-expressing cells in 90-week animals, both in the CA1 region of the hippocampus (P = 6.02 × 10−5) and in the frontal cortex (P = 0.004), compared with 20-week-old animals (Fig. 3N). This increase in expression was also visibly evident in the immunofluorescence images (Fig. 3H–M), especially in the CA1 region of the hippocampus (Fig. 3H and K). Furthermore, consistent with the quantification of transgenic lamin A/progerin expression, the number of cells expressing progerin in the CA1 region of the hippocampus was significantly higher than that of frontal cortex (P < 0.0001; Fig. 3N).
Figure 3.
Quantification of the transgenic lamin A/progerin and progerin expression in the brain and heart. (A–F) Immunofluorescent labeling of transgenic lamin A/progerin in three different regions of the brain, including the CA1 region of hippocampus, frontal cortex and the Purkinje cell layer of the cerebellum, of the HGPS animals. For both age groups, almost all cells were positive for the transgenic lamin A/progerin in the hippocampus, while in the cerebellum, there were very few positive cells. (G) Quantification of transgene-positive cells in the CA1 region of the hippocampus and the frontal cortex. (H–M) Immunofluorescent labeling of progerin in three different regions of the brain of HGPS animals. (N) Quantification of progerin-positive cells in the CA1 region of the hippocampus and the frontal cortex. (O–R) Immunofluorescent labeling of transgenic lamin A/progerin in the heart from HGPS animals at 20 or 74 weeks of age. (S and T) Quantification of cells expressing transgenic lamin A/progerin or progerin. (U–X) Immunofluorescent labeling of progerin in the ventricles and atria from the HGPS animals. Images of immunofluorescent staining were merged with DAPI. Values represent mean ± SEM (*P < 0.05, **P < 0.01–0.001, ***P < 0.001).
Both transgenic lamin A/progerin and progerin proteins were detected in the cardiomyocytes of the heart tissue (both atrium and ventricle regions) from 20- and 74-week-old HGPS animals (Fig. 3O–R and U–X, respectively). When the number of transgenic lamin A/progerin-positive cardiomyocytes was quantified, the number of positive cells significantly increased with age in both regions of the heart, ventricle and atrium (P = 0.0008 for ventricles, P = 0.030 for atria; Fig. 3S). There was a regional difference in the number of cardiomyocytes expressing transgenic lamin A/progerin, in which the level was higher in the atrium compared with the ventricle (P = 0.009; Fig. 3S). However, there were no significant differences in the number of cells expressing progerin between 20- and 74-week animals, nor between ventricle and atrium regions of the heart (Fig. 3T).
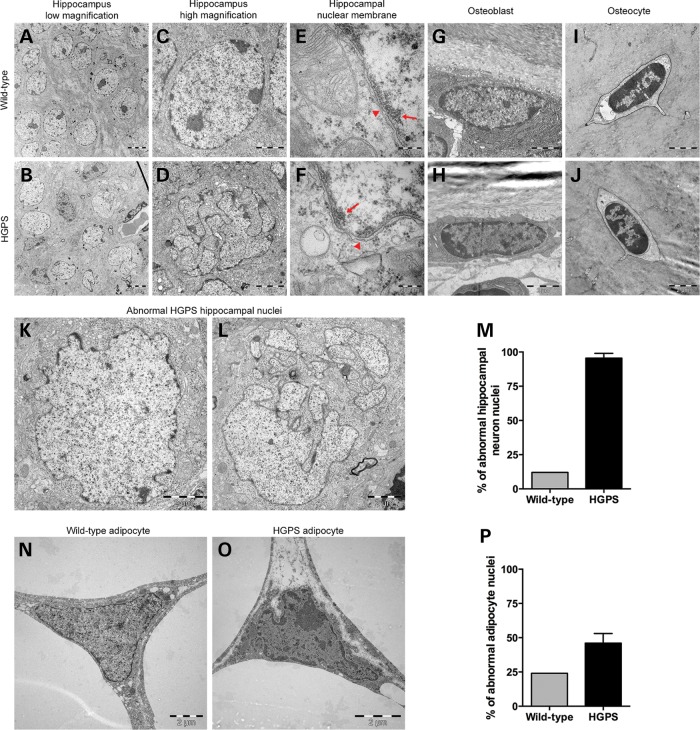
Severe neuronal nuclear distortions were observed in the hippocampus at the ultrastructural level
From the immunofluorescence analyses, it was evident that the transgenic human lamin A/progerin- and progerin-positive cells in HGPS brain had abnormal nuclear morphology, with many folds and blebs (Fig. 3A–F and H–M). In order to obtain higher resolution images of neuronal nuclei, the CA1 region of the hippocampus from 70-week HGPS and wild-type animals were analyzed using transmission electron microscopy (Fig. 4). Femur and white adipose tissue from the same group of animals were also analyzed (Fig. 4). The hippocampal nuclei of HGPS animals were severely distorted, with multiple lobulations and irregular extensions, which resulted in deep invaginations of the nuclear envelope (Fig. 4B, D, K and L) compared with the normal round nuclei of the wild-type animal (Fig. 4A and C). On average, 130 hippocampal neurons were counted for each animal to quantify the number of cells with abnormal nuclei. Notably, 95.5% of hippocampal neurons of HGPS animals had either irregular shape or severe invaginations that resulted in a fragmented appearance, which was significantly higher than that of the wild-type hippocampus (Fig. 4M). It has been shown that in HGPS skin fibroblasts, peripheral heterochromatin that normally lies adjacent to the nuclear envelope is lost (5,12). Therefore, it was important to search for this loss in the hippocampal neurons of the HGPS animals. However, there was no apparent loss of peripheral heterochromatin (arrows) in HGPS hippocampal neurons compared with wild-type (Fig. 4E and F).
Figure 4.
Electron microscopic analyses of nuclei of various cell types from 70-week HGPS and wild-type animals. While the wild-type hippocampus nuclei from the CA1 region consistently had a smooth round shape (A and C), the HGPS hippocampal nuclei of the CA1 region had a very irregular shape (B and K) and presented with extensive folding, blebbing and lobulation of the nuclear envelope, which was so severe that the nuclei looked fragmented (D and L). The images from the nuclear membranes (arrowheads; E and F) of the hippocampal neurons in the wild-type (E) and HGPS (F) animals showed no apparent loss of heterochromatin (arrows). (G–J) There were no significant differences in the nuclear structure of osteoblasts or osteocytes. (M) Quantification of abnormal nuclei in the hippocampus showed that 95.5% of hippocampal neurons had abnormal nuclei, compared with 11% in wild-type animals. (N and O) Nuclei of adipocytes from HGPS animals showed moderate folding and irregularity in nuclear morphology compared with wild-type animals. (P) Quantification of the percentage of adipocytes with abnormal nuclei morphology.
Ultrastructural changes in white adipocytes of HGPS animals
Fifty osteoblasts, osteocytes and adipocytes from each animal were analyzed for the presence of folds and/or blebs of the nucleus, and the number of cells with these abnormalities was quantified. There were no apparent differences in nuclear structure in osteoblasts or in osteocytes of HGPS animals compared with wild-type animals (Fig. 4G–J). However, in 46% of adipocytes from HGPS animals, there were noticeable folds and irregularity in the nucleus compared with the relatively smooth wild-type adipocyte nucleus (Fig. 4N–P).
No evidence of inclusions in the brain from HGPS animals
It has been well documented that expression of mutant proteins in post-mitotic neurons is often accompanied by deposition of the aberrant proteins in ubiquitin-positive inclusion bodies (13). Therefore, we explored whether the forced overexpression of human lamin A and progerin in neurons of HGPS mice resulted in the formation of ubiquitin inclusions. However, detailed analysis of the cortex and hippocampus in 90-week HGPS animals did not reveal evidence of such structures in brain sections of these mice (Supplementary Material, Fig. S5). We also analyzed the brain for protein aggregates associated with Alzheimer's disease, but found no evidence for aberrant Tau in the brain sections of HGPS animals (Supplementary Material, Fig. S6).
Expression of abnormal proteins can also result in impairment of the ubiquitin/proteasome system (14,15), which is the primary intracellular proteolytic system that eliminates misfolded proteins (16). Moreover, it has been previously reported that progerin can inhibit the catalytic activity of the proteasome (17). To explore whether progerin causes functional impairment of ubiquitin-dependent proteasomal degradation, we transiently expressed wild-type lamin A or progerin, or a minigene that expresses both lamin A and progerin, in a cell line that stably expresses a green fluorescent protein (GFP)-based reporter substrate of the ubiquitin/proteasome system (Supplementary Material, Fig. S7). Expression of the lamin A and/or progerin variants did not cause accumulation of the reporter substrate of the ubiquitin/proteasome system, suggesting that the system remains sufficiently able to facilitate degradation of ubiquitin-dependent substrates, regardless of its expression of progerin. Accordingly, treatment with a small compound proteasome inhibitor resulted in accumulation of the GFP substrate in progerin-expressing cells, underscoring that the system is operative even under conditions of overexpressed progerin.
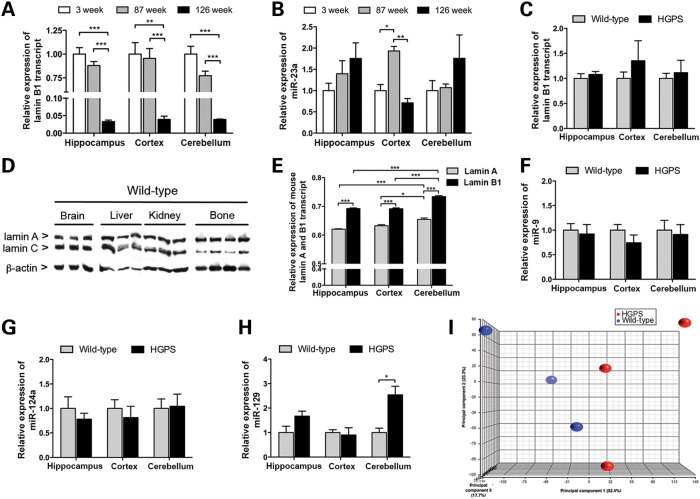
Lamin B1 and miRNA expression in the brain
Recent studies have suggested that a decrease in lamin B1 levels could serve as a marker for senescence (18–20). However, it has been shown that overexpression of LMNB1 results in abnormal nuclear morphology such as extensive folding, blebbing and lobulation in the nuclear envelope of neuronal cell lines (21). In this study, analysis of lamin B1 expression in different regions of the brain in wild-type animals showed that the level of Lmnb1 transcript decreased significantly in an age-dependent manner, in which the level was lowest at 126 weeks compared with 3 and 87 weeks in all regions of the brain (P = 1.45 × 10−5 for hippocampus, P = 0.0007 for cortex and P = 5 × 10−5 for cerebellum; Fig. 5A). However, there was no difference between 3 and 87 weeks (Fig. 5A). Prior studies have shown that miR-23a targets the LMNB1 gene and regulates lamin B1 expression (20,21). For this reason, miR-23a expression in various regions of the wild-type brain was analyzed at 3, 87 and 126 weeks of age (Fig. 5B). Relative expression levels of miR-23a showed an increasing trend with age. Nevertheless, there was a significant increase in the level of miR-23a in the cortex at 87 weeks (P = 0.002), but this level decreased again by 126 weeks (P = 0.0005), suggesting that there may be regulators other than miR-23a that may govern the expression of lamin B1. Relative Lmnb1 transcript expression was also measured in the brain tissue of 20-week HGPS and wild-type animals. However, there were no significant changes in the level of Lmnb1 transcript in all regions of the brain of HGPS animals compared with wild-type animals (Fig. 5C).
Figure 5.
Gene expression and miRNA analyses of different regions of the brain in HGPS and wild-type animals. (A and B) Graphs showing the relative expression of lamin B1 transcript (A) and miR-23a (B) in various regions of the brain from wild-type animals at 3, 83 and 126 weeks of age. (C) Lamin B1 transcript levels in HGPS animals compared with wild-type animals. (D) Western blot showing the expression of mouse lamin A and C in brain, liver, kidney and tail-bone tissues from adult wild-type animals. The sc-6215 antibody was used to detect the lamin A and C proteins. (E) Relative expression level of mouse lamin A and B1 transcripts in various regions of the brain in 20-week-old wild-type animals. (F–H) Relative expression of miR-9 (F), miR-124a (G) and miR-129 (H) in various regions of the brain of 20-week HGPS and wild-type animals. (I) Principal component analysis of the top 1000 genes with the highest average expression levels revealed very small changes in gene expression in the hippocampi of HGPS animals (red) compared with wild-type (blue). Values represent mean ± SEM (*P < 0.05, **P < 0.01–0.001, ***P < 0.001).
Recent studies have suggested that the lack of progeroid phenotype in neuronal cells is due to down-regulation of lamin A by a neural-specific microRNA, miR-9 (22,23). In this study, the relative levels of lamin A and C proteins in the wild-type brain, liver, kidney and bone tissues were analyzed by western blot (Fig. 5D). The results showed that the lamin A and C proteins were expressed at different levels in all investigated tissues. In brain, the lamin A to C ratio was 0.65 (SD = 0.04); in kidney, 1.25 (SD = 0.20); in liver, 0.82 (SD = 0.13) and in bone, 3.47 (SD = 1.18). To further understand the relative expression of lamin A and B1 in the brain, relative levels of mouse lamin A and B1 transcripts were quantified in different regions of the brain in wild-type animals (Fig. 5E). In agreement with a prior study (22), lamin B1 was more highly expressed in all regions of the brain compared with lamin A (P < 0.0001 for all regions; Fig. 5E). Furthermore, the cerebellum had the highest level of expression for both lamin A and B1 compared with hippocampus and cortex (P = 0.0007, P = 0.015, respectively; Fig. 5E).
The database of predicted and published microRNAs (miRWalk) suggested that in addition to miR-9, miR-124a and miR-129 regulate the expression of lamin A. To determine whether expression of the HGPS mutation results in changes in the expression of lamin B1 or miRNAs that target lamin A, the levels of lamin B1, miR-9, miR-124a and miR-129 were measured in 20-week HGPS and wild-type animals (Fig. 5F–H). However, there were no obvious changes in the levels of lamin B1 transcript or in any miRNAs upon transgenic expression of the HGPS mutation (Fig. 5C and F–H).
Negligible global changes in gene expression in the hippocampus
A number of studies have identified changes in gene expression in cells from HGPS patients and in tissues from animal models of HGPS (24,25). To investigate whether the severe nuclear distortions seen in 95.5% of the hippocampal neurons in HGPS animals (Fig. 4B, D, K and L) resulted in changes in gene expression, global genome transcript analysis was performed using Affymetrix exon arrays with RNA extracted from the hippocampus of 63-week-old HGPS and wild-type animals. Surprisingly, despite severe distortions in the nuclei of hippocampal neurons of HGPS animals (Fig. 4D, K and L), there were only negligible changes in global gene expression. Only five genes, out of the 16 572 RefSeq genes from the main meta-probe list, showed a statistically significant 2-fold change (Supplementary Material, Table S1). Principal component analysis (PCA) of samples grouped by genotype, using the Partek Genomic Suite software, did not result in any specific clustering, again indicating that no major differences in hippocampal gene expression between wild-type and HGPS animals (Fig. 5I).
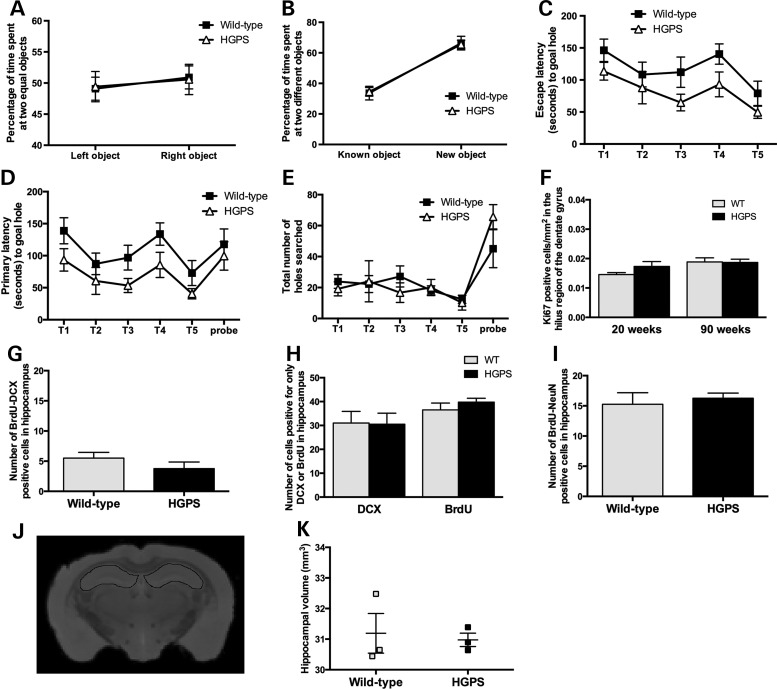
Behavioral assays reveal no effect on memory in HGPS animals
To functionally evaluate a possibly significant pathology in the hippocampi of the HGPS animals, we performed behavioral assays. The novel object recognition (NOR) test revealed no difference in object memory consolidation between HGPS and wild-type animals, and both groups had a significant preference for the novel object in the probe trial (Fig. 6A and B). In the short-protocol of the Barnes maze test (Fig. 6C–E), we detected changes by training trails (within group effect) but not by genotype (between-group effect).
Figure 6.
Functional analysis of hippocampus and neurogenesis in HGPS animals. (A and B) Object memory consolidation. (A) Pre-trial with two identical objects (left and right). Fraction of the time spent at each object on the ordinate, while object position is displayed on the abscissa. Key to genotype can be found in A. (B) Probe trial with familiar and novel objects. Both groups show a preference for the novel object (paired t-test; P < 0.001). The mean and SEM have been indicated. (C–E) Barnes maze test on spatial memory and learning. (C) The escape latency decreases across trials (T1–T5) in parallel with an improvement in primary latency (first encounter with correct hole) shown in (D) and the decrease in total number of holes searched in each trial (E). The improvement in escape latency across trials was evident within animals (F = 6.3; P < 0.001) but not between animal groups (F = 4.0; P = 0.07); thus, the slightly better performance of the HGPS line did not reach statistical significance. (F) No difference was detected between HGPS and wild-type animals at both 20 and 90 weeks of hippocampal cell proliferation. (G–I) Adult neurogenesis in HGPS and wild-type animals measured by double labeling with BrdU/DCX (G and H), BrdU/NeuN (I). (J) Hippocampal volumes were manually outlined using the ITK-SNAP software. A representative MRI image illustrating segmentation of the hippocampus from HGPS mice is outlined in the figure. (K) No significant difference, indicative of hippocampal atrophy, was seen between HGPS and wild-type mice. The mean and SEM have been indicated.
Adult neurogenesis appears unaffected by progerin expression
The subgranular zone of the hippocampal dentate gyrus is one area of the brain with postnatal production of neurons (26). To analyze the effect of progerin on cell proliferation, we stained hippocampal sections from 20- and 90-week-old HGPS and wild-type animals and counted the Ki67-positive cells in the hilus of the dentate gyrus. The result showed no significant difference in the number of proliferating cells between HGPS and wild-type mice at 20 or 90 weeks of age (Fig. 6F). To determine if the expression of progerin had an effect on adult neurogenesis, we performed repeated bromodeoxyuridine (BrdU) labeling of dividing cells for five consecutive days, in 54- or 57-week-old HGPS and wild-type mice (n = 4 per sample group). Following a chase period of 16 days, animals were sacrificed and hippocampi from each animal were collected and sectioned. Eight microtome sections, collected every tenth section, from each hippocampus were double-labeled with BrdU and doublecortin (DCX, a marker for immature neurons) or BrdU and NeuN (a marker for mature neurons). Counting of double-positive cells for BrdU and DCX showed no significant difference (P = 0.28), indicating that the fraction of newly formed and immature neurons was similar between HGPS and wild-type animals (Fig. 6G). To assess the difference in regenerative patterns between HGPS and wild-type animals, the cells positive for DCX or only positive for BrdU were also counted. Although the fraction of cells that were DCX positive but BrdU negative were larger than expected, the results did not reveal a significant difference between wild-type and HGPS animals (P = 0.94) (Fig. 6H). Additionally, the fraction of cells that were only positive for BrdU did not show a significant difference between the HGPS and wild-type animals (P = 0.37), which suggested that other proliferating cells in the hippocampus were unaffected (Fig. 6H). Counting of double-positive cells for BrdU and NeuN showed no significant differences between the HGPS and wild-type animals (P = 0.65; Fig. 6I), suggesting that the fraction of neurons that have reached maturity was similar in both populations.
Ex vivo magnetic resonance imaging reveals no effect on hippocampal volume in HGPS animals
Ex vivo magnetic resonance imaging (MRI) was performed to assess changes in hippocampal volume indicative of hippocampal atrophy (27,28). Ninety-five- or 109-week-old HGPS and wild-type mice (n = 3 per sample group) were examined using a 9.4 T Varian MRI system. Hippocampal volumes were outlined manually using the software ITK-SNAP (Fig. 6J). There was no significant difference (P = 0.77) in hippocampal volume between the HGPS and wild-type sample groups (Fig. 6K).
Discussion
Although lamin A is expressed in terminally differentiated cells, the phenotype of HGPS is restricted to certain tissues, usually of mesenchymal origin. Several suggestions have been proposed to explain the apparent lack of phenotype in the brain (22,23). However, it is important to add that, to our knowledge, no results of systematic neuropathologic evaluations of brain samples from HGPS subjects have been published. In this study, we analyzed the effects of long-term neural expression of the common progeroid lamin A mutation, LMNA c.1824C>T; p.G608G, to test the hypothesis that neuropathological changes would likely occur in children with HGPS if they were to live longer. The results presented in this work indicate that the detrimental effects of progerin expression were less than anticipated in the brain, even after 90 weeks of transgenic expression. Careful pathological examination of HGPS brain tissue did not indicate any significant changes, except for neuronal nuclear distortions, compared with wild-type mice of the same age.
Accumulation of progerin was previously suggested to contribute to the phenotype development in HGPS (5,10). In this study, we analyzed the accumulation of transgenic lamin A/progerin and progerin using immunofluorescence with antibodies specific to human lamin A/progerin or progerin in the brain, bone and heart. Our results suggested that progerin accumulates in neuronal cells of the hippocampus and the frontal cortex of the brain, but not in cardiomyocytes of the atrium and ventricle of the heart. However, accumulation of lamin A/progerin was seen in ventricles and atria. This discrepancy may have been due to sensitivity differences in the antibodies. In bone tissue, there was a decrease in the number of transgene-positive cells. To rule out the possibility of increased loss of osteocytes in HGPS animals, as previously reported in HGPS (11), the number of empty osteocyte lacuna were counted. However, no evidence suggestive of early loss of osteocytes was observed. Difference in the accumulation of progerin between different types of tissues may be dependent on the initial level of progerin expression, which varied between different tissue types, or due to the detection range for the progerin antibody being limited.
Ultrastructural analysis of neurons, adipocytes, osteoblasts and osteocytes from HGPS mice showed that a significant fraction of the neurons and adipocytes showed increased nuclear distortion compared with the wild-type animals. It is important to note that the fraction of neuronal cells of the hippocampus that showed expression of transgenic lamin A and progerin was 91% after 90 weeks of expression. This is in contrast to results from quantification of the transgenic expression of human lamin A and progerin in the bone after 90 weeks of transgenic expression, in which only a very small fraction of osteocytes, <1%, expressed the transgene. Unfortunately, we were unable to quantify adipocytes with transgenic expression using immunofluorescence or immunohistochemistry. Analysis of the skin with antibodies to human lamin A and progerin only identified very scattered cells with transgenic expression in the dermis, the bulb region of the hair follicle and the hypodermis. The loss of subcutaneous fat in HGPS animals suggests that the proportion of transgene-positive adipocytes may be higher than identified, or that there may be a systemic response that causes this loss. Recently, Karakasilioti et al. (29) reported a mouse model for xeroderma pigmentosum with a DNA repair defect that resulted in a range of premature aging symptoms, including fat depletion. They showed that DNA damage triggered a chronic autoinflammatory reaction, which in turn caused fat depletion in their mouse model (29). Taking this into consideration, further analysis is needed to determine the origin of the fat depletion seen in our mouse model.
While post-mitotic cells have lost their ability to proliferate, mitotic cells are capable of proliferation (30). Both the brain and adipose tissue are mainly composed of post-mitotic cells. However, recent evidence suggests that limited repair and regeneration also occur within these tissues. In the hippocampus of the brain, it was shown that one-third of neurons are subject to change with an annual turnover of 1.75% of neurons within the renewing fraction (31). Approximately 10% of adipocytes are renewed annually (32). One explanation for the lack of neurodegeneration in HGPS may be that post-mitotic cells are less sensitive to structural defects, and the effect on adipocytes might be the result of a systemic response induced by chronic autoinflammatory reaction (29,33).
Our data show that the capacity of the ubiquitin/proteasome system in progerin-expressing cells is sufficient to safeguard degradation of ubiquitin-dependent substrate. This is in contrast to an earlier report, which indicated that the activity of the proteasome is reduced in fibroblasts of HGPS patients (17). However, it is noteworthy that cells contain excessive proteasome activity because under normal conditions, the activity of the dominant chymotrypsin-like activity has to be inhibited >80% to induce accumulation of ubiquitylated substrate (34). Even more striking is that the caspase-like activity can be completely inhibited by selective inhibitors without affecting the functionality of the proteasome in ubiquitin-dependent proteasomal degradation (35).
A recent publication suggested that the expression of lamin A in the brain is regulated by miR-9 and that this miRNA maintains the expression of lamin A and progerin in a HGPS mouse model at undetectable levels (22). In our model system, we saw expression of the HGPS mutation in the hippocampus, frontal cortex and cerebellum. However, we did not identify any significant changes in the miRNA levels in HGPS animals compared with the wild-type animals in any regions of the brain. The reason for this result could be because during the construction of the target transgenic mouse, the 3′ untranslated region (UTR) included in the LMNA gene was very short (36), and therefore our model may be less sensitive to miRNA regulation.
A previous study that used whole-genome expression analysis in cells from HGPS patients identified >300 genes that showed a significant difference of at least 2-fold (25). Whole-genome exon array analysis of neuronal cells from the hippocampus in our HGPS mice and wild-type controls showed that only five genes demonstrated a significant 2-fold change. This is surprising because one might imagine that severe nuclear distortion would have a more significant effect on the gene expression. This is in agreement with a previous study that reported nuclear distortion of keratinocytes without effects on the skin in a transgenic mouse model. However, this study assessed expressed progerin fused with a flag epitope tag and did not analyze changes in gene expression (37). The behavioral tests showed that object memory consolidation was unaffected by the expression of progerin (Fig. 6A and B) and that both genotypes behaved similar to aged-matched C57BL/6J (38). The preserved object memory consolidation process is relevant for the results in Barnes maze because the spatial navigation strategy to find the escape hole depends on recollection of the static object queues present around the arena (39). The results recorded in the Barnes maze, used here to assess spatial memory and learning, detected no significant difference between genotype. Taken together, our results support the hypothesis that neuronal cells are less sensitive to progerin expression, regardless of its sustained expression in adulthood.
Experimental Procedures
Transgenic mice
Animals were housed in a 12-hour light/dark cycle at 20–22°C and 55–65% air humidity in a pathogen-free animal facility at the Karolinska University Hospital. The mice were fed irradiated mouse pellets RM3 (Scanbur BK AB) and water ad libitum. NSE-tTA (line A) (40) promoter mice were maintained on a CD1 background and were intercrossed with transgenic tetop-LAG608G mice, line VF1-07, generated and maintained on FVB/NCrl (36). HGPS mice referred to throughout the paper correspond to tetop-LAG608G+; NSE-tTA+ mice, and tetop-LAG608G−; NSE-tTA− mice are referred to as wild-type. Tetop-LAG608G+; NSE-tTA− mice were also included and are referred to as single transgenic animals. Ten 20-week-old wild-type animals (n = 7 females; n = 3 males) and 10 HGPS animals (n = 7 females; n = 3 males) were used for weight analysis. Likewise, eight 90-week-old animals (n = 4 for each group, all male) were used for weight analysis and other experiments, as stated below. NSE-tTA+ mice were intercrossed with transgenic tetop-LAG608G mice, of the line VF1-07, that was generated and maintained on C57BL/6J. Animal studies were approved by the Stockholm South Ethical Review board, Dnr S107-09, S12-11 and S101-12. Behavioral studies were approved by the Stockholm North Ethical Review board, Dnr N206/2012 and N84/2014.
Tissue collection and preparation
For RNA and protein extractions, animals were sacrificed by cervical dislocation without anesthesia. The whole brain was rapidly removed from the skull and immediately frozen in liquid nitrogen or placed into an ice-cold 0.9% saline solution. The brain (placed in ice-cold saline) was then dissected on a cold plate into three regions: cerebellum, cortex and hippocampus under the microscope. The dissection plate was placed on dry ice to keep the tissue cold. Other tissues, such as bone (femur), skin and heart, were collected as quickly as possible. The bone marrow was flushed out from the femur with phosphate-buffered saline (PBS). Dissected tissue samples of individual structures were immediately frozen in liquid nitrogen and stored separately at −80°C until use.
For histological staining, immunofluorescence and immunohistochemistry, animals were anesthetized with ketamine (90 mg/kg, i.p.) and xylazine (7.5 mg/kg i.p.) before undergoing transcardial perfusion. Each animal was first perfused with PBS to rinse out blood, followed by 4% paraformaldehyde (PFA) in PBS, pH 7.4. The perfusion solution was delivered at a constant volume of 10 ml/min using an automatic pump. After harvesting the brain, it was immersed in 4% PFA overnight. The carcass was also fixed overnight in 4% PFA. On the following day, the brain and dorsal skin tissues were sent for dehydration processing and paraffin embedding. The brain was divided in the sagittal plane, and femur bones were decalcified for 3 weeks using 12.5% ethylenediaminetetraacetic acid (EDTA) (pH 7.0) prior to paraffin embedding.
RNA analysis
Twenty-week HGPS and wild-type littermates (n = 3 for each group; one male and two females) were used for all RNA analyses. C57BL/6J animals that were never intercrossed with transgenic mice are referred to as ‘pure’ wild-type, and 3-, 87- and 126-week animals [n = 4 for 3-week (one male; three females) and 87-week (four females); n = 3 for 126-week (three males)] were used. Snap-frozen tissues were homogenized in ice-cold TRIzol reagent (Invitrogen), and total RNA was extracted according to the manufacturer's protocol. After resuspension of RNA in nuclease-free water (Gibco), the samples were measured and evaluated for concentration and purity (260 nm/280 nm ratio) using a BioPhotometer (Eppendorf), in which values between 1.7 and 2.0 for 260/280 nm ratio were considered optimal. Twenty micrograms of each RNA sample from skin, heart, kidney and lung was further purified with DNase I (RQ1 RNase-free DNase, Promega) and RNeasy mini columns (Qiagen) according to the manufacturer's instructions. Samples were stored at −80°C until use.
cDNA was synthesized from 1 μg of total RNA using random hexamers and SuperScript II reverse transcriptase (Invitrogen). To examine the presence of our transgene, RT-PCR for human lamin A and progerin was performed as previously described (36). β-actin was used as a control. Levels of human lamin A and progerin transcript were measured by quantitative RT-PCR (qRT-PCR) as previously described (41). Briefly, qRT-PCR reactions were prepared in triplicate for each sample with Power SYBR® Green PCR Master Mix (Life Technologies) and performed on a 7500 Fast Real-Time PCR system (Applied Biosystems). Transcript levels were determined by the comparative cycle threshold method and normalized to levels of β-actin. Specific primers for human lamin A and progerin were used for both RT-PCR and qRT-PCR. Primer sequences for the Lmnb1 gene have previously been published (22), and sequences for progerin, lamin A, and β-actin can be provided upon request.
Western blotting
Twenty-week HGPS (n = 3; two females and one male) and wild-type or single-transgenic littermates controls (n = 3; two females and one male) were used for western blotting. Tissues were homogenized in 5% radio-immunoprecipitation assay buffer containing 8 M Urea and proteinase inhibitor (Roche). Western blots were performed as previously described (36). Briefly, a mouse monoclonal anti-human lamin A/C antibody (mab3211, Chemicon) that does not cross-react with mouse lamin A/C protein was used to analyze transgenic expression at the protein level. To quantify the transgenic expression relative to wild-type mouse lamin A/C, we used an antibody raised against the N-terminal end of lamin A/C (sc-6215, Santa Cruz Biotechnology), which recognizes lamin A/C of both human and mouse origin. Secondary antibodies used were HRP-conjugated rabbit anti-goat (305-035-045, Jackson Immuno Research) and HRP-conjugated goat anti-mouse (115-036-062 and 115-035-062, Jackson Immuno Research). Protein extracts from HGPS patient cell line AG03506 were used as a control. The levels of protein expression for the mouse and human lamin A were normalized against β-actin (A5441, Sigma). Relative quantification using densitometry of the lamin A to lamin C proteins in brain (n = 3; three males), liver (n = 3; one female and two males), kidney (n = 6; two females and four males) and bone (n = 6; one female and five males) from adult wild-type mice was performed according to a previously described procedure (41). Leakiness expression of the transgene in the brain was quantified using the transgene-specific lamin A/C antibody (mab3211, Chemicon) on protein extracts of brain from HGPS and tetop-LAG608G+; NSE-tTA− single transgenic mice.
Bone measurements
The length and width of femur and tibia bones from 20-week HGPS and wild-type littermates (n = 3 for each group; one male and two females) were measured using an electronic digital caliper. The bone length was defined as the distance between the most distal part and the most proximal part of the bone; femur length was measured between the condyle and the greater trochanter, and tibia length was measured between the malleolus and the tibial tuberosity. Bone thickness was measured at the middle of the bone and was defined as the distance from the posterior to the anterior side of the bone.
Histological staining
Twenty- and 90-week HGPS and wild-type animals (n = 3 of each group for 20-week; one male and two females, n = 4 of each group for 90-week; four males) were used for all histological staining as well as immunofluorescence and immunohistochemistry. For heart tissue, 74-week HGPS (n = 3 females; n = 2 males) and wild-type littermates (n = 1 female; n = 2 males) were used instead of 90-week HGPS. Four-micrometer-thick sections of brain, skin, bone and heart tissues were cut using a microtome and collected on Superfrost glass slides (Thermo Scientific). Sections were incubated for 30 min at 60°C for adhesion of sections onto glass slides. These sections were processed for automated H&E and/or Masson's trichrome staining according to standard procedures.
Immunofluorescence
Four-micrometer-thick sections of paraffin-embedded brain, skin and heart were cut using a microtome and collected on Superfrost Plus glass slides (Thermo Scientific, Waltham, MA, USA), which were then incubated for 30 min at 60°C for adhesion. Each step was carried out at room temperature unless otherwise stated. For transgenic lamin A/progerin, progerin and keratin-5 labeling, sections underwent antigen retrieval in a pressure cooker, in which the sections were completely immersed in a 0.5 M EDTA, pH 8 solution. For BrdU, Ki67, NeuN and DCX labeling, sections underwent a double-antigen retrieval step including incubation in the microwave (700 W for 8 min in 10 mm citrate buffer at pH 8) that was followed by incubation in 2 m HCl (37°C water bath for 30 min). Following antigen retrieval, blocking was performed using 3% normal goat serum and then a mouse-to-mouse blocking reagent (Scytek, Logan, UT, USA). Primary antibodies included anti-human Lamin A/C (1 : 30, mab3211; Chemicon), anti-progerin (1:100, mAb13A4; Enzo Life Science), anti-Keratin5 (1:1000, PRB-160P; BioSite, San Diego, CA, USA), anti-ubiquitin (1:200, U5379; Sigma), anti-BrdU (1:25, 347580; BD Bioscience), anti-NeuN (1:100, ABN78; Millipore), anti-DCX (1:75, ab18723; Abcam) and anti-Ki67 (1:100, VP-K452; Vector Laboratories). Primary antibodies were incubated at 4°C overnight. Corresponding secondary antibodies were Alexa 488-conjugated donkey anti-goat (1:100, A-11055; Life Technologies), Alexa 488-conjugated goat anti-rabbit (1:100, A-11034, Life Technologies), Alexa 555-conjugated goat anti-mouse (1:100, A-2122, Life Technologies) and Fitc-conjugated goat anti-rabbit (1:300, ab6717, Abcam), which were incubated for 30 min at room temperature. Sections were counter-stained with DAPI (1:1000; Vector Laboratories) for 5 min prior to mounting (Vectashield mounting media, Vector Laboratories). Imaging was performed on a Nikon A1R, and an A1+ imaging system (Nikon Corporation, Japan).
Immunohistochemistry
Four-micrometer-thick sections of paraffin-embedded tissues, collected on Superfrost Plus glass slides (Thermo Scientific, Waltham, MA, USA), were analyzed for cleaved caspase-3 (for brain and heart tissues) GFAP and aberrant Tau (for brain tissue). Antigen retrieval for cleaved caspase-3 labeling required an incubation of sections in 10 mm citrate buffer, pH 6, at 95°C in water bath for 30 min, and for the aberrant Tau labeling, it required an incubation in formic acid at room temperature for 30 min. Following antigen retrieval, sections were blocked with 3% normal goat serum prior to anti-cleaved Caspase 3 (1:200, cab 9664, cell signaling), anti-GFAP (1:25000, MAB360, Millipore) and aberrant Tau (1:100, MC-1, (42)) primary antibody incubation at 4°C. The following day, biotinylated goat anti-rabbit secondary antibody (1:200, 65-6140, Zymed) or goat anti-mouse secondary antibody (1:300, 62-6540, Zymed) was applied for 30 min at room temperature, followed by the label antibody (ABC Elite; Vector Laboratories, Burlingame, CA, USA) for 30 min. DAB chromagen (Dako Cytomation, Carpinteria, CA, USA) was applied for 3 min, followed by rinsing with distilled water. Sections were then counter-stained with Mayers hematoxylin (Histolab), then mounted with mounting medium for light microscopy (Pertex, Histolab).
For bone tissues, 4-μm-thick sections of paraffin-embedded femur, collected on Superfrost Plus glass slides (Thermo Scientific, Waltham, MA, USA), were labeled for transgenic lamin A/progerin (1:3000, mab3211, Chemicon). All procedures were identical to caspase-3 labeling except that antigen retrieval was performed at 97.5°C and DAB was exposed for 2 min. A biotinylated goat anti-mouse secondary antibody (1:400, 62-6540, Zymed) was used.
Quantification of immunofluorescent staining
Using confocal microscopy, immunofluorescent images were taken from 20- and 90-week HGPS brain tissues (n = 3 (two females, one male) for 20-week HGPS, n = 4 (four males) for 90-week HGPS), and from 20- and 74-week HGPS heart tissues [n = 3 (2 females, 1 male) for each age group] that were labeled for transgenic lamin A/progerin or progerin. From these images, 400 DAPI-positive cells were counted, and among these, 400 DAPI-positive cells, transgenic lamin A/progerin- or progerin-positive cells were counted to calculate the frequency of cells expressing transgenic lamin A/progerin or progerin. Quantification was conducted in two different brain regions: frontal cortex and CA1 region of hippocampus, and in the ventricle and atrium of the heart. The number of transgenic lamin A/progerin- or progerin-positive cells was too low in the cerebellum for quantification, and the area was excluded.
Quantification of immunohistochemical staining in bone
Quantification of cells positive for transgenic lamin A/progerin expression was conducted in the diaphyseal cortical bone region of femur from 20- (n = 4; three females and one male) and 90-week (n = 4; four males) HGPS animals using a Nikon Eclipse E1000 microscope. A minimum of 2000 osteocytes were counted for each animal, and then the number of transgenic lamin A/progerin-positive cells were counted to calculate the frequency of osteocytes expressing transgenic lamin A/progerin. We also counted osteocytes in 3-week femurs from Tetop-LAG608G+; Sp7-tTA+, F1-line VF1-07, expressing the HGPS mutation in bone tissue (n = 3; three females), and wild-type littermate controls (n = 3; 2 females and 1 male) (11).
Quantification of empty osteocyte lacuna
Quantification of the presence of empty osteocyte lacuna was conducted in H&E-stained sections of femur from 20-week HGPS (n = 3; three females) and wild-type animals (n = 3; one female and two males) and 90-week HGPS and wild-type male animals (n = 3 per genotype). Five hundred osteocyte lacuna in the diaphyseal cortical bone region were counted in each animal.
Transmission electron microscopy
Seventy-week animals [n = 2 (one male and one female) for HGPS; n = 1 (female) for wild-type] were sacrificed by cervical dislocation, and the hippocampus, femur and abdominal white fat were dissected. These tissues were fixed in 2% glutaraldehyde + 1% PFA in 0.1 m phosphate buffer, pH 7.4 at room temperature for 30 min and stored at 4°C. Specimens were rinsed in 0.1 m phosphate buffer, pH 7.4 and post fixed in 2% osmium tetroxide 0.1 m phosphate buffer, pH 7.4 at 4°C for 2 h, dehydrated in ethanol followed by acetone, and embedded in LX-112 (Ladd, Burlington, Vermont, USA). Semi-thin sections were cut and stained with toluidine blue O and used for light microscopic analysis. Ultrathin sections (∼40–50 nm) were cut by a Leica EM UC 6 (Leica, Wien, Austria) and contrasted with uranyl acetate followed by lead citrate and examined using a Tecnai 12 Spirit Bio TWIN transmission electron microscope (FEI Company, Eindhoven, The Netherlands) at 100 kV. Digital images were randomly taken with a Veleta camera (Olympus Soft Imaging Solutions, GmbH, Münster, Germany). A total of 130 hippocampal neurons, 50 osteoblasts, 50 osteocytes and 50 adipocytes were assessed for nuclear shape. To be counted as a misshapen, the nucleus had to present with lobulations and folds that altered the normal round shape of the nucleus.
Ubiquitin-dependent proteasomal degradation reporter assay
Human melanoma cell MelJuSo line stably expressing UbG76V-GFP, which has been previously described (43), was grown in DMEM GlutaMAX™ medium (Life Technologies) complemented with 10% FCS at 37°C under 5% CO2. To generate the pHcRed1-C1_mycLAΔ150 expression vector, the pET3-LAΔ150 (5) was digested with BamHI and XbaI, and the mycLAΔ150 insert fragment was gel-purified and ligated to a pHcRed1-C1 vector (BD Biosciences Clontech), previously digested with BamHI and XbaI. Additional vectors were generated from the pHcRed1-C1_mycLAΔ150 vector. The vector was first digested with XbaI, then filled in with T4 DNA polymerase and gel-purified to remove the LAΔ150 insert. The previously published pIRES2-eGFP-tetop-LAwt and pIRES2-eGFP-tetop-LAG608G vectors (LA-77 and LA-75, respectively) (36) were digested with BamHI before being filled in with T4 DNA polymerase and digested with AscI to release the LA minigenes for gel purification. The LA minigenes were then ligated to the gel-purified pHcRed1-C1_myc vector fragment to generate the pHcRed1-C1_LA-75, and pHcRed1-C1_LA-77 vectors. Transient transfection with cellular expression vectors was performed using Lipofectamine® 2000 transfection reagent (Life Technologies) according to the manufacturer's protocol. MelJuSo UbG76V-GFP stable human melanoma cells were grown on 18 mm coverslips in six-well plates for 24 h after transfection. MG132 was added 5 h before fixation in a 10 µm concentration. Cells were then fixed for 20 min in PBS with 4% PFA and mounted with 10% Mowiol®; 2.5% 1,4-diazabicyclo[2.2.2]octane, 25% glycerol in 0.1 m Tris, pH 8.5. Images were acquired on a Zeiss LSM510 META confocal laser scanning microscope with a Zeiss Plan Apochromat 63x/1.40 oil DIC objective (Carl Zeiss, Jena, Germany) operated by LSM5 software. The images were processed using the Zeiss LSM Image Browser software.
MicroRNA analysis
Twenty-week HGPS and wild-type littermates (n = 3 for each group; one male and two females) were used for miRNA-9, miRNA-124 and miRNA-129 analyses. Three-, 87- and 126-week pure C57BL/6J wild-type animals [n = 4 for 3- (one male; three females) and 87-week (females); n = 3 for 126-week (males)] were used for miRNA-23a analysis. Total RNA was isolated from snap-frozen mouse brain tissues using a mirVANA miRNA isolation kit (Ambion), and RNA samples were quantified and evaluated for purity (260/280 nm ratio) using a spectrophotometer (BioPhotometer, Eppendorf). miRNA detection was performed using TaqMan microRNA assays (Applied Biosystems) with a no-UNG no-Amperase master mix (Applied Biosystems) according to manufacturer's instruction. Briefly, 10 ng of total RNA was reverse-transcribed using the TaqMan miRNA reverse transcription kit (Applied Biosystems) and PCR amplified using an Applied Biosystems 7500 Real-Time PCR system. As an internal control, miRNA expression was normalized against snoRNA202. All protocols were performed according to the manufacturer's instruction.
Statistical analysis
Statistical analyses were performed using one- and two-way ANOVAs, unpaired Student’s t-test and a Chi-squared for life span. A two-tailed P-value of 0.05–0.01 was considered significant (*), a P-value of 0.01–0.001 is indicated as ** and a P-value smaller than 0.001 is indicated as ***, unless otherwise indicated.
Exon array analysis
Sixty-three-week-old HGPS (n = 3; females) and wild-type animals (n = 3; one female and two males) were sacrificed by cervical dislocation before the hippocampus was dissected and snap-frozen in liquid nitrogen as described in the previous section. Total RNA was extracted using the mirVANA miRNA isolation kit (Ambion), which was then quantified using a spectrophotometer (BioPhotometer, Eppendorf), followed by a quality check with the Agilent Tapestation 2200. The criteria for high-quality RNA were an RNA Integrity Number (RIN) above 8.0 and ribosomal ratio (28S/18S) above 1.5. One hundred fifty nanograms of RNA from each animal were used in preparation for Affymetrix whole transcriptome (WT) microarray analysis using an Ambion WT Expression Kit (Affymetrix, Santa Clara, CA, USA). Samples were then hybridized to mouse Affymetrix GeneChip Sense Target (ST) Arrays and incubated in a rotating hybridization 640 oven (Affymetrix) at 45°C for 16 h at 60 rpm. Following hybridization, the arrays were washed and stained on an GeneChip® Fluidics 450 workstation (Affymetrix, Santa Clara, USA) using the FS450_0001 and FS450_0007 fluidics script. The arrays were scanned using the 3000 7G Affymetrix GeneChip Scanner (Affymetrix, Santa Clara, CA, USA). Labeling and hybridization of all exon arrays were performed at the Bioinformatics and Expression Analysis (BEA) core facility (Karolinska Institutet, Sweden). Affymetrix CEL files with exon array raw data were imported into Partek Genomic Suite software (Partek Inc., St. Louis, MO, USA) and filtered to include only those probe sets in the ‘main’ meta-probe list, which represents 16,572 RefSeq genes and full-length GenBank mRNAs. The main data include target-sequence, perfect-match, unique probe sets. Robust multi-array average is not an algorithm from Affymetrix but has been implemented into the Affymetrix Expression Console (EC). Robust multi-array average include methods for summarization, background correction and normalization. For robust multi-array average (RMA) references, see http://bmbolstad.com/misc/ComputeRMAFAQ/ComputeRMAFAQ.html, last accessed on 30 October 2014. Normalized probe signals were converted into a log 2 scale, which was a default setting in the RMA method. Annotations were provided by Affymetrix (NetAffx build 32). Sample data quality was assessed by the EC report, the quality control steps in Partek and visual inspection of the images from scanning. All arrays met all quality parameters established by Affymetrix, and no outliers were detected. Detection of differentially expressed genes was performed by following the pre-defined exon work flow menu in Partek. Exons were summarized into genes and analysis of variance was performed. A list of changed genes was chosen on the basis of a 2-fold increase or decrease in the expression level in HGPS hippocampal samples compared with the wild-type, together with an FDR P-value of <0.05.
Behavioral studies
Eight HGPS and eight wild-type male littermate mice between 55 and 60 weeks old were subjected to behavioral analysis of object and spatial memory. After arrival at the behavioral suite, the animals were allowed to acclimatize for 1 week. In the second week, object (reference) memory was examined by means of the NOR test, as described in detail elsewhere (44). After a pause of 1 week, the same animals were tested in the Barnes maze (39) using an arena measuring 124 cm in diameter with 36 holes each with a width of 4.5 cm. In this test, we used a shortened protocol developed by Attar et al. (45) with five training trials over 2 days followed by the probe trial on the third day. This protocol emphasizes the early learning phase and has been shown to be more sensitive with respect to discrete spatial learning difficulties/memory consolidation than the standard protocol with 15 training trials (45). The only aversive stimulus used was light (indirect light providing an illumination of the arena of 600 Lux from above and 400 Lux from below). All tests were recorded with a high-resolution video camera at 15 frames per second and fed into a computer where the records were analyzed with the Ethovision XT10 software (Noldus, The Netherlands). Software settings were not changed between groups and all analyses were executed at least twice. A paired Student's t-test was used to assess results from the NOR test while repeated measures used ANOVA (with genotype as in-between factor and trials as within-subject variable). In case of significance, a post-hoc Bonferroni analysis was performed. Statistical significance was set to P < 0.05. For all analyses, the Statistical V6 software was used.
Adult neurogenesis
BrdU labeling of dividing cells in the dentate gyrus of the hippocampus was used to monitor neurogenesis (26) in HGPS and wild-type female mice. Fifty-four- or 57-week-old bitransgenic (n = 4) and wild-type (n = 4) mice were injected intraperitoneally every 24 h during five consecutive days with BrdU (50 mg/kg bodyweight). The mice were sacrificed 16 days after the first day of injection. The whole hippocampus was cut into 4-μm-thick sections. The sections were dried at 60°C for 1 h and processed for double-labeling experiments with BrdU/DCX and BrdU/NeuN. Every tenth section was stained, and positive neuronal cells, for a total of eight sections/animal, were counted.
Ex vivo MRI
Whole-brain samples from HGPS (n = 3; one female and two males) and wild-type (n = 3; one female and two males) mice 95- or 109-week-old were analyzed using MRI. MRI data were acquired using 9.4 T scanner (Agilent Technologies, Santa Clara, CA, USA) with a millipede coil of 30 mm inner diameter. PFA fixed brains were positioned in a syringe filled with Fomblin YL VAC 06/6 (Solvay Specialty Polymers, Italy), to provide a cylindrical shape of homogenous magnetic susceptibility. A 3D multi-echo sequence was acquired where 10 spin echos were acquired with 6.37 ms separation with a TR time of 800 ms. The matrix size was 512 × 256 × 256 covering a field of view of 51.2 × 25.6 × 25.6 mm3. A single exponential decay was fitted to each pixel and an image corresponding to a TE time of 25 ms was calculated from the T2 and initial intensity. The hippocampi were manually segmented using the software ITK-SNAP version 3.2.0 (46).
Supplementary Material
Funding
This work was supported by grants from the Karolinska Institutet (to J.-H.B., B.U., M.E.); the K.A. Wallenberg Foundation KAW 2010.0127 (to B.U.); the Swedish Research Council (to N.P.D., M.E.); the Swedish Foundation for Strategic Research (to M.E.); the Torsten and Ragnar Söderberg Foundation (to M.E.); the Loo and Hans Osterman Foundation (to M.E.); the Progeria Research Foundation (to M.E.) and Marie Curie Actions within The European Community's Seventh Framework Programme, FP7/2012 No. 264508 (to N.P.D.). Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Supplementary Material
Acknowledgements
The pHcRed1-C1_mycLMNAdel150, pHcRed1-C1_LA-75 and pHcRed1-C1_LA-77 vectors were kind gifts from Michael R. Erdos. Postmortem human brain tissue samples were used as positive control tissue samples and were obtained from the Netherlands Brain Bank to F.W.v.L. for the ISAO project #06502 and #09514. We would like to thank Raoul Kuiper for pathological consultation, Sylvie Le Guyader for microscopy consultation, Malin Lijebäck and David Brodin for exon array analysis and Tomas McKenna, Gwladys Revechon, Xiuzhe Wang, Johanna Olsson, Hasina Nasser and Juliane Schwaderer for technical assistance. This study was in part performed at the Live Cell Imaging unit, Department of Biosciences and Nutrition, Karolinska Institutet, Huddinge, Sweden. The MRI scanning was performed at the Department of Comparative Medicine/Karolinska Experimental Research and Imaging Centre at Karolinska University Hospital, Solna, Sweden.
Conflict of Interest statement. None declared.
References
- 1.Baek J.-H., McKenna T., Eriksson M. Hutchinson–Gilford progeria syndrome. In: Puiu M., editor. Genetic Disorders. Vol. 1. Croatia: InTech, Rijeka; 2012. pp. 65–87. [Google Scholar]
- 2.Merideth M.A., Gordon L.B., Clauss S., Sachdev V., Smith A.C., Perry M.B., Brewer C.C., Zalewski C., Kim H.J., Solomon B., et al. Phenotype and course of Hutchinson–Gilford progeria syndrome. N. Engl. J. Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., et al. Lamin a truncation in Hutchinson–Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 4.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaffidi P., Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. [Google Scholar]
- 8.McClintock D., Ratner D., Lokuge M., Owens D.M., Gordon L.B., Collins F.S., Djabali K. The mutant form of lamin A that causes Hutchinson–Gilford progeria is a biomarker of cellular aging in human skin. PLoS One. 2007;2:e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez S., Coppede F., Sagelius H., Eriksson M. Increased expression of the Hutchinson–Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur. J. Hum. Genet. 2009;17:928–937. doi: 10.1038/ejhg.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olive M., Harten I., Mitchell R., Beers J.K., Djabali K., Cao K., Erdos M.R., Blair C., Funke B., Smoot L., et al. Cardiovascular pathology in Hutchinson–Gilford progeria: correlation with the vascular pathology of aging. Arterioscler. Thromb. Vasc. Biol. 2010;30:2301–2309. doi: 10.1161/ATVBAHA.110.209460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt E., Nilsson O., Koskela A., Tuukkanen J., Ohlsson C., Rozell B., Eriksson M. Expression of the Hutchinson–Gilford progeria mutation during osteoblast development results in loss of osteocytes, irregular mineralization, and poor biomechanical properties. J. Biol. Chem. 2012;287:33512–33522. doi: 10.1074/jbc.M112.366450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCord R.P., Nazario-Toole A., Zhang H., Chines P.S., Zhan Y., Erdos M.R., Collins F.S., Dekker J., Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson–Gilford progeria syndrome. Genome Res. 2013;23:260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciechanover A., Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 14.Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 15.Lindsten K., de Vrij F.M., Verhoef L.G., Fischer D.F., van Leeuwen F.W., Hol E.M., Masucci M.G., Dantuma N.P. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J. Cell Biol. 2002;157:417–427. doi: 10.1083/jcb.200111034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman M.Y., Goldberg A.L. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 17.Viteri G., Chung Y.W., Stadtman E.R. Effect of progerin on the accumulation of oxidized proteins in fibroblasts from Hutchinson Gilford progeria patients. Mech. Ageing Dev. 2010;131:2–8. doi: 10.1016/j.mad.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., Goldman R.D. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freund A., Laberge R.M., Demaria M., Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreesen O., Chojnowski A., Ong P.F., Zhao T.Y., Common J.E., Lunny D., Lane E.B., Lee S.J., Vardy L.A., Stewart C.L., et al. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J. Cell Biol. 2013;200:605–617. doi: 10.1083/jcb.201206121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S.T., Fu Y.H. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model Mech. 2009;2:178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung H.J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S., Yang S.H., Barnes R.H., 2nd, Hong J., Sun T., Pleasure S.J., et al. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. USA. 2012;109:E423–E431. doi: 10.1073/pnas.1111780109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissan X., Blondel S., Navarro C., Maury Y., Denis C., Girard M., Martinat C., De Sandre-Giovannoli A., Levy N., Peschanski M. Unique preservation of neural cells in Hutchinson–Gilford progeria syndrome is due to the expression of the neural-specific miR-9 microRNA. Cell Rep. 2012;2:1–9. doi: 10.1016/j.celrep.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Ly D.H., Lockhart D.J., Lerner R.A., Schultz P.G. Mitotic misregulation and human aging. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- 25.Csoka A.B., English S.B., Simkevich C.P., Ginzinger D.G., Butte A.J., Schatten G.P., Rothman F.G., Sedivy J.M. Genome-scale expression profiling of Hutchinson–Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–243. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 26.Wojtowicz J.M., Kee N. BrdU assay for neurogenesis in rodents. Nat. Protocols. 2006;1:1399–1405. doi: 10.1038/nprot.2006.224. [DOI] [PubMed] [Google Scholar]
- 27.Malaeb S.N., Davis J.M., Pinz I.M., Newman J.L., Dammann O., Rios M. Effect of sustained postnatal systemic inflammation on hippocampal volume and function in mice. Pediatr. Res. 2014;76:363–369. doi: 10.1038/pr.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persson A., Sim S.C., Virding S., Onishchenko N., Schulte G., Ingelman-Sundberg M. Decreased hippocampal volume and increased anxiety in a transgenic mouse model expressing the human CYP2C19 gene. Mol. Psychiatry. 2014;19:733–741. doi: 10.1038/mp.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karakasilioti I., Kamileri I., Chatzinikolaou G., Kosteas T., Vergadi E., Robinson A.R., Tsamardinos I., Rozgaja T.A., Siakouli S., Tsatsanis C., et al. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 2013;18:403–415. doi: 10.1016/j.cmet.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campisi J., d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 31.Spalding K.L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H.B., Bostrom E., Westerlund I., Vial C., Buchholz B.A., et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spalding K.L., Arner E., Westermark P.O., Bernard S., Buchholz B.A., Bergmann O., Blomqvist L., Hoffstedt J., Naslund E., Britton T., et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 33.Rosengardten Y., McKenna T., Grochova D., Eriksson M. Stem cell depletion in Hutchinson–Gilford progeria syndrome. Aging Cell. 2011;10:1011–1020. doi: 10.1111/j.1474-9726.2011.00743.x. [DOI] [PubMed] [Google Scholar]
- 34.Dantuma N.P., Lindsten K., Glas R., Jellne M., Masucci M.G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 35.Myung J., Kim K.B., Lindsten K., Dantuma N.P., Crews C.M. Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Mol. Cell. 2001;7:411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 36.Sagelius H., Rosengardten Y., Hanif M., Erdos M.R., Rozell B., Collins F.S., Eriksson M. Targeted transgenic expression of the mutation causing Hutchinson–Gilford progeria syndrome leads to proliferative and degenerative epidermal disease. J. Cell Sci. 2008;121:969–978. doi: 10.1242/jcs.022913. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Panteleyev A.A., Owens D.M., Djabali K., Stewart C.L., Worman H.J. Epidermal expression of the truncated prelamin A causing Hutchinson–Gilford progeria syndrome: effects on keratinocytes, hair and skin. Hum. Mol. Genet. 2008;17:2357–2369. doi: 10.1093/hmg/ddn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fahlström A., Zeberg H., Ulfhake B. Changes in behaviors of male C57BL/6J mice across adult life span and effects of dietary restriction. Age (Dordr) 2012;34:1435–1452. doi: 10.1007/s11357-011-9320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnes C.A. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J. Comp. Physiol. Psychol. 1979;93:74–104. doi: 10.1037/h0077579. [DOI] [PubMed] [Google Scholar]
- 40.Chen J., Kelz M.B., Zeng G., Sakai N., Steffen C., Shockett P.E., Picciotto M.R., Duman R.S., Nestler E.J. Transgenic animals with inducible, targeted gene expression in brain. Mol. Pharmacol. 1998;54:495–503. doi: 10.1124/mol.54.3.495. [DOI] [PubMed] [Google Scholar]
- 41.McKenna T., Rosengardten Y., Viceconte N., Baek J.-H., Grochova D., Eriksson M. Embryonic expression of the common progeroid lamin A splice mutation arrests postnatal skin development. Aging Cell. 2014;13:292–302. doi: 10.1111/acel.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jicha G.A., Bowser R., Kazam I.G., Davies P. Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res. 1997;48:128–132. doi: 10.1002/(sici)1097-4547(19970415)48:2<128::aid-jnr5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 43.Menendez-Benito V., Verhoef L.G., Masucci M.G., Dantuma N.P. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum. Mol. Genet. 2005;14:2787–2799. doi: 10.1093/hmg/ddi312. [DOI] [PubMed] [Google Scholar]
- 44.Fahlström A., Yu Q., Ulfhake B. Behavioral changes in aging female C57BL/6 mice. Neurobiol. Aging. 2011;32:1868–1880. doi: 10.1016/j.neurobiolaging.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Attar A., Liu T., Chan W.T., Hayes J., Nejad M., Lei K., Bitan G. A shortened Barnes maze protocol reveals memory deficits at 4-months of age in the triple-transgenic mouse model of Alzheimer's disease. PLoS One. 2013;8:e80355. doi: 10.1371/journal.pone.0080355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.