Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder characterized by the selective death of motor neurons. Causative mutations in the global RNA-processing proteins TDP-43 and FUS among others, as well as their aggregation in ALS patients, have identified defects in RNA metabolism as an important feature in this disease. Lethal congenital contracture syndrome 1 and lethal arthrogryposis with anterior horn cell disease are autosomal recessive fetal motor neuron diseases that are caused by mutations in another global RNA-processing protein, hGle1. In this study, we carried out the first screening of GLE1 in ALS patients (173 familial and 760 sporadic) and identified 2 deleterious mutations (1 splice site and 1 nonsense mutation) and 1 missense mutation. Functional analysis of the deleterious mutants revealed them to be unable to rescue motor neuron pathology in zebrafish morphants lacking Gle1. Furthermore, in HeLa cells, both mutations caused a depletion of hGle1 at the nuclear pore where it carries out an essential role in nuclear export of mRNA. These results suggest a haploinsufficiency mechanism and point to a causative role for GLE1 mutations in ALS patients. This further supports the involvement of global defects in RNA metabolism in ALS.
Introduction
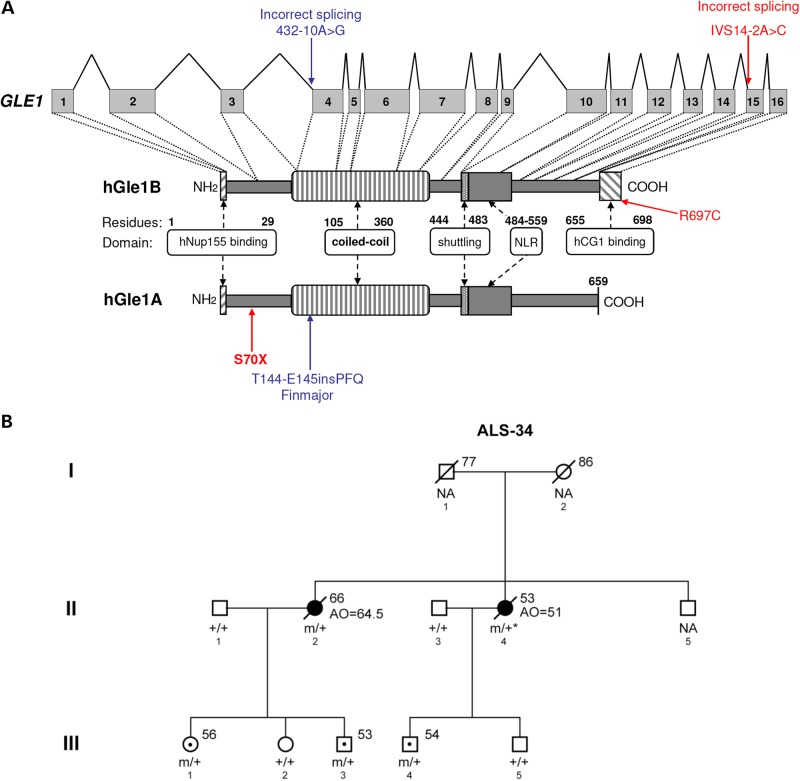
Amyotrophic lateral sclerosis (ALS) is a fatal, late onset motor neuron disease, characterized by selective motor neuron loss. ALS is increasingly becoming recognized as a RNA metabolism disorder where aggregates containing the RNA-binding proteins TDP-43 or FUS/TLS are observed in the motor neurons of most ALS patients (1). In addition, ALS causative mutations have been identified in the genes encoding these proteins as well as in genes encoding several other RNA-binding proteins (2). Interestingly, ALS causative mutations have now been observed to affect various stages of the RNA life-cycle, including gene transcription, pre-mRNA splicing, ribonucleoprotein complex formation, mRNA transport and translation (2). Gle1 (NM_001003722) is an essential multi-functional modulator of DEAD-box RNA helicases, and it has critical roles in the nuclear export of mRNA (3–5), as well as in the initiation (6,7) and termination of translation (3). These cellular roles are well conserved from yeast to mammals. In humans, two hGle1 isoforms (hGle1A and hGle1B) are expressed and they are identical except for their C-termini (Fig. 1A). By comparison to hGle1B, the hGle1A isoform is 43 amino acids (aa) shorter and ends with a unique 4-aa segment. hGle1A is localized diffusely throughout the nucleus and cytoplasm, whereas the longer hGle1B isoform is localized predominantly at the nuclear envelope (NE), more specifically to the nuclear pore complex (NPC) (8). The NPC localization of hGle1B requires interaction of its unique C-terminal region with the nucleoporin NUPL2 (NM_007342.2) (here referred to as hCG1) (9) and its N-terminal region with hNup155 (NM_153485.1) (Fig. 1A) (10). Interestingly, mutations in GLE1 were shown to cause two closely related disorders: lethal congenital contracture syndrome 1 (LCCS1; OMIM #253310) and lethal arthrogryposis with anterior horn cell disease (LAAHD; OMIM #611890). Both LCCS1 and LAAHD are autosomal recessive conditions that lead to death of fetuses before the 32nd week of gestation or soon after birth, respectively (11). LCCS1 and LAAHD were originally reported in Finnish families (12,13), and both disorders are characterized by fetal akinesia, joint contractures and skeletal muscle underdevelopment speculated to be the consequence of a near-complete absence of motor neurons in the developing anterior horn of the spinal cord. The majority of LCCS1 cases are homozygous for the GLE1 FinMajor mutation. This is a single-nucleotide substitution that generates an illegitimate splice acceptor site in GLE1 intron 3 and a 3-aa insertion in the coiled coil domain of hGle1 (Fig. 1A), while leaving the rest of the protein unchanged. Individuals with LAAHD are usually compound heterozygotes with the FinMajor mutation and another mutation in GLE1. Considering the striking shared feature of selective motor neuron loss/absence in ALS, LCCS1 and LAAHD as well as the critical multi-functional roles of TDP-43, FUS/TLS and hGle1 in RNA metabolism, we hypothesized that mutations in GLE1 might be causative for ALS.
Figure 1.
Identification of GLE1 mutations in ALS patients. (A) Structure of GLE1 gene and hGle1A and hGle1B proteins. Amino acid residues of known functional domains are indicated [adapted from Kendirgi et al. (8)]. Locations of identified changes are shown in red for ALS patients and blue for LCCS1 GLE1-FinMajor mutation. (B) Familial ALS-34 pedigree containing the GLE1-c.1965-2A>C mutation that encodes the hGle1-IVS14-2A>C protein. Genotypes of analyzed family members are indicated (squares, males; circles, females; black symbols, affected individuals; symbols with central dot, unaffected mutation carriers; slash marks, deceased individuals; ‘+’, WT; ‘m’, mutant; ‘NA’, no DNA sample available; asterisk, inferred genotype; numbers to the upper right of individual family members indicate age at death/current age).
Results
Identification of novel GLE1 variants in ALS patients
To test for the presence of ALS causative mutations in GLE1, we designed primers and used the Sanger method to sequence the 16 coding exons and exon–intron junctions of GLE1 in 1123 individuals [173 familial ALS (FALS) cases, 760 sporadic ALS (SALS) cases and 190 controls]. We also looked at GLE1 across an independent whole-exome sequencing (WES) data set of 485 control individuals with no known neurodegenerative disorders; GLE1 had a high exome coverage (≥30×). All patients and controls used in the study were Caucasian with a mixture of French and French-Canadian origin. A total of 11 rare [minor allele frequency (MAF) <1%] non-synonymous variations were identified, including 3 specific to ALS patients, 3 shared by ALS patients and controls and 5 specific to controls (see Table 1). Although no excess of rare GLE1 variations was found in ALS patients over controls (or vice versa), we observed an absence of deleterious (nonsense, splice site or frame-shift) variations in controls. In contrast, we identified two deleterious mutations in ALS patients: a nonsense mutation (c.209C>A that encodes p.S70X) in an SALS patient that was diagnosed limb onset ALS at the age of 61 and died at the age of 63 (Fig. 1A) and a splice site mutation (c.1965-2A>C whose protein product will be referred to as hGle1-IVS14-2A>C) in an FALS case (II-2) from pedigree ALS-34 (see Fig. 1A and B). The other variation identified exclusively in ALS patients was a missense variation (c.2089C>T encoding p.R697C) that exhibited high Grantham Matrix and Genomic Evolutionary Rate Profiling (GERP) scores (see Table 1 and Materials and Methods), suggesting a damaging effect on the function of the protein. All variations identified exclusively in ALS patients were absent from controls initially investigated by Sanger sequencing and WES, as well as from 285 additional controls screened via Sanger sequencing specifically for these variations (nTotal of Controls = 960). They were also absent from the publically available data of the Exome Variant Server (EVS). For all the ALS patients in which GLE1 variations were found, there were no mutations found in other commonly mutated ALS genes including C9orf72, SOD1, TARDBP and FUS/TLS. The splice site variation was first identified in individual (II-2) from pedigree ALS-34, who developed bulbar onset ALS at the age of 64.5 years and died at the age of 66. Following the identification of the splice site variation in individual II-2, an additional three individuals from the third generation of this family were found to carry the variation (III-1, III-3 and III-4) (Fig. 1B). This allowed us to infer that individual II-4 who developed limb onset ALS at the age of 51 and died at age 53 (for whom we did not have DNA available for testing) also carried the splice site variation. Individuals III-1, III-3 and III-4 were not affected at the time of this study; however, they were all at a younger age (56, 53 and 54, respectively) than the age of onset observed for ALS II-2 (64.5 years).
Table 1.
Rare (minor allele frequency <1%) GLE1 variations identified in ALS patients and controls
| Detected variants |
Frequency |
Prediction scores |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon | hg19/dbSNP | Coding DNA variant | Amino acid | FALS (n = 173) | SALS (n = 760) | Controls Sanger (n = 190) | Controls WES (n = 485) | Controls EVS (MAF %) | Grantham | GERP | |
| ALS only | 2 | g.131271264C>A | c.209C>A | p.S70X | 0 | 1 | 0a | 0 | No | NA | 3.650 |
| Intron 14 | g.131302552A>C | c.1965-2A>C | IVS14-2A>C | 1 | 0 | 0a | 0 | No | NA | 4.190 | |
| 16 | g.131303441C>T | c.2089C>T | p.R697C | 0 | 1 | 0a | 0 | No | 180 | 4.650 | |
| ALS and controls | 2 | g.131271171G>A RS138871311 | c.116G>A | p.C39Y | 0 | 1 | 2a | 1 | Yes (0.0581) | 194 | 4.350 |
| 7 | g.131287573G>A RS138310419 | c.1000G>A | p.E334K | 3 | 10 | 3 | 2 | Yes (1.0116) | 56 | 3.060 | |
| 13 | g.131300296G>T | c.1808G>T | p.R603L | 0 | 1 | 0a | 0 | Yes (0.0116) | 102 | 4.770 | |
| Controls only | 1 | g.131267089C>G RS150246404 | c.5C>G | p.P2R | 0 | 0 | 0 | 1 | Yes (0.0698) | 103 | 4.310 |
| 4 | g.131285910G>A | c.682G>A | p.V228M | 0 | 0 | 0 | 3 | No | 21 | −2.060 | |
| 6 | g.131287520G>A RS147943229 | c.947G>A | p.R316Q | 0 | 0 | 0 | 2 | Yes (0.1744) | 43 | 5.350 | |
| 7 | g.131287572C>G | c.999C>G | p.C333W | 0 | 0 | 1 | 0 | No | 215 | −3.330 | |
| 12 | g.131298693G>A RS121434407 | c.1706G>A | p.R569H | 0 | 0 | 1 | 2 | Yes (0.0465) | 29 | 5.920 | |
For these variations, 285 additional controls were tested by Sanger sequencing. Genomic positions are according to NM_001003722 from NCBI37/hg19 on the positive DNA strand. Reference SNP ID numbers (RS) were taken from the dbSNP database. Grantham matrix and GERP scores were obtained using SeattleSeq Annotation 137. FALS, familial amyotrophic lateral sclerosis; SALS, sporadic amyotrophic lateral sclerosis; WES, whole exome sequencing; EVS, Exome Variant Server (NHLBI GO Exome Sequencing Project, http://evs.gs.washington.edu/EVS/); MAF, minor allele frequency.
In vitro analysis of deleterious GLE1 mutants
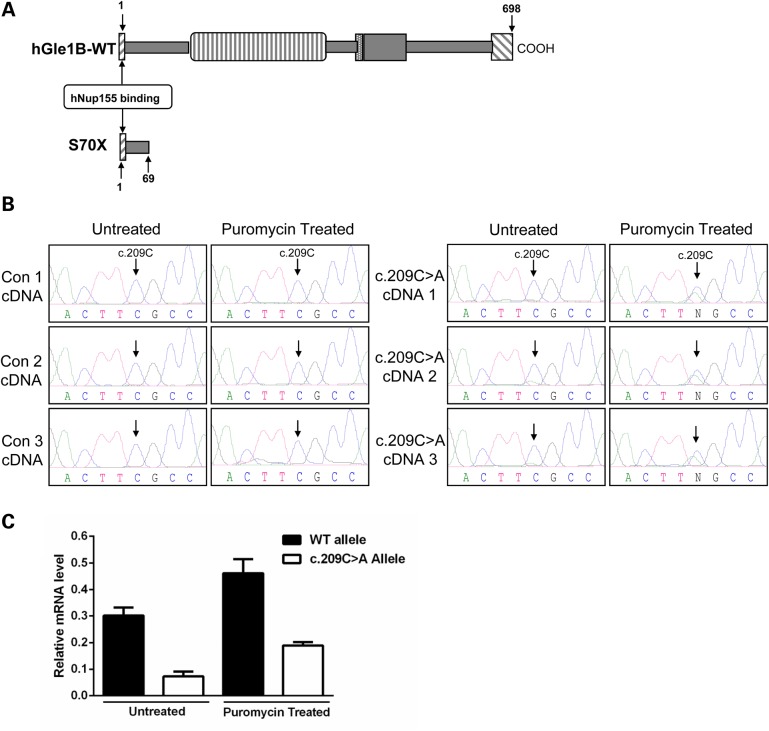
To gain insight into how deleterious mutations in GLE1 may lead to ALS pathology, we investigated the cellular effects of the c.209C>A and c.1965-2A>C mutations. The first introduces a premature stop codon in exon 2 of GLE1 that would be predicted to result in a severely truncated hGle1-S70X protein of only 69-aa residues (Fig. 2A). We observed very low sequencing peaks for the c.209C>A allele (Fig. 2B) in cDNA prepared using a lymphoblastoid cell line (LCL) from the c.209C>A ALS case and therefore hypothesized that it was processed by the nonsense mediated decay (NMD) pathway (14). We tested this by treating patient and control LCL with puromycin [an NMD pathway inhibitor (15)] (Fig. 2B) and using reverse transcription–multiplex ligation-dependent probe amplification (RT-MLPA) (16) to make quantitative measures of allelic mRNA expression. The level of the mutant allele was significantly lower than the WT allele in both untreated and puromycin-treated LCL (P < 0.0001—main effect of allele) (Fig. 2C). Predictably, the level of both alleles increased significantly with puromycin treatment (P = 0.0011—main effect of puromycin treatment). Although the interaction between the two main effects was not significant, we did see a greater effect size of puromycin treatment on the mutant allele relative to WT allele when the data were analyzed by Cohen's d (17) (Cohen's d values r = 0.907 mutant versus 0.728 WT), suggesting reduced stability of mutant mRNA over WT mRNA (Fig. 2B and C). The instability of the c.209C>A mRNA was further supported by a complete absence of the truncated hGle1-S70X protein (predicted molecular weight ∼8.3 kDa) in western blots prepared using the same LCL or HeLa cells transiently expressing an N-terminally FLAG-tagged hGle1–S70X construct (Supplementary Material, Fig. S3). Western blot conditions were optimized for the detection of small proteins and were probed using a rabbit polyclonal antibody (Sigma) recognizing the N-terminus of hGle1 (for which the epitope is within the 66 aa that are present in the truncated hGle1-S70X protein), or an antibody for the N-terminal FLAG tag in the case of transfected HeLa cells.
Figure 2.
Analysis of the GLE1-c.209C>A nonsense mutation. (A) Schematic comparison of the truncated hGle1-S70X and hGle1B-WT proteins. (B) Sequencing chromatograms for cDNAs prepared from puromycin treated/untreated LCL of three control individuals and the ALS case presenting the GLE1-c.209C>A mutation (note: three cDNAs produced from three separate RNA extractions were analyzed for the GLE1-c.209C>A LCL). Arrows indicate the c.209C residue. (C) Quantitative allele-specific expression measurements for WT and c.209C>A GLE1 mRNA in puromycin-treated and untreated GLE1-c.209C>A LCL. Expression levels calculated relative to the WT allele in four control LCL; error bars represent SEM.
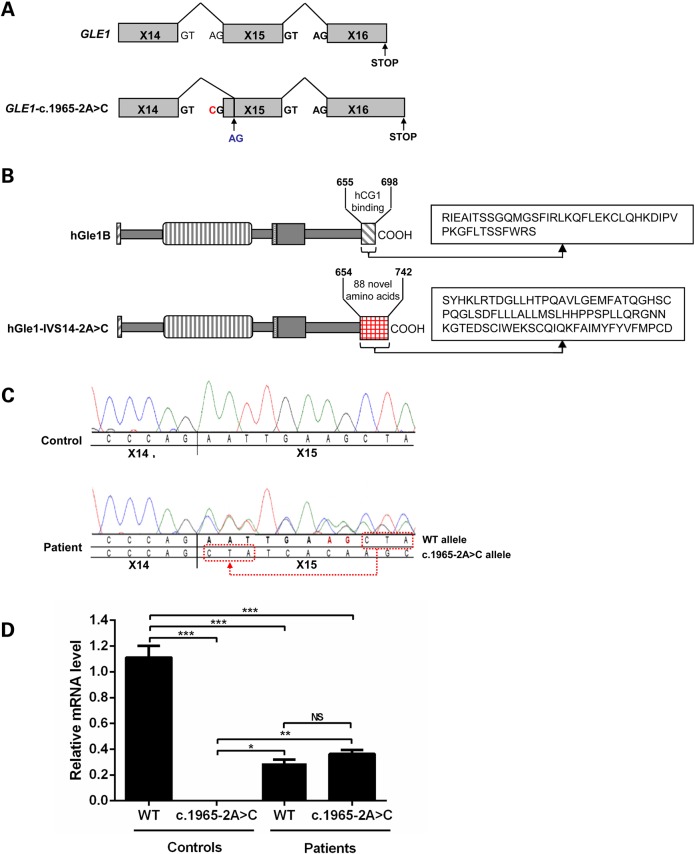
The deleterious GLE1-c.1965-2A>C splice site mutation in intron 14 was predicted by Mutation Taster (18) to result in the loss of a WT splice acceptor site and the use of an alternative splice site 8 bp further downstream at the beginning of exon 15 (Fig. 3A). This alternative splice site results in a shift in the reading frame and a replacement of the last 44 aa of WT hGle1B with 88 different aa in the hGle1-IVS14-2A>C protein product (Fig. 3B). Polymerase chain reaction (PCR) amplification and sequencing of cDNA prepared from a c.1965-2A>C LCL and a control LCL confirmed this prediction (Fig. 3C). To investigate whether this mutation influenced the stability of the mRNA transcript, we performed allele-specific quantitative PCR on cDNA from two c.1965-2A>C and two control LCLs, but no significant difference was observed between the WT and the mutant c.1965-2A>C alleles (Fig. 3D). The levels of expression of both the WT and the c.1965-2A>C alleles were significantly lower (P ≤ 0.001) than the level of expression of the homozygous WT allele in control LCLs (0.29 ± 0.03 for WT allele and 0.36 ± 0.03 for splice mutant allele versus 1.11 ± 0.09 for WT allele in controls) as would be expected in the heterozygous state.
Figure 3.
Analysis of the GLE1-c.1965-2A>C splice site mutation. (A) Schematic of predicted GLE1 exon 14–15 alternative splicing pattern in individuals expressing the heterozygous GLE1-c.1965-2A>C splice site mutation. The c.1965-2A>C mutant residue is indicated in red and the alternative splice site in blue. (B) Schematic comparison of the hGle1-IVS14-2A>C and hGle1B-WT proteins. The 88 aa replacing the terminal 44 aa of hGle1B in the hGle1-IVS-2A>C mutant are indicated. (C) cDNA-sequencing traces from LCL derived from an individual with the GLE1-c.1965-2A>C splice site mutation, and a control individual confirms the loss of the WT splice acceptor site in intron 14. (D) Quantitative expression comparisons between WT and c.1965-2A>C GLE1 in cDNA prepared from the LCLs of two control individuals and two individuals expressing this mutation. Expression levels were all calculated relative to WT allele in controls; error bars represent SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
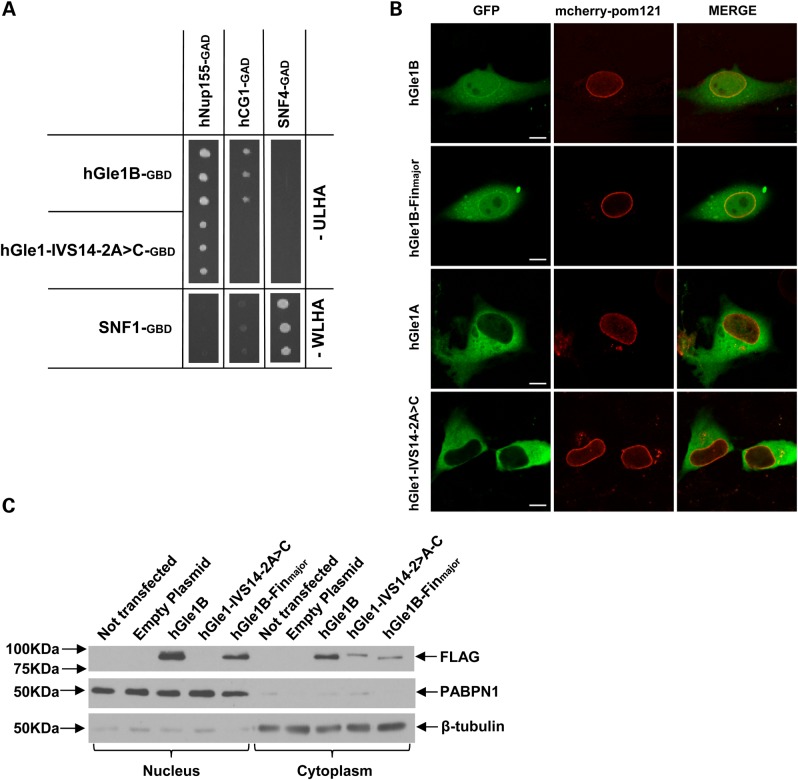
The unique C-terminal 43-aa span of hGle1B is necessary and sufficient for its interaction with the nucleoporin hCG1 (9), and siRNA-mediated knockdown of hCG1 in HeLa cells alters the steady-state localization of hGle1 (9). Moreover, the hGle1A isoform lacking the unique C-terminal hCG1-binding region does not localize predominantly at the NE or NPC (8). We therefore hypothesized that the replacement of the last 44 aa of hGle1B in the hGle1-IVS14-2A>C mutant protein would result in failed hCG1 binding and a resultant loss of NE/NPC localization. In support of this, the hGle1-IVS14-2A>C mutant protein was observed to be unable to bind hCG1, whereas it retained its ability to bind Nup155 in yeast-2-hybrid assays (Fig. 4A). When we transiently co-expressed GFP-tagged hGle1 constructs and a mcherry-tagged Pom-121 [a marker of the NE (19)] in HeLa cells, we observed the hGle1-IVS14-2A>C mutant protein to be absent from the nucleus with no co-localization with mcherry-Pom-121 at the NE (Fig. 4B). This cellular distribution was very similar to that observed for the WT hGle1A isoform (Fig. 4B) (8). The absence of the hGle1-splice protein in the nucleus and the NE was confirmed by performing additional immunocytochemistry detections (Supplementary Material, Fig. S1) and by immunodetecting western blots on which nuclear and cytoplasmic cell fractions were loaded (Fig. 4C). In contrast to hGle1-IVS14-2A>C mutant protein and hGle1A, the localization of the hGle1-FinMajor protein was similar to WT hGle1 (being present at the NE, nucleus and cytoplasm), as previously reported (11) (Fig. 4B and C; Supplementary Material, Fig. S1).
Figure 4.
Cellular localization and interactions of the hGle1-IVS14-2A>C mutant protein. (A) Yeast-2 hybrid analysis for the ability of the hGle1-IVS14-2A>C mutant protein to interact with hNup155 and hCG1. Interactions were detected by growth on media lacking uracil, leucine, histidine and adenine (-ULHA) (for hGle1B-GBD and IVS14-2A>C-GBD) or lacking tryptophan, leucine, histidine and adenine (-WLHA) (for SNF1-GBD control). (B) Live-cell imaging of HeLa cells transfected with WT or mutant GFP-tagged GLE1 constructs and mcherry-tagged Pom-121 (nuclear envelope/NPC marker); scale bar: 10 μm. (C) Western blot of nuclear and cytoplasmic proteins from HeLa cells transfected with FLAG-tagged WT or mutant GLE1 constructs, probed with antibody against FLAG. PABPN1 and β-tubulin represent nuclear and cytoplasmic loading controls, respectively.
In vivo analysis of deleterious GLE1 mutants
Having investigated the effects of the c.209C>A and c.1965-2A>C mutations in HeLa cells, we next tested their impact using an animal model. We previously have documented that knocking down the GLE zebrafish orthologue (zGle1) via the injection of antisense morpholinos (AS-MO) in one-cell stage embryos recapitulates LCCS1 pathology (20). Specifically, zGle1 morphants show a pronounced reduction in motor neuron number, accompanied by multiple LCCS1-like functional/morphological defects including immobility, diminished pharyngeal arches, a curved body axis and edema (20). Intriguingly, the reduction in motor neurons in zGle1 morphants is due to the apoptosis of neuronal precursors rather than degeneration of differentiated motor neurons (20). Moreover, the motor neurons that manage to differentiate in the zGle1 morphants display marked arborization defects (20). These defects appear to occur through a non-cell autonomous mechanism involving the absence of zGle1 in surrounding cells, not only in the motor neurons themselves (20). Thus, as reported for GLE1 (12,13), it appears that zGle1 also plays a critical role in the development and maturation of motor neurons.
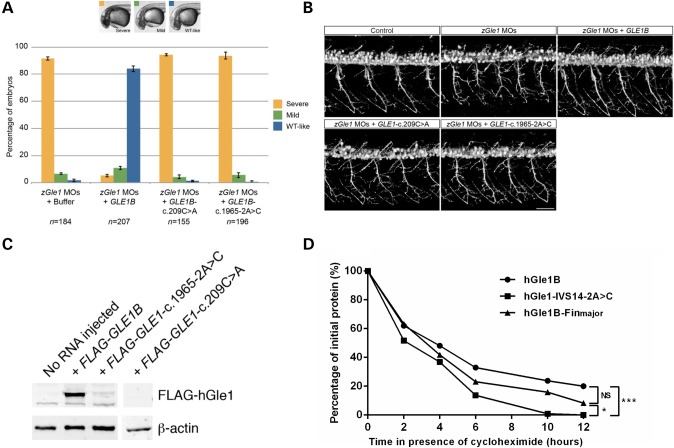
In our previous studies, we observed that the motor neuron defects observed in zGle1 morphants could be rescued by co-injecting the zGle AS-MO with in vitro-transcribed GLE1B-WT mRNA but not GLE1B mRNA containing the LCCS1 FinMajor mutation (20). In these experiments, the rescue of head and spinal neuronal death of zGle1 morphants correlated with the rescue of motor neuron defects (20). Therefore, we concluded that the extent of head/spinal neuronal death could serve as a readout of motor neuron defects. In the current study, we performed similar rescue experiments to assess the functionality of the hGle1-S70X and hGle1-IVS14-2A>C mutants by quantifying their ability to rescue zGle1 morphant head neuronal cell death (categorized as severe, mild or WT-like). Here, we found that unlike GLE1B-WT mRNA, mRNA containing the c.209C>A and c.1965-2A>C mutations could not rescue the zGle1 morphant head neuronal cell death (Fig. 5A) or the motor neuron defects (Fig. 5B). Given that the hGle1-IVS14-2A>C mutant protein behaved in a similar fashion to hGle1A, in terms of its localization and lack of hCG1 interaction, we also assessed the ability of hGle1A to rescue the morphant phenotype and determined that it did not (Supplementary Material, Fig. S2).
Figure 5.
In vivo effects of hGle1-S70X and hGle1-IVS14-2A>C mutants. (A) Graph showing the extent of head cell necrosis at 32 h post-fertilization (scored as severe, mild or WT-like) in zebrafish embryos that were injected with two non-overlapping zGle1 translation-blocking morpholinos (zGle1 MOs) followed by either buffer alone or 150 pg of FLAG-tagged human GLE1B-WT, GLE1-c.209C>A or GLE1-1965-2A>C mRNA at the one-cell stage. Values graphed as ‘mean ± SEM’, n = number of embryos analyzed in each condition. (B) Confocal images of live 2-day-old control zebrafish and zGle1 morphants (zGle1 MOs) that were injected with buffer alone (zGle1 MOs) or with 150 pg of FLAG-tagged human GLE1B-WT, GLE1-c.209C>A or GLE1-1965-2A>C mRNA at the one-cell stage. Motoneurons were labeled by the mnx1:TagRFP-T transgene, and images are lateral views of the trunk spinal cord with the dorsal side to the top and the anterior side to the left. Scale bars: 50 µm. (C) Western blot analysis of hGle1 expression in zGle1 morphants injected with buffer alone (no RNA) or 150 pg of FLAG-tagged GLE1B-WT, GLE1-c.1965-2A>C or GLE1-c.209C>A mRNA. Blots probed with rabbit anti-hGle1 antibody (which does not recognize the zebrafish Gle1 protein). Actin was used as a loading control. (D) Graph showing rate of degradation of hGle1B-WT versus hGle1-IVS14-2A>C and hGle1-FinMajor Flag-tagged proteins in HeLa cells in the presence of the translation inhibitor cycloheximide (*P ≤ 0.05, ***P ≤ 0.001).
To gain further insight into the lack of rescue observed with our ALS mutants, we looked at the expression level of the hGle1-S70X and hGle1-IVS14-2A>C mutant proteins in the injected zGle1 morphants. As would be predicted from our cell culture experiments, we did not detect the truncated hGle1-S70X protein in injected morphants (Fig. 5C). However, we were surprised to observe very low levels of hGle1-IVS14-2A>C mutant protein expression compared with hGle1B-WT. We are confident that this was not a result of reduced levels of injected mRNA, as in all cases, the RNA concentration and integrity were confirmed by electrophoresis on formaldehyde RNA gels and, before each injection, the volume was carefully measured by a micro-ruler. Thus, we instead speculated that the hGle1-IVS14-2A>C mutant protein was unstable and degraded quicker than hGle1B-WT. To assess this stability issue, we followed the transient protein expression of FLAG-tagged, hGle1B-WT, hGle1-FinMajor and hGle1-IVS14-2A>C in HeLa cells. This was done by preparing protein lysates from HeLa cells expressing these proteins for increasing periods of time in the presence of a protein synthesis inhibitor (cyclohemide) (21). The hGle1-IVS14-2A>C mutant protein was observed to have a significantly faster turnover rate than both the hGle1B-WT and hGle1B-FinMajor proteins (*P ≤ 0.05 and P ≤ 0.01, respectively) (Fig. 5D). By comparison, there was no significant difference between the turnover rates of the hGle1B-WT and hGle1B-FinMajor proteins.
Discussion
In this study, we report the identification of the first heterozygous mutations in GLE1 ever found to be associated with ALS. Three rare variations (two deleterious and one missense) were observed in ALS patients, and these were absent from all controls initially investigated by Sanger sequencing and WES, as well as from 285 additional controls screened via Sanger sequencing specifically for these variations (nTotal of Controls = 960). They were also absent from the EVS. Our investigations of the two deleterious mutations, c.209C>A and c.1965-2A>C, suggest that they operate via a haploinsufficiency mechanism. Specifically, the c.209C>A nonsense mutation results in degradation of c.209C>A mRNA by the NMD pathway and, therefore, reduces the levels of hGle1 both at the NPC (Isoform B) and in the cytoplasm (Isoform A). Whereas the c.1965-2A>C splice site mutation removes the domain within hGle1B required for interaction with NPC-scaffolding protein hCG1. Similar to hGle1A, the hGle1-IVS14-2A>C-mutant protein therefore does not localize at steady state to the NPC and is biased toward the cytoplasm. Thus, the c.1965-2A>C splice site mutation effectively alters the balance between the hGle1 protein pools in the cytoplasm versus at the NPC. Surprisingly, expression of either hGle1A or hGle1-IVS14-2A>C alone is not sufficient to rescue the zGle1 morphant phenotype, suggesting a specific requirement for hGle1B activity in motor neuron function.
In comparison with the ALS-GLE1 mutations, the main FinMajor causative mutation for LCCS1 and LAAHD results in the insertion of a proline–phenylalanine–glutamine peptide in the essential hGle1 coiled coil domain for Isoforms A and B. Our studies document that the FinMajor PFQ-insertion specifically disrupts Gle1-Gle1 self-association (5). Further, this defective self-association results in Gle1 dysfunction at the NPC during mRNA export but not during translation. Interestingly, the FinMajor protein localizes at steady state to the NPC, and its levels are not significantly altered when compared with WT hGle1. Instead, dysfunctional self-association at the NPC contributes directly to LCCS-1/LAAHD pathology (5) In comparison, importantly, the ALS-linked hGle1-S70X and hGle1-IVS14-2A>2 proteins studied here have perturbed steady-state localization to the NPC. Both mutations appear to result in the reduction of WT hGle1 levels at the NPC and in the cytoplasm. Thus, we propose that the ALS-linked mutations operate via a distinct mechanism from FinMajor in LCCS1. Specifically, altering the overall hGle1 levels and/or shifting the subcellular pools of hGle1B versus hGle1A potentially impacts the ALS disease pathology in these patients. These results highlight an intriguing disease paradigm whereby differentially altering the activity of the essential RNA metabolism factor Gle1 may contribute to distinct pathological outcomes in adults and in utero.
It is intriguing that hGle1, like TDP-43 and FUS, plays roles at multiple stages of the mRNA life-cycle and that all three are linked to devastating human diseases with motor neuron pathologies. Previous studies have not reported any functional links between hGle1 and either TDP-43 or FUS. However, hGle1 could for example be required for the nuclear export of the same mRNAs that need TDP-43 and FUS function. Future studies will shed light on this and further characterize the role of global defects in RNA metabolism in motor neuron health and in ALS pathogenesis.
Materials and Methods
Standard protocol approvals, registrations and patient consent
Protocols were approved by the Centre Hospitalier de l′Université de Montréal ethics committee on human experimentation and patients gave written informed consent.
Gene screening
One hundred and seventy-three unrelated FALS cases, 760 SALS cases and 190 matched controls, of European descent, were screened for mutations in GLE1 by PCR amplification and Sanger sequencing of the 16 coding exons (plus 50 bp of flanking introns) of GLE1. Sequencing of the exons in which variants were identified was then carried out in 285 further controls. Primer sequences are listed in Supplementary Material, Table S1, and amplification conditions were as follows: following 5 min of denaturation at 95°C, eight cycles were performed with 20 s at 94°C, 20 s at 65°C (decreasing by 1°C with each cycle) and 30 s at 72°C. A further 34 cycles of 20 s at 94°C, 20 s at 58°C and 30 s at 72°C were performed before a final extension step of 5 min at 72°C. PCR products were sequenced at the Genome Quebec Innovation Center (Montréal, QC, Canada) using a 3730XL DNA analyzer. Mutations were detected using mutation surveyor (version 3.10, SoftGenetics, PA, USA) and analyzed using Mutation Taster (18), GERP (22), the Grantham matrix (23) and alternative splice site predictor (24). In silico predictions for the c.209C>A and c.1965-2A>C mutations were confirmed through sequencing of patient cDNA generated using the SuperScript VILO cDNA Synthesis Kit and M-MLV RT enzyme, respectively, (both Invitrogen) as per the manufacturer's instructions. cDNA primers used were as follows:
c.209C>A Fwd-CGGCGCGAGGATGTTTTAGAAGAA
c.209C>A Rev-CTTTGCCCTTGGTTCCATTTGGTG
c.1965-2A>C Fwd-ATGCTAATTCTCATCAAAGAGGAC
c.1965-2A>C Rev-CCCTTGGGGACAGGAATGT
Plasmids
Details of all plasmids used are given in Supplementary Methods and Supplementary Material, Table S2.
Yeast two-hybrid analyses
PJ69-4A yeast cells (25) were grown to an OD600 of 0.6 and co-transformed via the lithium acetate method with bait plasmids carrying a leucine (L) marker and a Gal4 transactivation domain (GAD) and prey plasmids carrying either a tryptophan (W) marker (SFN4 control) or a uracil (U) marker (hGle1 variants) and a Gal4 DNA-binding domain (GBD) (Supplementary Material, Table S2). Co-transformation of SNF1-GBD with SNF4-GAD [two interacting yeast proteins (26)] and hGle1variant–GBD constructs with SNF4-GAD provided positive and negative controls, respectively. Cells were plated on synthetic minimal media (SD) supplemented with 2% glucose and lacking leucine and uracyl (SD/–LU) or leucine and tryptophan (SD/–LW) and then incubated at 30°C for 3–5 days. Surviving colonies were replica plated on SD media lacking histidine (H) and adenine (A) (SD/–LUHA or SD/–LWHA). 3-mm 3-Amino-1,2,4-triazole (3-AT) was included in SD/–LUHA and SD/–LWHA media to remove nonspecific interactions in negative controls. Plates were assessed for growth after 5 days of incubation.
Lymphoblastoid cell lines
LCLs were established and maintained as described (27). Total RNA was extracted using Trizol (Invitrogen) as per the manufacturer's instructions. For puromycin treatments, 5 × 106 cells per LCL were incubated with 300 μg/ml puromycin (Sigma) for 6 h before harvesting. Non-treated cells received an equivalent volume of dimethyl sulfoxide (DMSO).
HeLa cells
HeLa cells were cultured as described (28). Cells were transfected with FLAG-tagged GLE1 constructs (in pcDNA 3.1(+) vectors—see Supplementary Material, Table S2 for vector details) using jetPRIME reagent (Polypus transfection, Inc.) as per the manufacturer's instructions. Transfections were carried out 24 h after cell seeding, and cells were harvested or processed further 24 h after transfection.
Quantitative reverse transcriptase–PCR (RT-PCR)
LCL cDNA was generated from RNA extractions prepared from two individuals with the GLE1- c.1965-2A>C mutation and two control individuals using the QuantiTect Reverse Transcription Kit (Qiagen). qRT-PCR was performed using the QuantiFast SYBR Green PCR Kit (as per manufacturer's instructions). Primers were as follows:
GLE1-WT fwd-TCATCAAAGAGGACTACTTTCCCAGAATTG
GLE1-WT rev-TTTTGTGACCCATGAAAATCTCCTCTCCTA
GLE1- c.1965-2A>C fwd-TCATCAAAGAGGACTACTTTCCCAGCTAT
GLE1- c.1965-2A>C rev-TTTTGTGACCCATGAAAATCTCCTCTCCTA
RNA polymerase II (Pol-II) fwd-CCCCGCTCCATTGCTGCCAA
Pol-II rev-GTGCAGAGTTGGCTGCCGGT
PCR conditions were as follows: 95°C for 5 min, then 40 cycles at 95°C for 10 s and 60°C for 30 s. Fluorescent signals were captured using the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems). Expression levels of GLE1-WT and c.1965-2A>C alleles were normalized to the Pol II housekeeping gene and calculated in comparison with the mean level of expression of the WT allele in controls, using the 2-ΔΔCt method (29). Three experiments (each including three cDNA samples per LCL) were averaged. Statistics were performed by one-way ANOVA with post hoc Bonferroni tests.
Reverse transcription–multiplex ligation-dependent probe amplification
RT-MLPA was carried out using an SALSA MLPA RNA reagent kit (MRC-Holland) as per the manufacturer's instructions with the following exceptions: cDNA was generated using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). RT-MLPA probes for GLE1-WT, GLE1-c.209C>A and Pol-II were custom designed with M13 tails (see Supplementary Material, Table S3). PCR amplification of bound, ligated probes was performed using VIC-labeled M13 primers (Invitrogen). PCR products were run on an ABI 3730 DNA Analyzer (Applied Biosystems) and analyzed using GeneMapper software version 4.0 (Applied Biosystems). Peak heights for WT and c.209C>A alleles were normalized to Pol-II. Relative levels of WT and c.209C>A GLE1 mRNAs in patient cells were then assessed by normalization to the WT allele in controls. Four experiments, each involving four controls and three sets of c.209C>A patient RNA were averaged. Statistics were performed by two-way ANOVA, and effect sizes were calculated via Cohen's d (17).
Live-cell imaging
HeLa cells were plated in 35 mm No. 1.5 glass bottom dishes (Mattek) and co-transfected with plasmids expressing indicated GFP-hGle1 variants and mcherry-pom121. 12–18 h cells were imaged using a confocal microscope (LSM510, Zeiss) using a Zeiss 63X/1.2 water objective.
Nuclear and cytoplasmic protein fractionation
Fractionation of HeLa cells was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) as per the manufacturer's instructions. Western blots were performed as described (28) using antibodies against FLAG (F3165; Sigma), Poly(A)-Binding Protein Nuclear 1 [A gift from Dr Bear, described in (30)] and α-tubulin (ab4074; Abcam). Results are representative of three experiments.
Cycloheximide protein degradation assay
Transfected HeLa cells were treated with 100 μg/ml cycloheximide (Sigma) for 2, 4, 6, 10 or 12 h and then homogenized in SDS–UREA buffer (0.5% SDS, 8 M Urea, 2% β-mercaptoethanol). Control cells (0 time point) received an equivalent volume of DMSO. Extracted proteins were immunoblotted as described (28) using primary antibodies against FLAG (F3165; Sigma) and β-actin (MAB1501; Chemicon). WT/mutant hGle1 levels at each time point were measured by quantitative densitometry (Image J software; http://rsbweb.nih.gov/ij/), normalized to β-actin and expressed as a percentage of the level in control cells. Five experiments were averaged, and statistics were performed via repeated-measures ANOVA with post hoc Bonferroni tests.
Zebrafish rescue experiments
Zebrafish rescue experiments were performed as described (20). Briefly, WT embryos injected with two non-overlapping zebrafish (z)Gle1 translation-blocking morpholinos were sequentially injected with buffer alone or with 150 pg of in vitro-transcribed FLAG- or EGFP-tagged wild-type, c.209C>A or c.1965-2A>C human GLE1B mRNA at the one-cell stage. Extent of head necrosis, categorized as severe, mild or WT-like, was scored at 2 days of age. To examine the rescue of motor neuron defects, embryos with motoneurons labeled by mnx1:TagRFP-T transgene were used in the rescue experiments. The spinal motor neurons and their axon arborization were examined by live confocal microscopy at 2 days of age. To verify the protein expression levels of injected human GLE1B mRNAs, injected embryos were lysed at the bud stage and subjected to western blot analyses using a rabbit anti-human GLE1 antibody, which does not recognize the zebrafish Gle1 protein and was generated as follows: bacterially expressed recombinant GST-tagged hGle1(aa 1–362) (pSW1423) proteins were purified and injected into rabbits to generate a polyclonal antiserum (Covance, Inc., Princeton, NJ, USA). The production bleeds were collected from animal (VU#178) and affinity purified against the same GST-hGle1(aa 1–362).
Statistics
Statistics were performed using Graphpad Prism Software Version 5. The threshold for significance was set at P ≤ 0.05.
Supplementary Material
Funding
V.V.B., H.D. and G.A.R. are supported by the Canadian Institutes of Health Research (CIHR). A.W.F., L.-E.J. and S.R.W. are supported by the National Institutes of Health (R37 GM51219 to S.R.W., F31 NS070431 to A.W.F.) and the March of Dimes (S.R.W.). G.A.R. holds the Canada's Research Chair in Neurogenetics and the Wilder Penfield Chair in Neuroscience.
Supplementary Material
Acknowledgements
We thank the patients involved in this study as well as Annie Raymond, Pascale Thibodeau, Anne Desjarlais and Pierre Provencher for technical support, sample collection and organization and Christine Vande Velde for her advice and for reading the manuscript. We also acknowledge the support of the Canadian Institutes for Health Research (CHIR), the ALS division of the Muscular Dystrophy Association (ALS-MDA) and the US ALS Association (ALSA).
Conflict of Interest statement. None declared.
References
- 1.Mackenzie I.R.A., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 2.Strong M.J. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS) J. Neurol. Sci. 2010;288:1–12. doi: 10.1016/j.jns.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 3.Alcázar-Román A.R., Bolger T.A., Wente S.R. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 2010;285:16683–16692. doi: 10.1074/jbc.M109.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy R., Wente S.R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 5.Folkmann A.W., Collier S.E., Zhan X., Aditi, Ohio M.D., Wente S.R. Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease. Cell. 2013;155:582–593. doi: 10.1016/j.cell.2013.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolger T.A., Wente S.R. Gle1 is a multifunctional DEAD-box protein regulator that modulates Ded1 in translation initiation. J. Biol. Chem. 2011;286:39750–39759. doi: 10.1074/jbc.M111.299321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolger T.A., Folkmann A.W., Tran E.J., Wente S.R. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kendirgi F., Barry D.M., Griffis E.R., Powers M.A., Wente S.R. An essential role for hGle1 nucleocytoplasmic shuttling in mRNA export. JCB. 2003;160:1029–1040. doi: 10.1083/jcb.200211081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kendirgi F., Rexer D.J., Alcázar-Román A.R., Onishko H.M., Wente S.R. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol. Biol. Cell. 2005;16:4304–4315. doi: 10.1091/mbc.E04-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayala H.J., Kendirgi F., Barry D.M., Majerus P.W., Wente S.R. The mRNA export factor human Gle1 interacts with the nuclear pore complex protein Nup155. Mol. Cell. Proteomics. 2004;3:145–155. doi: 10.1074/mcp.M300106-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Nousiainen H.O., Kestila M., Pakkasjarvi N., Honkala H., Kuure S., Tallila J., Vuopala K., Ignatius J., Herva R., Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuopala K., Ignatius J., Herva R. Lethal arthrogryposis with anterior horn cell disease. Hum. Pathol. 1995;26:12–19. doi: 10.1016/0046-8177(95)90109-4. [DOI] [PubMed] [Google Scholar]
- 13.Herva R., Leisti J., Kirkinen P., Seppanen U. A lethal autosomal recessive syndrome of multiple congenital contractures. Am. J. Med. Genet. 1985;20:431–439. doi: 10.1002/ajmg.1320200303. [DOI] [PubMed] [Google Scholar]
- 14.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noensie E.N., Dietz H.C. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 2001;19:434–439. doi: 10.1038/88099. [DOI] [PubMed] [Google Scholar]
- 16.Rotman J., van Gils W., Butler D., Spaink H.P., Meijer A.H. Rapid screening of innate immune gene expression in zebrafish using reverse transcription—multiplex ligation-dependent probe amplification. BMC Res. Notes. 2011;4:196. doi: 10.1186/1756-0500-4-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen J. Statistical Power Analysis for the Behavioral Sciences. Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 18.Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Meth. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell J.M., Mansfeld J., Capitanio J., Kutay U., Wozniak R.W. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J. Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jao L.-E., Appel B., Wente S.R. A zebrafish model of lethal congenital contracture syndrome 1 reveals Gle1 function in spinal neural precursor survival and motor axon arborization. Development. 2012;139:1316–1326. doi: 10.1242/dev.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chini C.C.S., Wood J., Chen J. Chk1 is required to maintain Claspin stability. Oncogene. 2006;25:4165–4171. doi: 10.1038/sj.onc.1209447. [DOI] [PubMed] [Google Scholar]
- 22.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., Marín A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 25.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang R., Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson M.A., Gusella J.F. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984;20:856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 28.Catoire H., Dion P.A., Xiong L., Amari M., Gaudet R., Girard S.L., Noreau A., Gaspar C., Turecki G., Montplaisir J.Y., et al. Restless legs syndrome-associated MEIS1 risk variant influences iron homeostasis. Ann. Neurol. 2011;70:170–175. doi: 10.1002/ana.22435. [DOI] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Hosoda N., Lejeune F., Maquat L.E. Evidence that poly(a) binding protein c1 binds nuclear pre-mRNA poly(a) tails. Mol. Cell. Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.