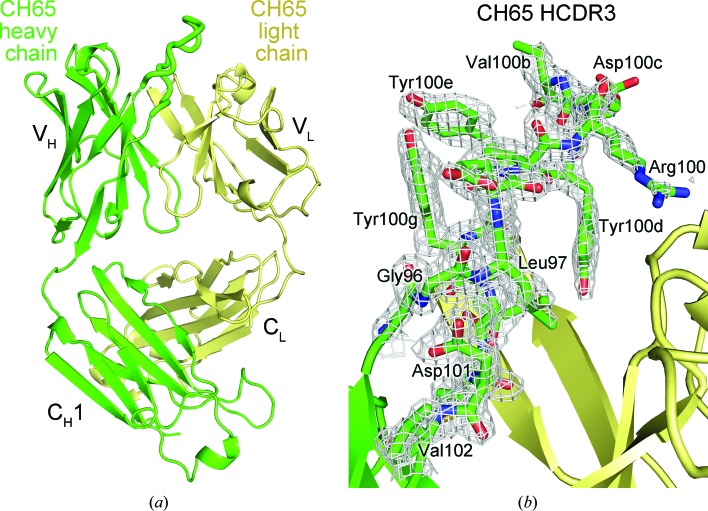
The crystal structure of anti-influenza antibody CH65 at 1.70 Å resolution reveals that the affinity-matured antibody evolved to preconfigure the antibody loops to increase binding affinity to the hemagglutinin.
Keywords: influenza, antibody, adaptive immunity, affinity maturation
Abstract
Influenza viruses remain a persistent challenge to human health owing to their inherent ability to evade the immune response by antigenic drift. However, the discovery of broadly neutralizing antibodies (bnAbs) against divergent viruses has sparked renewed interest in a universal influenza vaccine and novel therapeutic opportunities. Here, a crystal structure at 1.70 Å resolution is presented of the Fab of the human antibody CH65, which has broad neutralizing activity against a range of seasonal H1 isolates. Previous studies proposed that affinity maturation of this antibody lineage pre-organizes the complementarity-determining region (CDR) loops into an energetically favorable HA-bound conformation. Indeed, from the structural comparisons of free and HA-bound CH65 presented here, the CDR loops, and in particular the heavy-chain CDR3, adopt the same conformations in the free and bound forms. Thus, these findings support the notion that affinity maturation of the CH65 lineage favorably preconfigures the CDR loops for high-affinity binding to influenza hemagglutinin.
1. Introduction
The recent explosion in the identification and characterization of broadly neutralizing antibodies (bnAbs) against influenza viruses has bolstered enthusiasm and efforts towards the design of universal flu vaccines and novel therapeutics. These bnAbs bind the influenza hemagglutinin glycoprotein (HA; Wilson et al., 1981 ▶) and target functionally conserved epitopes on the HA head and stem regions (Lee & Wilson, 2015 ▶). In conjunction with these advances in antibody discovery, the developmental pathways that the HA stem-targeted VH1–69-encoded antibodies take to convert from germline to affinity-matured forms are an active area of investigation (Lingwood et al., 2012 ▶; Avnir et al., 2014 ▶; Pappas et al., 2014 ▶). These studies show that somatic mutations that arise during the affinity-maturation process can affect the conformations of the antibody complementarity-determining region (CDR) loops. As such, the identification and characterization of bnAbs against the HA have afforded exquisite insight into the recognition and neutralization of influenza viruses.
The affinity-maturation pathway of an HA head-targeted antibody lineage has also been characterized (Whittle et al., 2011 ▶; Schmidt et al., 2013 ▶). These antibodies were isolated from an individual immunized with the 2007 trivalent influenza vaccine. The affinity-matured antibodies CH65 and CH67, which derive from the same antibody precursors, have a broad spectrum of recognition against seasonal H1 isolates and insert their heavy-chain CDR loop 3 (HCDR3) into the receptor-binding site. One of the key recognition elements is mimicry of the interactions of the endogenous sialic acid receptor, where an Asp residue in HCDR3 corresponds to the carboxylate of sialic acid. This feature has also been found in other HA receptor binding site-targeted bnAbs (Hong et al., 2013 ▶; Lee et al., 2014 ▶). However, the unmutated common ancestor (UCA) and the intermediate (I-2) precursor antibodies in the CH65–CH67 lineage have significantly weaker affinity for the H1 HAs (Schmidt et al., 2013 ▶). These studies have suggested that the mutations acquired through affinity maturation dictate the structural conformation of the antibody CDR loops, as for the VH1–69 stem-targeted antibody CR6261 (Lingwood et al., 2012 ▶). Molecular-dynamics simulations and crystal structure comparisons of UCA, I-2, CH65–HA, apo CH67 and CH67–HA have suggested that the maturation pathway leads to a preconfiguration of the antibody HCDR3 conformation that is optimal for antigen recognition and which is observed in the Fab–HA complexes (Schmidt et al., 2013 ▶). Therefore, we determined the apo CH65 Fab crystal structure to test this hypothesis.
2. Materials and methods
2.1. Protein expression and purification
The CH65 Fab was cloned into the pFastBac Dual vector (Invitrogen) with N-terminal gp67 and honeybee melittin signal peptides fused to the heavy chain and light chain, respectively, and a C-terminal His6 tag fused to the heavy chain. Recombinant bacmid DNA was generated using the Bac-to-Bac system (Invitrogen) and baculovirus was generated by transfecting purified bacmid DNA into Sf9 cells using Cellfectin II (Invitrogen). The CH65 Fab was expressed by infecting suspension cultures of High Five cells (Invitrogen) with high-titer baculovirus for 3 d at 28°C. The protein was purified from the supernatant by Ni–NTA (Qiagen), Protein G and Mono S chromatography (GE Healthcare), followed by a final step of gel filtration (GE Healthcare) in a buffered solution consisting of 50 mM NaCl, 10 mM Tris pH 8.
2.2. Crystallization, data collection, structure determination and refinement
The CH65 Fab was initially screened for crystallization using the Robotics Core at the Joint Center for Structural Genomics (JCSG). Crystal hits were optimized by mixing 1 µl concentrated protein solution (11.8 mg ml−1) with 1 µl mother liquor (1.8 M ammonium sulfate, 0.1 M CHES pH 10) at room temperature. The crystals were cryoprotected with mother liquor supplemented with 20%(v/v) glycerol, flash-cooled and stored in liquid nitrogen until data collection. Diffraction data were collected on the GM/CA CAT 23ID-B beamline at the Advanced Photon Source (APS) to 1.70 Å resolution and were processed in the orthorhombic space group P212121 using the XDS package (Kabsch, 2010 ▶). The structure was determined by molecular replacement with Phaser (McCoy et al., 2007 ▶) using CDR-trimmed CH65 Fab variable and constant domains from the CH65–HA complex as the search model (PDB entry 3sm5 chains H and L; Whittle et al., 2011 ▶). The apo CH65 Fab was iteratively built using Coot (Emsley et al., 2010 ▶) and was refined in PHENIX (Adams et al., 2010 ▶). The refinement parameters included an initial round of simulated annealing using the default phenix.refine parameters and rigid-body refinement (set for each Ig domain) of the molecular-replacement solution model, followed by restrained refinement including TLS refinement (set for each Ig domain). The Fab was renumbered to the Kabat scheme. Ramachandran statistics were calculated using MolProbity (Chen et al., 2010 ▶).
3. Results and discussion
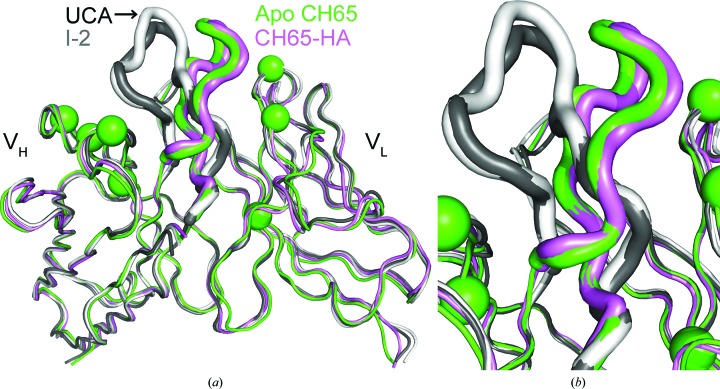
The crystal structure of the apo CH65 Fab was determined at 1.70 Å resolution (Fig. 1 ▶ a) and the asymmetric unit contains one copy of the Fab (Table 1 ▶). The structure was determined by molecular replacement using a CDR-trimmed model of the HA-bound CH65 Fab (PDB entry 3sm5) and the loops were iteratively rebuilt after an initial round of simulated annealing. The final Fab model is well ordered and the maps show clear electron density for the rebuilt CDR loops, including HCDR3 (Fig. 1 ▶ b). While the elbow angles of the free and the bound antibodies differ owing to the inherent flexibility of the loops connecting the variable and constant domains (Stanfield et al., 2006 ▶), the variable heavy (VH) and variable light (VL) Ig domains of the apo CH65 Fab align well with those of the HA-bound Fab (Cα r.m.s.d. of 0.7 Å), but not as well with those of the UCA or I-2 (Cα r.m.s.d. of 1.8 Å) (Fig. 2 ▶ a). Furthermore, the apo CH65 CDR loops adopt similar conformations as in the HA-bound Fab (Fig. 2 ▶ b). Structural deviations of UCA and I-2 in comparison to CH65 result from different conformations of the HCDR3 loop, despite very similar VH and VL structures excluding HCDR3 (Cα r.m.s.d. of 0.8 Å). This observation supports the notion that affinity maturation configures the HCDR3 of the CH65–CH67 antibody lineage to adopt an energetically favorable conformation to bind the HA with high affinity (Schmidt et al., 2013 ▶).
Figure 1.
Crystal structure of apo CH65 Fab. (a) Overall Fab structure with the heavy chain and light chain colored green and yellow, respectively. The HCDR3 is shown as thick tubes. The variable and constant Ig domains for each antibody chain are labeled. (b) The 2F o − F c electron-density simulated-annealing OMIT map of the CH65 HCDR3 loop is shown as a gray mesh and is contoured at 1.0σ.
Table 1. Data-collection and refinement statistics for apo CH65 Fab.
Values in parentheses are for the outer shell.
| Data collection | |
| Beamline | 23ID-B, APS |
| Wavelength () | 1.0332 |
| Space group | P212121 |
| Unit-cell parameters (, ) | a = 57.1, b = 67.0, c = 130.6, = = = 90.0 |
| Resolution () | 501.70 (1.731.70) |
| No. of observations | 391436 |
| No. of unique reflections | 55829 (2739) |
| Multiplicity | 7.0 (6.5) |
| Completeness (%) | 99.6 (93.3) |
| I/(I) | 14.7 (2.5) |
| R merge | 0.08 (0.82) |
| R p.i.m. | 0.03 (0.34) |
| Refinement statistics | |
| Resolution () | 46.81.70 |
| No. of reflections (work set) | 55419 |
| No. of reflections (test set) | 2802 |
| R cryst (%) | 18.1 |
| R free (%) | 20.7 |
| No. of protein atoms | 3318 |
| No. of waters | 348 |
| Average B factor (2) | 27.4 |
| Wilson B factor (2) | 21.6 |
| R.m.s.d. from ideal geometry | |
| Bond lengths () | 0.009 |
| Bond angles () | 1.21 |
| Ramachandran statistics (%) | |
| Favored | 97.1 |
| Outliers | 0 |
| PDB code | 4wuk |
Figure 2.
Comparison of apo and HA-bound structures of the CH65–CH67 antibody lineage. (a) The heavy variable (VH) and light variable (VL) Ig domains are depicted and the HCDR3 loops of the UCA (PDB entry 4hk0; Schmidt et al., 2013 ▶), I-2 (PDB entry 4hk3; Schmidt et al., 2013 ▶), apo CH65 (PDB entry 4wuk) and HA-bound CH65 (PDB entry 3sm5) are shown as thick tubes. Spheres indicate amino-acid positions that differ between the UCA and the CH65 and CH67 antibodies. (b) Enlarged view of the HCDR3 loops.
Interestingly, CH65 has the same HCDR3 amin-acid sequence as the inferred UCA and I-2 precursors (Schmidt et al., 2013 ▶), which implies that the conformation of this loop is regulated by affinity maturation in other regions of the variable domain, such as the HCDR1, HCDR2 and framework region 3 (FR3), as well as the LCDR loops. Seven changes arise between the UCA and the mature CH65 and CH67 antibodies at heavy-chain positions 31, 34, 52 and 56 as well as light-chain positions 30, 36 and 50 (Fig. 2 ▶ a). As these changes cluster on either side of HCDR3 (Fig. 2 ▶ a), systematic somatic mutagenesis of these positions in the UCA via affinity maturation may preconfigure the HCDR3 loop into the HA-bound conformation.
This dependency on the affinity-maturation state of the antibody, independent of the HCDR3 loop, has also been observed for the esterolytic antibody 48G7 (Patten et al., 1996 ▶; Wedemayer et al., 1997 ▶). These studies showed that the affinity-matured 48G7 antibody has a 30 000-fold higher affinity for haptens compared with its germline precursor. Just as for CH65, affinity maturation of 48G7 pre-organizes its CDR loops into a preferable configuration to bind its substrate, whereas the germline 48G7 CDR loops have different conformations. Thus, these findings further support the notion that affinity maturation can stabilize a conformation of the HCDR3 loop that enables high-affinity binding to the antigen (Babor & Kortemme, 2009 ▶).
Altogether, the findings of this study reveal that the CDR loops of the apo CH65 Fab adopt a similar conformation to the affinity-matured, HA-bound Fab. The structural characterization of the apo CH65 Fab is in agreement with the suggestion that this affinity-matured antibody arranges its CDR loops into a favorable conformation to recognize the HA, thereby accounting for the increased binding affinity in comparison to its germline precursors (Schmidt et al., 2013 ▶). These results confirm the notion that somatic hypermutation not only changes the identity of the amino acids that contact its antigen but can also pre-organize the CDR loops into a more stable and favorable conformation to bind their cognate antigens (Wedemayer et al., 1997 ▶; Yin et al., 2003 ▶; Babor & Kortemme, 2009 ▶; Sela-Culang et al., 2012 ▶).
Supplementary Material
PDB reference: apo CH65 Fab, 4wuk
Acknowledgments
We thank Henry Tien of the Robotics Core at the JCSG for automated crystal screening, Robert Kirchdoerfer for reagents, the staff of the Advanced Photon Source (APS) GM/CA CAT beamline 23ID-B for support and Marc Elsliger as well as the instructors from the 2012 CCP4 School held at APS for computational and crystallographic support. The work was funded by NIH R56 AI099275 and T32AI007244 (to PSL). The JCSG robotic crystallization facility is supported by U54 GM094586. The GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under contract No. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH. The authors declare no competing financial interests. This is The Scripps Research Institute manuscript No. 28090.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Avnir, Y. et al. (2014). PLoS Pathog. 10, e1004103. [DOI] [PMC free article] [PubMed]
- Babor, M. & Kortemme, T. (2009). Proteins, 75, 846–858. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Hong, M., Lee, P. S., Hoffman, R. M. B., Zhu, X., Krause, J. C., Laursen, N. S., Yoon, S.-I., Song, L., Tussey, L., Crowe, J. E. Jr, Ward, A. B. & Wilson, I. A. (2013). J. Virol. 87, 12471–12480. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lee, P. S., Ohshima, N., Stanfield, R. L., Yu, W., Iba, Y., Okuno, Y., Kurosawa, Y. & Wilson, I. A. (2014). Nature Commun. 5, 3614. [DOI] [PMC free article] [PubMed]
- Lee, P. S. & Wilson, I. A. (2015). Curr. Top. Microbiol. Immunol. 386, 323–341. [DOI] [PMC free article] [PubMed]
- Lingwood, D., McTamney, P. M., Yassine, H. M., Whittle, J. R. R., Guo, X., Boyington, J. C., Wei, C.-J. & Nabel, G. J. (2012). Nature (London), 489, 566–570. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Pappas, L., Foglierini, M., Piccoli, L., Kallewaard, N. L., Turrini, F., Silacci, C., Fernandez-Rodriguez, B., Agatic, G., Giacchetto-Sasselli, I., Pellicciotta, G., Sallusto, F., Zhu, Q., Vicenzi, E., Corti, D. & Lanzavecchia, A. (2014). Nature (London), 516, 418–422. [DOI] [PubMed]
- Patten, P. A., Gray, N. S., Yang, P. L., Marks, C. B., Wedemayer, G. J., Boniface, J. J., Stevens, R. C. & Schultz, P. G. (1996). Science, 271, 1086–1091. [DOI] [PubMed]
- Schmidt, A. G. et al. (2013). Proc. Natl Acad. Sci. USA, 110, 264–269.
- Sela-Culang, I., Alon, S. & Ofran, Y. (2012). J. Immunol. 189, 4890–4899. [DOI] [PubMed]
- Stanfield, R. L., Zemla, A., Wilson, I. A. & Rupp, B. (2006). J. Mol. Biol. 357, 1566–1574. [DOI] [PubMed]
- Wedemayer, G. J., Patten, P. A., Wang, L. H., Schultz, P. G. & Stevens, R. C. (1997). Science, 276, 1665–1669. [DOI] [PubMed]
- Whittle, J. R. R., Zhang, R., Khurana, S., King, L. R., Manischewitz, J., Golding, H., Dormitzer, P. R., Haynes, B. F., Walter, E. B., Moody, M. A., Kepler, T. B., Liao, H.-X. & Harrison, S. C. (2011). Proc. Natl Acad. Sci. USA 108, 14216–14221. [DOI] [PMC free article] [PubMed]
- Wilson, I. A., Skehel, J. J. & Wiley, D. C. (1981). Nature (London), 289, 366–373. [DOI] [PubMed]
- Yin, J., Beuscher, A. E., Andryski, S. E., Stevens, R. C. & Schultz, P. G. (2003). J. Mol. Biol. 330, 651–656. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: apo CH65 Fab, 4wuk