Summary
Exogenous melatonin application confers abiotic stress resistance in bermudagrass through modulation of antioxidants and metabolic homeostasis, and extensive transcriptional reprogramming such as the reorientation of photorespiratory, carbohydrate, and nitrogen metabolism.
Key words: Abiotic stress, antioxidant, bermudagrass, melatonin, metabolites, reactive oxygen species, transcriptomic.
Abstract
Melatonin (N-acetyl-5-methoxytryptamine), a well-known animal hormone, is also involved in plant development and abiotic stress responses. In this study, it is shown that exogenous application of melatonin conferred improved salt, drought, and cold stress resistances in bermudagrass. Moreover, exogenous melatonin treatment alleviated reactive oxygen species (ROS) burst and cell damage induced by abiotic stress; this involved activation of several antioxidants. Additionally, melatonin-pre-treated plants exhibited higher concentrations of 54 metabolites, including amino acids, organic acids, sugars, and sugar alcohols, than non-treated plants under abiotic stress conditions. Genome-wide transcriptomic profiling identified 3933 transcripts (2361 up-regulated and 1572 down-regulated) that were differentially expressed in melatonin-treated plants versus controls. Pathway and gene ontology (GO) term enrichment analyses revealed that genes involved in nitrogen metabolism, major carbohydrate metabolism, tricarboxylic acid (TCA)/org transformation, transport, hormone metabolism, metal handling, redox, and secondary metabolism were over-represented after melatonin pre-treatment. Taken together, this study provides the first evidence of the protective roles of exogenous melatonin in the bermudagrass response to abiotic stresses, partially via activation of antioxidants and modulation of metabolic homeostasis. Notably, metabolic and transcriptomic analyses showed that the underlying mechanisms of melatonin could involve major reorientation of photorespiratory and carbohydrate and nitrogen metabolism.
Introduction
Melatonin (N-acetyl-5-methoxytryptamine) is a well-known animal hormone that is involved in many biological processes including sleep, mood, circadian rhythms, retinal physiology, seasonal reproductive physiology, temperature homeostasis, sexual behaviour, antioxidative activity, and immunological enhancement (Galano et al., 2011; Venegas et al., 2012; Calvo et al., 2013). However, melatonin is not only found exclusively in animals, but is ubiquitously present in almost all forms of life including protists, prokaryotes, eukaryotic unicells, algae, fungi, and plants (Dubbels et al., 1995; Hattori et al., 1995; Kolář and Macháčkova, 2005; Arnao and Hernández-Ruiz, 2006, 2007; Tan et al., 2007a, b, 2012).
In 1995, two reports first identified melatonin in higher plants (Dubbels et al., 1995; Hattori et al., 1995). In the last 20 years, additional research found that melatonin is universally distributed in leaves, roots, stems, fruits, and seeds of all plant species examined (Manchester et al., 2000; Reiter et al., 2001; Kolář and Macháčkova, 2005; Hernández-Ruiz and Arnao, 2008; Murch et al., 2009; Zhao et al., 2013). Interestingly, remarkably high concentrations of melatonin have been identified and quantified in popular beverages (beer, tea, coffee, and wine), crops (barley, corn, grape, wheat, rice, tobacco, and oats), and Arabidopsis in comparison with those in animals (Manchester et al., 2000; Kolář and Macháčkova, 2005; Arnao and Hernández-Ruiz, 2006, 2009b , 2013a , b; Tan et al., 2007a, 2012; Ramakrishna et al., 2012). Additionally, the melatonin content of tomato and rice has been modified by genetic engineering (Okazaki and Ezura, 2009; Okazaki et al., 2009, 2010; Byeon et al., 2012, 2013, 2014; Byeon and Back, 2014a , b). The well-known beneficial effects of melatonin on human health and the abundance of melatonin in popular beverages and crops may encourage the daily consumption of these products (Tan et al., 2012).
To date, melatonin has also been found to be a ubiquitous modulator in multiple plant developmental processes and various stress responses (Kolář and Macháčkova, 2005; Arnao and Hernández-Ruiz, 2006; Tan et al., 2007a, 2012). Changes in melatonin in plants may be involved in circadian rhythms, flowering, promotion of photosynthesis, preservation of chlorophyll (Arnao and Hernández-Ruiz, 2009a ; Tan et al., 2012), stimulation and regeneration of root system architecture (Hernández-Ruiz et al., 2005; Pelagio-Flores et al., 2012; Zhang et al., 2014), delayed senescence of leaves (Byeon et al., 2012; Wang et al., 2012, 2013 a, b), and alleviation of oxidative damage mediated by reactive oxygen species (ROS) and reactive nitrogen species (RNS) burst (Tan et al., 2012) Moreover, melatonin protects against multiple abiotic processes such as cold stress (Posmyk et al., 2009a ; Kang et al., 2010; Bajwa et al., 2014), copper stress (Posmyk et al., 2008, 2009b), high temperature (Byeon and Back 2014b ), salt stress (Li et al., 2012), osmotic stress (Zhang et al., 2013), drought stress (Wang et al., 2014), and pathogen infection (Yin et al., 2013). The mechanisms were partially characterized after the direct exogenous application of melatonin to plants (Posmyk et al., 2008, 2009a, b; Zhao et al., 2011; Li et al., 2012; Pelagio-Flores et al., 2012; Wang et al., 2012, 2013a , b; Yin et al., 2013; Bajwa et al., 2014) or the creation of transgenic plants that produced either more or less melatonin through modulating its metabolic pathway (Kang et al., 2010; Byeon et al., 2013, 2014; Park et al., 2013; Byeon and Back, 2014a ; Wang et al., 2014). Finally, the recent studies which showed the protective roles of melatonin in response to abiotic stress indicate that this indole might be a potentially ideal target for future genetic engineering technology to improve abiotic stress resistance in plants. Thus, transgenic plants with higher melatonin concentration might lead to breakthroughs to improve crop production in agriculture as well as the general health of humans (Tan et al., 2012).
Bermudagrass [Cynodon dactylon (L). Pers.] is a warm-season turfgrass for lawns, parks, and sport fields cultivated worldwide (Shi et al., 2012, 2013a, b, 2014 b, c, d). In response to global changed environmental stresses, improvement of abiotic stress resistance is very important for grass engineering (Shi et al., 2012, 2013a, b, 2014 b, c, d). As mentioned above, melatonin might be an ideal target for future genetic engineering of some plant species. However, the endogenous melatonin concentration and the possible role of melatonin in response to abiotic stress in bermudagrass is largely unknown. In this study, endogenous melatonin was examined after abiotic stress treatments in bermudagrass plants, and exogenous melatonin treatment was applied to investigate the in vivo role of melatonin in the response of bermudagrass to abiotic stress. In addition, the effects of exogenous melatonin treatment on ROS accumulation and cell damage, as well as underlying antioxidant responses, were determined. Moreover, comparative metabolomic and transcriptomic analyses were performed to identify differentially expressed metabolites and genes after exogenous melatonin treatment. This study provided some insights into the physiological and molecular mechanisms of melatonin in bermudagrass responses to multiple abiotic stresses.
Materials and methods
Plant materials and growth conditions
Newly harvested common bermudagrass seeds were used in this study. After stratification in deionized water at 4 °C for 4 d in darkness, the bermudagrass seeds were sown in soil in the growth room, which was controlled at 28 °C, with an irradiance of ~150 μmol quanta m–2 s–1, 16h light and 8h dark cycles, and ~65% relative humidity.
Plant abiotic stress treatment
To test the effect of exogenous melatonin on plant physiological responses and abiotic stress resistance, 21-day-old bermudagrass plants were irrigated with water or with different concentrations of melatonin solutions for 7 d, respectively. After melatonin pre-treatment, all control and melatonin-pre-treated 28-day-old bermudagrass plants were subjected to subsequent salt, drought, or cold stress treatments. For salt stress treatment, 28-day-old bermudagrass plants were irrigated with NaCl solutions for 25 d; the NaCl concentration was increased stepwise by 50mM every 5 d to a final concentration of 300mM. For drought stress treatment, 28-day-old plants were subjected to a drought condition by withholding water for 21 d and then re-watered for 4 d. For cold stress treatment, 28-day-old bermudagrass plants were subjected to 4 °C treatment for 21 d, and then transferred to –10 °C for 8h. The freezing stress-treated plants were then recovered overnight at 4 °C and transferred to a standard growth room (28 °C) for 4 d. In each independent experiment, three pots with ~40 plants in each pot were used for each treatment in one concentration of melatonin, and at least three independent experiments were performed to obtain the results.
The survival rate of the salt-, drought-, or freezing-stressed plants was recorded at 25 d after stress treatments. The plant leaf samples from melatonin-pre-treated 28-day-old plants were collected at the indicated time points after salt, drought, or cold treatment for the assays of multiple of physiological parameters.
Quantification of melatonin by enzyme-linked immunosorbent assay (ELISA)
Melatonin from plant leaves was extracted using the acetone–methanol method as described by Pape and Lüning (2006). Briefly, 1g of plant leaf samples was ground in liquid nitrogen, and then transferred to 5ml of extraction mixture (acetone:methanol:water=89:10:1) and homogenized extensively on ice, and the homogenate was centrifuged at 4500 g for 5min at 4 °C. The supernatant was moved to a new centrifuge tube containing 0.5ml of 1% trichloric acid for protein precipitation. After centrifugation at 12 000 g for 10min at 4 °C, the extract was used for quantification of melatonin using the Melatonin ELISA Kit (EK-DSM; Buhlmann Laboratories AG, Schonenbuch, Switzerland) according to the manufacturer’s instruction as described in Shi and Chan (2014a ).
Quantifications of chlorophyll content
Plant leaf chlorophyll was extracted using 80% (v/v) acetone for 6h with shaking in the dark. The concentration of chlorophyll was then calculated by examining the absorbance at 645nm and 663nm of the centrifuged supernatant.
Quantification of electrolyte leakage (EL)
The EL of plant leaves under control and abiotic stress conditions was assayed using a conductivity meter (Leici-DDS-307A, Shanghai, China) as previously described (Shi et al., 2012, 2013a, b, 2014 b, c, d). The relative EL was expressed as the ratio of initial conductivity to the conductivity after boiling.
Determination of malondialdehyde (MDA) content
The MDA content was extracted using chilled thiobarbituric acid (TBA) reagent, and was quantified via determining the absorbance of the supernatant at 450, 532, and 600nm as previously described (Shi et al., 2012, 2013a, b, 2014 b, d).
Determination of ROS accumulation and antioxidants
As two major indicators of ROS accumulation, hydrogen peroxide (H2O2) and superoxide radical (O2·–) contents were quantified using the titanium sulphate method and the Plant O2·– ELISA Kit (10-40-488, Bejing Dingguo, Beijing, China) as previously described (Shi et al., 2012, 2013a, b, 2014 b, d).
The activities of three antioxidant enzymes, namely superoxide dismutase (SOD; EC 1.15.1.1), catalase (CAT; EC 1.11.1.6). and peroxidase (POD; EC 1.11.1.7), were assayed using a Total SOD Assay Kit (S0102; Haimen Beyotime, Haimen city, China), a CAT Assay Kit (S0051; Haimen Beyotime), and a Plant POD Assay Kit (A084-3; Nanjing Jiancheng, Nanjing city, China), respectively, as described by Shi et al., 2012, 2013a, b, 2014 b, d). The concentrations of reduced glutathione (GSH) and oxidized glutathione (GSSG) were determined using the GSH and GSSG Assay Kit (S0053; Haimen Beyotime) as described by Shi et al. (2014b, d), and the GSH redox state was calculated as the ratio of GSH concentration to the concentration of GSH plus GSSG.
Extraction, identification, and quantification of metabolites
Extraction, identification, and quantification of metabolites from plant leaves were performed as in Shi et al. (2014d ). Briefly, the metabolite extraction and sample derivatization were performed as in Lisec et al. (2006), then the derivatizated extract was injected into a DB-5MS capillary cloumn (30 m×0.25 mm×0.25 μm; Agilent J&W GC column, California, USA) using gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) (Agilent 7890A/5975C) according to the protocol described by Shi et al. (2014d ). After the GC-TOF-MS assay, the various metabolites were identified via comparing every retention time index-specific mass with reference spectra in mass spectral libraries (NIST 2005, Wiley 7.0). The numerous metabolites were then quantified based on the pre-added internal standard (ribitol) in the process of metabolite extraction.
Hierarchical cluster analysis
The hierarchical cluster analysis of several metabolites was performed using the CLUSTER program (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/) and Java Treeview (http://jtreeview.sourceforge.net/) as in Shi et al., 2012, 2013a, b, 2014 b, d). For cluster analysis, all metabolites were quantified as fold change relative to the wild-type bermudagrass plants under control conditions, which was set as 1.0.
RNA extraction, library construction, and sequencing
For RNA extraction, 28-day-old bermuagrass plants in pots that were irrigated with water or 20 μM melatonin for 7 d were used. Each treatment was represented by two replicate leaf samples, and each sample contained leaves from at least 30 seedlings. Total RNA was extracted with TRIzol (Invitrogen) and was quantified as previously described (Shi et al., 2013c ). RNA quality was determined using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol. The cDNA libraries were constructed with the mRNA-Seq Sample Preparation Kit™ (Illumina, San Diego, CA, USA) and the DNA yield and fragment insert size distribution of each library were determined on the Agilent Bioanalyzer. The cDNA libraries were then sequenced on an Illumina HiSeq2500 sequencing instrument using the 100bp single end protocol.
Quantitative real-time PCR
The above RNA samples were also used for synthesis of first-strand cDNA with reverse transcriptase (BIO-RAD, Hercules, CA, USA), and the cDNAs were used for quantitative real-time PCR using a CFX96™ Real Time System (BIO-RAD) as previously described (Shi et al., 2013c ). The specific primers of the analysed genes for real-time PCR are listed in Supplementary Table S1 available at JXB online, and the housekeeping genes have been described in Hu et al. (2012).
Bioinformatics analyses of RNA-Seq data
Raw RNA-Seq reads were first trimmed for low quality regions using clean reads with length longer than 25bp, obtained after trimming low quality bases (Q<17) using the SolexQA tool (v2.2) and removing adaptor sequences using the cutadapt tool (v1.3) (Martin, 2011). A total of 679 million clean RNA-Seq reads from 20 libraries, and four libraries were used for transcriptome profiling in this study. Transcriptome analyses of RNA-Seq data were used for transcriptome assembly using Trinity software (v r20131110) (Haas et al., 2013). The resulting pre-assembled transcriptome were refined according to the methods described by Ranjan et al. (2014). After transcripts expressed at a low level and redundant sequences were removed, 28 456 high quality transcripts were retained as the final reference transcriptome for bermudagrass.
To obtain putative annotations, the final transcriptome sequences were compared with the NCBI nr protein database by BlastX using an E-value of 1e-5 as the cut-off. Blast2GO (v 2.5.0) (Gotz et al., 2008) was used to assign GO terms to each transcript. The final transcriptome sequences were also compared with Arabidopsis (TAIR10) and rice (MSU release7) protein database using BlastX with an E-value cut-off of 1e-5. The best hit from these two well-annotated species was used to annotate each bermudagrass transcript.
To evaluate the abundance of each transcript, reads from individual libraries were mapped to the final reference transcripts using Bowtie, and the read counts on each transcript were estimated by the software RSEM with default parameters (Li and Dewey, 2011). Differentially expressed transcripts were identified by the R package edge R (Robinson et al., 2010.).
GO term enrichment analysis of differentially expressed genes was carried out using the topGO Bioconductor package (v 2.16.0) (Alexa et al., 2006) for up- and down-regulated genes, respectively. The Classification SuperViewer Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) was used to generate an overview of the enriched pathways (Provart and Zhu, 2003), and MapMan was used as the classification source to assign functional pathways for each gene (Thimm et al., 2004; Yang et al., 2013). The normalized frequency (NF) of each functional category was calculated as described in Chan et al. (2011): NF=sample frequency of each category in this experiment/background frequency of each category in the Arabidopsis genome.
Statistical analysis
The experiments in this study were repeated three times and the data shown are the means ±SEs. The means are the average of three independent experiments. Each independent experiment was a pooled sample from at least 30 bermudagrass plants. Bars with different letters above the columns in the figures indicate significant differences at P<0.05 (Duncan’s range test).
Results
Abiotic stress induced the endogenous melatonin level in bermudagrass
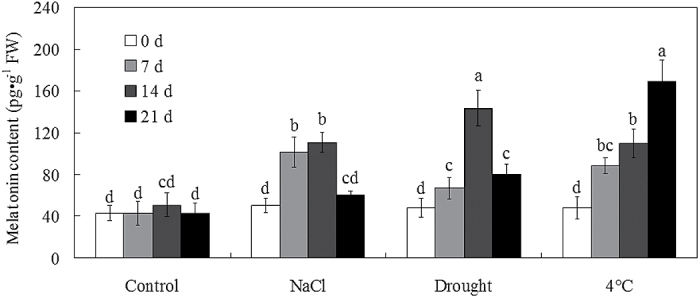
To investigate how abiotic stress affected the melatonin content, endogenous melatonin levels in bermudagrass leaves were quantified after treatments with 300mM NaCl, drought, or cold (4 °C) stresses for 0, 7, 14, and 21 d. Melatonin content remained steady at ~50 pg g–1 fresh weight (FW) in non-treated control plants (Fig. 1). After abiotic stress treatments, the melatonin content in bermudagrass leaves significantly increased (Fig. 1). The induction of melatonin content by multiple abiotic stress treatments indicated the in vivo role of melatonin in bermudagrass response to abiotic stress.
Fig. 1.
The endogenous melatonin level was induced by salt, drought, and cold stresses in bermudagrass. Twenty-eight-day-old bermudagrass plants were treated with control, 300mM NaCl, drought, and cold (4 °C) stresses for 0, 7, 14, and 21 d, respectively. Bars with different letters above the columns of figures indicate significant differences at P<0.05 (Duncan’s range test).
Exogenous melatonin improved abiotic stress resistance in bermudagrass
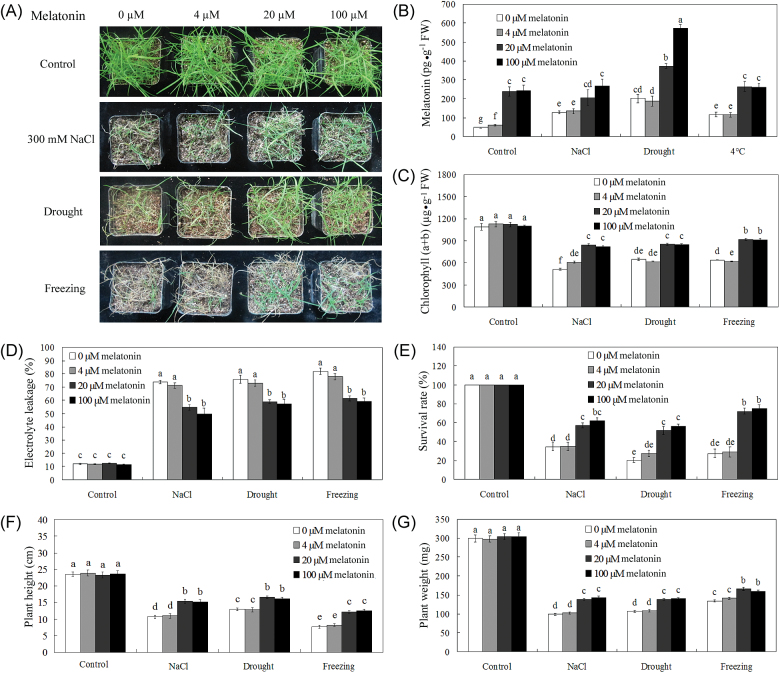
After 7 d pre-treatment with different concentrations of melatonin (0, 4, 20, and 100 μM melatonin, respectively), no significant differences were observed between non-treated and melatonin pre-treated plants (28 d old) (Fig. 2A). When salt, drought, or cold (4 °C) stresses were applied, the endogenous melatonin levels were activated, and 20 μM and 100 μM melatonin-pre-treated bermudagrass had significantly higher levels than non-melatonin-treated plants (Fig. 2B). Growth and physiological parameters including chlorophyll, EL, survival rate, plant height, and plant biomass (weight) of melatonin-pre-treated 28-day-old bermudagrass plants were generally equivalent to those of non-treated plants under well-watered conditions for the following 25 d (Fig. 2C–G). After salt, drought, or freezing stress treatments, growth of both melatonin-pre-treated and non-treated plants was inhibited, but 20 μM and 100 μM melatonin-pre-treated plants had greener leaf tissues than those of non-treated bermudagrass plants (Fig. 2A). Consistently, 20 μM or 100 μM melatonin-pre-treated plants exhibited a significantly higher chlorophyll content, lower EL, and higher survival rate than did non-treated bermudagrass plants (Fig. 2C–E). Moreover, 20 μM and 100 μM melatonin-pre-treated plants exhibited healthy growth in comparison with non-treated and 4 μM melatonin-pre-treated plants, with significantly higher plant height and weight (Fig. 2F, G). These results indicate that exogenous melatonin application improved salt, drought, and freezing stress resistance in bermudagrass.
Fig. 2.
Application of exogenous melatonin improved abiotic stress resistance in bermudagrass. (A) Growth of 28-day-old plants with different melatonin treatments and under control, salt, drought, or freezing stress conditions. The picture is representative of one pot for every treatment in one concentration of melatonin, and at least nine pots with ~120 lines were used for the stress treatments with similar results. (B) Melatonin content of 28-day-old plants with different melatonin treatments and under control, 300mM NaCl, drought stress, and cold (4 °C) stress conditions for 14 d. (C, D) Chlorophyll (C) and EL (D) of 28-day-old plants with different treatments (0, 4, 20, and 100 μM melatonin, respectively) under control, 300mM NaCl, drought stress, and freezing stress conditions for 21 d. (E) The survival rate of bermudagrass plants after 25 d of treatments of control, 300mM NaCl, re-watered from drought, and freezing stresses. (F, G) Plant height (F) and fresh weight (G) of bermudagrass plants after 24 d of treatments of control, 300mM NaCl, re-watered from drought, and freezing stresses. The results shown are the means ±SE (n=3 for B, n=4 for C–E, and n=12 for F and G), and bars with different letters above the columns of figures indicate significant differences at P<0.05 (Duncan’s range test). (This figure is available in colour at JXB online.)
Exogenous melatonin alleviated abiotic stress-induced ROS accumulation in bermudagrass
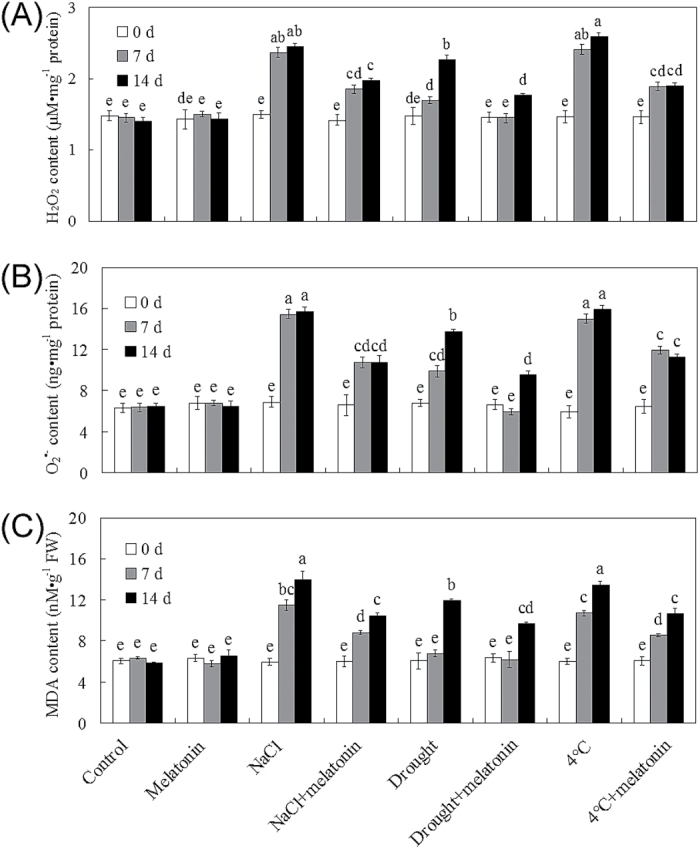
As the major indicators of the stress-triggered ROS level and oxidative damage, H2O2, O2·–, and MDA contents were assayed among control and 20 μM melatonin-pre-treated plants during abiotic stress treatments. Under control conditions, melatonin had no significant effect on H2O2, O2·–, and MDA contents (Fig. 3A–C). When salt, drought, and cold (4 °C) stresses were applied, melatonin-pre-treated plants showed significantly lower levels of H2O2, O2·–, and MDA in comparison with non-treated bermudagrass plants, conferring less oxidative damage (Fig. 3A–C). These results indicated that exogenous application of melatonin could modulate abiotic stress-triggered ROS accumulation and related oxidative damage in bermudagrass.
Fig. 3.
Abiotic stress-induced ROS accumulation and MDA content were alleviated by exogenous melatonin in bermudagrass. (A–C) Quantifications of H2O2 content (A), O2·– content (B), and MDA content (C) of 28-day-old plants with different treatments (control and 20 μM melatonin) under control, 300mM NaCl, drought, and cold (4 °C) stress conditions on the designated days. The results shown are the means ±SEs, and bars with different letters above the columns of figures indicate significant differences at P<0.05 (Duncan’s range test).
Effects of exogenous melatonin on ROS-related antioxidants in bermudagrass response to abiotic stress
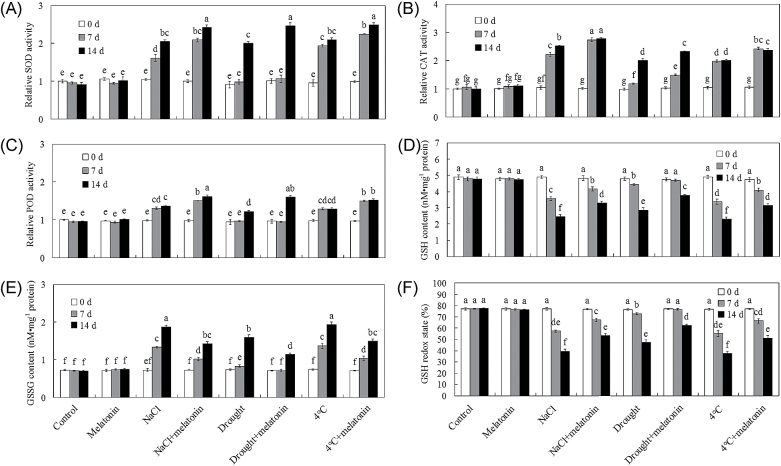
To alleviate abiotic stress-triggered ROS burst, plants have developed complex antioxidant defence system, including several antioxidant enzymes and non-enzymatic glutathione antioxidant pool. Under control conditions, no significant differences in antioxidant enzymes and the non-enzymatic glutathione antioxidant pool were found between non-treated and melatonin-pre-treated bermudagrass (Fig. 4A–F). Under abiotic stress conditions, the activities of antioxidant enzymes (SOD, CAT, and POD) and the GSSG content were greatly induced, while GSH content was significantly decreased (Fig. 4A–F). Additionally, melatonin-pre-treated plants showed significantly higher activities of antioxidant enzymes (SOD, CAT, and POD) and a higher GSH redox state in comparison with non-treated plants, conferring more effective antioxidants (Fig. 4A–F). These results indicated that melatonin had significant effects on both antioxidant enzymes and the non-enzymatic glutathione antioxidant pool, which might be consistent with alleviated abiotic stress-induced ROS accumulation and related oxidative damage in bermudagrass.
Fig. 4.
Effects of exogenous melatonin on ROS-related antioxidants in bermudagrass response to abiotic stress. (A–F) Quantifications of SOD activity (A), CAT activity (B), POD activity (C), GSH (D), GSSG (E), and GSH redox state (F) of 28-day-old plants with different treatments (control and 20 μM melatonin) under control, 300mM NaCl, drought, and cold (4 °C) stress conditions on the designated days. The results shown are the means ±SEs, and bars with different letters above the columns of figures indicate significant differences at P<0.05 (Duncan’s range test).
Modulation of metabolic homeostasis by exogenous melatonin treatment under control and abiotic stress conditions
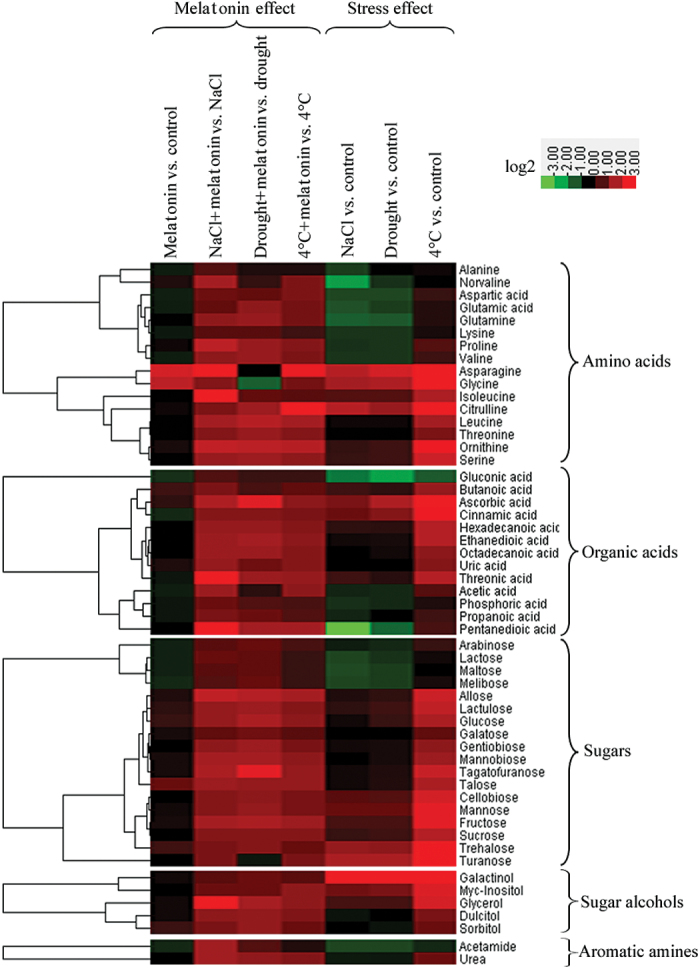
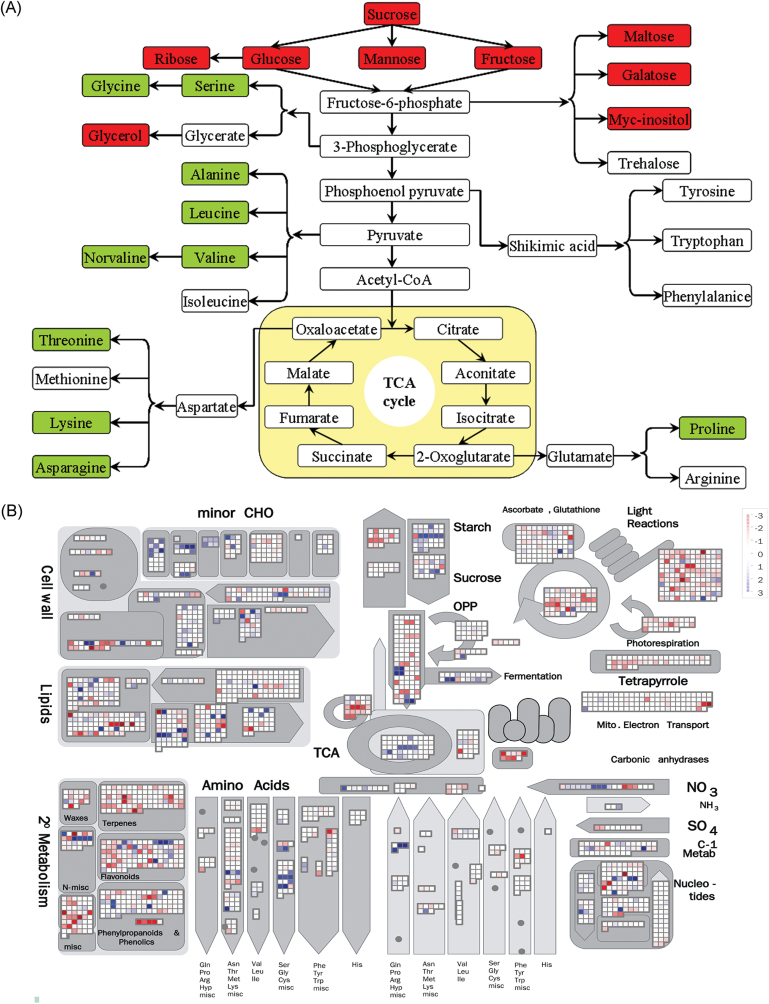
To gain more insights into the modulation of metabolic homeostasis by exogenous melatonin treatment under control and abiotic stress conditions, GC-TOF-MS was performed to identify differentially expressed metabolites. In total, 54 metabolites, comprising 16 amino acids, 13 organic acids, 18 sugars, five sugar alcohols, and two aromatic amines, were reproducibly examined in non-treated and melatonin-pre-treated plants under control and abiotic stress conditions (Fig. 5; Supplementary Table S2 at JXB online). Under control conditions, no significant regular pattern of these metabolites was shown in non-treated and melatonin-pre-treated plants (Fig. 5; Supplementary Table S2). When salt, drought, and cold (4 °C) stresses were applied, melatonin-pre-treated plants exhibited higher concentrations of almost all the 54 metabolites than non-treated plants (Fig. 5; Supplementary Table S2). Additionally, many of these metabolites were commonly regulated by salt, drought, and cold (4°C) stresses (Fig. 5; Supplementary Table S2). Interestingly, 18 metabolites, comprising 10 amino acids, six sugars, and two sugar alcohols, were assigned to the carbon metabolic pathway comprising glycolysis, oxidative pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle, indicating the direct link between melatonin and the carbon metabolic pathway in bermudagrass response to abiotic stress. Melatonin-pre-treated plants exhibited significantly higher levels of these metabolites than non-treated plants under abiotic stress conditions (Fig. 6A).
Fig. 5.
Hierarchical cluster analysis of metabolites modulated by exogenous melatonin in bermudagrass response to abiotic stress. Hierarchical cluster analysis of 54 metabolites of 28-day-old plants by melatonin effect (melatonin versus control, NaCl+melatonin versus NaCl, drought+melatonin versus drought, 4 °C+melatonin versus 4c°C) and by stress effect (NaCl versus control, drought versus control, 4 °C versus control). The log2 ratios and scale bars are shown in the resulting tree figure, which was obtained using the CLUSTER software package and the Java Treeview. The concentrations of these metabolites are listed in Supplementary Table S2 at JXB online. (This figure is available in colour at JXB online.)
Fig. 6.
The effects of melatonin on the metabolites that were involved in the carbon metabolic pathway. (A) Assignment of the 18 metabolites from 54 assayed metabolites to the carbon metabolic pathway. The concentrations of these 18 metabolites are shown in Supplementary Table S2 at JXB online. (B) Effect of exogenous melatonin treatment on primary metabolism in bermudagrass. Transcripts participating in the same pathway or homologues are represented by a set of closely connected cubes, and the 2-based logarithm of fold change value (log2FC) is denoted. (This figure is available in colour at JXB online.)
Transcriptome analysis: GO annotation and enrichment analysis
Since melatonin pre-treatment increased salt, drought, and freezing stress resistance in bermudagrass, the 28-day-old plants (without and with 7 d of pre-treatment) were used for transcriptomic analysis to reveal the effect of melatonin pre-treatment on global transcriptional reprogramming. Approximately 679 million RNA-Seq reads were used for de novo assembly of the bermudagrass transcriptome. After removing poorly expressed and redundant transcripts, 28 456 high quality transcripts were retained as the final reference transcriptome. These annotated transcripts were used to search various protein databases. In total, 22 137 (78%) had BLASTX hits in the well-annotated Arabidopsis and/or rice protein database (E-value >1e-5). GO annotation was performed using the Blast2GO pipeline, and 18 701 (66%) transcripts were assigned with at least one GO term. Among the three GO categories, 13 402 transcripts were annotated in Biological Process, 14 052 transcripts in Molecular Function, and 14 685 in Cellular Component.
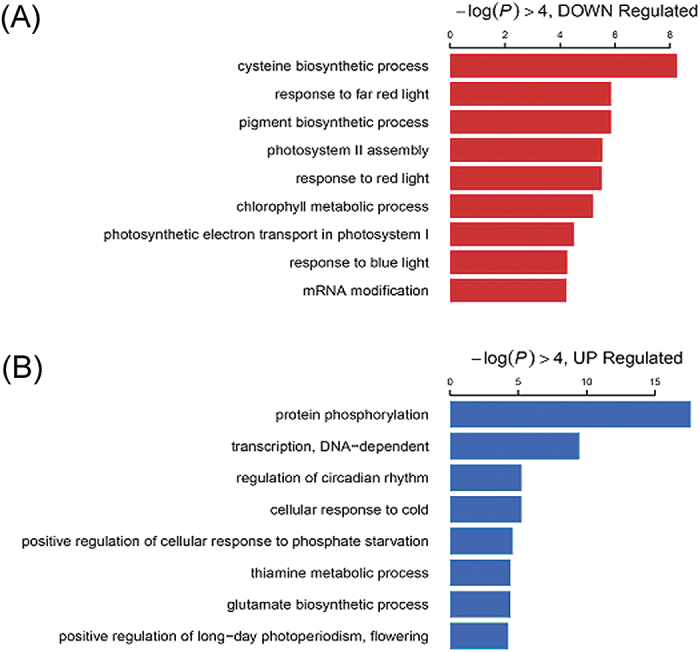
Using fold change >2 and false discovery rate (FDR) <0.05 as thresholds, 3933 transcripts (2361 up-regulated and 1572 down-regulated by exogenous melatonin treatment) were identified as differentially expressed genes (Supplementary Tables S4, S5 at JXB online). Many stress-responsive genes were highly induced by exogenous melatonin treatment in bermudagrass (Table 1). Interestingly, several C-REPEAT-BINDING FACTORS/DEHYDRATION-responsive ELEMENT-BINDING PROTEIN (CBF/DREB) genes and target genes, heat shock transcription factors (TFs), zinc finger TFs, WRKY, MYB, bHLH genes, and hormone-related genes were highly induced >16-fold after melatonin treatment (Table 2). GO enrichment analysis in the biological process domain suggested that genes related to the cysteine biosynthetic process, response to light signal, and the photosynthetic process were down-regulated. In particular, the studies of Wang et al. (2012) showed that melatonin can lower ROS damage of many photosynthetic components. Therefore, the expression of genes involved in the photosystem might been suppressed through a negative feedback mechanism. The up-regulated genes were greatly enriched with the GO terms involved in gene expression regulatory process, such as protein phosphorylation, DNA-dependent transcription, regulation of circadian rhythm, etc. (Fig. 7).
Table 1.
Pathway enrichment analysis of genes whose expression was significantly affected by exogenous melatonin pre-treatment in bermudagrass
| Group | MapMan pathway | Up-regulation | Down-regulation | ||
|---|---|---|---|---|---|
| NF | P-value | NF | P-value | ||
| I | N metabolism | 5.71 | 0.0000 | 4.00 | 0.0140 |
| Major CHO metabolism | 3.30 | 0.0000 | 2.31 | 0.0100 | |
| TCA/org transformation | 2.58 | 0.0023 | 2.63 | 0.0076 | |
| Transport | 2.05 | 0.0000 | 2.42 | 0.0000 | |
| Hormone metabolism | 2.02 | 0.0000 | 1.44 | 0.0110 | |
| Metal handling | 1.78 | 0.0440 | 2.19 | 0.0260 | |
| Redox | 1.59 | 0.0160 | 2.60 | 0.0000 | |
| Secondary metabolism | 1.42 | 0.0094 | 2.75 | 0.0000 | |
| II | Gluconeogenesis/ Glyoxylate cycle | – | – | 24.02 | 0.0000 |
| Glycolysis | – | – | 2.30 | 0.0210 | |
| Photosystem | – | – | 8.21 | 0.0000 | |
| Tetrapyrrole synthesis | – | – | 4.88 | 0.0001 | |
| Fermentation | 6.63 | 0.0005 | – | – | |
| Minor CHO metabolism | 2.99 | 0.0000 | – | – | |
| Signalling | 2.53 | 0.0000 | 0.95 | 0.0550 | |
| C1 metabolism | 2.32 | 0.0430 | – | – | |
| III | RNA | 1.84 | 0.0000 | 1.04 | 0.0340 |
| Cofactor and vitamin metabolism | 1.83 | 0.0400 | – | – | |
| Amino acid metabolism | 1.71 | 0.0036 | 1.70 | 0.0120 | |
| Lipid metabolism | 1.64 | 0.0009 | 1.69 | 0.0023 | |
| Nucleotide metabolism | 1.54 | 0.0280 | 1.87 | 0.0120 | |
| Stress | 1.46 | 0.0000 | – | – | |
| Development | 1.41 | 0.0016 | – | – | |
| Miscellaneous | 1.27 | 0.0015 | 1.71 | 0.0000 | |
| Protein | 1.11 | 0.0031 | 0.73 | 0.0000 | |
| IV | Cell | 0.80 | 0.0260 | – | – |
| Not assigned | 0.45 | 0.0000 | 0.76 | 0.0000 | |
| Mitochondrial electron transport/ATP synthesis | 0.24 | 0.0085 | – | – | |
| DNA | 0.12 | 0.0000 | 0.18 | 0.0000 | |
| MicroRNA, natural antisense, etc | 0.00 | 0.0000 | 0.00 | 0.0000 | |
Twenty-one-day-old bermuagrass plants in pots were irrigated with water or 20 μM melatonin for 7 d, then the 28-day-old plants (without and with 7 d of pre-treatment) were used for transcriptomic analysis. Differentially expressed genes (i.e. with P-value ≤0.05 and log2 fold change ≥1 or log2 fold change ≤ –1) were annotated using the Classification SuperViewer Tool and with MapMan. MapMan was used as the classification source.
Group I indicates highly enriched pathways of both up- and down-regulated genes; group II indicates highly enriched pathways of either up- or down-regulated genes; group III indicates slightly enriched pathways; and group IV indictes under-represented pathways. Scales of normalized frequency (NF) are as follows:
| ≥4 | 2–4 | 1–20. | 5–1 | ≤0.5 |
Table 2.
List of getnes highly induced (>16-fold) by exogenous melatonin pre-treatment in bermudagrass
| Seq_ID | logFC | P-value | FDR | Putative annoation in Arabdopsis | E-valuea | Putative annoation in rice | E-valuea |
|---|---|---|---|---|---|---|---|
| comp58508_c2_seq12 | 8.47 | 5.35E-247 | 2.50E-245 | CBF4, DREB1D | 2e-07 | DRE | 1e-35 |
| comp58508_c2_seq7 | 7.49 | 0 | 0 | DREB1A, CBF3 | 9e-15 | DRE | 2e-25 |
| comp57631_c0_seq7 | 2.98 | 0 | 0 | DREB2A | 2e-23 | AP2 domain containing protein | 2e-62 |
| comp55697_c1_seq4 | 4.92 | 0 | 0 | DNA-binding protein | 1e-23 | AP2 domain containing protein | 6e-34 |
| comp49674_c0_seq1 | 4.49 | 0 | 0 | HRE1 | 1e-14 | AP2 | 1e-44 |
| comp52672_c0_seq8 | 5.57 | 0 | 0 | ERD15 | 5e-17 | Expressed protein | 2e-46 |
| comp51935_c1_seq1 | 5.89 | 0 | 0 | Cold-regulated gene 27 | 3e-13 | Expressed protein | 4e-74 |
| comp59530_c3_seq1 | 5.50 | 4.31E-51 | 4.22E-50 | HSFA6B | 5e-61 | HSF | 4e-133 |
| comp51719_c2_seq14 | 4.61 | 0 | 0 | HSFC1 | 1e-57 | HSF | 1e-113 |
| comp56606_c0_seq9 | 5.48 | 9.30E-52 | 9.25E-51 | HSP20-like | 2e-17 | hsp20 | 3e-27 |
| comp60859_c0_seq13 | 5.13 | 1.51E-14 | 5.65E-14 | SLT1 | HSP20-like | 4e-172 | SLT1 protein | 0 |
| comp53718_c0_seq3 | 6.51 | 0 | 0 | DnaJ | 2e-36 | Heat shock protein DnaJ | 1e-78 |
| comp51393_c1_seq14 | 5.36 | 2.40E-58 | 2.67E-57 | DnaJ | 6e-16 | Heat shock protein DnaJ | 1e-36 |
| comp23443_c0_seq1 | 7.89 | 6.84E-42 | 5.67E-41 | DNAJ heat shock protein | 1e-35 | Expressed protein | 4e-61 |
| comp53904_c0_seq1 | 4.32 | 0 | 0 | WRKY25 | 4e-35 | WRKY53 | 4e-51 |
| comp55751_c0_seq1 | 6.18 | 0 | 0 | WRKY40 | 2e-53 | WRKY71 | 1e-132 |
| comp55960_c0_seq3 | 4.57 | 1.84E-212 | 7.05E-211 | WRKY46 | 4e-35 | WRKY74 | 1e-93 |
| comp54790_c0_seq12 | 5.79 | 1.72E-246 | 7.99E-245 | WRKY51 | 1e-31 | WRKY67 | 6e-37 |
| comp57307_c2_seq3 | 5.57 | 0 | 0 | AZF1 | 8e-12 | C2H2 zinc finger protein | 1e-23 |
| comp54697_c5_seq1 | 4.22 | 1.63E-163 | 4.59E-162 | ATL6 | 3e-42 | Zinc finger, C3HC4 type | 1e-55 |
| comp57307_c1_seq2 | 5.61 | 0 | 0 | STZ, ZAT10 | 4e-25 | C2H2 zinc finger protein | 3e-36 |
| comp55694_c0_seq3 | 5.27 | 0 | 0 | NIP2 | 1e-51 | RING-H2 finger protein ATL2B | 2e-90 |
| comp57199_c1_seq6 | 4.07 | 1.01E-155 | 2.69E-154 | AtMYB78 | 6e-14 | MYB | 2e-14 |
| comp60805_c1_seq6 | 6.20 | 0 | 0 | RVE1 | 5e-36 | MYB | 7e-97 |
| comp60805_c1_seq4 | 5.70 | 0 | 0 | LHY | 2e-13 | MYB | 2e-92 |
| comp58066_c0_seq2 | 4.59 | 8.75E-312 | 5.22E-310 | BHLH | 3e-38 | Ethylene-responsive protein | 1e-95 |
| comp54812_c1_seq1 | 6.33 | 0 | 0 | BHLH92 | 3e-19 | BHLH | 2e-79 |
| comp56171_c3_seq1 | 5.02 | 0 | 0 | anac032, NAC032 | 6e-07 | NAC | 3e-54 |
| comp54298_c0_seq8 | 4.56 | 0 | 0 | AF2 | 7e-73 | NAC | 2e-145 |
| comp54697_c3_seq1 | 4.60 | 2.23E-132 | 5.10E-131 | RING/U-box protein | 1e-28 | Zinc finger | 4e-49 |
| comp53888_c2_seq1 | 4.15 | 9.66E-50 | 9.21E-49 | CMPG2 | 1e-15 | U-box | 7e-20 |
| comp53888_c0_seq1 | 4.74 | 1.02E-122 | 2.15E-121 | PUB29 | 2e-31 | U-box | 9e-90 |
| comp48740_c0_seq1 | 6.07 | 0 | 0 | Calcium-binding EF-hand protein | 3e-35 | EF hand family protein | 8e-68 |
| comp48880_c0_seq1 | 5.33 | 0 | 0 | Calcium-binding EF-hand protein | 1e-28 | OsCML31 | 4e-60 |
| comp61173_c2_seq4 | 5.64 | 0 | 0 | Calcium-binding protein | 3e-34 | OsCML10 | 1e-57 |
| comp42867_c0_seq1 | 5.25 | 6.59E-310 | 3.93E-308 | Calcium-binding protein | 1e-43 | OsCML14 | 7e-72 |
| comp50275_c0_seq5 | 4.36 | 1.47E-284 | 7.97E-283 | Calcium-binding protein | 5e-25 | EF hand | 1e-40 |
| comp60797_c0_seq1 | 5.12 | 0 | 0 | Calmodulin-binding protein | 2e-160 | Calmodulin-binding protein | 0 |
| comp55169_c0_seq2 | 4.50 | 0 | 0 | Calmodulin-binding protein | 8e-85 | Calmodulin-binding protein | 3e-145 |
| comp51845_c0_seq1 | 4.67 | 0 | 0 | CML43 | 3e-39 | OsCML27 | 6e-78 |
| comp55740_c0_seq1 | 4.20 | 0 | 0 | TCH2, CML24 | 2e-38 | OsCML16 | 1e-80 |
| comp57565_c0_seq8 | 9.08 | 7.50E-220 | 3.05E-218 | CKA1 | 0 | Casein kinase II | 0 |
| comp54827_c0_seq1 | 6.78 | 6.62E-48 | 6.09E-47 | Kinase | 2e-20 | Kinase | 3e-26 |
| comp55548_c0_seq1 | 4.39 | 0 | 0 | Kinase | 6e-148 | Phosphotransferase | 0 |
| comp55703_c1_seq1 | 4.29 | 7.18E-159 | 1.96E-157 | Kinase | 7e-65 | Tyrosine protein kinase | 2e-124 |
| comp60546_c0_seq1 | 4.11 | 1.75E-75 | 2.41E-74 | Kinase | 0 | Leucine-rich repeat protein | 0 |
| comp54489_c2_seq1 | 5.52 | 3.60E-185 | 1.19E-183 | JMJD5 | 8e-139 | jmjC protein 5 | 9e-146 |
| comp48478_c1_seq1 | 4.45 | 1.09E-76 | 1.52E-75 | HDA18 | 2e-64 | Histone deacetylase | 2e-71 |
| comp57103_c1_seq1 | 4.45 | 0 | 0 | ERF-1 | 3e-22 | ERF | 1e-43 |
| comp51949_c0_seq2 | 4.49 | 0 | 0 | JAZ2, TIFY10B | 2e-10 | ZIM protein | 8e-31 |
| comp59913_c1_seq8 | 7.52 | 0 | 0 | CYP707A1 | 8e-50 | Cytochrome P450 | 1e-64 |
| comp50455_c0_seq1 | 3.42 | 0 | 0 | PYL5, RCAR8 | 7e-55 | Cyclase/dehydrase | 1e-87 |
| comp55459_c2_seq1 | 4.20 | 0 | 0 | PP2C | 6e-101 | PP2C | 1e-164 |
| comp55059_c0_seq4 | 4.33 | 2.77E-269 | 1.43E-267 | SHY2, IAA3 | 2e-47 | OsIAA24 | 2e-64 |
| comp56859_c0_seq6 | 5.51 | 3.14E-169 | 9.21E-168 | Alcohol dehydrogenase | 8e-100 | Dehydrogenase | 9e-100 |
| comp55475_c1_seq1 | 4.41 | 0 | 0 | WCRKC thioredoxin 1 | 4e-45 | Thioredoxin | 4e-80 |
| comp49522_c0_seq2 | 4.02 | 9.96E-36 | 7.38E-35 | Oxidoreductase | 2e-88 | Dehydrogenase | 4e-136 |
| comp57222_c0_seq2 | 5.09 | 0 | 0 | FMO1 | 2e-91 | Monooxygenase | 0 |
| comp56296_c0_seq6 | 5.02 | 2.03E-49 | 1.92E-48 | Copper transport protein | 2e-09 | Heavy metal-associated protein | 3e-26 |
| comp46851_c0_seq1 | 4.92 | 5.38E-117 | 1.07E-115 | Heavy metal transport | 1e-06 | Expressed protein | 4e-21 |
| comp51430_c0_seq1 | 4.45 | 7.94E-278 | 4.22E-276 | Heavy metal transport | 1e-15 | Heavy metal-associated protein | 3e-41 |
Twenty-one-day-old bermuagrass plants in pots were irrigated with water or 20 μM melatonin for 7 d, then the 28-day-old plants (with and without 7 d of pre-treatment) were used for transcriptomic analysis.
logFC, log2 fold change; FDR, false discovery rate.
eE-value, expected value for putative annotation in Arabidopsis or rice.
Fig. 7.
The Biological Process GO terms enrichment of down-regulated (A) and up-regulated (B) genes between control and melatonin-pre-treated bermudagrass. The horizontal axis shows –log10 of the P-value. Twenty-one-day-old bermuagrass plants in pots were irrigated with water or 20 μM melatonin for 7 d, then the 28-day-old plants (with and without 7 d of pre-treatment) were used for transcriptomic analysis. (This figure is available in colour at JXB online.)
To confirm the reliability of the RNA-Seq data, the expression of 18 genes (nine up-regulated and nine down-regulated by exogenous melatonin treatment) that were differentially expressed between control and melatonin-treated plants was assessed via quantitative real-time PCR. Consistently, the results of the real-time PCR assay exhibited the same trend and were correlated with the RNA-Seq data (Supplementary Fig. S1 at JXB online), confirming the reproducibility of RNA-Seq data.
Pathway and GO term enrichment analyses
The transcriptome data were submitted to the Mercator web tool to align with the public protein database, and 14 288 transcripts were located in at least one point in plant biological pathways. As shown in Table 1, pathway analysis revealed that melatonin affected the expression of many genes involved in N metabolism, minor carbohydrate metabolism, TCA/org transformation, transport, hormone metabolism, metal handling, redox, and secondary metabolism (Table 1, group I). Other transcripts involved in stress response and metabolism were extensively changed after melatonin treatment (Fig. 6B; Supplementary Fig. S2 at JXB online). These results indicated that melatonin treatment might induce a stress response in bermudagrass. The pathway analysis results were consistent with the study carried out by Weeda et al. (2014), which showed that melatonin altered many genes involved in plant defence (Supplementary Fig. S2), and these changes might contribute to the enrichment of stress-related GO terms (Fig. 7).
Discussion
As sessile organisms, plants have developed sophisticated strategies to respond to diverse environmental stresses. The stress signals are perceived by several receptors at the cell membrane level, followed by their transduction to multiple second messengers such as abscisic acid (ABA), H2O2, nitric oxide (NO), etc. These activate downstream stress-responsive genes and physiological responses, eventually leading to protective responses at the whole-plant level (Shi et al., 2012, 2013a, b, 2014 b, c, d; Shi and Chan, 2014a , b). Although no direct evidence had indicated that melatonin could serve as a second messenger, the induction of melatonin by multiple stress treatments (Fig. 1) indicates an in vivo role for melatonin in bermudagrass response to abiotic stress (Arnao and Hernández-Ruiz, 2006, 2009b , 2013a , b; Tan et al., 2007a, 2012).
In this study, the protective role of melatonin on the response of bermudagrass to abiotic stress was revealed. Under control conditions, melatonin had no significant effect on bermudagrass growth or its physiological responses (Fig. 2). Under abiotic stress conditions, however, melatonin-pre-treated plants exhibited significantly higher chlorophyll content, lower EL, and higher survival rate than did non-treated bermudagrass plants (Fig. 2C–E). After recovery from abiotic stress treatments, melatonin-pre-treated plants exhibited better growth status than non-treated plants, with higher biomass (plant height and weight) (Fig. 2F, G). These results indicate that exogenous melatonin application improved salt, drought, or freezing stress resistance in bermudagrass, in accordance with the enhanced resistance to cold stress (Posmyk et al., 2009a ; Kang et al., 2010; Bajwa et al., 2014), copper stress (Posmyk et al., 2008, 2009b), high temperature (Byeon and Back, 2014b ), salt stress (Li et al., 2012), osmotic stress (Zhang et al., 2013), drought stress (Wang et al., 2014), and pathogen infection (Yin et al., 2013) due to melatonin in various plant species.
As an antioxidant in animals, melatonin scavenges free radicals directly, stimulates the activities of antioxidant enzymes including CAT, SOD (both MnSOD and CuSOD), glutathione reductase (GR), and glutathione peroxidase (GPX), and increases the efficiency of mitochondrial oxidative phosphorylation (Tan et al., 1993, 1999, 2003, 2007a, b; Reiter et al., 2000; Kolář and Macháčkova, 2005). In plants, melatonin is also an important antioxidant and a radical scavenger (Arnao et al., 1996, 2001; Cano et al., 2003, 2006). Li et al. (2012) also found that exogenous melatonin modulates salinity-induced oxidative damage by directly scavenging H2O2 and enhancing the activities of antioxidative enzymes in Malus hupehensis. Consistently, exogenous application of melatonin significantly activated ROS detoxification of antioxidants, including enzymatic antioxidant enzymes (SOD, CAT, and POD) and non-enzymatic glutathione (GSH redox state), to maintain cellular ROS (mainly including H2O2 and O2·–) at a relatively low level. This results in the alleviation of abiotic stress-induced oxidative damage and further conferred improved abiotic stress resistance (Figs 3, 4). Consistently, RNA-Seq found that many genes involved in redox, many POD genes and glutathione S-transferases (GST) genes were significantly modulated by exogenous melatonin treatment (Table 1; Supplementary Fig. S2 at JXB online). In summary, the positive modulation by exogenous melatonin of the ROS detoxification system might contribute greatly to enhanced abiotic stress resistance in bermudagrass.
Moreover, comparative metabolomic analysis showed the actions of melatonin treatment on carbon metabolites and amino acid metabolism under abiotic stress conditions. Notably, melatonin-pre-treated plants exhibited higher concentrations of 54 metabolites compared with non-melatonin-treated plants (Fig. 5; Supplementary Table S2 at JXB online). Among these metabolites, proline and some carbohydrates (fructose, sucrose, glucose, maltose, cellobiose, trehalose, galactose, and galactinol) are important compatible solutes to respond to abiotic stress for osmotic adaptation (Krasensky and Jonak, 2012). Thus, higher levels of proline and carbohydrates (glucose, maltose, fructose, sucrose, and trehalose) in melatonin-pre-treated plants provided beneficial effects under abiotic stress conditions. In addition, higher levels of other metabolites including multiple amino acids, organic acids, and sugars in melatonin-pre-treated plants indicate the beneficial physiological processes in melatonin-pre-treated plants during abiotic stress treatment; the data further confirm the protective role of melatonin in response to abiotic stress. Notably, 18 metabolites comprising 10 amino acids, six sugars, and two sugar alcohols of the carbon metabolic pathway exhibited significantly higher levels in melatonin-pre-treated plants under abiotic stress conditions (Fig. 6A). These results indicate the widespread effects of melatonin treatment in carbon metabolism and amino acid metabolism; these metabolites might play some role in melatonin-mediated abiotic stress resistance in bermudagrass.
Additionally, comparative transcriptomic analysis identified 2361 up-regulated and 1572 down-regulated transcripts as a consequence of exogenous melatonin treatment. Quantitative real-time PCR of 18 genes supported the reliability of the RNA-Seq data (Supplementary Fig. S1 at JXB online). Pathway enrichment analysis indicated that eight pathways were over-represented among differentially expressed genes between control and melatonin-treated bermudagrass plants, including N metabolism, major carbohydrate metabolism, TCA/org transformation, transport, hormone metabolism, metal handling, redox, and secondary metabolism (Table 1, group I). The enrichment of redox-related genes affected by melatonin (Table 1, group I) was consistent with the effects of exogenous melatonin on ROS detoxification in bermudagrass (Figs 4, 5). In animals, melatonin is known to be involved in circadian rhythms (Tal et al., 2011). Interestingly, the GO enrichment analysis also showed that transcripts that function in regulation of rhythms and flowering were over-represented (Fig. 7). Notably, the pathway analysis results were consistent with those of the study carried out in Arabidopsis by Weeda et al. (2014). Thus, melatonin altered many genes involved in plant defence in bermudagrass (Supplementary Fig. S2); these changes probably contributed to the enrichment of stress-related GO terms (Fig. 7). Additionally, exogenous melatonin treatment had significant effects on various signalling pathways including primary metabolism, secondary metabolism, photosynthesis, large enzyme families, receptor-like kinases, proteolysis, and autophagy pathways in bermudagrass as determined using MapMan software (Fig. 6B; Supplementary Fig. S2). Those genes modulated by exogenous melatonin treatment might also contribute to melatonin-enhanced abiotic stress resistance in bermudagrass.
Asparagine accumulation shows that nitrogen re-distribution and mobilization were important features of the melatonin response (Lea et al., 2007; Maaroufi-Dguimi et al., 2011). Jia et al. (2001) also suggested that asparagine may be a signalling molecule involved in sensing the nitrogen status. In addition, asparagine is an amino group donor for the synthesis of the photorespiratory intermediate glycine. Nagy et al. (2013) and Shi et al. (2014) found that this is also a good indicator of drought stress in drought-tolerant and sensitive wheat and bermudagrass cultivars. At the same time, some carbohydrate metabolism compounds increased: these included fructose, glucose, and trehalose, but not sucrose. Such differential dynamics of carbohydrates could reflect modifications of carbon balance and carbon utilization. Moreover, asparagine synthetase genes that are involved in asparagine synthesis are regulated by the level of carbohydrates (Lam et al., 1998; Foito et al., 2009). TCA/org transformation is important for the Calvin cycle for CO2 assimilation and separation of initial carbon fixation by contact with air and secondary carbon fixation into sugars (Selwood and Jaffe, 2011). Glycolysis is an important metabolic pathway in carbohydrate metabolism, and the central role of glycolysis in the plant metabolic pathway is to provide energy such as ATP and generates precursors for anabolism such as fatty acids and amino acids (Plaxton, 1996). In accordance with the metabolic profiles, transcriptomic analysis found that many genes involved in TCA/org transformation and N metabolism were modulated by melatonin treatment (Table 1; Fig. 6B). Genes which functioned in sucrose and amino acid metabolism were also greatly changed after melatonin treatment (Fig. 6B), leading to altered sucrose and amino acid contents revealed by metabolite analysis (Figs 5, 6). These results showed that the underlying mechanisms related to melatonin may involve major reorientation of photorespiratory and carbohydrate and nitrogen metabolism.
To date, various TFs have been shown to be involved in plant stress responses via activating stress-responsive gene expression, such as BASIC LEUCINE ZIPPER PROTEINS (bZIPs), CBF/DREBs, ETHYLENE-RESPONSIVE ELEMENT-BINDING FACTORS (ERFs), MYBs, WRKYs, and zinc finger proteins (ZFPs) (Shi et al., 2014a, e). In the current study, many TFs were significantly regulated by exogenous melatonin treatment (Table 2; Supplementary Tables S4, S5 at JXB online), and these TFs might contribute to the enhanced stress tolerance of melatonin-treated plants, thus indicating that melatonin might pre-condition to be resistant to abiotic stresses. Some protein kinases [such as mitogen-activated protein kinase (MAPK)] and calcium signalling kinases [including calcium-dependent protein kinases (CDPKs), calcineurin B-like (CBL)-interacting protein kinases (CIPKs), and calcium-related protein kinases (CRKs)] were also transcriptionally regulated by exogenous melatonin treatment (Table 1; Supplementary Tables S4, S5). This suggests that kinase signalling might play critical roles in melatonin-mediated stress responses. As sessile organisms, plants cannot avoid unfavourable stress conditions by adjusting their location; thus, they have evolved complex strategies to perceive stress signals and further translate the perception into effective responses, which might largely depend on various protein kinases and TFs (Shi et al., 2014a, e).
Recently, it was found that one cysteine2/histidine2-type zinc finger TF, zinc finger of Arabidopsis thaliana 6 (ZAT6), is involved in melatonin-mediated freezing stress response, and the AtZAT6-activated CBF pathway was essential for melatonin-mediated freezing stress response in Arabidopsis (Shi and Chan, 2014a ). This study together with others in sunflower (Helianthus annuus) (Mukherjee et al., 2014) and in Arabidopsis (Shi and Chan, 2014a ; Weeda et al., 2014) indicate that melatonin is involved in long-distance signal transduction in plants. Moreover, some important genes in plant hormone signalling [RCAR/PYR/PYL, SNF1-related protein kinases 2 (SnRK2), and nine-cis-epoxycarotenoid dioxygenase (NCED) genes in ABA signalling, and jasmonate (JA)-JIM-domain proteins (JAZs) in JA signalling] that were significantly regulated by exogenous melatonin treatment (Table 1; Supplementary Tables S4, S5 at JXB online) might also have some function in melatonin-mediated cross-talk among plant hormones, as well as in stress responses. Kolář and Macháčkova (2005) also found that melatonin might function as an auxin to promote vegetative growth. These results suggested that melatonin might serve as a plant hormone that cross-talks with other plant hormones. Thus, melatonin triggered extensive transcriptional reprogramming and pre-conditioned resistance to multiple abiotic stresses. Further investigation of the in vivo roles of these genes will shed additional light on melatonin-mediated stress responses in bermudagrass.
Taken together, this study provides the first evidence of the protective roles of exogenous melatonin in bermudagrass response to multiple abiotic stresses. This involved the activation of antioxidants, modulation of metabolic homeostasis, and extensive transcriptional reprogramming.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Validation of differentially expressed genes by quantitative real-time PCR.
Figure S2. Effect of exogenous melatonin treatment on stress-related pathways in bermudagrass.
Table S1. The specific primers used for real-time PCR.
Table S2. Concentrations of 54 metabolites in 28-day-old bermudagrass plants under control conditions and different treatments [20 μM melatonin, 300mM NaCl, drought, and cold (4 °C)] stress conditions for 14 d.
Table S3. Summary of RNA-Seq data.
Table S4. List of up-regulated genes in melatonin-treated bermudagrass plants.
Table S5. List of down-regulated genes in melatonin-treated bermudagrass plants.
Acknowledgements
We thank Professor Pingfang Yang, Dr Yanqiong Jiang, and Dr Longxin Hu for their help with this study. This research was supported by the Knowledge Innovative Key Program of the Chinese Academy of Sciences (No. 55Y455446O0544), Youth Innovation Promotion Association of the Chinese Academy of Sciences (No. 29Y429371O0437) to HS, by funding from the Chinese Academy of Sciences to RL, and by ‘the Hundred Talents Program’ of the Chinese Academy of Sciences (Nos 54Y154761O01076 and 29Y329631O0263) to ZC. HS designed and performed the experiments, analysed the data, and wrote and revised the manuscript; CJ analysed the transcriptomic data; TY performed the metabolic experiment; HZ performed the transcriptomic experiment; RL analysed the transcriptomic data and revised the manuscript; DT and RR revised the manuscript; ZC designed the experiments and revised the manuscript; and all authors approved the manuscript.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. [DOI] [PubMed] [Google Scholar]
- Arnao MB, Cano A, Alcolea JF, Acosta M. 2001. Estimation of free radical-quenching activity of leaf pigment extracts. Phytochemical Analysis 12, 138–143. [DOI] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. 2006. The physiological function of melatonin in plants. Plant Signaling and Behavior 1, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. 2007. Melatonin in plants: more studies are necessary. Plant Signaling and Behavior 2, 381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. 2009a. Protective effect of melatonin against chlorophyll degradation during the senescence of barley leaves. Journal of Pineal Research 46, 58–63. [DOI] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. 2009b. Chemical stress by different agents affects the melatonin content of barley roots. Journal of Pineal Research 46, 295–299. [DOI] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. 2013a. Growth conditions determine different melatonin levels in Lupinus albus L. Journal of Pineal Research 55, 149–155. [DOI] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. 2013b. Growth conditions influence the melatonin content of tomato plants. Food Chemistry 138, 1212–1214. [DOI] [PubMed] [Google Scholar]
- Arnao MB, Sanchez-Bravo J, Acosta M. 1996. Indole-3-carbinol as a scavenger of free radicals. Biochemistry and Molecular Biology International 39, 1125–1134. [DOI] [PubMed] [Google Scholar]
- Bajwa VS, Shukla MR, Sherif SM, Murch SJ, Saxena PK. 2014. Role of melatonin in alleviating cold stress in Arabidopsis thaliana . Journal of Pineal Research 56, 238–245. [DOI] [PubMed] [Google Scholar]
- Byeon Y, Back K. 2014a. An increase in melatonin in transgenic rice causes pleiotropic phenotypes, including enhanced seedling growth, delayed flowering, and low grain yield. Journal of Pineal Research 56, 408–414. [DOI] [PubMed] [Google Scholar]
- Byeon Y, Back K. 2014b. Melatonin synthesis in rice seedlings in vivo is enhanced at high temperatures and under dark conditions due to increased serotonin N-acetyltransferase and N-acetylserotonin methyltransferase activities. Journal of Pineal Research 56, 189–195. [DOI] [PubMed] [Google Scholar]
- Byeon Y, Park S, Kim YS, Back K. 2013. Microarray analysis of genes differentially expressed in melatonin-rich transgenic rice expressing a sheep serotonin N-acetyltransferase. Journal of Pineal Research 55, 357–363. [DOI] [PubMed] [Google Scholar]
- Byeon Y, Park S, Kim YS, Park DH, Lee S, Back K. 2012. Light-regulated melatonin biosynthesis in rice during the senescence process in detached leaves. Journal of Pineal Research 53, 107–111. [DOI] [PubMed] [Google Scholar]
- Byeon Y, Park S, Lee HY, Kim YS, Back K. 2014. Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. Journal of Pineal Research 56, 275–282. [DOI] [PubMed] [Google Scholar]
- Calvo JR, González-Yanes C, Maldonado MD. 2013. The role of melatonin in the cells of the innate immunity: a review. Journal of Pineal Research 55, 103–120. [DOI] [PubMed] [Google Scholar]
- Cano A, Hernández-Ruiz J, Arnao MB. 2006. Changes in hydrophilic antioxidant activity in Avena sativa and Triticum aestivum leaves of different age during de-etiolation and high-light treatment. Journal of Pineal Research 119, 321–327. [DOI] [PubMed] [Google Scholar]
- Cano A, Alcaraz O, Arnao MB. 2003. Free radical-scavenging activity of indolic compounds in aqueous and ethanolic media. Analytical and Bioanalytical Chemistry 376, 33–37. [DOI] [PubMed] [Google Scholar]
- Chan Z, Grumet R, Loescher W. 2011. Global gene expression analysis of transgenic, mannitol-producing, and salt-tolerant Arabidopsis thaliana indicates widespread changes in abiotic and biotic stress-related genes. Journal of Experimental Botany 62, 4787–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbels R, Reiter RJ, Klenke E, Goebel A, Schnakenberg E, Ehlers C, Schiwara HW, Schloot W. 1995. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. Journal of Pineal Research 18, 28–31. [DOI] [PubMed] [Google Scholar]
- Foito A, Byrne SL, Shepherd T, Stewart D, Barth S. 2009. Transcriptional and metabolic profiles of Lolium perenne L. genotypes in response to a PEG-induced water stress. Plant Biotechnology Journal 7, 719–732. [DOI] [PubMed] [Google Scholar]
- Galano A, Tan DX, Reiter RJ. 2011. Melatonin as a natural ally against oxidative stress: a physicochemical examination. Journal of Pineal Research 51, 1–16. [DOI] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Research 36, 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8, 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori A, Migitaka H, Iigo M, Itoh M, Yamamoto K, Ohtani-Kaneko R, Hara M, Suzuki T, Reiter RJ. 1995. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochemistry and Molecular Biology International 35, 627–634. [PubMed] [Google Scholar]
- Hernández-Ruiz J, Arnao MB. 2008. Distribution of melatonin in different zones of lupin and barley plants at different ages in the presence and absence of light. Journal of Agricultural and Food Chemistry 56, 10567–10573. [DOI] [PubMed] [Google Scholar]
- Hernández-Ruiz J, Cano A, Arnao MB. 2005. Melatonin acts as a growth-stimulating compound in some monocot species. Journal of Pineal Research 39, 137–142. [DOI] [PubMed] [Google Scholar]
- Hu L, Huang Z, Liu S, Fu J. 2012. Growth response and gene expression in antioxidant related enzymes in two bermudagrass genotypes differing in salt tolerance. Journal of the American Society of Horticultural Science 137, 134–143. [Google Scholar]
- Jia M, Keutgen N, Matsuhashi S, Mitzuniwa C, Ito T, Fujimura T, Hashimoto S. 2001. Ion chromatographic analysis of selected free amino acids and cations to investigate the change of nitrogen metabolism by herbicide stress in soybean (Glycine max). Journal of Agricultural and Food Chemistry 49, 276–280. [DOI] [PubMed] [Google Scholar]
- Kang K, Lee K, Park S, Kim YS, Back K. 2010. Enhanced production of melatonin by ectopic overexpression of human serotonin N-acetyltransferase plays a role in cold resistance in transgenic rice seedlings. Journal of Pineal Research 49, 176–182. [DOI] [PubMed] [Google Scholar]
- Kolář J, Macháčkova I. 2005. Melatonin in higher plants: occurrence and possible functions. Journal of Pineal Research 39, 333–341. [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. 2012. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Journal of Experimental Botany 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G. 1998. Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana . The Plant Journal 16, 345–353. [DOI] [PubMed] [Google Scholar]
- Lea US, Slimestad R, Smedvig P, Lillo C. 2007. Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225, 1245–1253. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang P, Wei Z, Liang D, Liu C, Yin L, Jia D, Fu M, Ma F. 2012. The mitigation effects of exogenous melatonin on salinity-induced stress in Malus hupehensis . Journal of Pineal Research 53, 298–306. [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. 2006. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Maaroufi-Dguimi H, Debouba M, Gaufichon L, Clément G, Gouia H, Hajjaji A, Suzuki A. 2011. An Arabidopsis mutant disrupted in ASN2 encoding asparagine synthetase 2 exhibits low salt stress tolerance. Plant Physiology and Biochemistry 49, 623–628. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17, 10–12. [Google Scholar]
- Manchester LC, Tan DX, Reiter RJ, Park W, Monis K, Qi W. 2000. High levels of melatonin in the seeds of edible plants: possible function in germ tissue protection. Life Sciences 67, 3023–3029. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, David A, Yadav S, Baluška F, Bhatla SC. 2014. Salt stress-induced seedling growth inhibition coincides with differential distribution of serotonin and melatonin in sunflower seedling roots and cotyledons. Physiologia Plantarum 152, 714–728. [DOI] [PubMed] [Google Scholar]
- Murch SJ, Alan AR, Cao J, Saxena PK. 2009. Melatonin and serotonin in flowers and fruits of Datura metel L. Journal of Pineal Research 47, 277–283. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Németh E, Guóth A, Bona L, Wodala B, Pécsváradi A. 2013. Metabolic indicators of drought stress tolerance in wheat: glutamine synthetase isoenzymes and Rubisco. Plant Physiology and Biochemistry 67, 48–54. [DOI] [PubMed] [Google Scholar]
- Okazaki M, Ezura H. 2009. Profiling of melatonin in model tomato (Solanum lycopersicum L.) cultivar Micro-Tom. Journal of Pineal Research 43, 338–343. [DOI] [PubMed] [Google Scholar]
- Okazaki M, Higuchi K, Aouni A, Ezura H. 2010. Lowering intercellular melatonin by transgenic analysis of indoleamine 2,3-dioxygenase from rice in tomato plants. Journal of Pineal Research 49, 239–247. [DOI] [PubMed] [Google Scholar]
- Okazaki M, Higuchi K, Hanawa Y, Shiraiwa Y, Ezura H. 2009. Cloning and characterization of a Chlamydomonas reinhardtii cDNA arylalkylamine N -acetyltransferase and its use in the genetic engineering of melatonin content in the Micro-Tom tomato. Journal of Pineal Research 43, 373–382. [DOI] [PubMed] [Google Scholar]
- Pape C, Lüning K. 2006. Quantification of melatonin in phototrophic organisms. Journal of Pineal Research 41, 157–165. [DOI] [PubMed] [Google Scholar]
- Park S, Lee DE, Jang H, Byeon Y, Kim YS, Back K. 2013. Melatonin-rich transgenic rice plants exhibit resistance to herbicide-induced oxidative stress. Journal of Pineal Research 54, 258–263. [DOI] [PubMed] [Google Scholar]
- Pelagio-Flores R, Muňoz-Parra E, Ortiz-Castro R, López-Bucio J. 2012. Melatonin regulates Arabidopsis root system architecture likely acting independently of auxin signaling. Journal of Pineal Research 53, 279–288. [DOI] [PubMed] [Google Scholar]
- Plaxton WC. 1996. The organization and regulation of plant glycolysis. Annual Review of Plant Biology and Plant Molecular Biology 47, 185–214. [DOI] [PubMed] [Google Scholar]
- Posmyk MM, Bałabusta M, Wieczorek M, Sliwinska E, Janas KM. 2009a. Melatonin applied to cucumber (Cucumis sativus L.) seeds improves germination during chilling stress. Journal of Pineal Research 46, 214–223. [DOI] [PubMed] [Google Scholar]
- Posmyk MM, Janas KM, Kontek R. 2009b. Red cabbage anthocyanin extract alleviates copper-induced cytological disturbances in plant meristematic tissue and human lymphocytes. Biometals 22, 479–490. [DOI] [PubMed] [Google Scholar]
- Posmyk MM, Kuran H, Marciniak K, Janas KM. 2008. Presowing seed treatment with melatonin protects red cabbage seedlings against toxic copper ion concentrations. Journal of Pineal Research 45, 24–31. [DOI] [PubMed] [Google Scholar]
- Provart N, Zhu T. 2003. A browser-based functional classification superviewer for Arabidopsis genomics. Current Computer Molecular Biology 2003, 271–272. [Google Scholar]
- Ramakrishna A, Giridhar P, Sankar KU, Ravishankar GA. 2012. Melatonin and serotonin profiles in beans of Coffea species. Journal of Pineal Research 52, 470–476. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Ichihashi Y, Farhi M, Zumstein K, Townsley B, David-Schwartz R, Sinha NR. 2014. De novo assembly and characterization of the transcriptome of the parasitic weed Cuscuta pentagona identifies genes associated with plant parasitism. Plant Physiology 166, 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RJ, Tan DX, Burkhardt S, Manchester LC. 2001. Melatonin in plants. Nutrition Reviews 59, 286–290. [DOI] [PubMed] [Google Scholar]
- Reiter RJ, Tan DX, Osuna C, Gitto E. 2000. Actions of melatonin in the reduction of oxidative stress. A review. Journal of Biomedical Science 7, 444–458. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwood T, Jaffe EK. 2011. Dynamic dissociating homo-oligomers and the control of protein function. Archives of Biochemistry and Biophysics 519, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Chan Z. 2014. The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6-activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. Journal of Pineal Research 57, 185–191. [DOI] [PubMed] [Google Scholar]
- Shi H, Wang Y, Cheng Z, Ye T, Chan Z. 2012. Analysis of natural variation in bermudagrass (Cynodon dactylon) reveals physiological responses underlying drought tolerance. PLoS One 7, e53422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Ye T, Chen F, Deng J, Yang P, Zhang Y, Chan Z. 2014a. The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiology 165, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Chan Z. 2013a. Comparative proteomic and physiological analyses reveal the protective effect of exogenous polyamines in the bermudagrass (Cynodon dactylon) response to salt and drought stresses. Journal of Proteome Research 12, 4951–4964. [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chan Z. 2013b. Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermudagrass (Cynodon dactylon (L). Pers.). Plant Physiology and Biochemistry 71, 226–234. [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chan Z. 2014b. Nitric oxide-activated hydrogen sulfide is essential for cadmium stress response in bermudagrass (Cynodon dactylon (L). Pers.). Plant Physiology and Biochemistry 71, 226–234. [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chan Z. 2014c. Comparative proteomic responses of two bermudagrass (Cynodon dactylon (L). Pers.) varieties contrasting in drought stress resistance. Plant Physiology and Biochemistry 82, 218–228. [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chen F, Cheng Z, Wang Y, Yang P, Zhang Y, Chan Z. 2013c. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. Journal of Experimental Botany 64, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Zhong B, Liu X, Chan Z. 2014d. Comparative proteomic and metabolomic analyses reveal mechanisms of improved cold stress tolerance in bermudagrass (Cynodon dactylon (L). Pers.) by exogenous calcium. Journal of Integrative Plant Biology 71, 226–234. [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Zhong B, Liu X, Jin R, Chan Z. 2014e. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21 . New Phytologist 203, 554–567. [DOI] [PubMed] [Google Scholar]
- Tal O, Haim A, Harel O, Gerchman Y. 2011. Melatonin as an antioxidant and its semi-lunar rhythm in green macroalga Ulva sp. Journal of Experimental Botany 62, 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DX, Chen LD, Poeggeler B, Manchester L, Reiter RJ. 1993. Melatonin: a potent, endogenous hydroxyl radical scavenger. Endocrine Journal 1, 57–60. [Google Scholar]
- Tan DX, Hardeland R, Manchester LC, Korkmaz A, Ma S, Rosales-Corral S, Reiter RJ. 2012. Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. Journal of Experimental Botany 63, 577–597. [DOI] [PubMed] [Google Scholar]
- Tan DX, Hardeland R, Manchester LC, Poeggeler B, Lopez-Burillo S, Mayo JC, Sainz RM, Reiter RJ. 2003. Mechanistic and comparative studies of melatonin and classic antioxidants in terms of their interactions with the ABTS cation radical. Journal of Pineal Research 34, 249–259. [DOI] [PubMed] [Google Scholar]
- Tan DX, Manchester LC, Helton P, Reiter RJ. 2007a. Phytoremediative capacity of plants enriched with melatonin. Plant Signaling and Behavior 2, 514–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DX, Manchester LC, Reiter RJ, Plummer BF. 1999. Cyclic 3-hydroxymelatonin: a melatonin metabolite generated as a result of hydroxyl radical scavenging. Biological Signals and Receptors 8, 70–74. [DOI] [PubMed] [Google Scholar]
- Tan DX, Manchester LC, Terron MP, Flores LJ, Reiter RJ. 2007b. One molecule, many derivatives: a never-ending interaction of melatonin with reactive oxygen and nitrogen species? Journal of Pineal Research 42, 28–42. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Venegas C, García JA, Escames G, Ortiz F, López A, Doerrier C, García-Corzo L, López LC, Reiter RJ, Acuña-Castroviejo D. 2012. Extrapineal melatonin: analysis of its subcellular distribution and daily fluctuations. Journal of Pineal Research 52, 217–227. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao Y, Reiter RJ, He C, Liu G, Lei Q, Zuo B, Zheng XD, Li Q, Kong J. 2014. Changes in melatonin levels in transgenic ‘Micro-Tom’ tomato overexpressing ovine AANAT and ovine HIOMT genes. Journal of Pineal Research 56, 134–142. [DOI] [PubMed] [Google Scholar]
- Wang P, Sun X, Chang C, Feng F, Liang D, Cheng L, Ma F. 2013a. Delay in leaf senescence of Malus hupehensis by long-term melatonin application is associated with its regulation of metabolic status and protein degradation. Journal of Pineal Research 55, 424–434. [DOI] [PubMed] [Google Scholar]
- Wang P, Sun X, Li C, Wei Z, Liang D, Ma F. 2013b. Long-term exogenous application of melatonin delays drought-induced leaf senescence in apple. Journal of Pineal Research 54, 292–302. [DOI] [PubMed] [Google Scholar]
- Wang P, Yin L, Liang D, Li C, Ma F, Yue Z. 2012. Delayed senescence of apple leaves by exogenous melatonin treatment: toward regulating the ascorbate–glutathione cycle. Journal of Pineal Research 53, 11–20. [DOI] [PubMed] [Google Scholar]
- Weeda S, Zhang N, Zhao X, Ndip G, Guo Y, Buck GA, Fu C, Ren S. 2014. Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS One 9, e93462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Li Z, Bickel DR. 2013. Empirical Bayes estimation of posterior probabilities of enrichment: a comparative study of five estimators of the local false discovery rate. BMC Bioinformatics 14, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang P, Li M, et al. 2013. Exogenous melatonin improves Malus resistance to Marssonina apple blotch . Journal of Pineal Research 54, 426–434. [DOI] [PubMed] [Google Scholar]
- Zhang N, Zhang HJ, Zhao B, et al. 2014. The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. Journal of Pineal Research 56, 39–50. [DOI] [PubMed] [Google Scholar]
- Zhang N, Zhao B, Zhang HJ, Weeda S, Yang C, Yang ZC, Ren S, Guo YD. 2013. Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). Journal of Pineal Research 54, 15–23. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Qi LW, Wang WM, Saxena PK, Liu CZ. 2011. Melatonin improves the survival of cryopreserved callus of Rhodiola crenulata . Journal of Pineal Research 50, 83–88. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tan DX, Lei Q, Chen H, Wang L, Li QT, Gao Y, Kong J. 2013. Melatonin and its potential biological functions in the fruits of sweet cherry. Journal of Pineal Research 55, 79–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.