Highlight
Our study of Tap46 overexpression suggests that Tap46 enhances plant growth as a positive effector of the TOR signalling pathway. Furthermore, the abundance of Tap46/PP2Ac protein is regulated by TOR activity.
Key words: Kinematic analysis, phosphorylation of S6 kinase, PP2A catalytic subunit, rapamycin, real-time quantitative PCR, seed viability.
Abstract
Tap46, a regulatory subunit of protein phosphatase 2A (PP2A), plays an essential role in plant growth and development through a functional link with the Target of Rapamycin (TOR) signalling pathway. Here, we have characterized the molecular mechanisms behind a gain-of-function phenotype of Tap46 and its relationship with TOR to gain further insights into Tap46 function in plants. Constitutive overexpression of Tap46 in Arabidopsis resulted in overall growth stimulation with enlarged organs, such as leaves and siliques. Kinematic analysis of leaf growth revealed that increased cell size was mainly responsible for the leaf enlargement. Tap46 overexpression also enhanced seed size and viability under accelerated ageing conditions. Enhanced plant growth was also observed in dexamethasone (DEX)-inducible Tap46 overexpression Arabidopsis lines, accompanied by increased cellular activities of nitrate-assimilating enzymes. DEX-induced Tap46 overexpression and Tap46 RNAi resulted in increased and decreased phosphorylation of S6 kinase (S6K), respectively, which is a sensitive indicator of endogenous TOR activity, and Tap46 interacted with S6K in planta based on bimolecular fluorescence complementation and co-immunoprecipitation. Furthermore, inactivation of TOR by estradiol-inducible RNAi or rapamycin treatment decreased Tap46 protein levels, but increased PP2A catalytic subunit levels. Real-time quantitative PCR analysis revealed that Tap46 overexpression induced transcriptional modulation of genes involved in nitrogen metabolism, ribosome biogenesis, and lignin biosynthesis. These findings suggest that Tap46 modulates plant growth as a positive effector of the TOR signalling pathway and Tap46/PP2Ac protein abundance is regulated by TOR activity.
Introduction
Target of Rapamycin (TOR) is a Ser/Thr protein kinase that is evolutionarily conserved among all eukaryotes. The TOR signalling pathway integrates diverse signals, such as nutrients, energy, growth factors, and environmental conditions, to regulate cell growth and metabolism in yeast, plants, and mammals (Hay and Sonenberg, 2004; Wullschleger et al., 2006; Bögre et al., 2013; Xiong and Sheen, 2013). In yeast, TOR kinase becomes activated under favourable conditions to promote ribosome biogenesis, protein translation, transcription, and nutrient uptake, while rapamycin treatment or nutrient deficiency inactivates TOR, turning off the processes and activating autophagy, proteolysis, and stress responses instead (Loewith, 2010). In mammals, TOR signalling similarly regulates metabolic activity and cell growth in response to diverse external signals, and has been linked to amino acid sensing and the insulin signalling pathway (Zoncu et al., 2011; Laplante and Sabatini, 2012). TOR kinase forms two distinct multiprotein complexes in yeast and mammals, named TOR complex 1 (TORC1) and TOR complex 2 (TORC2), which contain both conserved and distinct protein components. Rapamycin-sensitive TORC1 is mainly involved in the control of metabolism, translation, and autophagy, while rapamycin-insensitive TORC2 regulates cytoskeleton structure, apoptosis, and transcription (Wullschleger et al., 2006; Zoncu et al., 2011; Shimobayashi and Hall, 2014).
In contrast to yeast and mammalian systems, molecular studies on the plant TOR signalling pathway have been impeded by its reduced sensitivity to rapamycin (Soulard et al., 2009). Moreover, disruption of plant TOR-pathway genes, such as TOR, Tap46, and RAPTOR, is lethal to the embryo (Menand et al., 2002; Deprost et al., 2005; Ahn et al., 2011). Expression of FKP12, which mediates rapamycin binding to TOR, renders Arabidopsis plants rapamycin sensitive, providing an invaluable tool for dissecting the signalling pathway in higher plants (Sormani et al., 2007; Ren et al., 2012; Xiong and Sheen, 2012). Using this system, Xiong et al. (2013) reported that TOR senses and transduces shoot photosynthesis-derived glucose signals that control cell proliferation in the root meristem through transcriptional reprogramming of numerous genes involved in metabolism and biosynthesis for energy production and growth. Furthermore, TOR directly phosphorylates the E2Fa transcription factor, which can then activate transcription of S-phase genes, a novel function for TOR in direct transcriptional regulation of the cell cycle (Xiong et al., 2013). The phytohormone auxin activates TOR kinase, promoting S6K1 phosphorylation at T449 and translation reinitiation of upstream open reading frame (uORF)-containing mRNAs via phosphorylation of eIF3h (eukaryotic initiation factor 3, subunit h) (Schepetilnikov et al., 2013).
Tap42/α4, a regulatory subunit of protein phosphatase 2A, is a downstream effector of TOR kinase in yeast and mammals. Tap42/α4 directly associates with the catalytic subunits of PP2A and the PP2A-related phosphatases PP4 and PP6 to form a heterodimer (Chen et al., 1998; Düvel and Broach, 2004). Yeast Tap42 is phosphorylated by TOR and acts downstream of TOR in many TOR-mediated responses (Jacinto et al., 2001; Dűvel and Broach, 2004). Loss of function of Tap42 mimics the effects of rapamycin treatment (Yan et al., 2006). α4, the mammalian homologue of Tap42, regulates dephosphorylation of the transcription factors c-jun and p53. Deletion of α4 rapidly initiates apoptosis in proliferating, as well as differentiated, cells in mice (Kong et al., 2004).
Tap46, a plant homologue of Tap42/α4, plays an essential role in plant cell growth and survival as a component of the TOR signalling pathway (Ahn et al., 2011). Like Tap42/α4, Tap46 interacts with catalytic subunits of PP2A, PP4, and PP6, and recombinant Tap46 is phosphorylated by immuno-precipitated TOR kinase in vitro. Tap46-deficient plants exhibit the signature phenotypes of TOR inactivation, including repression of global translation and activation of nitrogen mobilization and autophagy. Taken together, these results suggest that Tap46 may function as a downstream effector of TOR signalling in the regulation of plant survival, growth, and development (Ahn et al., 2011). In this study, we further investigated Tap46 functions in plants by characterizing its gain-of-function phenotype and its relationship with the TOR pathway. Tap46 overexpression in Arabidopsis stimulated overall plant growth resulting in enlarged plants with larger leaves and seeds, and increased nitrogen-assimilating activities. Increased phosphorylation of S6 kinase (S6K) was detected in Tap46 overexpression plants, suggesting activation of the TOR signalling pathway. Furthermore, TOR activity oppositely modulated protein abundance of Tap46 and the PP2A catalytic subunit (PP2Ac), suggesting their antagonistic relationship in the TOR signalling pathway.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana wild type (WT; ecotype Col-0) and the Tap46 overexpression transgenic plants in the Col-0 background were grown in a growth room at 23°C and 60 μmol m–2 s–1 light intensity under a 16h light/8h dark cycle. For rapamycin treatment, seeds were germinated and grown in six-well plates containing 1ml of liquid medium (0.5 X MS and 0.5% sucrose, pH 5.7) with 1 or 10 μM rapamycin.
Kinematic analysis of leaf growth
Kinematic analysis was performed as described previously (De Veylder et al., 2001; Gonzalez et al., 2010). For the leaf area analysis, first leaves were harvested from seven to eight plants grown in soil at 4 to 25 days after cotyledon emergence. The leaves were cleared with clearing solution [3:1 (v/v) ethanol:acetic acid] and mounted on microscope slides for photography, and the leaf area was determined with ImageJ software (http://rsb.info.nih.gov/ij/). Abaxial epidermal cells (40–100 cells) were drawn for four to five leaves with a light microscope (Olympus) fitted with a drawing tube and a differential interference contrast objective. Photographs of leaves and drawings were used to measure the leaf area and the average cell area with the ImageJ software. Epidermal cell numbers per leaf were calculated using the leaf area and average cell area.
Immunoblotting
Protein extracts (30 µg) were subjected to SDS-PAGE and immunoblotting as described previously (Ahn et al., 2011; Lee et al., 2013). Anti-Tap46 antibodies were generated in rabbits against an oligopeptide, NQPLIFGPASIVGGP, that corresponds to amino acid residues of Arabidopsis Tap46 at positions 289 to 303 using the antibody production services of Cosmogenetech (http://www.cosmogenetech.com). Immunoblotting was performed using mouse monoclonal antibodies against the haemagglutinin (HA) tag (1:10 000 dilution; Applied Biological Materials), the Myc tag (1:10 000 dilution; Applied Biological Materials), and the Flag tag (1:10 000; Sigma), or using rabbit polyclonal antibodies against α-tubulin (1:1000 dilution; Sigma), the PP2A catalytic subunit (1:1000; Cell Signalling), and Tap46 (1:5000; Cosmogenetech). The membranes were then treated with horseradish peroxidase-conjugated goat anti-mouse IgG antibodies (1:10 000; Invitrogen) or goat anti-rabbit antibodies (1:10,000; Invitrogen), respectively. Signals were detected on X-ray film (Kodak) using an ECL chemiluminescence kit (ELPIS-Biotech, Inc.).
Detection of S6K phosphorylation
Protoplast transient expression assays were carried out as described previously (Xiong and Sheen, 2012). Flag-tagged WT or mutant S6K1 (T449A) was transiently expressed in mesophyll protoplasts isolated from the transgenic Arabidopsis seedlings grown in (–)DEX or (+)DEX medium (DEX: dexamethasone). Protoplasts were harvested by centrifugation and the protein extract was subjected to immunoblotting with anti-phospho-p70 S6K (pThr-389) polyclonal antibody (1:1000; Cell Signalling) to detect phosphorylation of the Thr-449 residue in Arabidopsis S6K1 and Thr-455 residue in Arabidopsis S6K2 as described (Xiong and Sheen, 2012).
Other techniques
The following techniques are described in Supplementary Methods S1: generation of DEX-inducible Tap46 overexpression lines in Arabidopsis; Agrobacterium-mediated transient expression; real-time quantitative PCR; bimolecular fluorescence complementation (BiFC); co-immunoprecipitation; measurement of enzyme activities; accelerated-ageing treatment and tetrazolium staining of seeds; and statistical analyses.
Results
Overexpression of Tap46 leads to enhanced plant growth
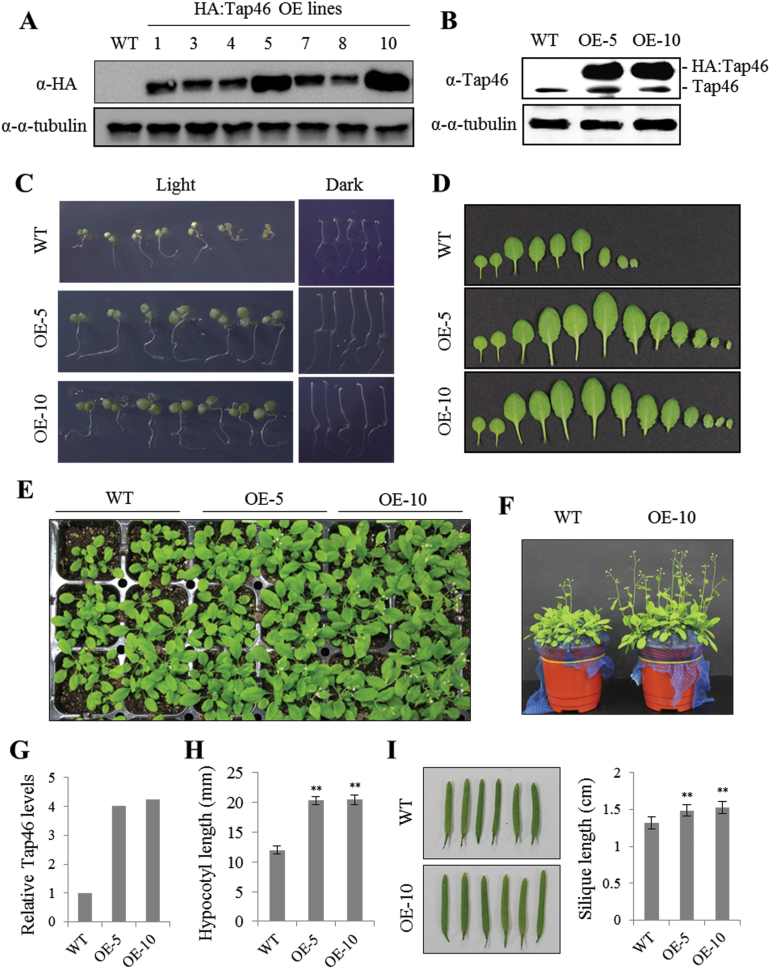
To investigate the role of Tap46 in plant growth, we generated transgenic Arabidopsis lines (Col-0 ecotype) that constitutively overexpress Tap46 fused to an HA tag under the control of a cauliflower mosaic virus 35S promoter. The presence of the HA:Tap46 transgene was confirmed by genomic PCR in >30 independent T1 transgenic lines. Ten T2 lines containing a single copy transgene were selected. T3 homozygous plants of most of these lines exhibited enhanced growth compared with the WT plants. Immunoblotting with anti-HA antibody using cell extracts from the T3 lines confirmed the expression of HA:Tap46 proteins (Fig. 1A). Two overexpression lines, OE-5 and OE-10, were selected for further study based on strong phenotypes and high HA:Tap46 expression indicated by immunoblotting with anti-HA and anti-Tap46 antibodies (Fig. 1A, B). Quantification of the immunoblot bands in Fig. 1B showed that total Tap46 protein levels in OE-5 and OE-10 leaves were ~4-fold higher than the levels in WT plants (Fig. 1G). The Tap46 overexpression seedlings exhibited enhanced cotyledon and root growth on Murashige and Skoog (MS) medium under light conditions (Fig. 1C). Dark-grown Tap46 overexpression seedlings also exhibited longer hypocotyls and roots (Fig. 1C): the hypocotyls of dark-grown OE-5 and OE-10 seedlings were ~1.8-fold longer than those of WT seedlings (Fig. 1H). Leaf series of plants grown in soil for 3 weeks showed an increase in size and number in OE-5 and OE-10 plants, suggesting enhanced plant growth (Fig. 1D). OE-5 and OE-10 plants grew bigger and faster, and reached maturity earlier, than WT plants (Fig. 1E, F). At maturity, the overexpression plants had enlarged leaves and siliques: for example, siliques of the overexpression plants were ~15–20% longer than those of WT plants (Fig. 1F, I). Thus, Tap46 overexpression affected the mechanisms that regulate plant growth rates and final organ size.
Fig. 1.
Enhanced growth of Arabidopsis Tap46 constitutive overexpression (OE) lines. (A) Immunoblotting with anti-HA antibodies to measure expression of HA:Tap46 fusion protein in WT (Col-0) and seven independent Tap46 OE lines. Thirty micrograms of total protein isolated from seedlings were subjected to immunoblotting. Anti-α-tubulin antibody was used as a loading control. (B) Immunoblotting with anti-Tap46 antibodies of protein extracts from WT and Tap46 OE (OE-5 and OE-10) seedlings. (C) Seedling growth of WT, OE-5, and OE-10 lines on MS medium under light and dark conditions 4 days after sowing. (D) Representative leaf series of WT, OE-5, and OE-10 plants grown for 3 weeks in soil. (E) WT, OE-5, and OE-10 plants grown for 3 weeks in soil. (F) WT and OE-10 plants grown for 4 weeks in soil. (G) Relative expression levels of Tap46 protein in WT, OE-5, and OE-10 seedlings based on quantification of the immunoblot band intensities in (B). Levels of endogenous and HA-tagged Tap46 were combined for the OE lines. (H) Average hypocotyl length of WT and Tap46 OE seedlings grown under dark conditions 4 days after sowing (n = 30). Asterisks denote the statistical significance of the differences between WT and Tap46 OE samples: *, P ≤ 0.05; **, P ≤ 0.01. (I) Average silique length of WT and Tap46 OE lines (n = 30). This figure is available in colour at JXB online.
Kinematic analysis of leaf growth
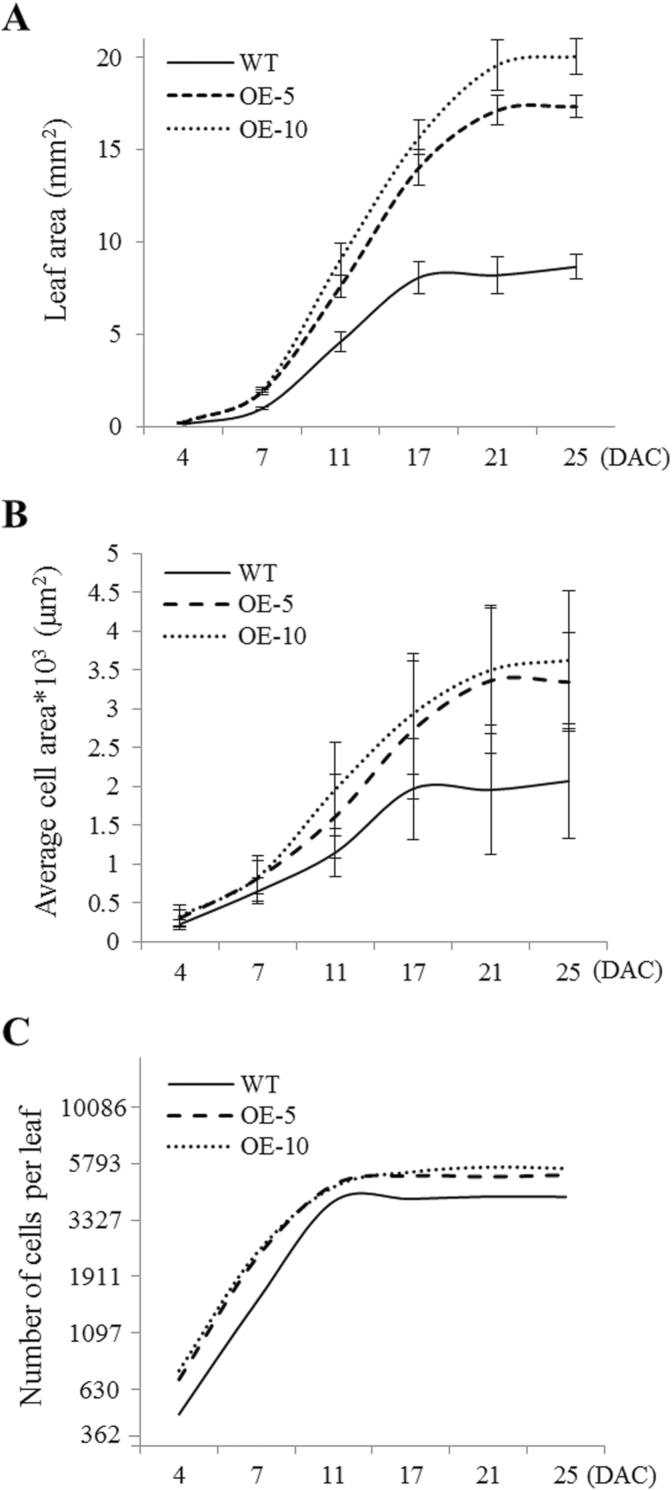
Final organ size is determined by two parameters: cell division and cell expansion (Horiguchi et al., 2006; Anastasiou and Lenhard 2007). To gain insight into the effect of Tap46 overexpression on the two parameters during leaf development, kinematic analysis was performed on the first leaves of OE-5 and OE-10 transgenic plants in comparison with those of WT plants (Fig. 2; Supplementary Table 1; Supplementary Figure S1). First leaves of seven to eight soil-grown plants were harvested at 4, 7, 11, 17, 21, and 25 days after cotyledon emergence (DAC) to assess leaf size and abaxial epidermal cell size, and to calculate the epidermal cell number using the two values. The leaf area of WT and Tap46 overexpression lines progressively increased to reach a plateau at 17 and 21 DAC, respectively. Increased leaf size of Tap46 overexpression plants was visible as early as 4 DAC, and the final leaf size was 2.0 to 2.31-fold larger than that of the WT at 25 DAC (Fig. 2A; Supplementary Table 1). Tap46 overexpression leaves had larger epidermal cells at 4 DAC compared with WT leaves, and the final cell size of Tap46 overexpression leaves at 25 DAC was 1.62- to 1.75-fold larger than that of WT leaves (Fig. 2B; Supplementary Table 1). Furthermore, the leaf epidermal cells of Tap46 overexpression lines reached their final sizes at 21 DAC, while those of the WT reached a plateau at 17 DAC, suggesting prolonged cell expansion in Tap46 overexpression leaves. When the first leaves of WT and Tap46 overexpression lines were observed at 2 DAC, there was no difference in epidermal cell sizes, suggesting that cell division occurs in a similar size in WT and Tap46 overexpression lines (Supplementary Figure S2). The epidermal cell number per leaf increased rapidly during the early stages and then remained nearly constant from 11 DAC onward for both WT and Tap46 overexpression plants. The estimated cell number of OE-5 and OE-10 leaves was 1.23-fold and 1.32-fold higher than that of WT plants at 25 DAC (Fig. 2C; Supplementary Table 1). Taken together, these results suggest that the leaf size enlargement of Tap46 overexpression plants is mainly caused by cell expansion, occurring more vigorously for an extended period of time, although increased cell division also contributes to the increased leaf size.
Fig. 2.
Kinematic analysis of leaf growth. Seeds were imbibed at 4°C for 24 hours before sowing. First leaves were collected from WT, OE-5, and OE-10 plants at 4, 7, 11, 17, 21, and 25 DAC. (A) Average areas of first leaves. (B) Average areas of epidermal cells on the abaxial side of first leaves. (C) Calculated numbers of epidermal cells per leaf.
Seed enlargement in Tap46 overexpression plants
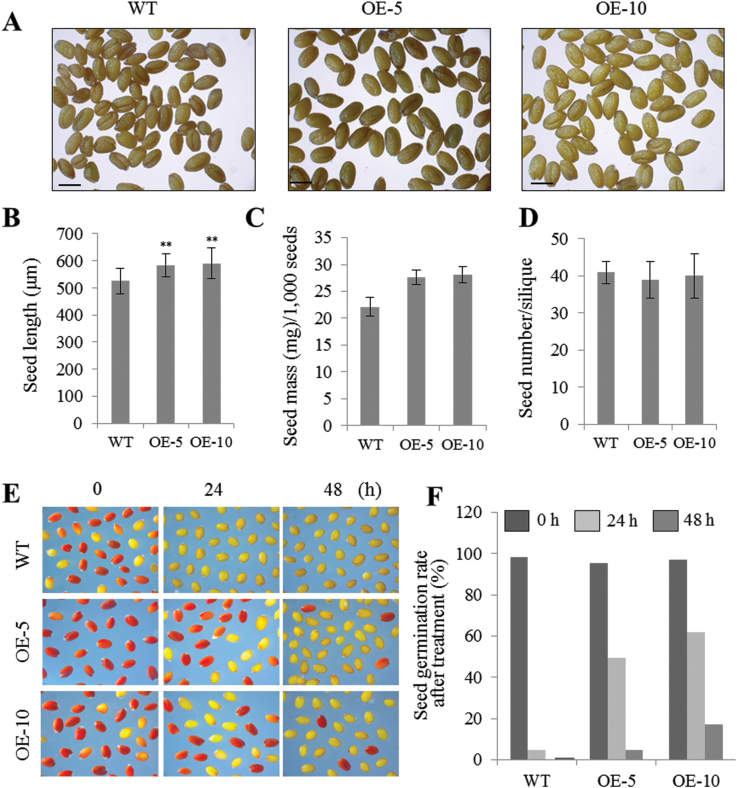
To investigate the effect of Tap46 overexpression on seed development, we determined the average length and mass of mature dry seeds from WT, OE-5, and OE-10 plants that had been grown in soil under the same conditions. OE-5 and OE-10 seeds were longer and heavier than WT seeds (Fig. 3A–C), although the seed number per silique was slightly reduced in Tap46 overexpression plants (Fig. 3D). To test seed quality, seed viability was monitored under accelerated-aging conditions (Fig. 3E, F). Seeds were incubated under high temperature (43°C) and high humidity (100% relative humidity) for 0–72 hours. Tetrazolium uptake assays were performed to determine seed viability. Upon incubation with the tetrazolium solution, viable seeds are stained red, while seeds with low viability remain white (Oge et al., 2008; Seo et al., 2011). OE-5 and OE-10 seeds survived and germinated at a higher frequency after 24 or 48 hours of treatment, compared with WT seeds (Fig. 3E, F). None of the WT or Tap46 overexpression seeds survived after 72 hours of treatment. Thus, Tap46 overexpression seeds appeared to be more resistant to environmental stresses that reduce seed viability.
Fig. 3.
Analysis of seed enlargement in Tap46 overexpression lines. (A) Mature seeds of WT, OE-5, and OE-10 plants. Scale bars, 500 µm. (B) Average seed length (n = 500). (C) Average seed mass per 1000 seeds (n = 3). (D) Average seed number per silique (n = 30). (E) Tetrazolium staining to determine seed viability. WT, OE-5, and OE-10 seeds were incubated under accelerated ageing conditions for 0–48 hours. Viable seeds are stained red (and appear here as the darker seeds). (F) Germination rate (%) of WT, OE-5, and OE-10 seeds after accelerated ageing treatment for 0–48 hours (n = 100). This figure is available in colour at JXB online.
DEX-inducible overexpression of Tap46 results in enhanced plant growth
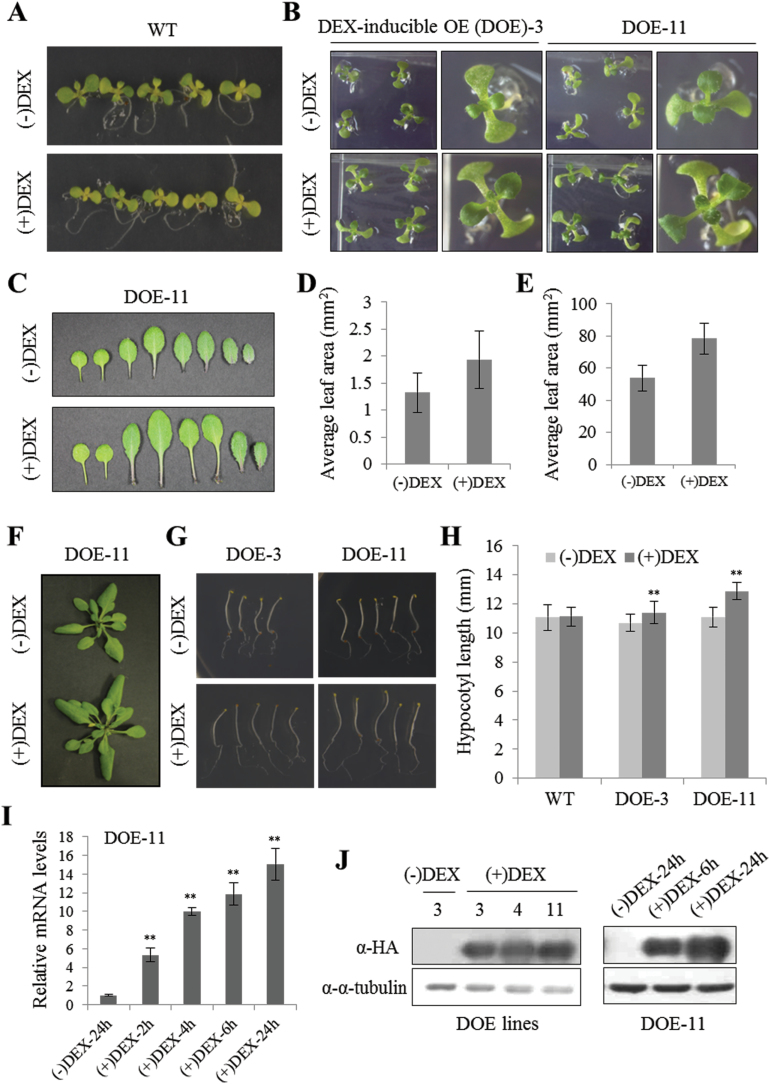
To confirm that Tap46 overexpression leads to enhanced seedling growth, we generated dexamethasone (DEX)-inducible Tap46 overexpression (DOE) lines in Arabidopsis that carry the HA:Tap46 transgene under the control of a DEX-inducible transcription system (Aoyama and Chua, 1997) (Fig. 4). WT and transgenic seedlings were grown in MS medium containing ethanol [(–)DEX] or 10 µM DEX [(+)DEX]. Upon DEX treatment, seedlings of two independent DOE lines (no. 3 and no. 11) showed enhanced shoot growth compared with (–)DEX DOE or WT seedlings with or without DEX treatment (Fig. 4A, B). Leaf series of soil-grown DOE-11 plants 16 days after germination (DAG) demonstrated that DEX spraying resulted in an increase in leaf size compared with (–)DEX plants (Fig. 4C). Quantification revealed that DEX spraying increased the average area of the first leaves (4 DAG) and the largest leaves (16 DAG) of DOE-11 plants, suggesting that Tap46 overexpression stimulates plant growth (Fig. 4D, E). Prolonged DEX spraying of soil-grown DOE lines resulted in enlarged plants (Fig. 4F). DEX-treated DOE seedlings also grew slightly faster than ethanol-treated seedlings under dark conditions, resulting in longer hypocotyls at 4 days after sowing (DAS; Fig. 4G, H). A time-dependent effect of DEX treatment on Tap46 mRNA levels was determined by real-time quantitative RT-PCR using Tap46-specific primers described in Supplementary Table S2 (Fig. 4I). DOE-11 seedlings were grown on MS medium for 10 days, transferred to new MS medium containing ethanol [(–)DEX] or 10 μM DEX [(+)DEX], and incubated for 2–24 hours. DOE-11 seedlings exhibited a progressive increase in Tap46 transcript levels upon DEX treatment compared with the (–)DEX 24-h sample (Fig. 4I). Immunoblotting with anti-HA antibodies detected ectopic expression of the HA:Tap46 fusion protein in the transgenic DOE-3, DOE-4, and DOE-11 lines upon DEX treatment for 24 hours (Fig. 4J, left). A time-dependent accumulation of HA:Tap46 protein in DOE-11 lines upon DEX treatment is shown in Fig. 4J (right). These results further support the hypothesis that Tap46 overexpression stimulates plant growth.
Fig. 4.
Enhanced growth of Arabidopsis DEX-inducible Tap46 overexpresion (DOE) lines. (A) WT seedling growth on MS medium containing ethanol [(–)DEX] or 10 μM DEX [(+)DEX] at 6 DAS. (B) Seedling growth of DOE-3 and DOE-11 lines on (–)DEX and (+)DEX medium 6 DAS. (C) Representative leaf series of soil-grown DOE-11 plants that were sprayed with ethanol or 30 µM DEX for 16 DAG. (D) Average area of the first leaves of soil-grown DOE-11 plants that were sprayed with ethanol or DEX for 4 days after germination. (E) Average area of the largest rosette leaves of soil-grown DOE-11 plants that were sprayed with ethanol or DEX for 16 days after germination. (F) Soil-grown DOE-11 seedlings sprayed with ethanol or DEX for three weeks. (G) Seedling growth 4 DAS on (–)DEX or (+)DEX medium under dark conditions. (H) Average hypocotyl length of dark-grown seedlings on (–)DEX and (+)DEX medium 4 DAS (n = 30). (I) Real-time quantitative RT-PCR analysis of Tap46 transcript levels. Transcript levels in (+)DEX samples are expressed relative to those in (–)DEX-24 hour samples. Values represent means ±SD of three replicates per experiment. (J) Immunoblotting to measure accumulation of HA:Tap46 fusion protein in DOE lines. Thirty micrograms of total protein isolated from different DOE lines (nos. 3, 4, and 11) with or without DEX treatment for 24 hours were subjected to immunoblotting with anti-HA antibodies (left). HA:Tap46 protein accumulation was detected by immunoblotting in DOE-11 seedlings after 6 and 24 hours of DEX treatment (right). Anti-α-tubulin antibody was used as a loading control. This figure is available in colour at JXB online.
Activities of nitrogen metabolic enzymes
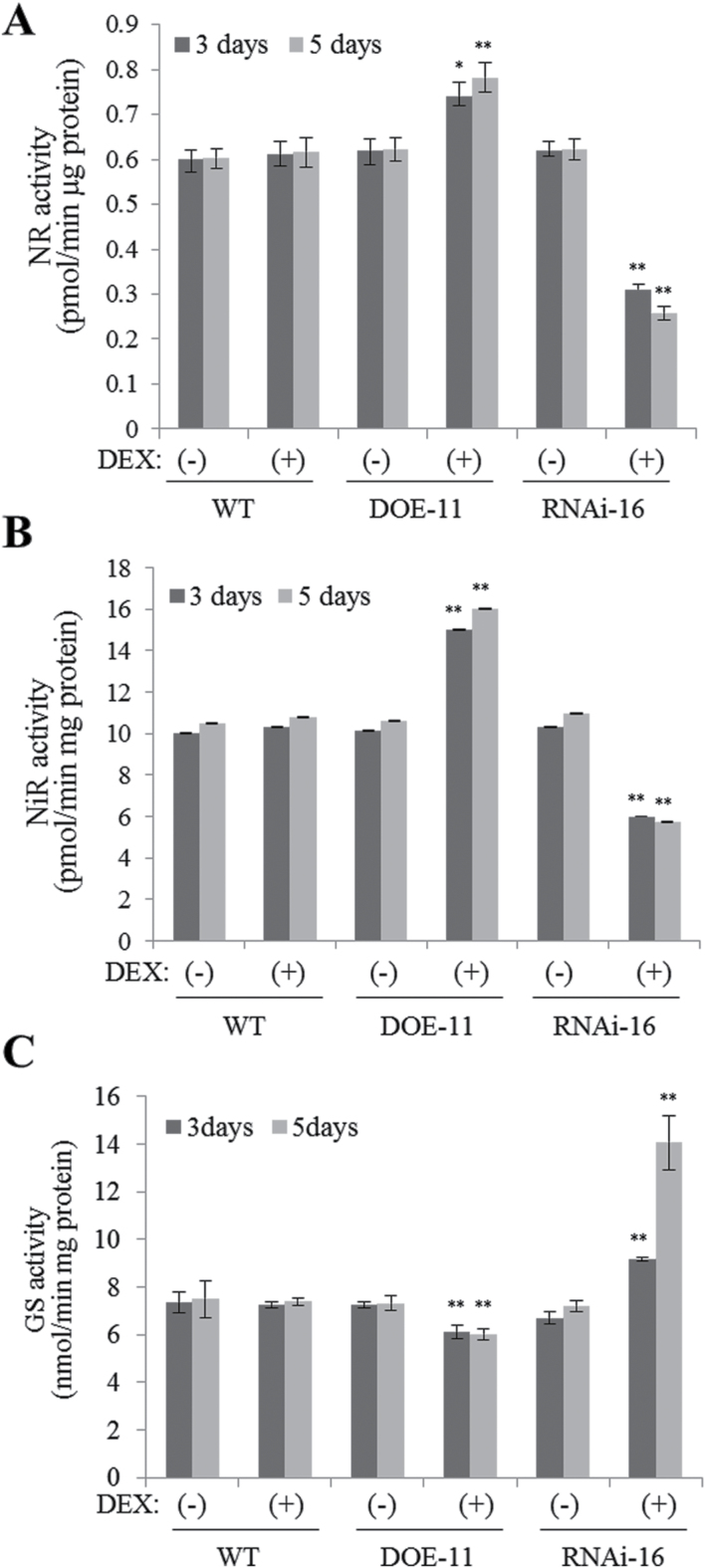
In aerobic soils, the major form of inorganic nitrogen is nitrate (Xu et al., 2012). A large portion of nitrate absorbed by the roots is transported to the shoot and reduced to nitrite by nitrate reductase (NR) and then to ammonium by nitrite reductase (NiR). Ammonium derived from nitrate or directly absorbed via ammonium transporters is further assimilated into amino acids by the glutamine synthetase (GS)/glutamine 2-oxoglutarate aminotransferase (GOGAT) cycle (Xu et al., 2012). Induction of TOR RNAi in Arabidopsis results in 2- to 3-fold higher levels of GS and glutamate dehydrogenase activities that are usually associated with leaf senescence and nutrient recycling (Deprost et al., 2007). Activation of Tap46 RNAi resulted in reduced NR activity (Ahn et al., 2011).
We examined the activities of several key nitrogen-assimilating enzymes in the DEX-inducible Tap46 overexpression (DOE-11) and DEX-inducible Tap46 RNAi (RNAi-16) lines (Ahn et al., 2011) (Fig. 5). Seedlings were grown for 2 weeks in MS medium and transferred to new MS medium containing ethanol [(–)DEX] or 10 µM DEX [(+)DEX] for further growth for 3 or 5 days. Increased total NR activities were detected in DOE-11 seedlings upon DEX treatment, while NR activities of DEX-treated RNAi-16 seedlings were lower than those of (–)DEX samples (Fig. 5A). There were no differences in the activity between WT (–)DEX and (+)DEX samples. To assess how much of the NR enzyme is in the non-phosphorylated active form, NR activation state was determined by comparing NR activity measured in the presence of Mg2+ with the activity measured in the presence of EDTA (total activity) (Supplementary Table S3; Lillo et al., 2003). NR activation state was only slightly increased in DOE-11 seedlings after DEX treatment for 3 and 5 days, while DEX treatment of WT seedlings did not change the activation state. These results suggest that the moderate increase (~25%) in total NR activity observed in DEX-treated DOE-11 seedlings is mainly caused by increased NR protein levels. NiR activities increased and decreased in DOE-11 and RNAi-16 lines, respectively, upon DEX treatment (Fig. 5B). These results suggest that the increased NR and NiR activities contributed to enhanced growth of the transgenic plants. GS activity, however, was slightly reduced in DOE-11 seedlings after 3 and 5 days of DEX treatment, while the activity significantly increased in DEX-treated RNAi-16 seedlings (Fig. 5C). GS functions as the major assimilatory enzyme for ammonium, but it also plays a role in re-assimilation of recycled nitrogen released from protein degradation during senescence (Pageau et al., 2006; Xu et al., 2012). Increased GS activity in the RNAi-16 lines is consistent with the early senescence phenotype and increased glutamine accumulation observed in the plants (Ahn et al., 2011).
Fig. 5.
Modulation of metabolic enzyme activities in DEX-inducible Tap46 overexpression and Tap46 RNAi (RNAi-16) plants. Enzyme activities were measured using protein fractions prepared from WT, DOE-11, and RNAi-16 seedlings that were grown in soil for 2 weeks and sprayed with ethanol [(–)DEX] or 30 μM DEX [(+)DEX] for 3 or 5 days. Data points represent mean ± SD of three experiments. Asterisks denote statistical significance of the differences between (–)DEX and (+)DEX samples on each day: *, P ≤ 0.05; **, P ≤ 0.01. (A) NR activities. (B) NiR activities. (C) GS activities.
Stimulated TOR signalling pathway in Tap46 DOE plants
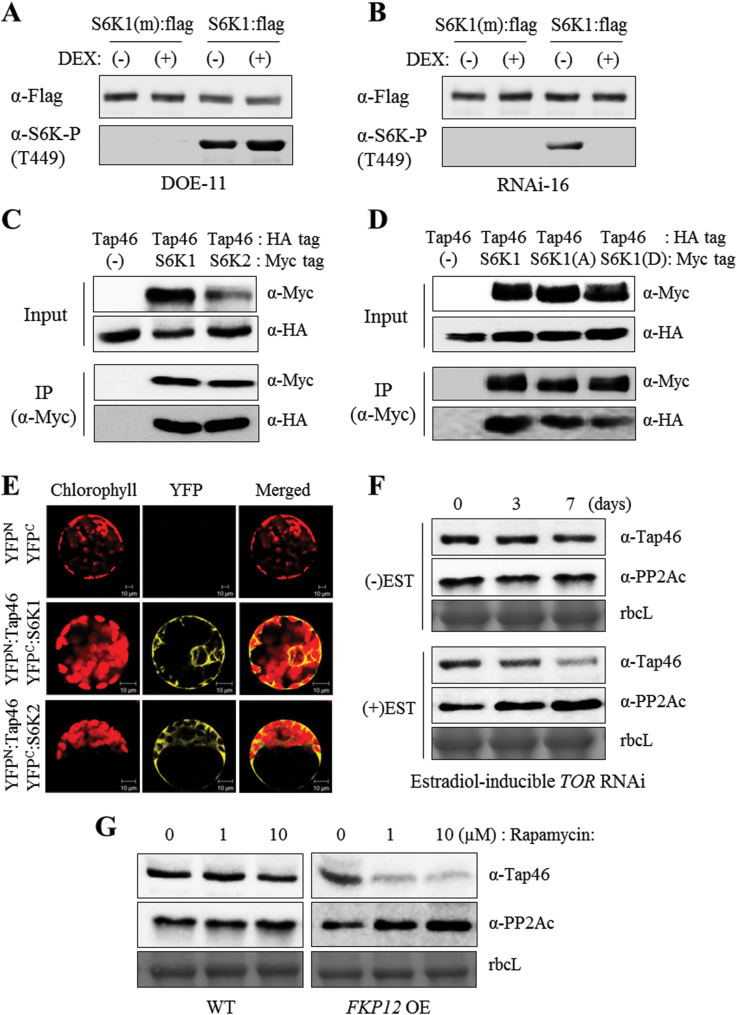
Mammalian TOR (mTOR) regulates cap-dependent translation machinery through phosphorylation of its downstream substrate S6K and 4E-BP in response to nutrients, energy, and environmental stresses (Ma and Blenis, 2009). Inactivation of mTOR kinase by rapamycin leads to dephosphorylation of S6K, and PP2A was reported to interact with and dephosphorylate S6K in mammalian cells (Peterson et al., 1999; Westphal et al., 1999). In Arabidopsis, TOR activation triggers phosphorylation of S6K at specific sites (S6K1 at T449 and S6K2 at T455), suggesting that S6K is a true substrate of TOR kinase in plants (Schepetilnikov et al., 2011; Xiong and Sheen, 2012). Since Tap46, a regulatory subunit of PP2A, functions as a downstream effector of plant TOR (Ahn et al., 2011), we investigated whether Tap46 overexpression or Tap46 RNAi affects S6K phosphorylation (Fig. 6A, B). DEX-inducible Tap46 overexpression (DOE-11) and Tap46 RNAi (RNAi-16) lines were grown for 2 weeks in MS medium and transferred to new MS medium containing ethanol [(–)DEX] or 10 µM DEX [(+)DEX] for 3 days. Flag-tagged WT S6K1 or mutant S6K1 [S6K1(m); T449A] were transiently expressed in protoplasts isolated from DOE-11 and RNAi-16 seedlings as previously described (Xiong and Sheen, 2012). Immunoblotting of the protein extracts with anti-Flag and anti-phospho-S6K antibodies, followed by quantification of the immunoblot band intensities using ImageJ (http://imagej.nih.gov/ij/), demonstrated that the levels of phosphorylated S6K1 at T449 increased and decreased in the DOE-11 and RNAi-16 plants, respectively, as compared with total S6K1 protein levels (Fig. 6A, B; Supplementary Figure S3A). The T449A mutation in S6K1 completely abrogated the phosphorylation (Figure 6A, B). Therefore, Tap46 expression positively correlated with S6K phosphorylation, supporting its function as a positive effector of the TOR signalling pathway.
Fig. 6.
Determination of the relationship between Tap46 and the TOR signalling pathway. (A) Phosphorylation of S6K1 and its mutant in DOE-11 seedlings upon DEX treatment. Flag-tagged WT S6K1 or mutant S6K1 [S6K1(m); T449A, non-phosphorylated form] were transiently expressed in protoplasts. Western blot analyses were performed with anti-Flag antibodies to detect total S6K1 protein and with anti-S6K1-P(T449) antibodies to detect S6K1 phosphorylated at T449. (B) Phosphorylation of S6K1 and its mutant in DEX-inducible Tap46 RNAi-16 seedlings upon DEX treatment. (C) Co-immunoprecipitation. Protein extracts were prepared from N. benthamiana leaves that expressed HA:Tap46 with S6K1:Myc or S6K2:Myc fusion proteins. After immunoprecipitation (IP) with anti-Myc antibodies, co-immunoprecipitated HA:Tap46 was detected by immunoblotting with anti-HA antibodies. To check IP efficiency, the precipitated fractions were also immunoblotted with anti-Myc antibodies (input). (D) Co-immunoprecipitation of HA:Tap46 with S6K1:Myc, S6K1(A):Myc, or S6K1(D):Myc fusion proteins. S6K1(T449A) is a non-phosphorylated form and S6K1(T449D) is a phospho-mimetic form. (E) Visualization of interactions of Tap46 with S6K1 and S6K2 using bimolecular fluorescence complementation (BiFC). YFPN:Tap46 was expressed together with YFPC:S6K1 or YFPC:S6K2 fusion proteins in N. benthamiana leaves by agroinfiltration for confocal laser scanning microscopy. (F) Immunoblot analysis to detect cellular Tap46 and PP2Ac protein levels in TOR RNAi lines. Thirty micrograms of total protein isolated from Arabidopsis estradiol-inducible TOR RNAi seedlings after 3 and 7 days of ethanol (–EST) or 10 µM estradiol (+EST) treatment were subjected to immunoblotting with anti-Tap46 and anti-PP2Ac antibodies. Coomassie-stained Rubisco large subunit (rbcL) is shown as a loading control. (G) Immunoblot analysis to detect cellular Tap46 and PP2Ac protein levels in WT and Arabidopsis FKP12 overexpression lines upon rapamycin treatment. Seedlings were treated with 1 or 10 µM rapamycin for 24 hours prior to immunoblotting. This figure is available in colour at JXB online.
To further examine the relationship between Tap46 and S6K, we examined the interaction of Tap46 with S6K using co-immunoprecipitation and BiFC (Fig. 6C–E). For co-immunoprecipitation, HA-fused Tap46 (HA:Tap46) was co-expressed with Myc-fused S6K1 (S6K1:Myc) or S6K2 (S6K2:Myc) in Nicotiana benthamiana leaves by agroinfiltration (Fig. 6C). Protein expression was detected by immunoblotting with anti-HA and anti-Myc antibodies (Input). The S6K1:Myc and S6K2:Myc proteins were immunoprecipitated from leaf extracts using anti-Myc antibodies. Following SDS-PAGE, western blotting was performed with anti-Myc antibodies to detect the immunoprecipitated S6K proteins and then with anti-HA antibodies to detect HA:Tap46 as a co-immunoprecipitant. As shown in Fig. 6C, HA:Tap46 was co-immunoprecipitated at similar levels by S6K1:Myc and S6K2:Myc, suggesting that Tap46 can interact with both S6K1 and S6K2. As a control, when expressed alone, HA:Tap46 was not immunoprecipitated by anti-Myc antibodies (Fig. 6C). To determine the effect of S6K1 phosphorylation on its association with Tap46, a similar co-immunoprecipitation assay was performed (Fig. 6D). HA:Tap46 co-expressed with Myc-fused WT S6K1, S6K1(T449A; non-phosphorylated form), or S6K1(T449D; phospho-mimetic form). S6K1:Myc protein and its mutants were immunoprecipitated from leaf extracts by anti-Myc antibodies. Immunoprecipitants were analysed by western blotting with anti-Myc and anti-HA antibodies. HA:Tap46 was co-immunoprecipitated at similar levels regardless of the S6K1 form, indicating that S6K1 may interact with Tap46 using a surface that excludes the T449 site (Fig. 6D).
We next performed BiFC assays to determine the interaction of Tap46 with S6K1 and S6K2 (Fig. 6E; Supplementary Figure S4). For BiFC, the pSPYNE-Tap46 construct was agroinfiltrated together with the pSPYCE-S6K1 or pSPYCE-S6K2 constructs into N. benthamiana leaves to co-express YFPN:Tap46 and YFPC:S6K1/2 fusion proteins. After 48 hours, mesophyll protoplasts prepared from the infiltrated leaves or epidermal cells of the infiltrated leaves were observed by confocal laser scanning microscopy. Co-expression of YFPN:Tap46 and YFPC:S6K1, and of YFPN:Tap46 and YFPC:S6K2, both resulted in strong cytosolic YFP fluorescence (Fig. 6E; Supplementary Figure S4). As a control, no fluorescence was detected in the leaf protoplasts co-expressing YFPN and YFPC (Fig. 6E), and in the epidermal cells co-expressing YFPN and YFPC, YFPN:Tap46 and YFPC, YFPN and YFPC:S6K1, or YFPN and YFPC:S6K2 (Supplementary Figure S4). Previously, GFP:S6K2 fusion protein that was transiently expressed in tobacco BY-2 cells by particle bombardment was mainly localized to the nucleus, but a minor portion was present in the cytosol (Mahfouz et al., 2006). We observed that GFP:S6K2 was localized to both the nucleus and the cytosol in N. benthamiana leaves (Supplementary Figure S5). These analyses suggest that Tap46 interacts with S6K1 and S6K2 predominantly in the cytosol.
We investigated how the protein levels of Tap46 and its partner, PP2Ac, are modulated upon TOR inactivation. Estradiol-inducible Arabidopsis TOR RNAi effectively downregulated TOR protein accumulation, leading to inhibition of seedling growth and phosphorylation of S6K1 and S6K2 (Xiong and Sheen, 2012). Furthermore, transgenic Arabidopsis plants overexpressing Arabidopsis FKP12 exhibited significantly enhanced sensitivity to rapamycin, a drug that inactivates TOR (Xiong and Sheen, 2012). These strategies were employed to modulate TOR activity. Estradiol-inducible TOR RNAi lines were grown for 10 days in MS medium and then transferred to new MS medium containing ethanol (–EST) or 10 µM estradiol (+EST) for further growth for 3 and 7 days. Immunoblotting was performed with the protein extracts using anti-Tap46 and anti-PP2Ac antibodies, and the immunoblot band intensities were measured using ImageJ. Protein levels of Tap46 and PP2Ac decreased and increased, respectively, in the TOR RNAi seedlings following estradiol treatment, whereas the control Rubisco large subunit (rbcL) levels remained constant (Fig. 6F; Supplementary Figure S3B). Thus, protein accumulation of Tap46 and PP2Ac are positively and negatively regulated, respectively, by TOR activity. Treatment of the FKP12 overexpression seedlings with 1 and 10 µM rapamycin also resulted in a progressive decrease and increase of Tap46 and PP2Ac protein levels, respectively (Fig. 6G; Supplementary Figure S3C). These results provide evidence that Tap46 and PP2A function as positive and negative effectors of TOR signalling, respectively, in plants.
Real-time quantitative RT-PCR analysis of gene expression
Transcriptome profiling of WT and tor mutant seedlings after a 2-hour glucose treatment at the photoautotrophic transition point identified numerous target genes of the glucose-TOR pathway in Arabidopsis (Xiong et al., 2013). The upregulated genes included those involved in root meristem activation, the cell cycle, DNA synthesis, ribosome biogenesis, protein translation, anabolic processes, cell wall biosynthesis, protein folding, and nitrate and lipid transport. The repressed genes encoded metabolic enzymes in catabolic pathways such as β-oxidation and the glyoxylate cycle, and proteins involved in protein/amino acid/lipid degradation or autophagy (Xiong et al., 2013). Comparative transcriptome analysis of rapamycin-treated WT and BP12-expressing Arabidopsis lines using RNA-seq also revealed a large number of differentially expressed genes (Ren et al., 2012). In rapamycin-treated BP12 lines, the downregulated genes were associated with anabolic pathways, such as cell wall biosynthesis, photosynthesis, and inorganic nutrient transport, whereas upregulated genes were related to catabolic pathways and stress pathways (Ren et al., 2012). In addition, TOR silencing by estradiol-inducible artificial microRNA resulted in transcript changes of the genes involved in the cell cycle, cell wall modification, stress response, senescence, and primary and secondary metabolism, accompanied by major changes in metabolite profiles (Caldana et al., 2013).
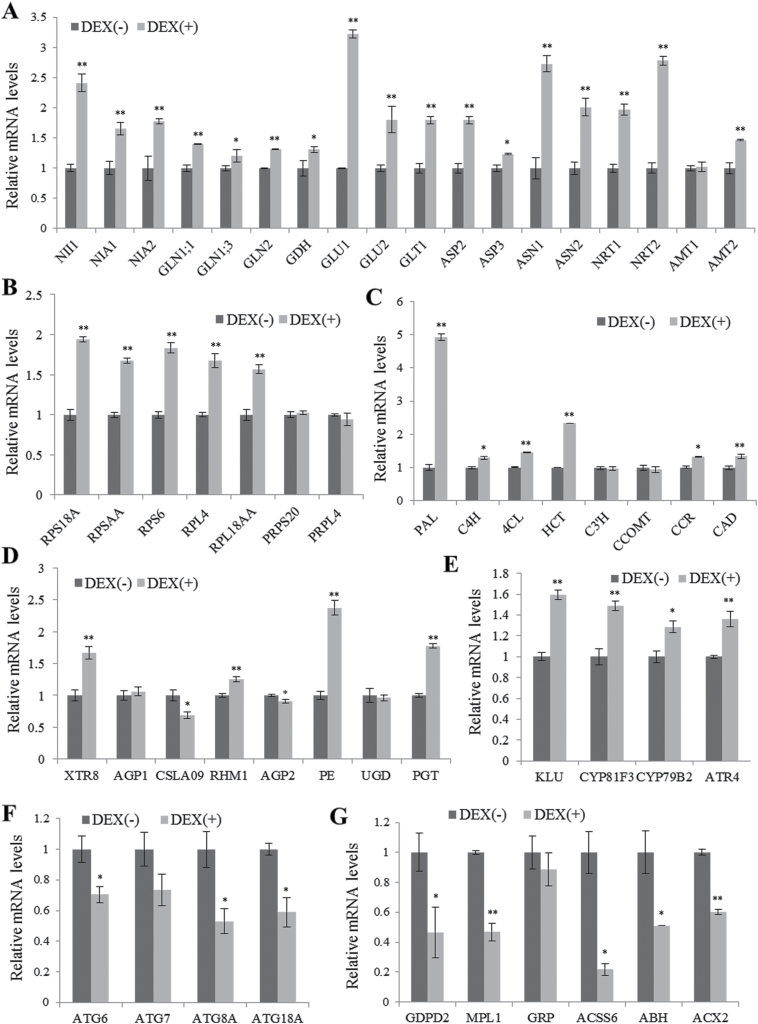
To examine changes in gene expression upon induction of Tap46 overexpression, real-time quantitative RT-PCR was performed using total RNA from DOE-11 seedlings after 3 days of ethanol [(–)DEX] or DEX [(+)DEX] treatment (Fig. 7; Supplementary Table S2). The focus of this analysis was on genes associated with nitrogen metabolism, ribosome biogenesis, lignin biosynthesis, cell wall biogenesis, cytochrome 450, autophagy, and lipid degradation, of which expression was modulated by the TOR pathway (Xiong et al., 2013). Among nitrogen metabolism genes, NII1 (nitrite reductase), GLU1 (glutamate synthase), ASN1 (asparagine synthetase), and NRT2 (nitrate transporter) genes were highly upregulated, suggesting increased nitrogen assimilation in (+)DEX seedlings (Fig. 7A). A few nuclear-encoded ribosomal protein genes were also upregulated upon DEX treatment, suggesting enhanced ribosome biogenesis processes, while plastid-encoded ribosomal protein genes (PRPS20 and PRPL4) were not affected (Fig. 7B). Among the lignin biosynthetic genes, transcript levels of PAL (phenylalanine ammonia-lyase), the first gene of the monolignol biosynthetic pathway, and HCT (hydroxycinnamoyl-CoA:shikimate hydroxylcinnamoyl transferase) significantly increased upon DEX treatment (Fig. 7C). Out of eight genes coding for synthesis and modification of cell walls, PE showed significantly increased transcript levels upon DEX treatment, while XTR8 and PGT were moderately induced (Fig. 7D). The transcript levels of four cytochrome 450 genes, KLU, CYP81F3, CYP79B2, and ATR4, were slightly higher in DEX-treated seedlings than in (–)DEX samples (Fig. 7E). KLU (CYP78A5) acts in the generation of a mobile growth-promoting signal, while CYP81F3, CYP79B2, and ATR4 (CYP83B1) are involved in auxin metabolism (Bak et al., 2011). In contrast, autophagy and lipid degradation-related gene transcripts were reduced in DEX-treated seedlings (Fig. 7F, G). These upregulated and downregulated genes in Tap46 overexpression seedlings mimic those induced by TOR activation (Xiong et al., 2013), further supporting the role of Tap46 in the TOR pathway.
Fig. 7.
Real-time quantitative RT-PCR analysis of gene expression in DOE-11 seedlings. DOE-11 seedlings grown on MS medium for one week were treated with ethanol [(–)DEX] or 10 µM DEX [(+)DEX] for 3 days for real-time quantitative RT-PCR. Data points represent mean ± SD of three experiments. Asterisks denote statistical significance of the differences between the (–)DEX and (+)DEX samples: *, P ≤ 0.05; **, P ≤ 0.01. (A) Nitrogen metabolism-related genes. (B) Ribosomal protein genes. (C) Lignin biosynthesis genes. (D) Cell wall biosynthesis genes. (E) Cytochrome 450 genes. (F) Autophagy genes. (G) Lipid degradation-related genes.
Discussion
Organ growth in plants is controlled by both genetic factors and environmental conditions that coordinate cell division and cell expansion in a spatial and temporal manner (Horiguchi et al., 2006; Anastasiou and Lenhard, 2007; Krizek, 2009). Arabidopsis leaf growth consists of proliferation, expansion, and maturation phases. In the proliferation phase, cell division and expansion occur simultaneously. In the expansion phase, cells cease to divide, but continue to expand. In the maturation phase, cells no longer divide or expand (De Veylder et al., 2001; Sonoda et al., 2009; Gonzalez et al., 2010). Genetic determinants of organ growth have been identified, and any perturbation in gene expression of one of these determinants may affect final organ size (Busov et al., 2008). Among the determinants, the plant hormones auxin and brassinosteroid (BR) are important plant growth regulators that stimulate both cell division and expansion. ARGOS and AINTEGUMENTA encode transcription factors involved in the auxin signalling pathway. Loss-of-function mutations of either gene result in smaller organs, while overexpression results in plant enlargement (Elliott et al., 1996; Mizukami and Fischer, 2000; Hu et al., 2003). ARF2, an auxin response factor, functions as a negative regulator of organ growth: arf2 mutant plants exhibit larger seeds, embryos, leaves, and flowers (Schruff et al., 2006). ARF2 mediates the crosstalk between auxin and BR signalling pathways that act synergistically in cell elongation (Vert et al., 2008). In addition, overexpression of gibberellin metabolism and response genes, TCP (TB1, CYC, and PCF) domain transcription factors, or growth-regulating factors (GRFs), also result in increased organ size (Busov et al., 2008).
The TOR pathway regulates cell growth in response to nutrient status and stress, thus linking final organ size and environmental conditions. Arabidopsis plants that overexpress TOR exhibit larger leaves and longer roots (Deprost et al., 2007), although expression of even higher levels of TOR results in pleiotropic defects in plant development (Ren et al., 2012). In contrast, RNAi of TOR or mutations of LST8/GβL, a partner of TOR, results in retarded seedling growth and decreased organ size (Deprost et al., 2007; Moreau et al., 2012; Xiong and Sheen, 2012). In this study, overexpression of Tap46 increased final organ size, including leaves, siliques, and seeds, without any negative pleiotropic effects under normal growth conditions. Analysis of cell number and cell area revealed that increased leaf size of Tap46 overexpression plants was mainly due to increased cell size, although cell number also increased to a small degree (Fig. 2). These results suggest that Tap46 plays a role in the regulation of both cell expansion and cell proliferation.
Xiong and Sheen (2012) recently reported a sensitive assay for monitoring endogenous TOR activity based on S6K phosphorylation. They found that TOR kinase phosphorylates the Thr-449 and Thr-455 residues of S6K1 and S6K2, respectively, in fully differentiated leaf cells. Additionally, either inactivation of TOR by rapamycin or TOR silencing by RNAi abrogated S6K phosphorylation. Considering that S6K is a key substrate and mediator of TOR kinase in yeast and mammals (Wullschleger et al., 2006; Ma and Blenis, 2009), the TOR-S6K1 paradigm appears to be conserved in plants as well. Furthermore, the phytohormone auxin triggers TOR activation leading to S6K1 phosphorylation on Thr-449 and efficient loading of uORF mRNAs onto polysomes, directly connecting the plant TOR/S6K pathway with translation (Schepetilnikov et al., 2013). Here, Tap46 overexpression and Tap46 RNAi increased and decreased S6K1 phosphorylation on Thr-449, respectively, supporting a role of Tap46 as a positive regulator of the TOR pathway (Fig. 6A, B).
Consistently, Tap46 protein levels decreased following estradiol-inducible TOR- RNAi or by rapamycin treatment (Fig. 6F, G). In contrast, PP2Ac protein levels increased when TOR activity was compromised by either method (Fig. 6F, G). Tap46 interacts with and sequesters PP2A catalytic subunits. Thus, Tap46 can modulate the formation of PP2A holoenzyme complexes comprised of the catalytic C subunit, the scaffolding A subunit, and the regulatory B subunit (B, B’, and B’’). Therefore, a decrease in Tap46 protein and an increase in PP2Ac in a cell may result in increased formation of PP2A holoenzyme complexes, subsequently stimulating cellular dephosphorylation activity. In yeast, Tap42 phosphorylation by TOR kinase correlates with Tap42 association with phosphatase catalytic subunits (Jiang and Broach, 1999). TOR inactivation induces dephosphorylation of Tap42 and the release of phosphatases from Tap46, followed by increased dephosphorylation of downstream targets of TOR signalling, supporting the hypothesis that Tap42 functions as a phosphatase inhibitor (Jacinto et al., 2001; Dűvel and Broach, 2004). However, mammalian α4 associated with different phosphatase catalytic subunits exhibits opposing allosteric effects on phosphatase activity depending on the substrate, suggesting that α4 can function as a phosphatase activator (Prickett and Brautigan, 2006; Nien et al., 2007). The PP2A family of phosphatases control diverse cellular processes, including cell cycle control, transcription, translation, and metabolism (Janssens and Goris, 2001; Farkas et al., 2007). This study suggests that Tap46 mainly acts as an inhibitor of PP2A activity in plants, but detailed mechanisms of Tap46 modulation of PP2A activity, both direct and indirect, require further study.
Using BiFC and co-immunoprecipitation, we demonstrated that Tap46 interacts with S6K1 and S6K2 in Arabidopsis (Fig. 6C-E). In mammalian cells, S6K is dephosphorylated by PP2A and interacts with PP2A (Ferrari et al., 1993; Peterson et al., 1999; Westphal et al., 1999). Furthermore, S6K1 is associated with the heterodimer of α4 and PP2Ac in mouse spleen B cells and α4 modulates S6K activity through negative regulation of PP2A activity upon B-cell activation (Yamashita et al., 2005). Whether Tap46 negatively regulates PP2Ac on plant S6Ks in a similar manner remains to be tested. Tap46 has multiple phosphatase partners, including catalytic subunits of PP2A, PP4, and PP6, and is a major regulator of cellular PP2A activities in plant cells (Ahn et al., 2011; Hu et al., 2014). Recently, Tap46 was shown to play a role in ABA-regulated seed germination through interaction with the ASCISIC ACID INSENSITIVE5 (ABI5) transcription factor in Arabidopsis (Hu et al., 2014). Constitutive overexpression of Tap46 and weak alleles of the tap46 mutation resulted in increased and decreased ABA sensitivity during seed germination, respectively. Tap46-overexpressing seeds contained increased transcript levels of late embryogenesis-abundant genes, AtEm1, AtEM6, and LeaD34, of which AtEm1 and AtEM6 are known to be ABI5-regulated genes (Hu et al., 2014). Furthermore, expression of stress-inducible genes such as RD29A, RD29B, and NCED3 was induced at higher levels in Tap46-overexpressing plants upon ABA treatment compared with WT plants, suggesting that Tap46 acts as a positive regulator of ABA-regulated gene expression (Hu et al., 2014). We observed in this study that the Tap46 OE-5 and OE-10 seeds are more resistant to environmental stresses such as high temperature and high humidity, based on the survival rate and germination rate after stress treatment (Fig. 3E, F). The enhanced resistance of the Tap46 overexpression seeds may be caused by their enhanced ABA sensitivity for regulation of ABA downstream genes under stress conditions as well as during seed maturation. It is noteworthy that Hu et al. (2014) did not observe enhanced growth in their Tap46 overexpression Arabidopsis lines, while immunoblotting with anti-Tap46 antibodies detected ~40% increases in the endogenous level of Tap46 proteins in the plants. We detected a ~4-fold increase in cellular Tap46 protein levels in seedlings of the overexpression lines (Fig. 1G), which may be responsible for the phenotype observed in this study.
Tap46 overexpression caused upregulation of a subset of genes related to nitrogen metabolism, ribosome biogenesis, and lignin biosynthesis, and downregulation of several genes related to autophagy and lipid degradation. This pattern of gene expression correlates with the pattern caused by TOR activation (Xiong et al., 2013), supporting a functional link to the TOR pathway. Transcriptome analysis of transgenic Arabidopsis lines overexpressing AVP1, GRF1, JAW, BRI1, or GA20OX1, all of which exhibit increased leaf size due to different mechanisms, previously showed that all five genes affect the transcriptome distinctly, suggesting that enhanced organ growth is controlled by multiple, possibly independent, pathways (Gonzalez et al., 2010). Nonetheless, kinematic analysis of leaf growth revealed that increased cell proliferation is mainly responsible for leaf-size enlargement in all of the five overexpression lines (Gonzalez et al., 2010). The leaf enlargement in Tap46 overexpression seedlings is predominantly caused by increased cell expansion (Fig. 2); also the Tap46 overexpression-induced gene expression profile, although analysed on a limited scale, did not seem to match any of the five lines. These results suggest that the Tap46-mediated growth-regulating pathway may function independently in organ-size regulation.
Supplementary material
Supplementary data can be found at JXB online.
Supplementary Table S1. Data points for the kinematic analysis shown in Fig. 2.
Supplementary Table S2. PCR primers used in this study.
Supplementary Table S3. Measurement of NR activation state.
Supplementary Figure S1. Representative epidermal cells of the first leaves of WT, OE-5, and OE-10 plants at 4 to 25 DAC.
Supplementary Figure S2. Sizes of the epidermal cells at 2 DAC.
Supplementary Figure S3. Quantification of the band intensity in western blots.
Supplementary Figure S4. Control experiments for BiFC.
Supplementary Figure S5. Subcellular localization of S6K2 in N. benthamiana leaves.
Supplementary Methods S1. Materials and methods.
Funding
This research was supported by the Cooperative Research Programme for Agriculture Science & Technology Development [Project numbers PJ009079 (PMBC) and PJ00952503 (SSAC)] from the Rural Development Administration of the Republic of Korea. CSA was supported by the Research Fellow Scholarship (NRF-2013R1A1A2062026) from the National Research Foundation of the Republic of Korea.
Supplementary Material
Acknowledgements
The authors wish to thank Dr Jen Sheen (Harvard Medical School, Boston, MA, USA) for providing seeds from the FKP12 overexpression lines and the estradiol-inducible TOR RNAi lines, and the S6K constructs for transfection assays.
References
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. 2011. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. The Plant Cell 23, 1185–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou E, Lenhard M. 2007. Growing up to one’s standard. Current Opinion in Plant Biology 10, 63–69. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. The Plant Journal 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, and Werck-Reichhart D. 2011. Cytochromes P450. Arabidopsis Book 9, e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Henriques R, Magyar Z. 2013. TOR tour to auxin. The EMBO Journal 32, 1069–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov VB, Brunner AM, Strauss SH. 2008. Genes for control of plant stature and form. New Phytologist 177, 589–607. [DOI] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. 2013. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana . The Plant Journal 73, 897–909. [DOI] [PubMed] [Google Scholar]
- Chen J, Peterson RT, Schreiber SL. 1998. Alpha 4 associates with protein phosphatases 2A, 4, and 6. Biochemical and Biophysical Research Communications 247, 827–832. [DOI] [PubMed] [Google Scholar]
- Deprost D, Truong H-N, Robaglia C, Meyer C. 2005. An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochemical and Biophysical Research Communications 326, 844–850. [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C. 2007. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports 8, 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D. 2001. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis . The Plant Cell 13, 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Broach JR. 2004. The role of phosphatases in TOR signaling in yeast. Current Topics in Microbiology and Immunology 279, 19–38. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. 1996. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C. 2007. Arabidopsis PPP family of serine/threonine phosphatases. Trends in Plant Science 12, 169–176. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Pearson RB, Siegmann M, Kozma SC, Thomas G. 1993. The immunosuppressant rapamycin induces inactivation of p70s6k through dephosphorylation of a novel set of sites. The Journal of Biological Chemistry 268, 16091–16094. [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, et al. 2010. Increased leaf size: different means to an end. Plant Physiology 153, 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes and Development 18, 1926–1945. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. 2006. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana . Journal of Plant Research 119, 37–42. [DOI] [PubMed] [Google Scholar]
- Hu R, Zhu Y, Shen G, Zhang H. 2014. TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis . Plant Physiology 164, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua N-H. 2003. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. The Plant Cell 15, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Molecular Cell 8, 1017–1026. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochemical Journal 353, 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Broach J. 1999. Tor proteins and protein phosphatase 2A reciprocally regulated Tap42 in controlling cell growth in yeast. The EMBO Journal 18, 2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M, Fox CJ, Mu J, Solt L, Xu A, Cinalli RM, Birnbaum MJ, Lindsten T, Thompson CB. 2004. The PP2A-associated protein alpha4 is an essential inhibitor of apoptosis. Science 306, 695–698. [DOI] [PubMed] [Google Scholar]
- Krizek BA. 2009. Making bigger plants: key regulators of final organ size. Current Opinion in Plant Biology 12, 17–22. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Lee H-S, Song J-Y, Jung YJ, Reinbothe S, Park Y-I, Lee SY, Pai H-S. 2013. Cell Growth Defect Factor1/CHAPERONE-LIKE PROTEIN OF POR1 plays a role in stabilization of light-dependent protochlorophyllide oxidoreductase in Nicotiana benthamiana and Arabidopsis . The Plant Cell 25, 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C, Lea US, Leydecker MT, Meyer C. 2003. Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in constitutive activation of the enzyme in vivo and nitrite accumulation. The Plant Journal 35, 566–573. [DOI] [PubMed] [Google Scholar]
- Loewith R. 2010. 9–TORC1 Signaling in budding yeast. The Enzymes 27, 147–175. [Google Scholar]
- Ma XM, Blenis J. 2009. Molecular mechanisms of mTOR-mediated translational control. Nature Reviews Molecular Cell Biology 10, 307–318. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DPS. 2006. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. The Plant Cell 18, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proceedings of the National Academy of Sciences, USA 99, 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. 2000. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences, USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, et al. 2012. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the Target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. The Plant Cell 24, 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien WL, Dauphinee SM, Moffat LD, Too CK. 2007. Overexpression of the mTOR alpha4 phosphoprotein activates protein phosphatase 2A and increases Stat1alpha binding to PIAS1. Molecular and Cellular Endocrinology 263, 10–17. [DOI] [PubMed] [Google Scholar]
- Oge L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. 2008. Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis . The Plant Cell 20, 3022–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageau K, Reisdorf-Cren M, Morot-Gaudry JF, Masclaux-Daubresse C. 2006. The two senescence-related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves. Journal of Experimental Botany 57, 547–557. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. 1999. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12–rapamycin associated protein. Proceedings of the National Academy of Sciences, USA 96, 4438–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Brautigan DL. 2006. The alpha4 regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. The Journal of Biological Chemistry 281, 30503–30511. [DOI] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, et al. 2012. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis . The Plant Cell 24, 4850–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. The EMBO Journal 32, 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA. 2011. Viral factor TAV recruits TOR/S6K1 signaling to activate reinitiation after long ORF translation. The EMBO Journal 30, 1343–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. 2006. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. [DOI] [PubMed] [Google Scholar]
- Seo YS, Kim EY, Kim WT. 2011. The Arabidopsis sn-1-specific mitochondrial acylhydrolase AtDLAH is positively correlated with seed viability. Journal of Experimental Botany 62, 5683–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. 2014. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nature Reviews Molecular Cell Biology 15, 155–162. [DOI] [PubMed] [Google Scholar]
- Sonoda Y, Sako K, Maki Y, Yamazaki N, Yamamoto H, Ikeda A, Yamaguchi J. 2009. Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. The Plant Journal 60, 68–78. [DOI] [PubMed] [Google Scholar]
- Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C. 2007. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biology 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Cohen A, Hall MN. 2009. TOR signaling in invertebrates. Current Opinion in Cell Biology 21, 825–836. [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proceedings of the National Academy of Sciences, USA 105, 9829–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal RS, Coffee RL, Marotta A, Pelech SL, Wadzinski BE. 1999. Identification of kinase-phosphatase signaling modules composed of p70 S6 kinase-protein phosphatase 2A (PP2A) and p21-activated kinase-PP2A. The Journal of Biological Chemistry 274, 687–692. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J, 2013. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2012. Rapamycin and glucose-Target of Rapamycin (TOR) protein signaling in plants. The Journal of Biological Chemistry 287, 2836–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2013. Moving beyond translation: Glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle 12, 1989–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Inui S, Maeda K, Hua DR, Takagi K, Sakaguchi N. 2005. The heterodimer of a4 and PP2Ac is associated with S6 kinase1 in B cells. Biochemical and Biophysical Research Communications 330, 439–445. [DOI] [PubMed] [Google Scholar]
- Yan G, Shen X, Jiang Y. 2006. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. The EMBO Journal 25, 3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Reviews Molecular Cell Biology 12, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.