Highlight
How the polyamine cadaverine alters plant development is poorly understood. Here it is shown that natural variation in ORGANIC CATION TRANSPORTER 1 affects root length sensitivity to exogenous cadaverine.
Key words: Arabidopsis, cadaverine, natural variation, polyamines, roots, transporter.
Abstract
Polyamines, including cadaverine, are organic cations that affect numerous biological processes including transcription, translation, cell signalling, and ion channel activity. They often function in biotic and abiotic stress responses in plants. Because little is known about how plants respond to cadaverine, a quantitative natural variation approach was used to identify genetic factors that contribute to this response. Here it is shown that Arabidopsis thaliana accessions have varying root length responses to exogenous cadaverine: Cape Verde Islands (Cvi) was one of the most resistant accessions tested, whereas Landsberg erecta (Ler) was one of the most sensitive. Recombinant inbred lines, near isogenic lines, and a microarray were used to show that variation in ORGANIC CATION TRANSPORTER 1 (OCT1) is at least partially responsible for this difference. OCT1 expression was higher in Cvi than in Ler, and oct1 mutants were more sensitive to cadaverine than wild-type plants. In oct1 mutants transformed with an ectopic copy of OCT1 originating from either Cvi or Ler, the expression level of the transgene, not its accession, correlated with the cadaverine response. These results suggest that decreased OCT1 expression confers cadaverine sensitivity in some accessions.
Introduction
Polyamines are organic cations that are prevalent in both prokaryotes and eukaryotes. Putrescine, spermidine, and spermine are the most common polyamines in plants, although thermospermine and cadaverine have also been found in many species including Arabidopsis thaliana (Shevyakova et al., 2001; Knott et al., 2007). Polyamines exist both as free molecules and as conjugates to other small molecules, mainly hydroxycinnamic acids, or proteins (Bagni and Tassoni, 2001). Both free and conjugated polyamines are important for cellular processes including cell growth, cell division, transcription, translation, and cell death. Their positive charges allow for interactions with nucleic acids, lipids, and proteins, which affect the activities and stabilities of these molecules (for a review, see Hussain et al., 2011).
Polyamines accumulate in certain plant tissues under many abiotic stresses including drought, salt, osmotic, and temperature stress, which all result in metabolic reprogramming and modifications to growth (Cuevas et al., 2008; Alet et al., 2011; Wang et al., 2011). Polyamines may protect against these stresses in several ways, including by altering gene expression, increasing the activities of antioxidant enzymes, and altering the cellular accumulation of ions (Liu et al., 2000; Takahashi et al., 2003; Tang and Newton, 2005). They may also function as signals that regulate cross-talk between hormonal pathways during stress responses (Obata and Fernie, 2012). Though essential for proper development, and beneficial at higher levels during stress, catabolism of polyamines produces H2O2 (Tisi et al., 2011). Although it is toxic, H2O2 also diffuses to nearby tissues where it activates various responses (Neill, 2002). To balance these effects, polyamine synthesis, degradation, localization, and conjugation must be tightly controlled.
Five potential Arabidopsis POLYAMINE UPTAKE TRANSPORTER (PUT) proteins were identified based on sequence similarity to known polyamine transporters in Leishmania major and Trympanosoma cruzi (Mulangi et al., 2012a, b ). When expressed in the polyamine uptake-deficient Δagp2 Saccharomyces cerevisiae mutant (Aouida et al., 2005), these proteins partially restored sensitivity to the polyamine analogue paraquat as well as to putrescine and spermidine. They also partially restored putrescine and spermidine uptake into the cells (Mulangi et al., 2012a ). PAR1/PUT2 has subsequently been shown to localize to the Golgi where it is required for the subcellular localization of paraquat in the chloroplast (Li et al., 2013). RMV1/PUT3 has been shown to be plasma membrane associated and to function in paraquat, putrescine, spermidine, and spermine uptake in Arabidopsis (Fujita et al., 2012). However, cadaverine transport by these proteins was not tested.
Less is known about cadaverine compared with the other polyamines. It is synthesized from lysine and therefore not directly derived from the polyamine putrescine, unlike spermidine, spermine, and thermospermine. Cadaverine is produced by some soil bacteria and yeast, possibly contributing to rhizosphere–root signalling (Perrig et al., 2007; Cloete et al., 2009). Exogenous cadaverine affects root development in several ways: it increases adventitious root growth in pine trees, lateral root formation in soybeans, and root weight in rice (Gamarnik and Frydman, 1991; Niemi et al., 2002; Cassán et al., 2009), whereas it shortens primary roots in soybeans (Gamarnik and Frydman, 1991).
Cadaverine levels and localization change upon exposure to stress. This molecule accumulates dramatically upon oxidative stress in rape leaves and upon drought stress in peppers (Aziz et al., 1997; Sziderics et al., 2010). In the ice plant, salt stress causes cadaverine to accumulate in true leaves (Kuznetsov et al., 2007), and heat shock stimulates cadaverine transport away from the heated tissue (Shevyakova et al., 2001). Cadaverine also confers protection against stress. It induces stomatal closure, a common stress response, in fava beans (Liu et al., 2000), and it prevents decreases in root and shoot weight upon oxidative stress in rice (Cassán et al., 2009).
Despite its profound impacts on overall cellular homeostasis and plant growth under stressful conditions, the underlying mechanisms that explain how cadaverine alters plant growth and development are still unknown. To address this gap, natural variation in root length sensitivity to exogenous cadaverine was examined, and ORGANIC CATION TRANSPORTER 1 (OCT1, AT1G73220) was identified as an important quantitative trait locus (QTL) involved in this response.
Materials and methods
Plant materials
QTL analysis used the following stocks from the Arabidopsis Biological Resource Center (Columbus, OH, USA): Cape Verde Islands (Cvi; CS8580), Landsberg erecta-2 Ler-2; CS8581), and a population of Cvi/Ler recombinant inbred lines (RILs; CS22000). This population consists of 162 individual RILs that were genotyped at 293 marker loci (Alonso-Blanco et al., 1998). The near isogenic lines (NILs) LCN1-12, -15, -14, -21, -22, and -28 (N717058, N17060, N17061, N17067, N17068, and N17074) (Keurentjes et al., 2007) were obtained from the Nottingham Arabidopsis Stock Center (Nottingham, UK). An-1 (CS22626), Bs-1 (CS6627), Knox-18 (CS22567), Sq-8 (CS22601), Ull2-3 (CS22587), and Wei-0 (CS22622) seed came from the Arabidopsis Biological Resource Center. The oct1-1 mutant and oct1-1 [35Spro:OCT1WS] transgenic lines were provided by Christine Lelandais-Briere (Lelandais-Brière et al., 2007).
Growth conditions
The seeds were sterilized by washing them with 95% ethanol. Except where noted in the text, they were plated on half-strength buffered Linsmaier and Skoog medium containing micro- and macronutrients, vitamins, and 1.5% sucrose (Caisson Laboratories, North Logan, UT, USA) that was supplemented with 1.5% agar type E (Sigma-Aldrich, St. Louis, MO, USA) and polyamines (Sigma-Aldrich) as required. The seedlings were stratified for 2–8 d and grown in either a Conviron TC16 growth chamber or an Enconair (now BioChambers, Winnipeg, Manitoba, Canada) Controller 6000 growth chamber for each experiment. Both chambers were set at 22 °C and a 16h light/8h dark cycle, and the light intensity was 50–70 μmol m–2 s–1 and provided by cool-white fluorescent bulbs (Grainger, Lake Forest, IL, USA).
Root length measurements
Seedling images were taken from the backs of the plates using a scanner (Epson, Suwa, Nagano Prefecture, Japan). Root length was measured using the segmented line tool in NIH Image (publically available at http://rsb.info.nih.gov/nih-image/) or the Neuron J plugin (Meijering et al., 2004) for Image J (Schneider et al., 2012).
Microscopy
To calculate cell length, the cell walls were stained by transferring the seedlings to 10 μg ml–1 propidium iodide (Sigma-Aldrich) for ~20min. Imaging was conducted using a Zeiss LSM 510 Meta Confocal microscope (Zeiss, Thornwood, NY, USA), and cell length measurements were performed in Adobe Photoshop.
QTL mapping
Plants were grown at a 30 ° tilt for 6 d on media with or without cadaverine. A concentration of 50 μM cadaverine was used in trial 1, and 100 μM cadaverine was used in trial 2. The mean root length value on medium containing cadaverine was subtracted from the mean value on control medium and then divided by the mean on control medium. This number was subtracted from 1, and this ratio, along with genotypic data for each RIL (Alonso-Blanco et al., 1998), was entered into WinQTL Cartographer version 2.5 (Wang et al., 2010) and the r/qtl package in R (http://www.r-project.org). For WinQTL Cartographer, maps were created using the Kosambi map function. During composite interval mapping, the model incorporated a 0.5 step rate with a 1.0 cM window size and 10 control markers. LOD thresholds were determined by 1000 permutations. In r/qtl, the function ‘scanone’ was used for interval mapping, and ‘scantwo’ was used to generate the two-dimensional plot. Significance levels were determined by 1000 permutations, implementing the EM algorithm. Fine mapping of the NILs was performed using standard methods and the primers listed in Supplementary Table S1 available at JXB online.
RT–PCR and qRT-PCR
Approximately 24 seedlings were grown at a 30 ° backward tilt at staggered times for each biological replicate on medium containing no cadaverine unless otherwise indicated. Whole roots from 7-day-old seedlings were dissected and immediately frozen in liquid nitrogen. RNA was prepared using an RNeasy Plant Mini Kit (QIAGEN, Venlo, The Netherlands) and stored at –80 ºC until use, typically within a few days. RNA samples were treated with RQ1 DNase (Promega, Madison, WI, USA) according to the instructions. The ratios at absorbencies of 260 to 280 and at 260 to 230 as well as gel electrophoresis were used to verify RNA quality. Poor quality samples were not used for subsequent experiments. RNA concentrations were determined using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA).
For reverse transcription–PCR (RT–PCR), cDNA was made using the iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). For quantitative real-time PCR (qRT-PCR), cDNA synthesis and qRT–PCR were done simultaneously using a qScript™ One-Step qRT-PCR Kit (Quanta Biosciences, Gaithersburg, MD, USA). The samples were run on a LightCycler® 480 Real Time-PCR System (Roche Applied Science, Basel, Switzerland). Each 6 μl reaction contained RNA at a final concentration of 2ng μl–1 and each primer at a concentration of 0.65 μM. Other components were included at the concentrations defined by the kit. Control reactions without reverse transcriptase were included and did not yield any detectable expression. Cycling conditions were 49 ºC for 10min, 95 ºC for 5min, and 45 cycles of 95 ºC for 5 s, 55 ºC for 15 s, and 72 ºC for 10 s. Melt curves were used to verify specificity. Four technical replicates were performed for each sample for each biological replicate, and three biological replicates were performed. LinRegPCR was used to analyse the data (Ramakers et al., 2003).
The primers used to amplify OCT1 were: 1F (5′ GTGGCTGTT CCTTCCACACT 3′), 1R (5′ AAAGGCCGTGACGAAAGTTA 3′), 2F (5′ CATCCTCGACAGCGTATGAC 3′), 2R (5′ CCTTC CTAACGCAACTAGCA 3′), 3F (5′ TTTGGTGTTGCATCAGTG CT 3′), and 3R (5′ CGGAGCGTTTCGAGTTTCT 3′). 4R (5′ CGTGAAATGCGTGTTGAAAG 3′) was used to detect possible transcript that extended into the T-DNA region. Primers 3F and 3R as well as 5′ CCTCTTGGATACGCGGTTT 3′ and 5′ CAAGA AGCCATCGAGGAGAC 3′ were used for OCT1 qRT–PCR. Primers used to amplify the reference gene At1g58050 (Czechowski et al., 2005) were: 5′ CCATTCTACTTTTTGGCGGCT 3′ and 5′ TCAATGGTAACTGATCCACTCTGATG 3′. This reference gene was chosen due to its consistent expression and its similar expression level to OCT1 (Czechowski et al., 2005)
Transgenic constructs
Genomic DNA from Cvi and Ler was used to amplify the region extending from 2079 bases before the start codon through to 417 bases after the stop codon of OCT1. This region was cloned in between the attL1 and attL2 sites in the Gateway entry vector pENTR/D-TOPO (Life Technologies, Madison, WI, USA). An LR reaction was performed to transfer this region into the binary vector pMDC99 (Curtis and Grossniklaus, 2003; Xu and Li, 2008). The constructs were sequenced and transformed into plants using the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). T3 seedlings likely to all carry the transgene as determined by antibiotic resistance were used in all experiments (Harrison et al., 2006).
Results
Arabidopsis accessions vary in their root length responses to cadaverine
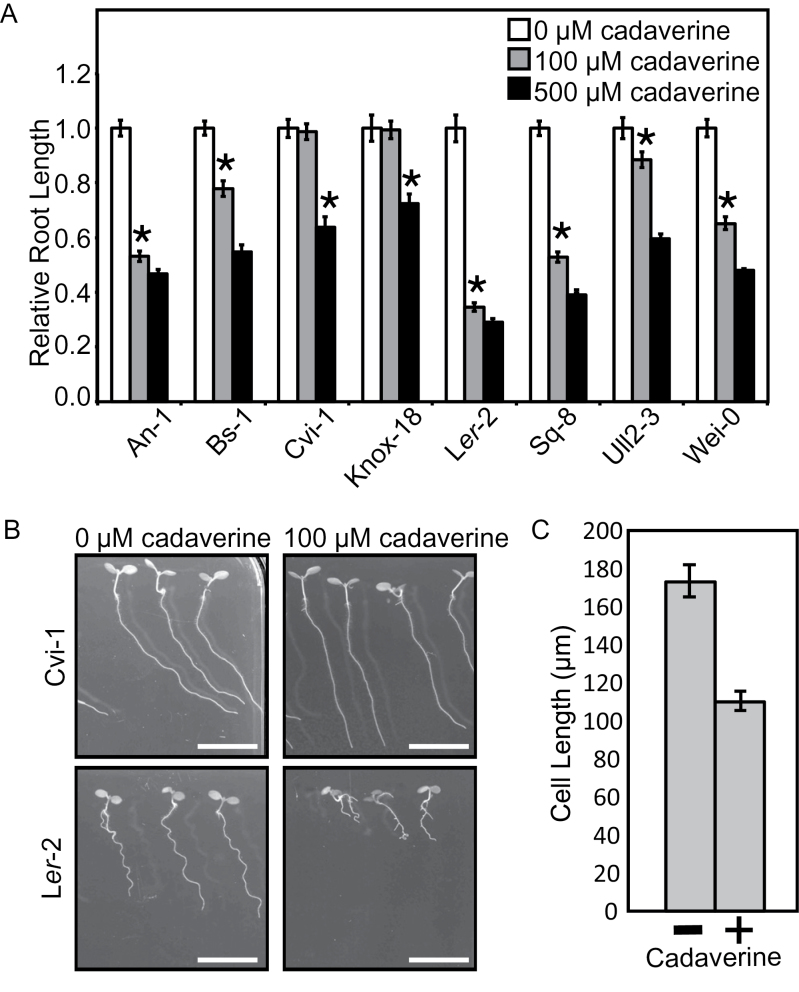
As a first step toward identifying genes involved in root growth responses to exogenous cadaverine, seedlings of eight Arabidopsis accessions were grown on media containing 0, 100, and 500 μM cadaverine. A decrease in root length between control and increasing cadaverine treatments was observed for all accessions tested (Fig. 1A). Alterations in root waving and skewing and an increase in adventitious and lateral root emergence were also noticed in most accessions (Supplementary Fig. S1 at JXB online). In terms of root length, Cvi-1 and Knox-18 were some of the most resistant accessions to cadaverine, whereas Ler-2 was one of the most sensitive (Fig. 1A, B).
Fig. 1.
Arabidopsis accessions show varying root length inhibition responses to exogenous cadaverine. (A) Seedlings were grown vertically for 3 d and then at a 30 ° backward tilt for another 3 d. Average root length on control medium was set to 1. Absolute average root lengths on control medium were 1.4, 1.6, 2.5, 2.1, 2.1, 2.3, 2.0, and 2.3cm in the accessions as shown from left to right. The asterisk indicates the first cadaverine concentration corresponding to a significantly shorter root compared with control medium (P<0.05, Student’s t-test). Error bars indicate ±SE, and n=12–27. (B) Images of Cvi-1 (top) and Ler-2 (bottom) seedlings growing on media without (left) and with (right) 100 μM cadaverine. The white scale bar is 1cm. (C) Average length of epidermal cells in the mature root zone from seedlings grown vertically for 7 d on either control medium or medium containing 100 μM cadaverine. Error bars indicate ±SE, and n=11–18.
To determine whether the effect of cadaverine on root length was due to reduced cell elongation or division, the length of the epidermal cells in the mature root zone of Ler seedlings grown on media containing 0 μM or 100 μM cadaverine was measured. It was found that although Ler roots were only ~35% as long on this concentration of cadaverine as on control medium (Fig. 1A), the average cell length of the seedlings on the medium containing cadaverine was ~64% of that of seedlings on control medium (Fig. 1C). This suggests that the reduction in root length is probably due to a combination of reduced cell elongation and division.
A QTL on chromosome 1 affects root length responses to cadaverine
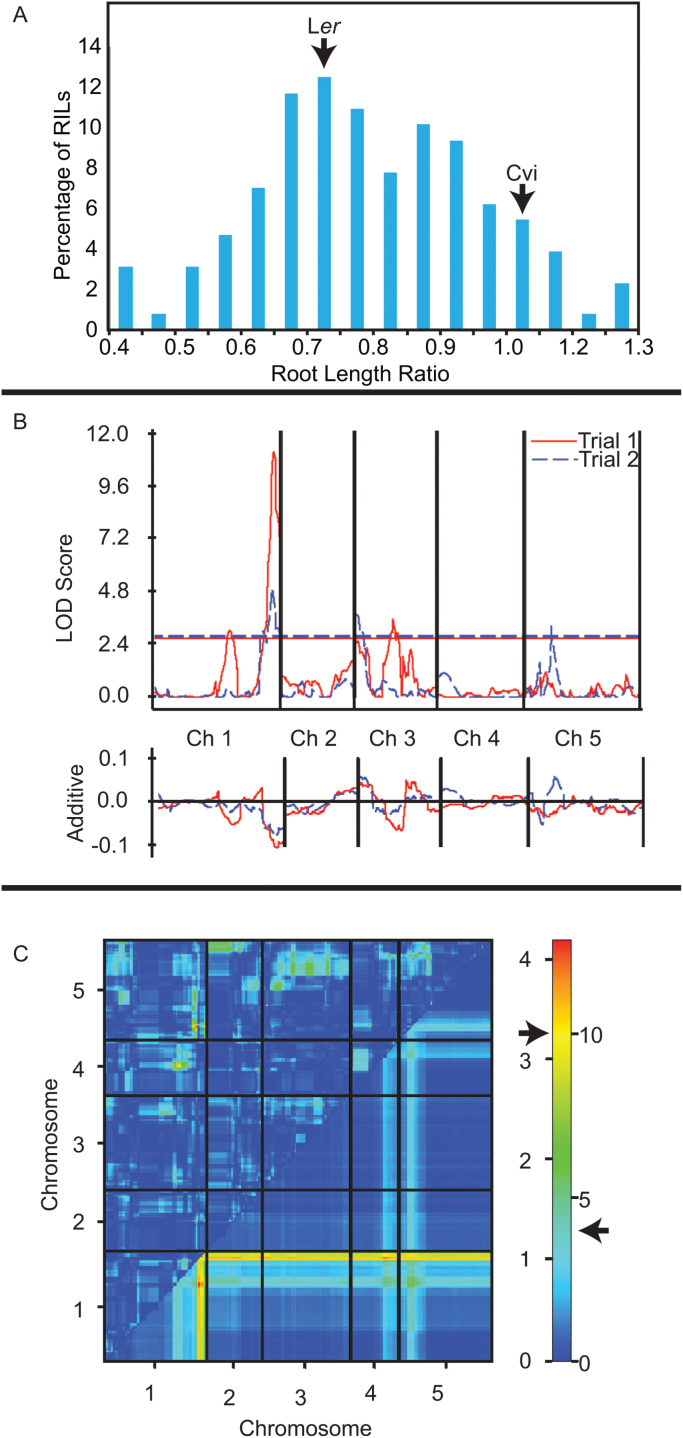
Due to the large difference between Cvi and Ler in root growth responses to exogenous cadaverine, a QTL study was performed with a group of RILs created from the two accessions. Other pairs of accessions would have been equally interesting to use for QTL analysis but, at the time this experiment was initiated, the Cvi×Ler population was the available population created from accessions with the most differing cadaverine responses. These RILs were previously characterized for genotypic contribution from the two parental lines along the five chromosomes (Alonso-Blanco et al., 1998). For both the parental lines and the RILs, root growth on medium containing cadaverine compared with growth on control plates lacking this compound was quantified. RIL mean root length ratios showed transgression with respect to the parental means (Fig. 2A). Furthermore, growth on 0 μM and 50 μM cadaverine displayed broad sense heritability values of 0.46 and 0.56, respectively. Taken together, these results indicate that this population is well suited for QTL mapping of root growth responses to exogenous cadaverine.
Fig. 2.
QTL analysis of root length responses to cadaverine in the Cvi/Ler RIL population. (A) Histogram showing root length ratios for the RILs on 100 μM cadaverine compared with control medium. (B) Composite interval mapping analysis for the root length ratio. The top portion of the graph gives the LOD score across each chromosome. The LOD significance threshold is shown as a horizontal line across the graph. The bottom graph shows the additive value toward the phenotype of each genomic region with respect to the Ler allele. (C) Two-dimensional QTL scan for the root length ratio from trial 1. The x- and y-axes represent positions along each chromosome. The region of the plot below the diagonal gives the additive QTL model, while the region above the diagonal shows epistatic interactions. A scale relating colour to LOD score is provided on the right of the heat map. Numbers on the left of this scale represent LOD scores for the epistatic portion of the plot, whereas those on the right represent scores for the additive model. The arrows indicate the significance thresholds for the epistatic (left) and additive (right) portions of the plot.
Using both composite interval mapping and a 2D scan approach, a strong QTL for relative root growth response to cadaverine was identified at the end of chromosome 1 in two independent trials (Fig. 2B). The additive portion of the QTL graph (Fig. 2B, bottom) predicts that having a Ler allele at this location results in a shorter root upon cadaverine treatment compared with a Cvi allele. No epistasis was observed between this QTL and other parts of the genome (Fig. 2C).
Near isogenic lines confirm the chromosome 1 QTL
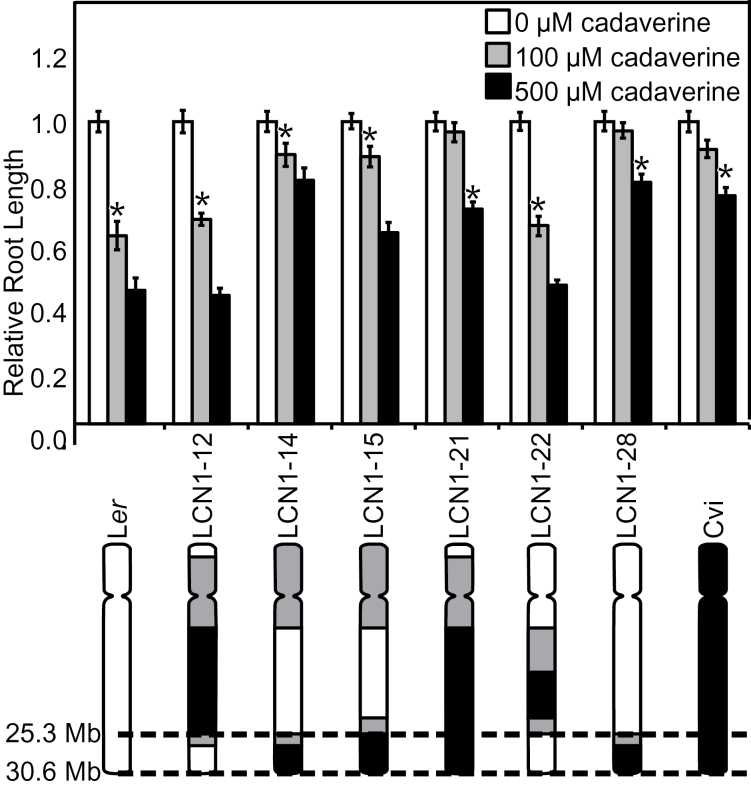
Six NILs with Cvi introgression of varying segments of chromosome 1 within an otherwise Ler background (Keurentjes et al., 2007) were used to confirm the root length QTL. The four LCN1 lines that contain a Cvi introgression at the end of chromosome 1 showed greater, more Cvi-like resistance to cadaverine than LCN1-12 and LCN1-22, the NILs that do not contain a Cvi introgression in this region (Fig. 3). The Ler to Cvi breakpoints in these lines were fine-mapped, and the candidate region was narrowed to a 5.3Mb interval at the end of chromosome 1 (primers shown in Supplementary Table S1 at JXB online).
Fig. 3.
Cadaverine root length responses of chromosome 1 NILs and fine mapping. Relative root lengths on media containing 0, 100, and 500 μM cadaverine are shown. Plants were grown at a 30 ° backward tilt for 6 d. The average root length on control medium was set to 1. Absolute average root lengths on control medium were 1.0, 1.2, 1.2, 1.2, 1.1, 1.2, 1.1, and 1.1cm in the lines as shown from left to right. The asterisk indicates the first concentration on which the root length was significantly different from the length on control medium (P<0.05, Student’s t-test). Error bars indicate ±SE, and n=18–34. In the diagram of chromosome 1 NILs (bottom), white bars are Ler chromosomal regions, black bars are Cvi regions, and grey bars indicate regions in which the breakpoint is undetermined. The parallel dashed lines flank the probable region of a locus controlling root length responses to cadaverine. Accession breakpoints were determined using primers based on polymorphisms described by Monsanto/CEREON between Columbia and Ler (Jander et al., 2002).
Identification of candidate genes for the chromosome 1 QTL
Because many of the causative loci behind the QTL are due to differences in gene expression (Alonso-Blanco et al., 2009), gene expression differences between Cvi and Ler were examined in the candidate region using previously published microarray data (Vaughn and Masson, 2011). The primary data are publicly available at Gene Expression Omnibus (NCBI; accession number GSE28275). From these arrays, 19 probe sets representing genes located in the candidate chromosomal segment give hybridization signals that differ significantly (95% confidence interval) by ≥2-fold between Cvi and Ler (Table 1). The largest predicted differentially expressed gene in this region was OCT1, which was estimated to show a 73-fold increase in Cvi compared with Ler.
Table 1.
Probe sets on chromosome 1 between 25.3Mb and 30.6Mb that show ≥2-fold differential expression between Cvi and Ler at the 95% confidence level
| Probe set ID | Locus ID | Ler over Cvi fold change | Log2 expression level | Description | |
|---|---|---|---|---|---|
| Ler | Cvi | ||||
| 260097_at | AT1G73220 | 72.7 down | 3.7 | 9.9 | Organic cation transporter (OCT1) |
| 264100_at | AT1G78970 | 48.5 down | 1.9 | 7.5 | Lupeol synthase (LUP1) |
| 245215_at | AT1G67830 | 6.0 down | 5.9 | 8.5 | α-Fucosidase (FXG1) |
| 245736_at | AT1G73330 | 5.1 down | 10.9 | 13.3 | Protease inhibitor (DR4) |
| 259941_s_at | AT1G71280 | 4.4 down | 5.2 | 7.4 | DEAD/DEAH box helicase |
| 260806_at | AT1G78260 | 2.9 down | 9.4 | 11.0 | RNA-binding protein |
| 260401_at | AT1G69840 | 2.5 down | 8.9 | 10.2 | Unknown protein |
| 264144_at | AT1G79320 | 2.2 down | 7.4 | 8.5 | Metacaspase (MC6) |
| 259705_at | AT1G77450 | 2.0 up | 12.1 | 11.1 | NAC transcription factor (NAC032) |
| 259977_at | AT1G76590 | 3.1 up | 8.3 | 6.7 | Zinc-binding protein |
| 261901_at | AT1G80920 | 3.4 up | 12.4 | 10.6 | DNA-J protein (J8) |
| 260411_at | AT1G69890 | 3.4 up | 7.6 | 5.9 | Unknown protein |
| 264957_at | AT1G77000 | 3.7 up | 10.1 | 8.2 | F-box protein (SKP2B) |
| 262362_at | AT1G72840 | 4.0 up | 7.4 | 5.4 | ATP-binding transmembrane receptor |
| 260207_at | AT1G70730 | 4.7 up | 10.8 | 8.6 | Phosphoglucomutase |
| 245731_at | AT1G73500 | 8.9 up | 10.1 | 6.9 | MAP kinase kinase (MKK9) |
| 260101_at | AT1G73260 | 12.2 up | 12.6 | 9.0 | Trypsin protease inhibitor |
| 259866_at | AT1G76640 | 14.3 up | 7.4 | 3.6 | Calmodulin-related protein |
| 264720_at | AT1G70080 | 20.8 up | 10.3 | 5.9 | Terpene synthase |
Abbreviations: NIL, near isogenic line; OCT, organic cation transporter; RIL, recombinant inbred line.
No other genes in Table 1 encode proteins that are directly relevant to polyamine metabolism or transport. Genes encoding a transglutaminase (At1g69820) and a cinnamyl alcohol dehydrogenase (At1g72680), both possibly linked to the formation of polyamine conjugates (Serafini-Fracassini and Del Duca, 2008; Fellenberg et al., 2009), are found in this QTL interval; however, they do not show ≥2-fold expression differences between Cvi and Ler.
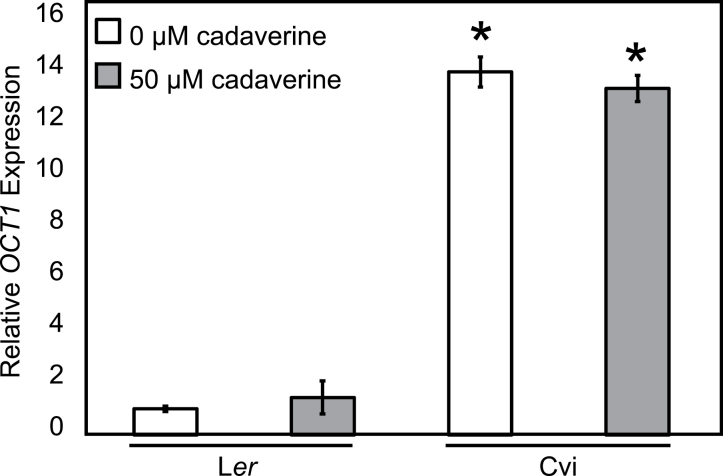
OCT1 from Cvi and Ler genomic DNA was sequenced. No large insertions or deletions were found, and only one amino acid change was found: residue 431 in the second exon is serine in Cvi and alanine in Ler (Supplementary Table S2 at JXB online). Analysing the 2kb of sequence upstream of the start codon revealed numerous single nucleotide polymorphisms and small insertions but no large insertions or deletions (Supplementary Table S2). Based on these data, it was hypothesized that OCT1 expression differences contribute to root length responses to exogenous cadaverine. qRT-PCR data confirmed the microarray result and showed that OCT1 expression was ~14 times as high in Cvi roots as in Ler roots (Fig. 4). Cadaverine treatment did not significantly change OCT1 expression in either accession (Fig. 4).
Fig. 4.
OCT1 expression relative to the reference gene At1G58050 is higher in Cvi roots than in Ler roots. Seedlings were grown on 0 μM or 50 μM cadaverine. Error bars indicate ±SE among biological replicates, and OCT1 expression for Ler on control medium was set to 1. The asterisk indicates a significant difference in expression compared with Ler for the given condition (P<0.05, Student’s t-test). Differences within each accession between cadaverine treatments are not significant.
oct1-1 mutants express a truncated OCT1 transcript
To evaluate a role for OCT1 in root length responses to cadaverine, the oct1-1 mutant, which was previously found to carry a single T-DNA insertion near the 3’ end of the gene, was obtained; both ends of the insertion were characterized (Lelandais-Brière et al., 2007). This mutant was also reported to not express any OCT1 RNA, as determined by RT–PCR using primers aligning on each side of the intron (Lelandais-Brière et al., 2007). To confirm this result, RT–PCR was performed on oct1-1 and (Wassilewskija) WS wild-type root RNA samples. As expected, PCR products were obtained from WS wild-type cDNA samples of the expected sizes using multiple primer sets (1F/1R, 2F/2R, and 3F/3R) (Supplementary Fig. S2 at JXB online). Also, no transcript was observed for oct1-1 mutants using primer sets near the 3′ end of the gene (2F/2R, 3F/3R, and 2F/4R) (Supplementary Fig. S2).
However, it was found that oct1-1 plants do express a partial OCT1 transcript using primers on each side of the intron (1F/1R) (Supplementary Fig. S2 at JXB online). This product size corresponds to approximately the predicted length if the intron is not spliced out of the RNA (238 bases instead of 135 bases), which would result in a premature stop codon in the intron. No product for any primer set was detected in control reactions without reverse transcriptase, which confirms the lack of genomic DNA contamination (Supplementary Fig. S2). Therefore, oct1-1 produces a truncated OCT1 transcript that is highly unlikely to encode a functional protein.
oct1-1 shows increased cadaverine sensitivity
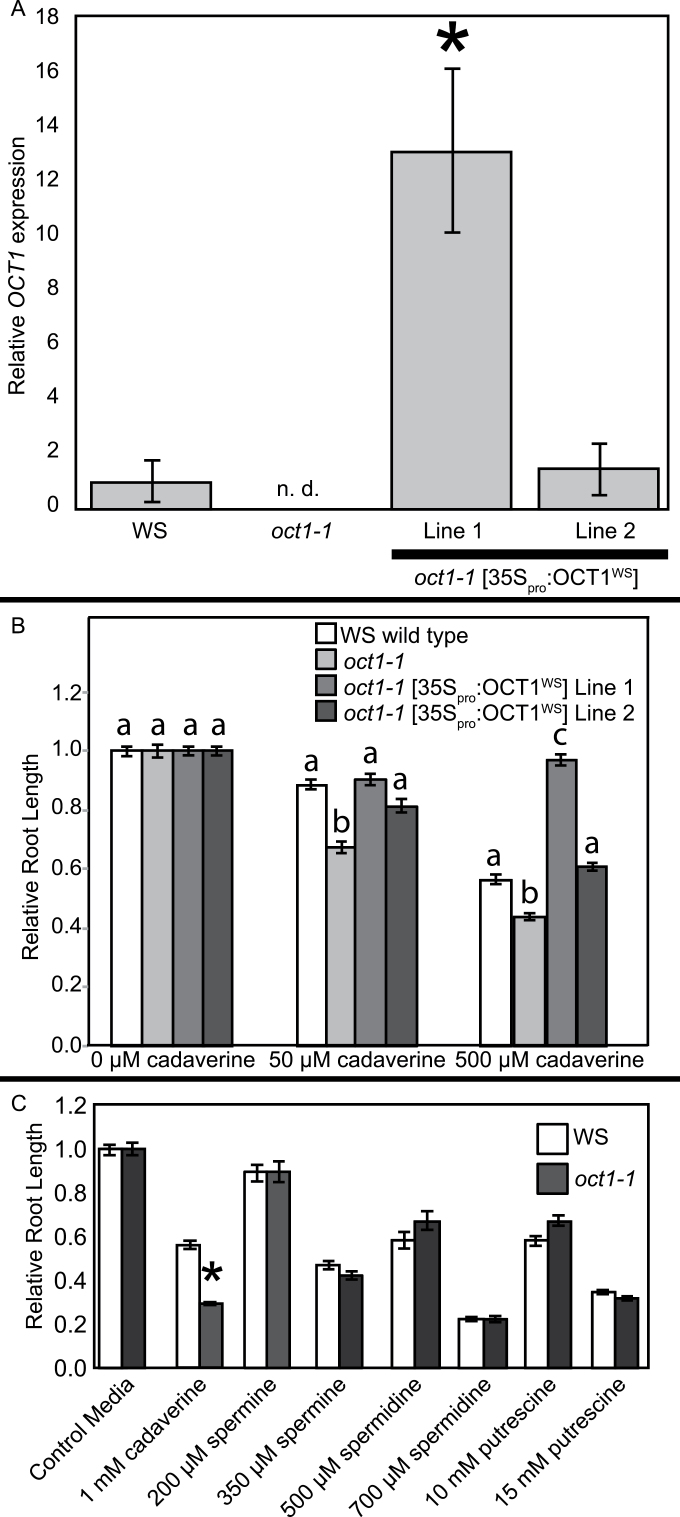
If Ler roots have higher sensitivity to cadaverine compared with Cvi roots because they have lower OCT1 expression, then oct1 mutant roots should also respond more strongly to cadaverine than wild-type plants. To test this, WS wild type, oct1-1, and two independent transgenic rescue lines (oct1-1 [35Spro:OCT1WS]) were grown on control medium and medium supplemented with cadaverine. Rescue line 1 had significantly higher OCT1 expression compared with the WS wild type, whereas rescue line 2 had a similar level of OCT1 expression (Fig. 5A). It was found that all of these plants had the same root length on control medium lacking cadaverine, but oct1-1 mutant roots were significantly shorter than wild-type roots on media containing 50 μM and 500 μM cadaverine (Fig. 5B). Robust root growth on medium containing cadaverine was restored in both independent transgenic rescue lines. Rescue line 2 showed a wild-type-like response to cadaverine, and rescue line 1 showed increased resistance to cadaverine (Fig. 5B). These results suggest that reduced OCT1 expression confers increased cadaverine sensitivity.
Fig. 5.
oct1-1 roots are more sensitive to exogenous cadaverine compared with wild-type plants but respond similarly to putrescine-derived polyamines. (A) OCT1 expression relative to the reference gene At1G58050 is shown. Error bars indicate ±SE among biological replicates, and OCT1 expression for the WS wild type on control medium was set to 1. The asterisk indicates a significant difference in expression compared with the WS wild type (P<0.05, Student’s t-test). The abbreviation n.d. indicates that expression was not detected. (B) Plants were grown for 6 d at a 30 ° backward tilt on media containing 0, 50, and 500 μM cadaverine. Root tips were marked after 3 d of growth, and the root growth over the following 3 d was measured for each treatment. Average root growth over the final 3 d on control medium was 1.2cm for the wild type, 1.4cm for oct1-1, 1.3cm for rescue line 1, and 1.4cm for rescue line 2; these lengths were set to 1. For each cadaverine concentration, the letters indicate significant differences in relative root length (P<0.01, pairwise Student’s t-tests). n=37–68, and error bars indicate ±SE. (C) Plants were grown for 6 d at a 30 ° backward tilt on media containing various concentrations of polyamines as indicated. Average root length on control medium was 1.3cm for the wild type and 1.4cm for oct1-1; these lengths were set to 1. n=33–44, and error bars indicate ±SE. No significant root length differences were observed for any treatment other than cadaverine (P<0.05, Student’s t-test).
The WS wild type and oct1-1 had similar root lengths on media containing the polyamines putrescine, spermidine, and spermine (Fig. 5C), which shows that this sensitivity is specific to cadaverine. Polyamines have been shown to shorten root growth through the generation of H2O2 by diamine and polyamine oxidases, which promotes premature cell differentiation and programmed cell death (Tisi et al., 2011). However, because the cadaverine-hypersensitive response of oct1-1 was specific to cadaverine (Fig. 5C), OCT1 probably functions in a different aspect of polyamine-mediated root growth regulation.
The expression level of OCT1, not its accession origin, correlates with cadaverine sensitivity
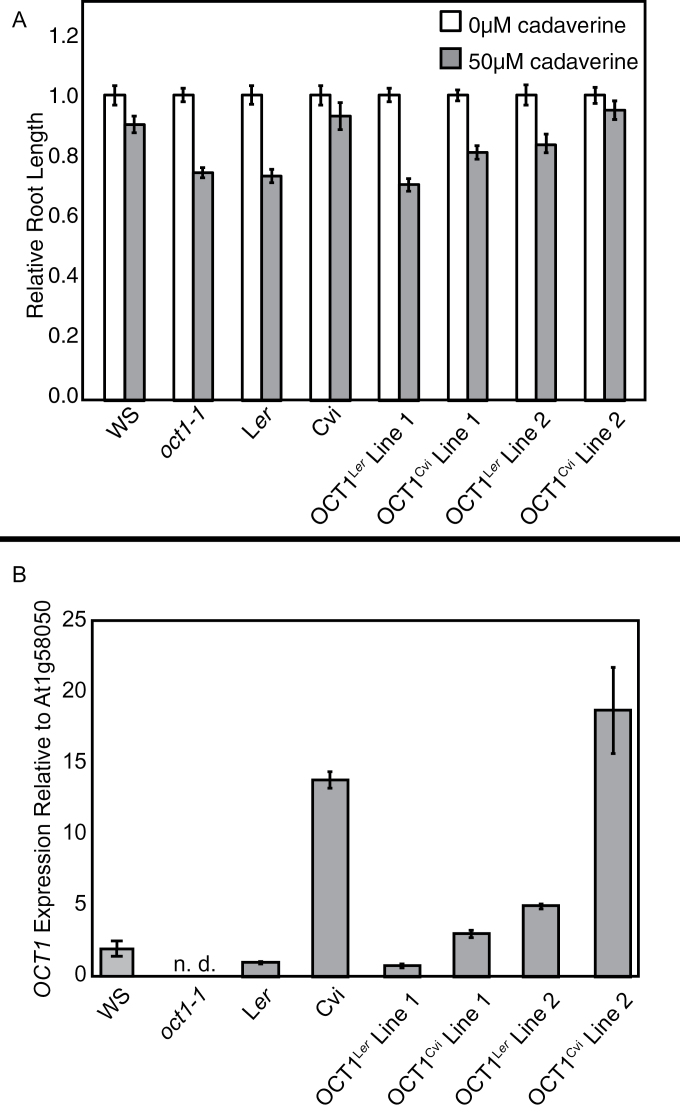
To determine if differences in the OCT1 sequence between Cvi and Ler or only OCT1 expression levels correlated with the cadaverine response, OCT1 was cloned from Cvi and Ler, and these constructs were introduced into oct1-1 mutants. These transgenic lines were then tested on media containing 0 μM and 50 μM cadaverine, and their relative root lengths were measured. A range of cadaverine resistance levels that did not correlate with the accession source of the transgene was found (Fig. 6A).
Fig. 6.
Higher OCT1 expression correlates with cadaverine resistance. OCT1Cvi and OCT1Ler indicate oct1-1 [OCT1pro:OCT1Cvi] and oct1-1 [OCT1pro:OCT1Ler], respectively. (A) Relative root lengths of plants on media containing 0 μM and 50 μM cadaverine. Plants were grown for 6 d at a 30 ° backward tilt. Average root length on control medium was set to 1. Absolute average root length on control medium was 2.2cm for Cvi, 1.9cm for Ler, 2.0cm for WS, 2.0cm for oct1-1, and 1.9, 2.0, 1.7, and 2.1cm for the transgenic lines as shown from left to right. Error bars indicate ±SE, and n=17–24. (B) Root OCT1 expression determined by qRT-PCR is shown relative to the reference gene At1G58050. OCT1 expression in Ler was set to 1. Error bars indicate ±SE among the biological replicates. The abbreviation n.d. indicates that expression was not detected.
Because the insertion location can affect transgene expression, it was predicted that the OCT1 expression levels would correlate with cadaverine sensitivity. To test this, OCT1 expression was measured in four of these lines by qRT-PCR. As expected, a range of OCT1 expression levels was found in the transgenic lines regardless of whether the OCT1 untranslated and coding regions came from Cvi or Ler. In general, lines with increased resistance to cadaverine had higher OCT1 expression (Fig. 6).
Genes other than OCT1 probably also contribute to the cadaverine response
The data suggest that differences in OCT1 expression contribute significantly to the different cadaverine root length responses in Cvi and Ler. To determine if differences in OCT1 expression among other natural accessions also correlate with variability in cadaverine sensitivity, OCT1 expression was measured in two other accessions, one resistant to cadaverine (Knox-18) and one sensitive to cadaverine (Sq-8). It was found that both lines had higher OCT1 expression than Ler even though only Knox-18 was more resistant to cadaverine than Ler. Furthermore, Sq-8 displayed higher OCT1 expression than Knox-18 despite being more sensitive to cadaverine (Supplementary Fig. S3 at JXB online). The OCT1 sequences of the eight natural accessions initially selected for the analysis of cadaverine response were also compared. Polymorphisms shared among the three most resistant accessions (Cvi-1, Knox-18, and Ull2-3) or the three most sensitive accessions (An-1, Ler-2, and Sq-8) were not seen (Supplementary Table S2). Together, these data suggest that factors other than OCT1 expression may also contribute significantly to differential cadaverine sensitivity among these accessions.
Ectopic copies of OCT1 derived from Cvi or Ler were also introduced into Columbia and Ler wild-type plants. Ler root length is highly sensitive to cadaverine, whereas Columbia root length is less sensitive to it. In general, the Columbia plants showed a range of responses to cadaverine regardless of the accession from which the transgene was derived (Supplementary Fig. S4 at JXB online). This result is similar to that obtained when these constructs were transformed into the oct1-1 mutant (Fig. 6). In contrast, when these constructs were transformed into Ler plants, the tested lines generally did not show increased resistance to cadaverine (Supplementary Fig. S4). Therefore, the ability of OCT1 to affect the cadaverine response may depend on the genetic background.
Discussion
It was found that Arabidopsis roots exhibited varying responses to exogenously applied cadaverine depending on the accession. In general, when cadaverine concentrations increased, a reduction in root length as well as alterations in skewing, lateral root branching, and root waving were seen. The reduction in root length is probably due to a combination of reduced cell division and expansion. A major QTL was identified on chromosome 1 that at least partially explains the root length difference in response to cadaverine between Cvi and Ler. By comparing phenotypes and gene expression in natural accessions, a mutant, and transgenic lines, OCT1 was identified as a key gene involved in this response. Restoring OCT1 expression in the mutant background resulted in a positive correlation between OCT1 expression levels and cadaverine resistance. Because the OCT1 expression levels in other natural accessions do not correlate well with cadaverine resistance, other genes are also likely to contribute to the variation of this trait between Arabidopsis accessions. These genes could contribute to endogenous cadaverine homeostasis, transport, and/or response. Unfortunately, the levels of endogenous cadaverine in Arabidopsis seedling roots and shoots were below the level of detection using HPLC methods, preventing a distinction from being made between these possibilities.
OCT1 encodes an organic cation transporter. In animals, OCT proteins take up a wide variety of endogenous compounds and xenobiotics, and, therefore, they affect substrate absorption, distribution, and excretion in many tissues. Most are polyspecific, and transport can occur in either direction (for a review, see Roth et al., 2012). Arabidopsis OCT1 is one of six OCT-like proteins in Arabidopsis (Lelandais-Brière et al., 2007). It was previously reported to localize to the plasma membrane, and promoter–β-glucuronidase (GUS) fusions showed activity in the vasculature of many organs including roots (Lelandais-Brière et al., 2007; Küfner and Koch, 2008). Because the other six OCT-like proteins localize to the tonoplast rather than the plasma membrane, it is likely that OCT1 has a different function.
The present work expands on these studies and suggests a role for OCT1 in affecting root growth responses to cadaverine. Future experiments will be aimed at clarifying the mechanism by which this occurs. One possibility is that OCT1 transports cadaverine away from elongating root cells and through the vasculature, causing it to accumulate in aerial tissues. Alternatively, cadaverine may alter the transport of another molecule through the membranes. Polyamines block many kinds of channels in multiple organisms. For example, spermine and spermidine directly block fast vacuolar cation channels in barley (Bruggemann et al., 1998), and all natural polyamines including cadaverine can regulate cation channel activity through cytoplasmic pathways (Liu et al., 2000; Shabala et al., 2007).
It has been demonstrated that exogenous cadaverine affects Arabidopsis root morphology and that there is natural variation in this response due in part to varying expression levels of OCT1. This work has potential implications for better understanding how polyamines contribute to development and mediate stress responses.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Arabidopsis natural accessions show varying root growth behaviours in response to cadaverine.
Figure S2. oct1-1 produces a truncated transcript.
Figure S3. OCT1 expression does not correlate with cadaverine sensitivity in all natural accessions.
Figure S4. OCT1 expression results in varying degrees of cadaverine sensitivity in Col and Ler.
Table S1. Primers for chromosome 1 fine mapping.
Table S2. Comparison of OCT1 sequences from eight Arabidopsis natural accessions.
Acknowledgements
This work was supported by grants from the University of Wisconsin–Madison College of Agricultural and Life Sciences Hatch program and the National Science Foundation (IOS-0821884 and IOS-1121694) to PHM, by a National Science Foundation Graduate Research Fellowship to AKS, and by a National Institutes of Health Training Grant in Genetics at the University of Wisconsin–Madison to LMV. We thank Hiroshi Maeda for critically reviewing the manuscript and Christine Lelandais-Brière for giving us the oct1-1 mutant and the oct1-1 [35Spro:OCT1WS] rescue lines. We also thank Katherine Baldwin and Shih-Heng Su for help with figures and technical assistance.
References
- Alet AI, Sánchez DH, Ferrando A, et al. 2011. Homeostatic control of polyamine levels under long-term salt stress in Arabidopsis: changes in putrescine content do not alleviate ionic toxicity. Plant Signaling and Behavior 6, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. 2009. What has natural variation taught us about plant development, physiology, and adaptation? The Plant Cell 21, 1877–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT. 1998. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. The Plant Journal 14, 259–271. [DOI] [PubMed] [Google Scholar]
- Aouida M, Leduc A, Poulin R, Ramotar D. 2005. AGP2 encodes the major permease for high affinity polyamine import in Saccharomyces cerevisiae . Journal of Biological Chemistry 280, 24267–24276. [DOI] [PubMed] [Google Scholar]
- Aziz A, Martin-Tanguy J, Larher F. 1997. Plasticity of polyamine metabolism associated with high osmotic stress in rape leaf discs and with ethylene treatment. Plant Growth Regulation 21, 153–163. [Google Scholar]
- Bagni N, Tassoni A. 2001. Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20, 301–317. [DOI] [PubMed] [Google Scholar]
- Bruggemann LI, Pottosin II, Schonknecht G. 1998. Cytoplasmic polyamines block the fast-activating vacuolar cation channel. The Plant Journal 16, 101–105. [Google Scholar]
- Cassán F, Maiale S, Masciarelli O, Vidal A, Luna V, Ruiz O. 2009. Cadaverine production by Azospirillum brasilense and its possible role in plant growth promotion and osmotic stress mitigation. European Journal of Soil Biology 45, 12–19. [Google Scholar]
- Cloete KJ, Valentine AJ, Stander MA, Blomerus LM, Botha A. 2009. Evidence of symbiosis between the soil yeast Cryptococcus laurentii and a sclerophyllous medicinal shrub, Agathosma betulina (Berg.) Pillans. Microbial Ecology 57, 624–632. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, Salinas J, Tiburcio AF, Ferrando A. 2008. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiology 148, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg C, Böttcher C, Vogt T. 2009. Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry 70, 1392–1400. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Iuchi S, Yamada K, Kobayashi Y, Urano K, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. 2012. Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 6343–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A, Frydman RB. 1991. Cadaverine, an essential diamine for the normal root development of germinating soybean (Glycine max) seeds. Plant Physiology 97, 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A. 2006. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SS, Ali M, Ahmad M, Siddique KHM. 2011. Polyamines: natural and engineered abiotic and biotic stress tolerance in plants. Biotechnology Advances 29, 300–311. [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. 2002. Arabidopsis map-based cloning in the post-genome era. Plant Physiology 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-de Vries H, Effgen S, Vreugdenhil D, Koornneef M. 2007. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175, 891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott JM, Römer P, Sumper M. 2007. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Letters 581, 3081–3086. [DOI] [PubMed] [Google Scholar]
- Küfner I, Koch W. 2008. Stress regulated members of the plant organic cation transporter family are localized to the vacuolar membrane. BMC Research Notes 1, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov V, Shorina M, Aronova E, Stetsenko L, Rakitin V, Shevyakova N. 2007. NaCl- and ethylene-dependent cadaverine accumulation and its possible protective role in the adaptation of the common ice plant to salt stress. Plant Science 172, 363–370. [Google Scholar]
- Lelandais-Brière C, Jovanovic M, Torres GAM, Perrin Y, Lemoine R, Corre-Menguy F, Hartmann C. 2007. Disruption of AtOCT1, an organic cation transporter gene, affects root development and carnitine-related responses in Arabidopsis. The Plant Journal 51, 154–164. [DOI] [PubMed] [Google Scholar]
- Li J, Mu J, Rai J, Fu F, et al. 2013. PARAQUAT RESISTANT1, a Golgi-localized putative transporter protein, is involved in intracellular transport of paraquat. Plant Physiology 162, 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Fu H, Bei Q, Luan S. 2000. Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiology 124, 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanl FA, Meng D, et al. 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nature Genetics 45, 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria J-CF, Steiner P, Hirling H, Unser M. 2004. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry 58A, 167–176. [DOI] [PubMed] [Google Scholar]
- Mulangi V, Chibucos MC, Phuntumart V, Morris PF. 2012a. Kinetic and phylogenetic analysis of plant polyamine uptake transporters. Planta 236, 1261–1273. [DOI] [PubMed] [Google Scholar]
- Mulangi V, Phuntumart V, Aouida M, Ramotar D, Morris P. 2012b. Functional analysis of OsPUT1, a rice polyamine uptake transporter. Planta 235, 1–11. [DOI] [PubMed] [Google Scholar]
- Neill S. 2002. Hydrogen peroxide signalling. Current Opinion in Plant Biology 5, 388–395. [DOI] [PubMed] [Google Scholar]
- Niemi K, Häggman H, Sarjala T. 2002. Effects of exogenous diamines on the interaction between ectomycorrhizal fungi and adventitious root formation in Scots pine in vitro . Tree Physiology 22, 373–381. [DOI] [PubMed] [Google Scholar]
- Obata T, Fernie AR. 2012. The use of metabolomics to dissect plant responses to abiotic stresses. Cellular and Molecular Life Sciences 69, 3225–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrig D, Boiero ML, Masciarelli OA, Penna C, Ruiz OA, Cassán FD, Luna MV. 2007. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Applied Microbiology and Biotechnology 75, 1143–1150. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Roth M, Obaidat A, Hagenbuch B. 2012. OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. British Journal of Pharmacology 165, 1260–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini-Fracassini D, Del Duca S. 2008. Transglutaminases: widespread cross-linking enzymes in plants. Annals of Botany 102, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Cuin TA, Pottosin I. 2007. Polyamines prevent NaCl-induced K+ efflux from pea mesophyll by blocking non-selective cation channels. FEBS Letters 581, 1993–1999. [DOI] [PubMed] [Google Scholar]
- Shevyakova NI, Rakitin VY, Duong DB, Sadomov NG, Kuznetsov VV. 2001. Heat shock-induced cadaverine accumulation and translocation throughout the plant. Plant Science 161, 1125–1133. [Google Scholar]
- Sziderics AH, Oufir M, Trognitz F, Kopecky D, Matusíková I, Hausman J-F, Wilhelm E. 2010. Organ-specific defence strategies of pepper (Capsicum annuum L.) during early phase of water deficit. Plant Cell Reports 29, 295–305. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Berberich T, Miyazaki A, Seo S, Ohashi Y, Kusano T. 2003. Spermine signalling in tobacco: activation of mitogen-activated protein kinases by spermine is mediated through mitochondrial dysfunction. The Plant Journal 36, 820–829. [DOI] [PubMed] [Google Scholar]
- Tang W, Newton RJ. 2005. Polyamines reduce salt-induced oxidative damage by increasing the activities of antioxidant enzymes and decreasing lipid peroxidation in Virginia pine. Plant Growth Regulation 46, 31–43. [Google Scholar]
- Tisi A, Federico R, Moreno S, Lucretti S, Moschou PN, Roubelakis-Angelakis KA, Angelini R, Cona A. 2011. Perturbation of polyamine catabolism can strongly affect root development and xylem differentiation. Plant Physiology 157, 200–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn LM, Masson PH. 2011. A QTL study for regions contributing to Arabidopsis thaliana root skewing on tilted surfaces. G3 1, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun P-P, Chen C-L, Wang Y, Fu X-Z, Liu J-H. 2011. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. Journal of Experimental Botany 62, 2899–2914. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. 2010. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC (http://statgen.ncsu.edu/qtlcart/).
- Xu R, Li QQ. 2008. Protocol: streamline cloning of genes into binary vectors in Agrobacterium via the Gateway(R) TOPO vector system. Plant Methods 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.