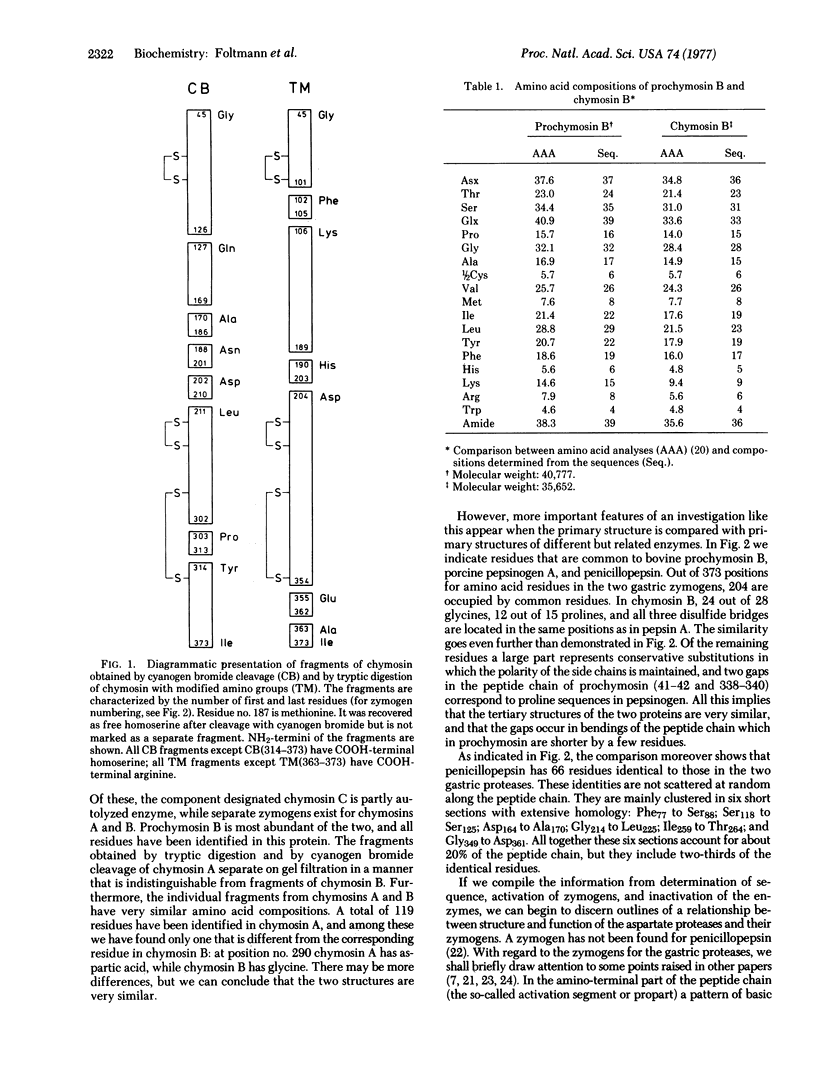
Abstract
The total sequence of 365 amino acid residues in bovine prochymosin is presented. Alignment with the amino acid sequence of porcine pepsinogen shows that 204 amino acid residues are common to the two zymogens. Further comparison and alignment with the amino acid sequence of penicillopepsin shows that 66 residues are located at identical positions in all three proteases. The three enzymes belong to a large group of proteases with two aspartate residues in the active center. This group forms a family derived from one common ancestor.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreeva N. S., Fedorov A. A., Gushchina A. E., Shutskever N. E., Riskulov R. R. Opredelenie trekhmernoi struktury pepsina pri razreshenii 2, 7 å. Dokl Akad Nauk SSSR. 1976;228(2):480–483. [PubMed] [Google Scholar]
- Chen K. C., Tang J. Amino acid sequence around the epoxide-reactive residues in pepsin. J Biol Chem. 1972 Apr 25;247(8):2566–2574. [PubMed] [Google Scholar]
- Cunningham A., Wang H. M., Jones S. R., Chiericato G., Rao L., Harris C. I., Rhee S. H., Hofmann T. Amino acid sequence of penicillopepsin. IV. Myxobacter AL-1 protease II and Staphylococcus aureus protease fragments and homology with pig pepsin and chymosin. Can J Biochem. 1976 Oct;54(10):902–914. doi: 10.1139/o76-128. [DOI] [PubMed] [Google Scholar]
- DJURTOFT R., FOLTMANN B., JOHANSEN A. STUDIES ON RENNIN. 8. ON THE MOLECULAR WEIGHT OF PRORENNIN AND RENNIN. C R Trav Lab Carlsberg. 1964;34:287–298. [PubMed] [Google Scholar]
- FOLTMANN B. STUDIES ON RENNIN. VII. ON THE AMINO ACID COMPOSITION OF PRORENNIN, RENNIN AND OF PEPTIDES LIBERATED DURING THE ACTIVATION OF PRORENNIN. C R Trav Lab Carlsberg. 1964;34:275–286. [PubMed] [Google Scholar]
- Foltmann B. A review on prorennin and rennin. C R Trav Lab Carlsberg. 1966;35(8):143–231. [PubMed] [Google Scholar]
- Foltmann B., Hartley B. S. The disulphide bridges and soluble tryptic peptides of calf rennin. Biochem J. 1967 Sep;104(3):1064–1074. doi: 10.1042/bj1041064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruton J. S. The mechanism of the catalytic action of pepsin and related acid proteinases. Adv Enzymol Relat Areas Mol Biol. 1976;44:1–36. doi: 10.1002/9780470122891.ch1. [DOI] [PubMed] [Google Scholar]
- Hapner K. D., Wilcox P. E. Fragmentation of bovine chymotrypsinogen A and chymotrypsin A-alpha. Specific cleavage at arginine and methionine residues and separation of peptides, including B and C chains of chymotrypsin. Biochemistry. 1970 Nov 10;9(23):4470–4480. doi: 10.1021/bi00825a002. [DOI] [PubMed] [Google Scholar]
- Harboe M. K., Foltmann B. Bovine pepsin: the sequence of the first 65 amino acid residues (completing the sequence of the first 110 residues of bovine pepsinogen). FEBS Lett. 1975 Dec 1;60(1):133–136. doi: 10.1016/0014-5793(75)80435-2. [DOI] [PubMed] [Google Scholar]
- Harboe M., Andersen P. M., Foltmann B., Kay J., Kassell B. The activation of bovine pepsinogen. Sequence of the peptides released, identification of a pepsin inhibitor. J Biol Chem. 1974 Jul 25;249(14):4487–4494. [PubMed] [Google Scholar]
- Hartley B. S. Strategy and tactics in protein chemistry. Biochem J. 1970 Oct;119(5):805–822. doi: 10.1042/bj1190805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagami T., Misono K., Michelakis A. M. Definitive evidence for similarity in the active site of renin and acidic protease. Biochem Biophys Res Commun. 1974 Jan 23;56(2):503–509. doi: 10.1016/0006-291x(74)90871-7. [DOI] [PubMed] [Google Scholar]
- Jenkins J. A., Blundell T. L., Tickle I. J., Ungaretti L. The low resolution structure analysis of an acid proteinase from Endothia parasitica. J Mol Biol. 1975 Dec 25;99(4):583–590. doi: 10.1016/s0022-2836(75)80173-2. [DOI] [PubMed] [Google Scholar]
- Keilová H. Inhibition of cathepsin D by diazoacetylnorleucine methyl ester. FEBS Lett. 1970 Feb 25;6(4):312–314. doi: 10.1016/0014-5793(70)80086-2. [DOI] [PubMed] [Google Scholar]
- Klemm P., Poulsen F., Harboe M. K., Foltmann B. N-terminal amino acid sequences of pepsinogens from dogfish and seal and of bovine pepsinogen B. Acta Chem Scand B. 1976;30(10):979–984. doi: 10.3891/acta.chem.scand.30b-0979. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. On the mechanism of action of pepsin. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):135–146. doi: 10.1098/rstb.1970.0016. [DOI] [PubMed] [Google Scholar]
- Knowles J. R., Wybrandt G. B. The sequence around an active-site aspartyl residue in pepsin. FEBS Lett. 1968 Sep;1(4):211–212. doi: 10.1016/0014-5793(68)80063-8. [DOI] [PubMed] [Google Scholar]
- Kovaleva G. G., Shimanskaya M. P., Stepanov V. M. The site of diazoacetyl inhibitor attachment to acid proteinase of Aspergillus awamori--an analog of penicillopepsin and pepsin. Biochem Biophys Res Commun. 1972 Nov 15;49(4):1075–1081. doi: 10.1016/0006-291x(72)90322-1. [DOI] [PubMed] [Google Scholar]
- Kulbe K. D. Micropolyamide thin-layer chromatography of phenylthiohydantoin amino acids (PTH) at subnanomolar level. A rapid microtechnique for simultaneous multisample identification after automated Edman degradations. Anal Biochem. 1974 Jun;59(2):564–573. doi: 10.1016/0003-2697(74)90310-8. [DOI] [PubMed] [Google Scholar]
- McPhie P. A spectrophotometric investigation of the pepsinogen-pepsin conversion. J Biol Chem. 1972 Jul 10;247(13):4277–4281. [PubMed] [Google Scholar]
- Morávek L., Kostka V. Complete amino acid sequence of hog pepsin. FEBS Lett. 1974 Jul 15;43(2):207–211. doi: 10.1016/0014-5793(74)81001-x. [DOI] [PubMed] [Google Scholar]
- Offord R. E. Electrophoretic mobilities of peptides on paper and their use in the determination of amide groups. Nature. 1966 Aug 6;211(5049):591–593. doi: 10.1038/211591a0. [DOI] [PubMed] [Google Scholar]
- Ong E. B., Perlmann G. E. The amino-terminal sequence of porcine pepsinogen. J Biol Chem. 1968 Dec 10;243(23):6104–6109. [PubMed] [Google Scholar]
- Pedersen V. B., Foltmann B. Amino-acid sequence of the peptide segment liberated during activation of prochymosin (prorennin). Eur J Biochem. 1975 Jun 16;55(1):95–103. doi: 10.1111/j.1432-1033.1975.tb02141.x. [DOI] [PubMed] [Google Scholar]
- Pedersen V. B., Foltmann B. The amino acid sequence of a hitherto unobserved segment from porcine pepsinogen preceeding the N-terminus of pepsin. FEBS Lett. 1973 Sep 15;35(2):255–256. doi: 10.1016/0014-5793(73)80298-4. [DOI] [PubMed] [Google Scholar]
- Pedersen V. B., Foltmann B. The amino acid sequence of the first 61 residues of chymosin (rennin EC 3.4.4.3). FEBS Lett. 1973 Sep 15;35(2):250–254. doi: 10.1016/0014-5793(73)80297-2. [DOI] [PubMed] [Google Scholar]
- Rajagopalan T. G., Stein W. H., Moore S. The inactivation of pepsin by diazoacetylnorleucine methyl ester. J Biol Chem. 1966 Sep 25;241(18):4295–4297. [PubMed] [Google Scholar]
- Sepulveda P., Jackson K. W., Tang J. The amino terminal sequences of acid proteases-human pepsin and gastricsin and the protease of Rhizopus chinensis. Biochem Biophys Res Commun. 1975 Apr 21;63(4):1106–1112. doi: 10.1016/0006-291x(75)90683-x. [DOI] [PubMed] [Google Scholar]
- Sepulveda P., Marciniszyn J., Jr, Liu D., Tang J. Primary structure of porcine pepsin. III. Amino acid sequence of a cyanogen bromide fragment, CB2A, and the complete structure of porcine pepsin. J Biol Chem. 1975 Jul 10;250(13):5082–5088. [PubMed] [Google Scholar]
- Smithies O., Gibson D., Fanning E. M., Goodfliesh R. M., Gilman J. G., Ballantyne D. L. Quantitative procedures for use with the Edman-Begg sequenator. Partial sequences of two unusual immunoglobulin light chains, Rzf and Sac. Biochemistry. 1971 Dec 21;10(26):4912–4921. doi: 10.1021/bi00802a013. [DOI] [PubMed] [Google Scholar]
- Subramanian E., Swan I. D., Davies D. R. The crystal at 5.5A resolution of an acid-protease from Rhizopus chinensis. Biochem Biophys Res Commun. 1976 Feb 9;68(3):875–880. doi: 10.1016/0006-291x(76)91226-2. [DOI] [PubMed] [Google Scholar]
- al-Janabi J., Hartsuck J. A., Tang J. Kinetics and mechanism of pepsinogen activation. J Biol Chem. 1972 Jul 25;247(14):4628–4632. [PubMed] [Google Scholar]


