Abstract
Hypoxia Inducible Factor (HIF) mediates a broad, conserved adaptive response to hypoxia, and the HIF pathway is a potential therapeutic target in cerebral ischemia. In this study, we investigated the mechanism by which in vitro ischemia (oxygen-glucose deprivation, OGD) affects canonical hypoxic HIF-1α stabilization. We validated the use of a reporter containing the oxygen dependent degradation domain of HIF-1α fused to firefly luciferase (ODD-luc) to quantitatively monitor distinct biochemical events leading to hypoxic HIF-1α expression or stabilization in a human neuroblastoma cell line (SH-SY5Y). When OGD was imposed following a 2 hour hypoxic stabilization of ODD-luc, the levels of the reporter were reduced, consistent with prior models proposing that OGD enhances HIF prolyl hydroxylase (PHD) activity. Surprisingly, PHD inhibitors and proteasome inhibitors do not stabilize ODD-luc in OGD. Further, OGD does not affect the half-life of ODD-luc protein following hypoxia, suggesting that OGD abrogates hypoxic HIF-1α induction by reducing HIF-1α synthesis rather than by enhancing its degradation. We observed ATP depletion under OGD versus hypoxia, and propose that ATP depletion enhances translational suppression, overcoming the selective synthesis of HIF concurrent with global decreases in protein synthesis in hypoxia. Taken together, these findings biochemically characterize a practical reporter for monitoring HIF-1α levels and support a novel model for HIF regulation in an in vitro model of human ischemia.
Keywords: HIF-1α, prolyl hydroxylase, oxygen and glucose deprivation, ischemia
Introduction
Hypoxia Inducible Factor (HIF) is a transcriptional activator that coordinates evolutionarily conserved adaptive responses to hypoxia (Wang and Semenza 1993). HIF is induced in cerebral ischemia (Stowe et al. 2008, Chavez and LaManna 2002), but perhaps insufficiently; augmenting HIF-mediated adaptive responses during cerebral ischemia may provide novel therapeutic strategies.
HIF is a heterodimer consisting of a constitutively present β subunit and a short-lived α subunit. HIF-1α protein half-life is regulated by oxygen-dependent degradation. Under normoxic conditions, prolyl hydroxylase domain enzymes (PHDs) hydroxylate Pro(402) and Pro(564) on HIF-1α, allowing it to be recognized by the E3 ubiquitin ligase Von Hippel-Lindau (pVHL) and targeted for proteasomal degradation (Jaakkola et al. 2001; Ivan et al. 2001; Bruick et al. 2001). Under hypoxia, a decrease in PHD activity leads to HIF-1α accumulation, heterodimerization with β-subunits, recruitment of p300/CBP, and transactivation of target gene expression (Jiang et al. 1996). HIF target genes include VEGF, EPO, GLUT1, PFK1, Neuroglobin and dozens of other genes that work in concert to restore oxygen delivery and to enhance glucose uptake and glycolysis in order to compensate for reduced oxidative phosphorylation.
HIF can be pharmacologically induced by structurally diverse small molecule PHD inhibitors. PHD inhibition consistently produces neuroprotection in diverse neurological disease models, including stroke (Siddiq et al. 2005; Nagel et al. 2010), Alzheimer’s Disease (McLachlan et al. 1991; Ritchie et al. 2003), allergic encephalomyelitis (Bowern et al. 1984), Parkinson’s Disease (Kaur et al. 2004), oxidative stress (Zaman et al. 1999), excitotoxicity (Li et al. 2011), trophic factor deprivation (Lomb et al. 2007) and mitochondrial dysfunction (Niatsetskaya et al. 2010). PHDs have been identified as the critical targets of iron chelators such as desferoxamine that are clinically beneficial in many neurological disorders and that recapitulate the neuroprotective effects of hypoxic preconditioning on later ischemic challenge (Siddiq et al. 2005). Novel PHD inhibitors poised for clinical trials have been identified through high-throughput screening (Smirnova et al. 2010).
Given the evidence for the regulation of HIF-1α by PHDs, many studies have inferred PHD activity from HIF levels in the absence of a direct assay of PHD enzyme activity in vivo. However, HIF stabilization can occur either via PHD inhibition or by impeding downstream parts of the ODD pathway, or through ODD-independent mechanisms such as p38-MAPK interaction (Emmerling et al. 2005) or NEDD8 conjugation to the PAS-B domain (Ryu et al. 2011). Likewise, PHD inhibition engages multiple downstream effector pathways in addition to HIF; some alternative substrates have been identified that show selectivity among the PHD isoforms, including β-(2) adrenergic receptor (Xie et al. 2009), and Rbp1 (Mikhaylova et al. 2008). Indeed, PHD inhibition produces neuroprotection via both HIF-dependent and HIF-independent mechanisms (Siddiq et al. 2009). Thus it is critical to decouple measures of PHD activity from HIF protein level in order to identify precisely how hypoxic adaptive responses are regulated in normal and pathological conditions and how they may be manipulated for therapeutic benefit.
In vitro studies using oxygen and glucose deprivation (OGD) have indicated that disruptions in glucose supply during cerebral ischemia may curtail endogenous neuroprotective programs. Glucose is required for hypoxic HIF stabilization in human mesangial and hepatoma (Hep3B) cells (Osada-Oka et al. 2010) and in human renal carcinoma (UOK262) cells (Sudarshan et al. 2009). These studies led to a model by which glucose fuels NADPH production through the pentose phosphate pathway to provide a substrate for NADPH-dependent oxidases, which in turn produce reactive oxygen species that inhibit prolyl hydroxylases. Accordingly, glucose deprivation enhances HIF-1α degradation by relieving a ROS-mediated inhibition on PHD enzyme activity. However, it remains unclear 1) whether PHDs in fact mediate the effects of glucose and 2) whether this model is applicable to neuronal cells.
Although the potential therapeutic value of HIF in cerebral ischemia has been widely appreciated for over a decade, prior studies of HIF regulation in neurons have been limited by the difficulty of performing quantitative analysis of immunoblotting employing commercially available HIF antibodies. Several HIF reporters containing luciferase and GFP have been developed and used for high-throughput screening for novel HIF activators (Smirnova et al. 2010) and imaging studies (Safran et al. 2006, Moroz et al. 2009). However, these studies have looked only at accumulation of the reporter without investigating the mechanism by which it accumulates. A full-length HIF reporter cannot distinguish which domains of the protein are important, and inclusion of the transactivation domain introduces the confound of exaggerating HIF-mediated gene transcription, thus engaging feedback loops that may alter HIF regulation. Luciferase has greater sensitivity and a wider dynamic range than GFP. Therefore, we used a reporter consisting of the oxygen-dependent degradation (ODD) domain of HIF-1α fused to firefly luciferase, stably expressed in SH-SY5Y human neuroblastoma cells.
The objectives of this study were: 1) to validate the use of ODD-luc as a quantitative biochemical reporter for HIF-1α stabilization specifically via the PHD-, VHL- and proteasome-dependent pathway, and 2) to apply this tool to test the hypothesis that glucose deprivation enhances HIF-1α degradation by increasing PHD enzyme activity in ischemic human neuronal cells.
Materials and Methods
Cell lines
SH-SY5Y human neuroblastoma cells were maintained in DMEM/F12+ Glutamax (Gibco 10565) with 10% Fetal Bovine Serum, 50 units Penicillin/mL, and 50μg Streptomycin/mL. The ODD-Luc construct encoded in pcDNA3.1 plasmid vector was constructed as described previously (Safran et al. 2006). QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technology) was used to introduce the Pro564Ala and Pro567Ala mutations for the AYIA control line. Cells were transfected with 1 mg plasmid DNA using Lipofectamine 2000 (Invitrogen). Transfected cell were grown in the presence of 0.5 mg/ml Geneticin (GIBCO-Invitrogen). Cell plating density was optimized for induction of ODD-luc activity after 2 hours; for all reported experiments, cells were plated at 5×10^5 cells/mL in volumes of 100μL/well in 96-well plates or 2.5mL/well in 6-well plates (Corning) 24 hours before treatments.
RCC4 cell lines were obtained from Health Protection Agency Culture Collections (cat#03112702 and 03112703) and maintained in DMEM + 10% FBS + 2 mM Glutamine + G418 0.5mg/ml (Gibco/Life Technologies.) Transient transfections of ODD-luc were performed using Lipofectamine 2000 (Invitrogen).
Hypoxic and Drug Treatments
Hypoxia (1% O2, 5% CO2, 94% N2) was induced using a custom-built hypoxic glovebox (Coy Biosystems). Media was equilibrated before treatments started, and depletion of O2 in the media was confirmed with a dissolved oxygen meter DO 6 (Oakton). Cells were washed with warm PBS, then treated with Glucose-free DMEM (Gibco 11966) supplemented with 10% FBS and Pen/Strep and to which glucose or drugs were added at noted concentrations. Cells were incubated for 2 hours in hypoxia or normoxia (21% O2, 5% CO2) at 37°C. Ciclopirox (CPO), 3,4 dihydroxybenzoate (DHB), dimethyloxaloylglycine (DMOG), were obtained from Sigma and solubilized in water or DMSO at 1000x concentrations. Leupeptin and NH4Cl were obtained from CalBiochem (EMD4 Biosciences).
Luciferase activity
Luciferase activity and Dual Luciferase activity assays (Promega) were performed according to manufacturer’s protocol in 96-well plates. Luminescence was read with an LMaxII384 luminometer (Molecular Devices) and analysed by SoftMax Pro software (Molecular Devices).
Immunoblots
After 2h treatments, cells were scraped in ice cold PBS and centrifuged at 1000g for 5 min. The pellet was lysed in RIPA buffer (1% Triton X-100, 1% SDS, 50 mM Tris-Cl, pH 7.4, 500 mM NaCl, 1 mM EDTA) plus 1:100 protease inhibitors cocktail (SIGMA), 10μM MG-132, and 2μL benzonase (Sigma). Protein concentration was quantified using BCA (Pierce) or QuickStart Bradford (Bio-Rad) assay. Samples were loaded on Nu-PAGE gels (Life Technologies), transferred to nitrocellulose membranes, incubated overnight at 4°C with primary antibodies diluted in Odyssey Blocking Buffer. Secondary fluorophore conjugated Odyssey InfraRedDye-680 and IRD-800 antibodies (LI-COR Biosciences) were added at 1:10,000 in Odyssey Blocking Buffer, and incubated for 1h at RT. Immunoreactive proteins were detected using Odyssey IR-imaging system (LI-COR Biosciences). Primary antibodies and dilutions used were: mouse anti-actin (Sigma) 1:10,000; mouse anti-luciferase sc-74548 (Santa Cruz) 1:1000; rabbit anti-HIF-1α NB100-479 (Novus) 1:1000; rabbit anti-Hydroxy-HIF-1α (Pro564) D43B5 (Cell Signaling).
Viability assays
Cell viability was assessed by the MTT (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Life Technologies) and confirmed by phase microscopy.
ATP assays
ATP concentration was measured using ATP Bioluminescent Kit (Sigma). In 96 well plates, cells were lysed with Cell Culture Lysis Reagent (Promega). 1:25 dilution of ATP assay was added, mixed, and luminescence was read immediately with an LMaxII384 luminometer (Molecular Devices).
Gene expression assays
Cells were lysed in Trizol (Invitrogen) and stored at −80°C. Total RNA was extracted using chloroform phase separation and isopropanol precipitation according to manufacturer protocol. Taqman RNA-to-Ct 1-step kit was used with gene expression assays for HIF-1α (Hs00153153_m1) and β-actin as endogenous control. Quantitative real-time PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems).
Statistics
Results are graphed as mean +/− SEM of at least three independent replicates. ANOVA and Tukey’s post-test were used to determine significance (p<0.05). Statistical analysis was performed using GraphPad Prism software.
Results
ODD-luc is a quantitative reporter for HIF stabilization and Prolyl Hydroxylase activity
HIF-1α protein levels are difficult to study quantitatively in neuronal cells due to the high coefficient of variation of immunoblots with commercially available antibodies. A fusion protein containing the oxygen dependent degradation domain of HIF-1α (a.a. 513–630) has been previously used as a reporter for HIF-1α stabilization, for high-throughput screening for novel activators of the hypoxic response (Smirnova et al. 2010), and for in vivo imaging (Safran et al. 2006, Goldman et al. 2011). We sought to use the ODD-luc construct as a biochemical reporter for HIF stabilization through the canonical PHD-dependent pathway in human neuroblastoma (SH-SY5Y) cells. We were interested in the mechanism by which HIF is engaged long before sustained stress leads to cell death, so we measured effects after no more than 2 hours of hypoxia (1% O2), oxygen and glucose deprivation (OGD), or drug treatments, a time window within which no cell death occurs. We first demonstrated that ODD-luc detection by western blot with a monoclonal antibody against firefly luciferase is highly specific, sensitive, and quantitative over a wide dynamic range (data not shown).
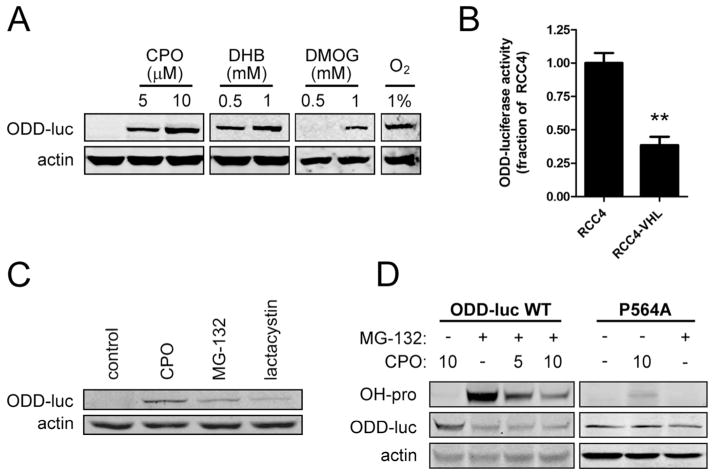
To ensure that the oxygen-dependent degradation of ODD-luc is achieved in a manner similar to that of endogenous full-length HIF-1α, we verified that ODD-luc activity and protein levels are dose-dependently stabilized within two hours by structurally diverse PHD inhibitors, including hypoxia (1% O2); ciclopirox (CPO), a specific inhibitor of the iron-binding site of the HIF prolyl hydroxylases; 3,4 di-hydroxybenzoate (DHB), a 2-oxoglutarate analog; and dimethyloxaloylglycine (DMOG) (Fig. 1A and Smirnova et al. 2010).
Figure 1.
ODD-luc is a biochemical reporter for HIF-1α. A. Luciferase immunoblot after treatment with hypoxia and structurally diverse PHD inhibitors: 5 or 10μM ciclopirox (CPO); 0.5 or 1mM 3,4 di-hydroxybenzoate (DHB), and 0.5 or 1mM dimethyloxaloylglycine (DMOG). B. ODD-luc activity in RCC4 cells null for, or ectopically expressing, VHL. C. ODD-luc immunoblot after treatment with proteasome inhibitors: 10uM MG-132 or 10uM lactacystin. D. Immunoblot for luciferase or hydroxy-proline(564) upon treatment with MG-132 (10μM) and diminished dose-dependently by CPO (μM doses as indicated) in wild-type ODD-luc or P564A mutant-expressing cells. **p<.01 in ANOVA with post-hoc Tukey test.
Canonical hydroxylation of the ODD degron during normoxia leads to the recruitment of the E3 Ubiquitin Ligase, Von Hippel Lindau protein. To determine whether ODD-luc is degraded in a VHL-dependent manner, we sought to measure ODD-luc stabilization upon knock-down of VHL in SH-SY5Y cells, however we could not confirm pVHL protein expression in SH-SY5Y cells with commercially available VHL antibodies. Instead, we transfected ODD-luc into VHL null human renal carcinoma cells expressing empty vector (RCC4-EV) or pVHL (RCC4-VHL). RCC4-VHL cells showed 60% less steady-state ODD-luc expression than RCC4-EV cells (Fig. 1B) indicating that normoxic degradation of ODD-luc is enhanced by expression of pVHL.
Ubiquitination of HIF-1α is followed rapidly by its degradation by the 26S proteasome. To verify that ODD-luc is degraded in a proteasome dependent manner, we used two structurally diverse proteasome inhibitors: MG-132, a competitive peptide inhibitor, and lactacystin, an irreversible alkylator. Both compounds were able to dose-dependently stabilize ODD-luc as measured by luciferase activity (data not shown) and western blot for luciferase (Fig. 1C).
As expected, proteasome inhibition caused accumulation of a 75kDa band corresponding to the ODD-luciferase. Of note, despite the ability of proteasome inhibition to elevate luciferase activity to a level similar to PHD inhibition, the protein levels of ODD-luc in the proteasome inhibitor-treated cells were lower. HIF-1α is ubiquitinated at several lysine residues (K532, K538, K547), all found within the ODD (Paltoglou and Roberts 2007). We therefore expected that proteasome inhibitors would cause accumulation of higher molecular weight bands corresponding to polyubiquitinated ODD-luc, but we were unable to detect these bands with the luciferase antibody, or with immunoblot for ubiquitin after immunoprecipitation with a luciferase antibody (IP:luc, IB:ubiquitin), or transfection with HA-ubiquitin followed by IP:HA, IB:luc or IP:luc, IB:HA, or with several other commercially available luciferase antibodies after IP:HA (data not shown). We conclude that ubiquitination of the ODD-luc construct partially masks the luciferase epitope but that ubiquitination does not affect luciferase activity.
We next verified that ODD-luc can be used to detect PHD-dependent hydroxylation at Pro(564). Using an antibody that recognizes HIF-1α hydroxylated at Pro(564), we observed that hydroxylation of ODD-luc is robustly observed upon stabilization with the proteasome inhibitor MG-132 and is dose-dependently diminished by the PHD inhibitor CPO (Fig. 1D). We further confirmed specificity of the hydroxy-proline immunoblotting by using a P564A mutant ODD-luc, which showed no change in luciferase levels with PHD or proteasome inhibition and which showed no hydroxylation under any condition (Fig. 1D).
Taken together, these findings confirm that the ODD-luc reporter is regulated by PHD, VHL, and the proteasome in a manner similar to endogenous HIF.
Oxygen-glucose deprivation decreases hypoxic accumulation of ODD-luc
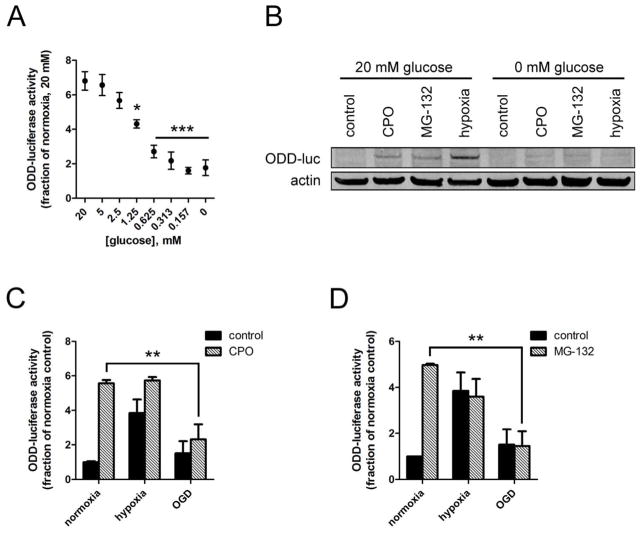
Hypoxia (1% O2, 2h) induced ODD-luc activity approximately 6-fold in normal glucose (20mM), but decreasing the glucose concentration dose-dependently abrograted the hypoxic induction of ODD-luc, to below 2-fold in the absence of glucose, as measured by luciferase activity (Fig. 2A). Hypoxia and OGD had no effect on cell viability within 2 hours (data not shown). Further, ODD-luc protein levels after 2h hypoxia were markedly decreased in the absence of glucose (Fig. 2B).
Figure 2.
Glucose deprivation abrogates hypoxic ODD-luc stabilization in a manner independent of PHD- or proteasome-mediated degradation. A. ODD-luc activity upon hypoxia (2h 1% O2) with varying glucose concentrations. B. ODD-luc protein stabilization by 2 hour treatment with 10μM CPO (C), 10uM MG-132 (M), or hypoxia (H) in the presence or absence of glucose. C. ODD-luc activity upon treatment with 10uM CPO in hypoxia or OGD D. ODD-luc activity upon treatment with 10uM MG-132 in hypoxia or OGD. **p<.01, ***p<.001 in ANOVA with post-hoc Tukey test.
This finding confirmed that oxygen-glucose deprivation abrogates hypoxic HIF-1α induction by acting on its ODD domain in human neuroblastoma cells, consistent with prior reports using other cell types. Therefore, we systematically investigated the mechanism by which glucose affects the ODD, starting with the hypothesis that glucose deprivation enhances PHD enzyme activity.
Glucose Deprivation does not enhance PHD activity or Proteasome-mediated ODD-luc degradation
To determine whether glucose deprivation enhances PHD enzyme activity in hypoxia, we sought to measure the effect of OGD on prolyl hydroxylation of ODD-luc by OH-Pro564 immunoblot when ODD-luc degradation was blocked with proteasome inhibition. Unexpectedly, proteasome inhibition with saturating doses of MG-132 (up to 20uM) or lactacystin (up to 10uM) was not able to accumulate ODD-luc protein in the absence of glucose, so we could not compare the hydroxylated fraction of ODD-luc across glucose concentrations (Fig 2B). Indeed, glucose deprivation completely abolished ODD-luc induction by CPO, MG-132, or Hypoxia as detected by luciferase immunoblot (Fig. 2B). Therefore, we turned to methods other than direct measurement of P564 hydroxylation to determine how oxygen -glucose deprivation negatively regulates HIF.
We reasoned that if enhanced PHD activity was responsible for the loss of hypoxic HIF induction in the absence of glucose, then PHD inhibition should rescue hypoxic stabilization of ODD-luc under oxygen and glucose deprivation (OGD). While ciclopirox (CPO, 10μM) induced ODD-luc activity approximately 5-fold in normoxia, it had no significant additive effect on hypoxic ODD-luc induction in normal glucose, and did not significantly increase ODD-luc levels under OGD (Fig. 2C), suggesting that PHD enzyme activity was not enhanced by glucose deprivation.
In light of these results, we hypothesized that glucose deprivation instead enhanced proteasomal degradation downstream of PHD activity, potentially by enhancing VHL-mediated ubiquitination or proteasomal recognition of ODD-luc. We reasoned that if glucose deprivation enhanced proteasome-dependent degradation, then proteasome inhibition should rescue hypoxic ODD-luc stabilization in OGD. MG-132 (10μM) induced ODD-luc activity approximately 5-fold in normoxia, had no significant additive effect on hypoxic ODD-luc induction in normal glucose, and did not significantly increase ODD-luc levels under OGD (Fig. 2D).
Taken together, these experiments indicated that glucose deprivation does not reduce hypoxic HIF induction through the enhancement of canonical PHD- and proteasome-dependent degradative pathway.
Glucose Deprivation does not enhance ODD-luc degradation
HIF-1α can be degraded not only by the 26S proteasome, but also by lysosomes; HIF-1α has been reported to be stabilized by lysosomal inhibition, independent of its stabilization by proteasome, HSP90 or calpain inhibition, downstream of 15-Deoxy-Delta(12,14)-prostaglandin-J(2) (Olmos et al. 2009). It is not known what domain(s) of HIF-1α are necessary for lysosomal degradation, and thus whether the ODD is sufficient for, or more susceptible than endogenous HIF-1α to, lysosomal degradation. We next investigated whether glucose deprivation enhances lysosomal degradation of ODD-luc in ischemic SH-SY5Y cells.
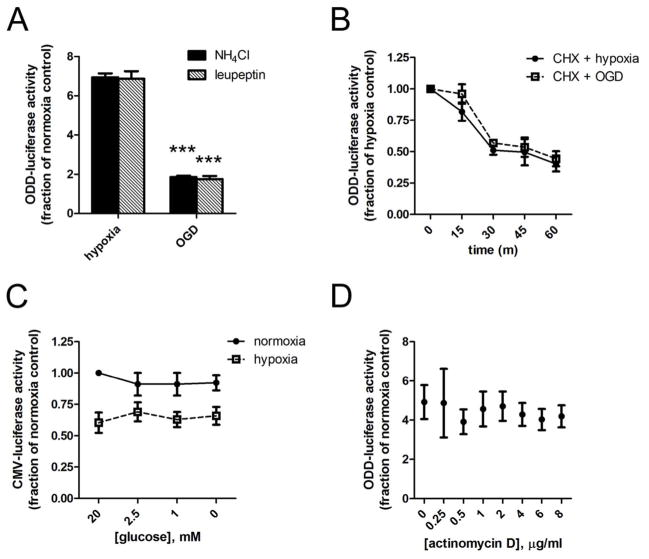
Structurally diverse lysosomal inhibitors NH4CL (5mM) and leupeptin (250 μM) were unable to stabilize ODD-luc under 2h OGD (Fig. 3A), and did not affect cell viability under these conditions. This finding is consistent with the previous finding that lysosomal degradation of HIF-1α occurs very slowly (Olmos et al. 2009), and suggests that enhanced lysosomal degradation cannot explain the loss of hypoxic ODD-luc induction in the absence of glucose over the short epoch of observation used in our analysis.
Fig 3.
Glucose Deprivation in hypoxia does not enhance ODD-luc degradation, or decrease global transcription, translation, or luciferase activity A. ODD-luc activity after 2h hypoxia or OGD in the presence of lysosomal inhibitors NH4Cl 5mM or leupeptin 250uM. B. ODD-luc activity after 2h pre-treatment with hypoxia followed by 0, 15, 30, 45, or 60 min treatment with Cyclohexamide (10ug/mL) in hypoxia with 20mM glucose (CHX) or 0mM glucose (CHX + GD); C. CMV-luc activity after 2h normoxia or hypoxia with glucose concentrations from 0–20mM; D. ODD-luc activity after 2h hypoxia and co-treatment with actinomycin D at 0.25–8ug/mL. ***p<.001, in ANOVA with posthoc Tukey test.
To further examine whether glucose deprivation enhances ODD-luc degradation, we measured the half-life of ODD-luc by blocking protein synthesis with cyclohexamide (CHX) in the presence or absence of glucose. In the presence of 10μg/mL CHX, glucose concentration had no significant effect on ODD-luc degradation, with an interpolated half-life of 45.1 minutes in normal glucose and 50.6 minutes in the absence of glucose (Fig. 3B).
Taken together, these data suggest that glucose deprivation does not enhance HIF-1α degradation through the ODD domain, but may instead suppress HIF-1α transcription or translation.
Glucose deprivation does not affect transcription of ODD-luc or HIF-1α
To determine whether glucose deprivation suppresses the transcription or translation of ODD-luc in hypoxia, we first examined the control construct that expresses firefly luciferase without the ODD in the same plasmid vector (CMV-Luc). CMV-Luc expression, as measured by luciferase activity assay or luciferase immunoblot, is constitutively much higher than ODD-luc, and is unaffected by hypoxia, PHD inhibition, or proteasome inhibition (data not shown). CMV-luc expression is reduced by hypoxia by about 30% (Fig. 3C). This is consistent with numerous reports of global inhibition of translation under hypoxia, which occurs rapidly through PERK and eIF2a phosphorylation and is maintained over a longer term through mTOR and EIF4e (Liu et al. 2006, Spriggs et al. 2010). In contrast to ODD-luc, CMV-luc expression is not affected by glucose concentration (Fig. 3C). This suggests that glucose deprivation does not abrogate hypoxic ODD-luc induction via non-selective suppression of transcription, translation, or luciferase activity.
We next examined whether transcriptional inhibition with actinomycin D could mimic the effect of glucose deprivation on ODD-luc levels. Actinomycin D did not abrogate ODD-luc levels induced by hypoxia within 2 hours (Fig. 3D), indicating that glucose deprivation acts post-transcriptionally to suppress ODD-luc in hypoxia.
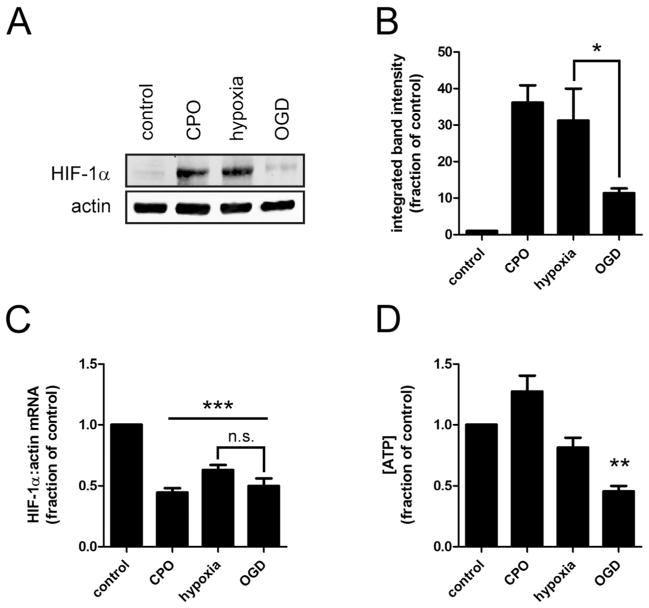
To confirm the relevance of our experiments with the ODD-luc reporter, we examined endogenous HIF-1α message and protein levels in wild-type SH-SY5Y cells treated with CPO, hypoxia, or OGD for 2h. HIF-1α mRNA levels were unchanged by OGD relative to hypoxia (Fig. 4C), indicating that glucose deprivation does not diminish HIF-1α transcription or mRNA stability. In contrast, HIF-1α protein levels were decreased in OGD relative to hypoxia (Fig. 4A and 4B), consistent with the findings using the ODD-luc reporter.
Figure 4.
OGD compared to hypoxia alone abrogates endogenous HIF-1α protein but not mRNA levels, and reduces cellular ATP levels. After 2 hour treatments with 10uM CPO, Hypoxia (1%O2, 20mM Glucose), or OGD (1%O2, 0mM Glucose): A. Representative immunoblot for HIF-1α; B. HIF-1α protein stabilization measured by densitometric analysis of three immunoblots; C. HIF-1α mRNA determined by qRT-PCR with beta-actin as endogenous control; D. ATP concentration, relative to untreated control. *p<.05, **p<.01, ***p<.001, n.s. no significant difference in ANOVA with post-hoc Tukey test.
Taken together, these data suggest that glucose deprivation decreases HIF-1α translation under hypoxia.
Glucose Deprivation decreases ATP levels
HIF-1α and other stress-responsive genes are preferentially translated even under the global suppression of protein synthesis that occurs in hypoxia (reviewed by Spriggs et al. 2010). We hypothesized that glucose deprivation produces an additional stress that overwhelms this selective translation of HIF-1α. Therefore we investigated how ATP levels are affected in SH-SY5Y cells in OGD compared to hypoxia alone.
We measured ATP concentration in cells treated with CPO, hypoxia, or OGD for 2 hours. ATP levels were not significantly affected by CPO or hypoxia, but were reduced by approximately 50% after 2h OGD compared to Hypoxia (Fig. 4D). As in previous experiments there was no difference in cell viability with any treatment within 2 hours (not shown).
Discussion
We have demonstrated for the first time that combined oxygen and glucose deprivation suppresses hypoxic HIF induction in human neuronal cells, consistent with findings in other cell types, including rat cortical neurons (Guo et al. 2008), human hepatoma (Hep3B) cells (Osada-Oka et al. 2010) and human renal carcinoma (UOK262) cells (Sudarshan et al. 2009). Using our ODD-luc reporter, we systematically investigated the mechanism by which glucose modulates HIF-1α protein levels, and found that glucose deprivation does not enhance PHD-dependent HIF-1α degradation; rather, we observe that glucose deprivation leads to ATP depletion and overcomes the selective synthesis of HIF-1α within the context of global translational suppression in hypoxia.
Several mechanisms have been proposed to explain the continued synthesis of HIF-1α during global translational suppression in hypoxia. HIF-1α mRNA is selectively stabilized in hypoxia by RNA binding proteins that recognize the 5′ and 3′ UTRs (Galbán et al. 2008). The HIF-1α 5′ UTR has been reported to contain an internal ribosome entry site (IRES), though this is controversial (reviewed by Spriggs et al. 2010). Other stress responsive proteins including ATF4 contain ‘decoy’ upstream open reading frames, which may also be present in HIF-1α (Spriggs et al. 2010). None of these mechanisms can explain our finding with the ODD-specific reporter. The fact that the ectopically expressed ODD-luc protein (but not the CMV-luc control protein) is regulated by glucose at the level of synthesis suggests that HIF-1α translation is regulated at least in part by the ODD portion of the coding region, a novel insight that should be verified with endogenous HIF-1α.
One way in which glucose deprivation could suppress HIF-1α synthesis in hypoxia is via activation of AMP-activated protein kinase (AMPK). AMPK is a key integrator of metabolic homeostasis. As part of its program to reduce ATP demand in times of scarcity, AMPK suppresses protein synthesis through two distinct mechanisms. First, AMPK activates TSC2, which inhibits mTOR, thus reducing activity of ribosomal S6 kinase and eukaryotic initiation factor-4 (eIF4). Second, AMPK activates eukaryotic elongation factor-2 (eEF2) kinase, leading to the phosphorylation and inhibition of eEF2. AMPK becomes activated by phosphorylation at threonine 172, which occurs in an LKB1-dependent manner upon ATP depletion or several other forms of cellular stress, including sustained hypoxia. Serum deprivation has been reported to accelerate hypoxic AMPK activation and subsequent inhibition of protein synthesis in hypoxia from 20h to 30 minutes (Liu et al. 2006). We propose that glucose deprivation has synergistic effects similar to serum deprivation in the context of hypoxia.
AMPK phosphorylation occurs in ischemia in vivo and contributes to injury; inhibiting pAMPK reduces infarct volume and behavioral deficits, and pAMPK downregulation is essential for neuroprotection by ischemic preconditioning (Li et al. 2010, Venna et al. 2012). This is consistent with the notion that neuronal resilience to ischemia can be enhanced by sustaining the synthesis of HIF-1α, so that stress-sensitive post-translational checkpoints allow it to engage a broad adaptive response when appropriate. On the other hand, increasing HIF-1α protein levels outright has produced conflicting effects. In vivo models show HIF can prevent injury (Baranova et al. 2007) and improve functional recovery (Wu et al. 2010), or increase infarct and edema volume (Chen et al. 2008; Higashida et al. 2011), or have opposing effects at progressive timepoints after stroke (Yeh et al. 2010). In vitro models also show HIF either protects or exacerbates cell death via expression of target genes that encode both pro-survival and pro-apoptotic proteins (Aminova et al. 2005, Guo et al. 2009), and demonstrate divergent roles of HIF in distinct CNS cell types (Vangeison et al. 2008). In contrast to direct HIF manipulation, PHD inhibition is broadly neuroprotective, as discussed above. Together these observations highlight the need to decouple measurements of PHD enzyme activity from HIF protein levels in order to identify the optimal therapeutic targets within hypoxia-responsive pathways. We validated the utility of the ODD-luc reporter as a quantitative assay for ODD-specific regulation of HIF protein levels. Although pathophysiological regulation of the reporter’s synthesis constrains its usefulness in ischemia, we validated that immunoblotting for hydroxy-P564 on ODD-luc can serve as a highly quantitative direct assay of PHD enzyme activity, a valuable resource for future studies of the mechanisms of HIF regulation.
NADPH supplementation has been reported to rescue hypoxic HIF accumulation under glucose deprivation, suggesting that glucose is required to replenish NADPH through the pentose phosphate pathway (Osada-Oka et al. 2010). In light of our findings, it is possible that NADPH bolsters HIF-1α accumulation in OGD not via ROS regulation of PHDs but instead by supporting energy homeostasis and preventing AMPK-mediated suppression of HIF-1α translation. In addition, Guo and colleagues have found in cortical neurons that glucose maintains a reducing environment (e.g. through NADPH facilitating GSH regeneration from GSSG via glutathione reductase), supporting a model by which ROS induces HIF-1α degradation under hypoxia, in contrast to findings in other systems that ROS can stabilize HIF-1α (Ryu et al. 2011), or that mitochondrial ROS production is in fact necessary for the hypoxic stabilization of HIF-1α (Guzy and Schumacker 2006). We saw no effect of structurally diverse antioxidants on hypoxic ODD-luc accumulation in normoxia or hypoxia in SH-SY5Y cells (data not shown), suggesting that any effect of ROS on HIF-1α protein levels in these cells occurs through ODD-independent mechanisms, e.g. not through ROS-mediated inhibition of PHDs.
Our finding of abrogated HIF-1α induction in OGD in vitro is not inconsistent with reports of HIF-1α protein accumulation in ischemia in vivo, because ischemic pathology unfolding in an intact brain involves diverse factors such as inflammation that may induce HIF-1α levels independent of mechanisms elucidated here.
Redundancy in the mechanisms by which HIF-1α may be induced highlights its importance and its potential power as a therapeutic target. The present study suggests that augmenting HIF signaling in ischemic neurons will not be achieved through antioxidant or PHD inhibition strategies, but rather by targeting ATP homeostasis and/or translational regulation, a premier candidate for which is AMPK. It remains to be determined whether HIF protein levels correlate with cellular health in the ischemic human brain. Additionally, our data suggest that any neuroprotective effects of prolyl hydroxylase inhibition in neuronal ischemia may be working through HIF-independent mechanisms given the scarcity of HIF when glucose is limiting.
Acknowledgments
The authors would like to thank B.J. Casey, David Eliezer, Hazel Szeto, and John Wagner for helpful discussions and Li Xia for technical assistance. This work was supported by NYS DOH C019772 (R.R.R.), the Thomas Hartman Foundation (R.R.R), the Miriam and Sheldon G. Adelson Medical Research Foundation (R.R.R) and the Burke Foundation.
Footnotes
The authors have no conflicts of interest to disclose.
References
- Aminova LR, Chavez JC, Lee J, Ryu H, Kung A, Lamanna JC, Ratan RR. Prosurvival and prodeath effects of hypoxia-inducible factor-1alpha stabilization in a murine hippocampal cell line. J Biol Chem. 2005;280(5):3996–4003. doi: 10.1074/jbc.M409223200. [DOI] [PubMed] [Google Scholar]
- Baranova O, Miranda LF, Pichiule P, Dragatsis I, Johnson RS, Chavez JC. Neuron-specific inactivation of the hypoxia inducible factor 1 alpha increases brain injury in a mouse model of transient focal cerebral ischemia. J Neurosci. 2007;27:6320–6332. doi: 10.1523/JNEUROSCI.0449-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowern N, Ramshaw IA, Clark IA, Doherty PC. Inhibition of autoimmune neuropathological process by treatment with an iron-chelating agent. J Exp Med. 1984;160(5):1532–43. doi: 10.1084/jem.160.5.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Chavez JC, LaManna JC. Activation of hypoxia-inducible factor-1 in the rat cerebral cortex after transient global ischemia: potential role of insulin-like growth factor-1. J Neurosci. 2002;22(20):8922–31. doi: 10.1523/JNEUROSCI.22-20-08922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jadhav V, Tang J, Zhang JH. HIF-1alpha inhibition ameliorates neonatal brain injury in a rat pup hypoxic-ischemic model. Neurobiol Dis. 2008;31(3):433–41. doi: 10.1016/j.nbd.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, Gorospe M. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol Cell Biol. 2008;28(1):93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SJ, Chen E, Taylor R, Zhang S, Petrosky W, Reiss M, Jin S. Use of the ODD-luciferase transgene for the non-invasive imaging of spontaneous tumors in mice. PLoS One. 2011;6(3):e18269. doi: 10.1371/journal.pone.0018269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Miyake M, Liu KJ, Shi H. Specific inhibition of hypoxia inducible factor 1 exaggerates cell injury induced by in vitro ischemia through deteriorating cellular redox environment. J Neurochem. 2009;108(5):1309–21. doi: 10.1111/j.1471-4159.2009.05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91(5):807–19. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- Higashida T, Peng C, Li J, Dornbos D, 3rd, Teng K, Li X, Kinni H, Guthikonda M, Ding Y. Hypoxia-inducible factor-1α contributes to brain edema after stroke by regulating aquaporins and glycerol distribution in brain. Curr Neurovasc Res. 2011;8(1):44–51. doi: 10.2174/156720211794520251. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271(30):17771–8. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- Li D, Bai T, Brorson JR. Adaptation to moderate hypoxia protects cortical neurons against ischemia-reperfusion injury and excitotoxicity independently of HIF-1α. Exp Neurol. 2011;230(2):302–10. doi: 10.1016/j.expneurol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Benashski SE, Siegel C, Liu F, McCullough LD. Adenosine monophosphate activated protein kinase inhibition is protective in both sexes after experimental stroke. Neurosci Lett. 2010;482(1):62–5. doi: 10.1016/j.neulet.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21(4):521–31. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomb DJ, Straub JA, Freeman RS. Prolyl hydroxylase inhibitors delay neuronal cell death caused by trophic factor deprivation. J Neurochem. 2007;103(5):1897–906. doi: 10.1111/j.1471-4159.2007.04873.x. [DOI] [PubMed] [Google Scholar]
- Kaur D, Andersen J. Does cellular iron dysregulation play a causative role in Parkinson’s disease? Ageing Res Rev. 2004;3(3):327–43. doi: 10.1016/j.arr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Mikhaylova O, Ignacak ML, Barankiewicz TJ, Harbaugh SV, Yi Y, Maxwell PH, Schneider M, Van Geyte K, Carmeliet P, Revelo MP, Wyder M, Greis KD, Meller J, Czyzyk-Krzeska MF. The von Hippel-Lindau tumor suppressor protein and Egl-9-Type proline hydroxylases regulate the large subunit of RNA polymerase II in response to oxidative stress. Mol Cell Biol. 2008;28(8):2701–17. doi: 10.1128/MCB.01231-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz E, Carlin S, Dyomina K, Burke S, Thaler HT, Blasberg R, Serganova I. Real-time imaging of HIF-1alpha stabilization and degradation. PLoS One. 2009;4(4):e5077. doi: 10.1371/journal.pone.0005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel S, Papadakis M, Chen R, Hoyte LC, Brooks KJ, Gallichan D, Sibson NR, Pugh C, Buchan AM. Neuroprotection by dimethyloxalylglycine following permanent and transient focal cerebral ischemia in rats. J Cereb Blood Flow Metab. 2011;31(1):132–43. doi: 10.1038/jcbfm.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niatsetskaya Z, Basso M, Speer RE, McConoughey SJ, Coppola G, Ma TC, Ratan RR. HIF prolyl hydroxylase inhibitors prevent neuronal death induced by mitochondrial toxins: therapeutic implications for Huntington’s disease and Alzheimer’s disease. Antioxid Redox Signal. 2010;12:435–443. doi: 10.1089/ars.2009.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos G, Arenas MI, Bienes R, Calzada MJ, Aragonés J, Garcia-Bermejo ML, Landazuri MO, Lucio-Cazaña J. 15-Deoxy-Delta(12,14)-prostaglandin-J(2) reveals a new pVHL-independent, lysosomal-dependent mechanism of HIF-1α degradation. Cell Mol Life Sci. 2009;66(13):2167–80. doi: 10.1007/s00018-009-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada-Oka M, Hashiba Y, Akiba S, Imaoka S, Sato T. Glucose is necessary for stabilization of hypoxia-inducible factor-1α under hypoxia: contribution of the pentose phosphate pathway to this stabilization. FEBS Lett. 2010;584(14):3073–9. doi: 10.1016/j.febslet.2010.05.046. [DOI] [PubMed] [Google Scholar]
- Paltoglou S, Roberts BJ. HIF-1a and EPAS ubiquitination mediated by the VHL tumour suppressor involves flexibility in the ubiquitination mechanism, similar to other RING E3 ligases. Oncogene. 2007;26:604–609. doi: 10.1038/sj.onc.1209818. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Li SH, Park HS, Park JW, Lee B, Chun YS. Hypoxia-inducible factor α subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J Biol Chem. 2011;286(9):6963–6970. doi: 10.1074/jbc.M110.188706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Kim WY, O’Connell F, Flippin L, Gunzler V, Horner JW, Depinho RA, Kaelin WG., Jr Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci USA. 2006;103:105–110. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq A, Aminova LR, Troy CM, Suh K, Messer Z, Semenza GL, Ratan RR. Selective inhibition of hypoxia-inducible factor (HIF) prolyl-hydroxylase 1 mediates neuroprotection against normoxic oxidative death via HIF- and CREB-independent pathways. J Neurosci. 2009;29:8828–8838. doi: 10.1523/JNEUROSCI.1779-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq A, Ayoub IA, Chavez JC, Aminova L, Shah S, LaManna JC, Patton SM, Connor JR, Cherny RA, Volitakis I, Bush AI, Langsetmo I, Seeley T, Gunzler V, Ratan RR. Hypoxia-inducible factor prolyl 4-hydroxylase inhibition. A target for neuroprotection in the central nervous system. J Biol Chem. 2005;280:41732–41743. doi: 10.1074/jbc.M504963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova NA, Rakhman I, Moroz N, Basso M, Payappilly J, Kazakov S, Hernandez-Guzman F, Gaisina IN, Kozikowski AP, Ratan RR, Gazaryan IG. Utilization of an in vivo reporter for high throughput identification of branched small molecule regulators of hypoxic adaptation. Chem Biol. 2010;17(4):380–91. doi: 10.1016/j.chembiol.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40(2):228–37. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Stowe AM, Plautz EJ, Nguyen P, Frost SB, Eisner-Janowicz I, Barbay S, Dancause N, Sensarma A, Taylor MD, Zoubina EV, Nudo RJ. Neuronal HIF-1 alpha protein and VEGFR-2 immunoreactivity in functionally related motor areas following a focal M1 infarct. J Cereb Blood Flow Metab. 2008;28(3):612–20. doi: 10.1038/sj.jcbfm.9600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshan S, Sourbier C, Kong HS, Block K, Valera Romero VA, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, Lee MJ, Gourlay CW, Trepel J, Linehan WM, Neckers L. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1alpha stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29(15):4080–90. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangeison G, Carr D, Federoff HJ, Rempe DA. The good, the bad, and the cell type-specific roles of hypoxia inducible factor-1 alpha in neurons and astrocytes. J Neurosci. 2008;28(8):1988–93. doi: 10.1523/JNEUROSCI.5323-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venna VR, Li J, Benashski SE, Tarabishy S, McCullough LD. Preconditioning induces sustained neuroprotection by downregulation of adenosine 5′-monophosphate-activated protein kinase. Neuroscience. 2012;201:280–7. doi: 10.1016/j.neuroscience.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268(29):21513–8. [PubMed] [Google Scholar]
- Xie L, Xiao K, Whalen EJ, Forrester MT, Freeman RS, Fong G, Gygi SP, Lefkowitz RJ, Stamler JS. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2(78):ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Ou LC, Gean PW, Hung JJ, Chang WC. Selective inhibition of early--but not late--expressed HIF-1α is neuroprotective in rats after focal ischemic brain damage. Brain Pathol. 2011;21(3):249–62. doi: 10.1111/j.1750-3639.2010.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman K, Ryu H, Hall D, O’Donovan K, Lin KI, Miller MP, Marquis JC, Baraban JM, Semenza GL, Ratan RR. Protection from oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21(waf1/cip1), and erythropoietin. J Neurosci. 1999;19:9821–9830. doi: 10.1523/JNEUROSCI.19-22-09821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]