Abstract
There is growing evidence that cancer-initiation could result from epigenetic changes. Y-box binding protein-1 (YB-1) is a transcription/translation factor that promotes the formation of tumors in transgenic mice; however, the underlying molecular events are not understood. To explore this in a human model system, YB-1 was expressed in mammary epithelial cells under the control of a tetracycline-inducible promoter. The induction of YB-1 promoted phenotypes associated with malignancy in three-dimensional breast acini cultures. This was attributed to YB-1 enhancing the expression and activity of the histone acetyltransferase p300 leading to chromatin remodeling. Specifically, this relaxation of chromatin allowed YB-1 to bind to the BMI1 promoter. The induction of BMI1 engaged the Polycomb complex resulting in histone H2A ubiquitylation and repression of the CDKN2A locus. These events manifested functionally as enhanced self-renewal capacity that occurred in a BMI1-dependent manner. Conversely, p300 inhibition with anacardic acid prevented YB-1 from binding to the BMI1 promoter and thereby subverted self-renewal. Despite these early changes, full malignant transformation was not achieved until RSK2 became overexpressed concomitant with elevated hTERT activity. The YB-1/RSK2/hTERT expressing cells formed tumors in mice that were molecularly subtyped as basal-like breast cancer. We conclude that YB-1 cooperates with p300 to allow BMI1 to over-ride p16INK4a-mediated cell cycle arrest enabling self-renewal and the development of aggressive breast tumors.
Keywords: Neoplastic cell transformation, Epigenetics, Breast cancer, Cancer stem cells, Human mammary epithelial cells (HMEC), Y-box binding protein-1 (YB-1)
Introduction
Y-box binding protein-1 (YB-1) is expressed in over 40% of breast cancers, but is most commonly associated with the estrogen receptor (ER)-negative subtypes [1, 2]. The activity of YB-1 is contingent upon phosphorylation of its serine-102 residue by p90 ribosomal S6 kinase (RSK) [3]. Most recently, it has been reported that activated pYB-1S102 is involved in maintaining tumor-initiating cell (TIC) populations via the transactivation of CD44 and CD49f [4]. Within the last decade, mounting evidence has supported the notion that cancer arises from a pool of stem/progenitor-like cells, colloquially referred to as TICs [5–8]. These cells share important properties with classical tissue stem cells including self-renewal and multi-lineage differentiation capacity [9]. Pioneering work in this field originated in studies of leukemia stem cells [10] and later was expanded to encompass solid tumors of the breast [11]. It has since been established that a subpopulation of mammary cells defined by CD44+/CD24− express putative stem cell markers and have the capacity to initiate tumors at limiting dilutions, a key characteristic of TICs [12].
Genes involved in cell fate determination, immortalization, and DNA repair are epigenetically deregulated in breast cancer, the most notable being the Polycomb group (PcG) target CDKN2A (encoding p16INK4a and p14ARF) [13]. Specifically, the PcG protein BMI1 transcriptionally represses this locus during the transformation of human mammary epithelial cells (HMECs) [14]. In addition, BMI1 has been linked to activation of human telomerase reverse transcriptase (hTERT) and induction of telomerase activity [14]. Taken together, this permits cells to escape from senescence, extend replicative life span, and acquire stem/progenitor cell properties such as enhanced self-renewal capacity. This rationalizes how BMI1 is able to provoke HMEC transformation [15, 16]. Notably, a subset of HMECs detected in disease free women exhibit silenced p16INK4a and genomic instability that could act as precursors to breast cancer [13, 17].
The expression of YB-1 in transgenic mice leads to the development of tumors with 100% incidence [18]. However the molecular events that lead up to tumor formation are unclear. In light of our previous finding that YB-1 functions as a cancer susceptibility gene [19], we wanted to address its potential role in transforming HMECs.
Materials and Methods
Cell lines and treatments
H16N2 HMECs with tetracycline-inducible YB-1 (HTRY) or LacZ (HTRZ) were cultured as previously described [20]. By culturing HTRY cells in doxycycline for 30 days two cell lines were established, referred to as HTRY-LT#1 and HTRY-LT#2. Transgene expression was achieved with 1 μg/ml doxycycline (Calbiochem, Gibbstown, NJ, USA) for 96 hours, unless otherwise noted. The normal HMECs 184hTERT and MCF10A (ATCC, Manassas, VA, USA) were cultured in HuMEC media (Invitrogen, Burlington, Canada). The breast cancer cell lines MDA-MB-231 (ATCC), SUM149 (Asterand, Detroit, MI, USA), and MDA-MB-435/LCC6 (a gift from Dr. Marcel Bally, BC Cancer Agency) were cultured as recommended. All cell lines tested negative for contamination with mycoplasma using a PCR-based detection assay (Agilent Technologies, Mississauga, Canada).
The inhibitors anacardic acid (Sigma-Aldrich, Oakville, Canada), BI-D1870 (synthesized by CDRD, Vancouver, Canada), LY-294002 (Calbiochem), and MG132 (Calbiochem) were dissolved in DMSO.
Quantitative real-time PCR
qRT-PCR was carried out with FAM-labeled Taqman Assays-on-Demand probes (Applied Biosystems, Carlsbad, CA, USA) on freshly isolated RNA as described [21]. Results were analyzed with the Δ-ΔCt method normalized to TBP and compared to a comparator sample.
Acini morphogenesis assay
Three-dimensional basement membrane cultures were setup using a well-established method [22] described in detail in Supplemental Methods.
Immunoblotting, immunofluorescence, and immunoprecipitation
Immunoblotting, immunofluorescence, and immunoprecipitation were performed as described previously [19]. Immunoblots were quantified using ImageJ densitometry software. Cyto-nuclear fractions were isolated using the NE-PER Extraction Kit (Thermo Fisher Scientific, Rockford, IL, USA). Images were acquired using an Olympus FV10i laser scanning confocal microscope. The specific antibodies are detailed in Supplemental Methods.
Mammosphere assay
Cells were plated at a density of 2 × 104 cells/well into ultra-low attachment 6-well plates (Corning, Corning, NY, USA) in MammoCult Basal media (StemCell Technologies). Spheres with a minimum diameter of 50 μm (or >15 cells) were counted at Day 7. For serial passaging, mammospheres were collected by centrifugation at 350g, dissociated with 0.25% trypsin, counted, and re-seeded.
Differentiation culture conditions
Secondary mammospheres were dissociated into a single cell suspension and plated at a density of 10 viable cells/well in a collagen-coated 8-well chamber slide. After 7 days, cells were fixed in 100% methanol at -20°C for 20 minutes and stained with CK14 and CK18 primary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Fluorescence-activated cell sorting
A single-cell suspension was double-stained with PE-conjugated CD44 (BD Biosciences) and FITC-conjugated CD49f (BD Biosciences) antibodies to sort out the CD44+/CD49f+ and CD44-/CD49f- populations as described previously [4]. For knockdown experiments, cells were treated with siBMI1 (10nM) for 96 hours, re-suspended in re-suspension buffer (2% FBS in 1x PBS), and stained with CD44-PE (1:20; BD Biosciences) and CD24-FITC (1:5; BD Biosciences) for 30 minutes at 4°C. Cells were washed with re-suspension buffer and stained with 7-AAD (1:20; BD Biosciences). Data was acquired using a FACSCanto flow cytometer and analyzed using FlowJo software.
Cell cycle analysis
Cells were fixed in 2% paraformaldehyde for 30 minutes and stained with Hoechst 33342 (10 μg/ml, Sigma-Aldrich) for 1 hour at room temperature. Cell cycle analysis was performed using a high-content screening platform (ArrayScan VTI) by scanning 20 fields/well using the Cell Cycle Bioapplication V3 software (Thermo Scientific) as described previously [23].
EdU quantification
Cells were plated at a density of 1000 cells/well in 96-well plates and treated with doxycycline 24 hours post-seeding. At 72 hours cells were pulsed with EdU for 2 hours before fixation and staining for EdU, according to the manufacturers instructions (Click-iT® EdU HCS kit (Invitrogen)). Nuclei were counterstained with Hoechst 33342. Cells were stained using YB-1 antibody (clone EP2706Y; 1:100; Abcam) and visualized with AF488 goat anti-rabbit secondary antibody (1:100; Invitrogen).
Histone acetyltransferase (HAT) activity assay
HAT activity in 3 μg of nuclear lysate was measured using the HAT Assay Kit (Active Motif, Carlsbad, CA, USA) according to manufacturer’s instructions. Nuclear protein was isolated and incubated in HAT assay buffer containing 0.5 mM acetyl-CoA and 50 μM histone H3 peptide for 15 minutes at room temperature. Fluorescence was measured at 460 nm with an excitation wavelength of 380 nm.
Chromatin immunoprecipitation
Promoter complexes were isolated using the EZ-ChIP Kit (Millipore, Billerica, MA, USA) according to manufacturer’s instructions described in Supplemental Methods.
siRNA and plasmid transfections
Cells were transfected with 20 nM siRNA using Lipofectamine RNAiMAX (Invitrogen). The siRNA target sequences are provided in Supplemental Experimental Procedures. Plasmid transfections (2 μg) were carried out using FuGENE HD (Roche, Laval, Canada). For selection of stable YB-1-expressing clones, culture media was supplemented with G418 (400 μg/ml). pLenti6/V5-RSK2 was constructed by inserting full length RSK2 into the pLenti6/V5-DEST vector. The pMIN-BMI1 construct was a gift from Dr. Sheila Singh (McMaster University).
Anchorage-independent growth assay
Soft agar assays were performed as previously described [24]. Cells were embedded in 0.6% agar at a density of 5 × 103 per well in a 24-well plate. Colonies were counted at 28 days.
Telomerase assay
The telomerase activity in 1 μg of total cell lysate was assessed using the Quantitative Telomerase Detection Kit (Allied Biotech, Vallejo, CA, USA) following the manufacturer’s instructions.
Tumor xenografts
HTRZ, HTRY-LT #1, and HTRY-LT #2 cells were injected into the fourth inguinal mammary gland of 8-week old female NOD scid gamma (NSG) mice (Charles River, Wilmington, MA, USA). Six mice/group were injected with two million cells suspended in Matrigel and collagen I (50:50). Doxycycline (625 mg/kg) was administered in food pellets (synthesized at Harlan Laboratories, Madison, WI, USA). For limiting dilution studies, five NOD/SCID mice/group were injected bilaterally with 105, 104, or 102 HTRZ and HTRY-LT #1 cells. Tumors were formalin-fixed, embedded in paraffin, and sections stained with hematoxylin and eosin (H&E) according to standard procedure. Immunohistochemical staining with Ki67 (Dako), YB-1 (1:1000; Epitomics), p300 (N-15; 1:200; Santa Cruz), and BMI1 (F-6; 1:100; Millipore) antibodies was carried out using conventional technique [25] and heat-induced epitope retrieval. All animal experiments were conducted in accordance with protocols approved by the University of British Columbia.
NanoString gene expression profiling
RNA (250 ng) was extracted from formalin-fixed paraffin embedded mammary glands from YB-1 transgenic mice [26] and analyzed using the nCounter Gene Expression Analysis system at the Centre for Translational and Applied Genomics (BC Cancer Agency, Vancouver, BC). A custom CodeSet was synthesized by NanoString Technologies (Seattle, WA, USA). Data were normalized according to NanoString recommendations and heatmaps were generated with median-centered genes and arrays and unsupervised hierarchical clustering with average linkage using Cluster 3.0 and Java Treeview 1.6 software.
Results
Ectopic YB-1 expression in HMECs promotes luminal filling of breast acini in vitro
To profile the molecular events that govern breast cancer progression, we engineered a Tet-On YB-1 expression system into non-malignant H16N2 HMECs. Our group has extensively characterized this system previously [19]. Cells conditionally expressing YB-1 under control of the tetracycline-inducible promoter were termed HMEC Tet-repressed YB-1 (HTRY), while a LacZ-expressing control cell line was designated HMEC Tet-repressed LacZ (HTRZ). Moreover, we developed two variant HTRY cells lines, HTRY-LT #1 and HTRY-LT #2, through sustained, long-term YB-1 expression. These cell lines expressed more YB-1 compared to HTRZ cells, as well as the normal mammary 184hTERT and MCF10A cell lines based on immunoblotting and qRT-PCR (Figure 1A and S1A). The level of YB-1 achieved in the Tet-On YB-1 over-expressing cell lines was 4–10 fold higher than in the HTRZ cells and was similar to the basal-like breast cancer (BLBC) cell lines SUM149, MDA-MB-231, and MDA-MB-435/LCC6 as measured by qRT-PCR (Figure 1B and S1B).
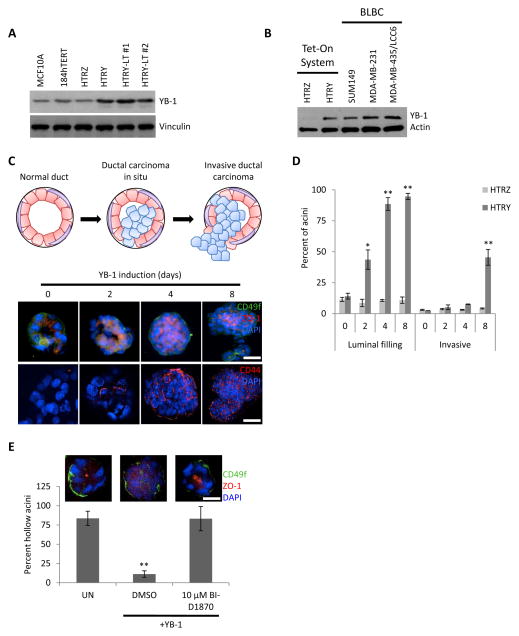
Figure 1. YB-1 over-expression in HMECs drove migration and luminal translocation in breast acinar structures.
(a) Immunoblot of YB-1 in the normal mammary epithelial MCF10A and 184hTERT cell lines compared to HTRZ and the YB-1-induced HTRY, HTRY-LT #1, and HTRY-LT #2 cell lines. (b) Immunoblot of YB-1 in HTRZ and HTRY cells compared to BLBC cell lines. (c) HTRY cells were grown as 3D acini on a reconstituted basement membrane. Following YB-1 induction, acinar structures were immunostained with ZO-1 (luminal), CD49f (basolateral), and CD44 antibodies. DAPI marked the nuclei. (d) Acinar structures (60 per time point) were evaluated for luminal filling and invasive outgrowths at the times indicated. (e) Acini were grown as above; however, YB-1 was induced along with the addition of DMSO or BI-D1870 (10 μM). At 96 hours luminal filling was evaluated. Scale bars represent 100 μm. UN, uninduced control. Data represented as mean ± SEM. P values were determined using t test. *, P <0.05; **, P < 0.01.
To question whether the over-expression of YB-1 in HMECs could drive phenotypic changes associated with breast cancer progression, such as luminal filling, we cultured HTRY cells on a reconstituted basement membrane. These cells became organized as three-dimensional polarized acini structures with a hollow lumen (Figure 1C). Subsequent induction of YB-1 led to luminal filling mirroring a ductal carcinoma in situ (DCIS). Notably, the earliest luminal outgrowths were found to express the TIC marker CD44, which, at later time points, was ubiquitously expressed by cells invading the luminal space (Figure 1C). By 8 days post-YB-1 induction, cells began to invade through the basement membrane and into the surrounding microenvironment similar to an invasive ductal carcinoma (IDC) (Figure 1C and 1D). In agreement, YB-1 expression caused HTRY and HTRY-LT cells to become invasive through Matrigel-coated Transwell chambers (Figure S2). As CD44 expression was previously found to be dependent upon YB-1 serine-102 phosphorylation by RSK [4], we questioned whether inhibiting this activation could prevent DCIS-like luminal outgrowths. Acini treated with the RSK inhibitor BI-D1870 failed to form luminal outgrowths following YB-1 induction and instead resembled the uninduced control (Figure 1E).
HMECs acquire characteristics of stem/progenitor-like tumor-initiating cells following YB-1 over-expression
Genome wide ChIP-on-chip analysis performed previously by our group identified a potential interaction between YB-1 and the BMI1 promoter [24]. As expected, expression of BMI1 was elevated in HTRY cells, relative to HTRZ cells, at the mRNA transcript level (Figure 2A), which correlated with increased protein abundance (Figure 2B). This stimulated histone H2A ubiquitylation and as a result the CDKN2A locus (encoding p16INK4a and p14ARF) was repressed. Consistent with our previous findings [4], the YB-1 transcriptional targets CD44 and CD49f were also more highly expressed in the HTRY cells. Notably, there was not a significant increase in BMI1, CD44, and CD49f expression in the long-term YB-1 expressing HTRY-LT #1 and HTRY-LT #2 cell lines relative to HTRY cells suggesting that the acquisition of a stem cell phenotype is an early event in the genesis of breast cancer.
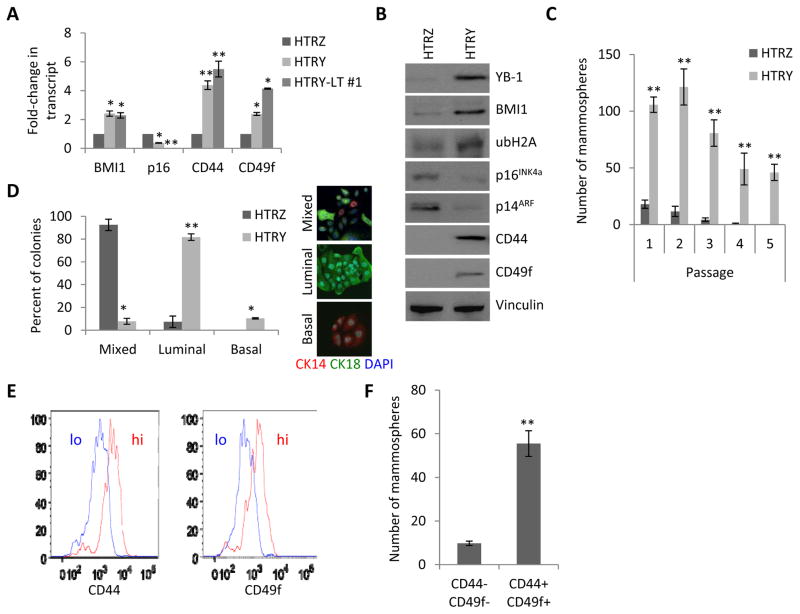
Figure 2. HMECs acquired TIC characteristics following short-term YB-1 over-expression.
(a) qRT-PCR analysis of TIC-associated genes in HTRZ, HTRY, and HTRY-LT #1 cells. (b) Immunoblot of BMI1, ubiquitinated histone H2A (ubH2A), p16INK4a, and p14ARF, in addition to the TIC markers CD44 and CD49f, in HTRZ and HTRY cells. (c) HTRZ and HTRY cells grown in mammosphere cultures and serially passaged. Doxycycline was replenished at each generation. (d) Differentiation culture of cells from secondary mammospheres. Colonies (x = 50) were evaluated for markers of luminal (CK18) and basal (CK14) differentiation using immunofluorescence. (e) FACS histograms depicting two HTRY cell populations defined as CD44/CD49f double-positive and CD44/CD49f double-negative. (f) Mammosphere assay of sorted HTRY cells. Data represented as mean ± SEM. P values were determined using t test. *, P <0.05; **, P < 0.01.
Accordingly, we questioned whether TIC marker expression correlated with stem cell properties. HTRZ and HTRY cells were plated into non-adherent mammosphere cultures as an in vitro surrogate assay for self-renewal capacity. In stark contrast to HTRZ cells, HTRY cells formed mammospheres that could subsequently be dissociated and single cells re-passaged as new spheres for at least five generations (Figure 2C). Moreover, we discovered that induction of YB-1 skewed cellular differentiation along the luminal lineage (Figure 2D). To validate that these molecular changes were not influenced by the H16N2 genetic background, we transiently transfected a FLAG:YB-1 expression vector into 184hTERT and MCF10A HMECs. Concomitant with YB-1 over-expression, both cell lines exhibited an increase in the expression of TIC-associated genes (Figure S3A), which translated into their ability to form mammospheres that could be serially passaged (Figure S3B) and an increased propensity for luminal differentiation (Figure S3C).
To address whether all HTRY cells or only a subpopulation acquired TIC properties, we analyzed the population by flow cytometry. Notably, we detected a fraction of cells enriched for CD44 and CD49f expression (Figure 2E). Sorting HTRY cells into two populations defined by high/high and low/low marker expression revealed that CD44+/CD49f+ cells had an enhanced ability to form mammospheres relative to the double-negative population (Figure 2F). These results suggest that YB-1 confers TIC properties to a small subpopulation of HTRY cells.
Chromatin remodeling by p300 underlies the reprogramming of HMECs into TICs
Work from other groups has highlighted the importance of the HAT protein p300 in maintaining the stem cell compartment [27, 28]. Coupled with the fact that p300 is up-regulated in CD44-positive breast cancer cells [29], we questioned whether its expression altered histone acetylation patterns to facilitate the reprogramming of HMECs into TICs. Compared to the normal mammary 184hTERT and HTRZ cells, p300 protein was strongly expressed in the YB-1 over-expressing cell lines as well as MDA-MB-231 cells (Figure 3A), which corresponded to higher HAT activity (Figure S4A). Anacardic acid, a potent inhibitor of p300 [30], decreased the activity of recombinant p300 (Figure S4B) as well as HAT activity in total cell lysate by over 80% (Figure S4C).
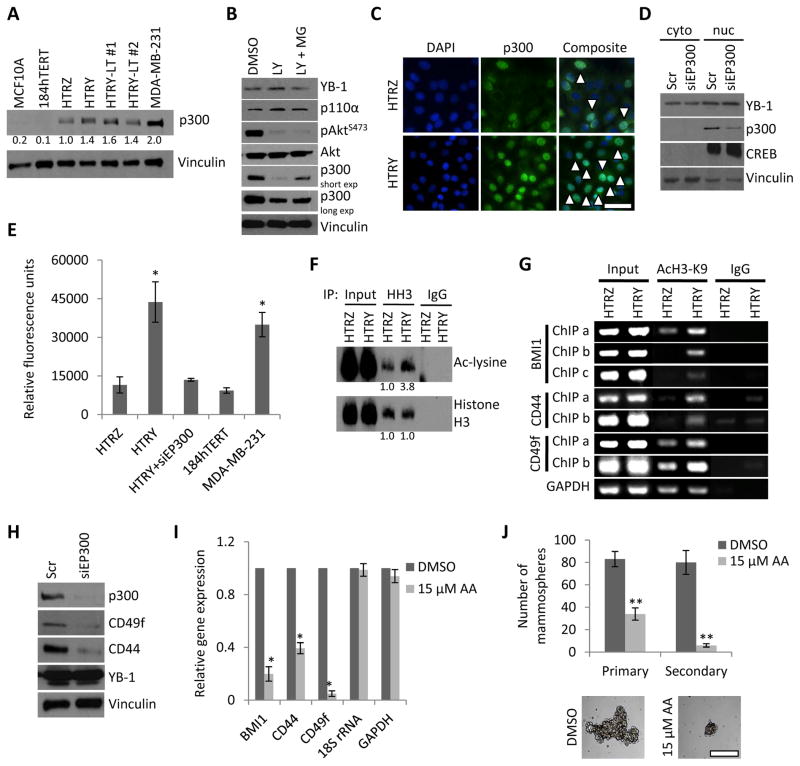
Figure 3. Increased p300 HAT activity underlies the reprograming of HMECs into TICs.
(a) Immunoblot analysis of p300 in normal mammary MCF10A and 184hTERT cell lines compared to HTRZ, HTRY, HTRY-LT #1, and HTRY-LT #2 cells and the BLBC cell line MDA-MB-231. (b) Immunoblot analysis of p300 and PI3K signaling components in HTRY cells treated for 24 hours with DMSO, LY294002 (LY, 20 μM), or LY in combination with MG132 (MG, 5 μM). (c) Immunofluorescence of p300 localization in HTRZ and HTRY cells. Arrows denote p300-positive nuclei (visualized by DAPI). Scale bar represents 100 μm. (d) Immunoblot analysis of cytoplasmic and nuclear fractions following siRNA-mediated p300 knockdown (5 nM) in HTRY cells. CREB and vinculin were used to assess the purity of the nuclear and cytoplasmic fractions, respectively. (e) HAT activity in nuclear lysate from HTRZ, HTRY, and HTRY cells treated for 96-hours with siRNA targeting EP300. 184hTERT and MDA-MB-231 were a negative and positive control for p300 activity, respectively. (f) Histone H3 (HH3) immunoprecipitation followed by immunoblotting to measure acetylated lysine residues. Protein levels were evaluated by densitometry (normalized to HTRZ). (g) ChIP targeting the promoters of BMI1, CD44, and CD49f in induced HTRZ and HTRY cells. DNA templates were pulled down with acetyl-histone H3 (Lys9) or nonimmune IgG antibody. GAPDH served as a control. (h) Immunoblot analysis of TIC markers in HTRY cells treated with EP300 siRNA for 96 hours. (i) qRT-PCR analysis of mRNA transcript in HTRY cells pre-treated with DMSO or anacardic acid (AA) for 4 hours prior to induction. (j) HTRY cells serially passaged as mammospheres in the presence of DMSO or AA. Primary mammospheres are shown. Scale bar represents 200 μm. Data represented as mean ± SEM. P values were determined using t test. *, P <0.05; **, P < 0.01.
To deduce the mechanism responsible for p300 upregulation, we discovered that the catalytic subunit of phosphoinositide 3-kinase (PI3K), p110α, was expressed in the HTRY cells and treatment with the PI3K inhibitor LY294002 suppressed AKT activation and decreased p300 expression (Figure 3B). Further, we noted that the deleterious effect of LY294002 on p300 expression could be partially rescued by concomitant treatment with the proteasome inhibitor MG132 (Figure 3B). This is consistent with reports that phosphorylation by AKT protects p300 from proteasome-mediated degradation [31]. We therefore attribute the upregulation of p300 to AKT-driven protein stabilization. The finding that YB-1 over-expression ensures p300 stability is further supported by a significant decrease in HAT activity following siRNA-mediated inhibition of YB-1 or its upstream kinases RSK1 and RSK2 (Figure S4D).
We next questioned whether YB-1 over-expression in HMECs influenced p300 activity. The number of p300-positive nuclei and the intensity of staining was much greater in HTRY cells relative to HTRZ cells as visualized by immunofluorescence (Figure 3C). In support, cytonuclear fractionation confirmed that both YB-1 and p300 were localized to the nuclear compartment in HTRY (Figure 3D) and MDA-MB-231 cells (Figure S4E). As a consequence of increased p300 expression HAT activity was nearly four-fold higher in the HTRY cells compared to the HTRZ cells (Figure 3E). Silencing EP300 (encoding p300) in HTRY cells significantly decreased HAT activity suggesting that p300 is the predominant HAT enzyme present in these cells (Figure 3E). As a control we report that 184hTERT cells have low p300 expression and low HAT activity compared to MDA-MB-231 cells, which express more p300 and have much higher HAT activity (Figure 3E).
The p300 protein epigenetically regulates gene expression by acetylating lysine residues on histone proteins. Therefore, as a direct measure of p300 activity we assessed the extent of histone acetylation by immunoprecipitation of histone H3. Using an anti-acetylated-lysine antibody we detected enrichment in the pool of acetylated histone H3 in HTRY cells relative to HTRZ cells (Figure 3F). To refine this analysis we next evaluated acetylation of histone H3 at lysine 9 (AcH3-K9) along promoter-centered chromatin of TIC-associated genes. The 9th lysine residue (H3K9) of histone H3 is a preferential substrate of p300 [27]. DNA purified after immunoprecipitation with anti-AcH3-K9 antibody was evaluated by PCR using primers targeting the length of the BMI1, CD44, and CD49f promoters. Higher levels of promoter-associated histone H3-K9 acetylation were observed in HTRY cells compared to HTRZ cells (Figure 3G).
To demonstrate that inhibition of p300 could prevent the reprogramming of HMECs into TICs, we silenced its expression and/or activity using an siRNA and pharmacologic approach. Silencing EP300 yielded a decrease in CD44 and CD49f expression in both HTRY cells (Figure 3H) and MDA-MB-231 cells (Figure S4F). We further demonstrate that inhibition of p300 activity using anacardic acid correlated with loss of BMI1, CD44, and CD49f mRNA expression (Figure 3I). Quenching p300 activity resulted in fewer, and smaller, primary mammospheres that could not be serially passaged (Figure 3J).
Chromatin remodeling permits YB-1 to transcriptionally regulate BMI1
We next questioned whether p300-mediated chromatin relaxation was a pre-requisite for YB-1 to bind TIC-associated gene promoters. In addition to CD44 and CD49f, we now show that YB-1 binds to the BMI1 promoter in both HTRY and MDA-MB-231 breast cancer cell lines using conventional ChIP (Figure S5A). Consistent with the role of BMI1 in the development of cancer its expression was inversely correlated with p16INK4a when normal mammary epithelial cells were compared to the BLBC cell lines SUM149 and MDA-MB-231 (Figure S5B). Furthermore, in ChIP assays using an anti-YB-1 antibody, pre-treating HTRY cells with AA to inhibit p300 activity prevented YB-1 binding to the BMI1 and CD49f promoters as well as one region on the CD44 promoter (Figure 4A). This suggests that p300-mediated chromatin remodeling is upstream of YB-1 promoter binding and gene transcription.
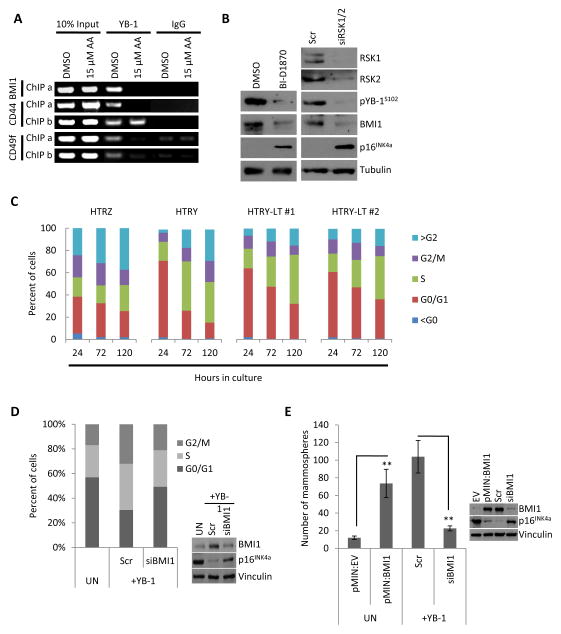
Figure 4. YB-1 transcriptionally regulated BMI1 to enhance self-renewal capacity.
(a) ChIP analysis of HTRY cells pre-treated with DMSO or anacardic acid (AA) for 4 hours prior to induction. DNA templates were pulled down with YB-1 or nonimmune IgG antibody and different promoter regions (ChIP a and ChIP b) were amplified using primers flanking YB-1 binding sites in the BMI1, CD44, and CD49f promoters. (b) Immunoblot analysis of HTRY cells treated with RSK1/2 siRNA for 96 hours or BI-D1870 for 24 hours. (c) The percentage of cells in each phase of the cell cycle at the indicated time points after plating was quantified by DNA content based on Hoechst 33342 intensity using an ArrayScan VTI. (d) Cell cycle profile of HTRY cells treated with scrambled control (scr) or BMI1 siRNA for 96 hours was measured using an ArrayScan VTI. Uninduced (UN) cells served as a control. Immunoblotting confirmed BMI1 knockdown. (e) Uninduced (UN) HTRY cells transfected with empty vector (EV) or BMI expression plasmid and induced HTRY cells treated with scrambled (scr) or BMI1 siRNA for 96 hours were grown in mammosphere cultures. BMI1 over-expression and knockdown was confirmed by immunoblotting. Data represented as mean ± SEM. P values were determined using t test. **, P < 0.01.
Consistent with the novel finding that the induction of YB-1 increased BMI1 expression as described above, we also report that siRNA-mediated loss of YB-1 significantly decreased the expression of BMI1 in MDA-MB-231 as well as HTRY-LT #2 cells (Figure S5C). As we have previously demonstrated that YB-1 Ser-102 phosphorylation by RSK is a prerequisite for its DNA binding and transcriptional activity [2, 32, 33], we inhibited the kinase using siRNA or the small molecule inhibitor BI-D1870. In HTRY cells, suppression of RSK1/2 expression and/or activity led to a decrease in activated pYB-1S102 resulting in loss of BMI1 and rescue of p16INK4a (Figure 4B).
BMI1-mediated silencing of p16INK4a has been associated with a loss of G1/S checkpoint fidelity [34]. In accordance, analysis of cell cycle kinetics revealed that the proportion of cells in G0/G1 was shifted in the YB-1 over-expressing cell lines compared to HTRZ cells (Figure 4C). This could explain why cellular doubling time decreased from 118 hours in the HTRZ cells to 101-, 72-, and 88-hours in the HTRY, HTRY-LT #1, and HTRY-LT #2 cells, respectively (Figure S6A). Consistent with this finding, HTRY cells had a higher proliferative rate relative to HTRZ cells based on EdU incorporation (Figure S6B). We were able to partially restore G1/S checkpoint activity following knockdown of BMI1 in HTRY cells using siRNA (Figure 4D). In support, loss of BMI1 suppressed the growth of MDA-MB-231 cells over 72 hours (Figure S6C). This was due, in part, to an increase in nuclear p16INK4a accumulation (Figure S7A and S7B). These observations suggest that YB-1 expression potentiated G1/S checkpoint slippage via a BMI1-dependent mechanism.
The abovementioned findings prompted us to question whether the enhanced self-renewal potential of HTRY cells was a direct consequence of BMI1 expression. The mammosphere forming capacity of uninduced HTRY cells was increased from 12 ± 2 to 73.7 ± 16.1 following transient transfection with a pMIN:BMI1 expression vector (Figure 4E). Conversely, siRNA-mediated silencing of BMI1 in HTRY cells decreased their ability to form mammospheres by nearly 80% (from 104 ± 18.5 to 22.7 ± 2.8) (Figure 4E). Notably, loss of BMI1 reduced the CD44+/CD24- stem cell population by ~25% in MDA-MB-231 cells (Figure S8). Perplexingly, no change was observed in CD44 or CD49f mRNA expression; however, CD44 protein level decreased (data not shown). Thus, we conclude that YB-1 cooperates with p300 and BMI1 to promote and sustain a population of cells with tumor-initiating potential.
Sustained up-regulation of YB-1 leads to full transformation and tumor-initiation
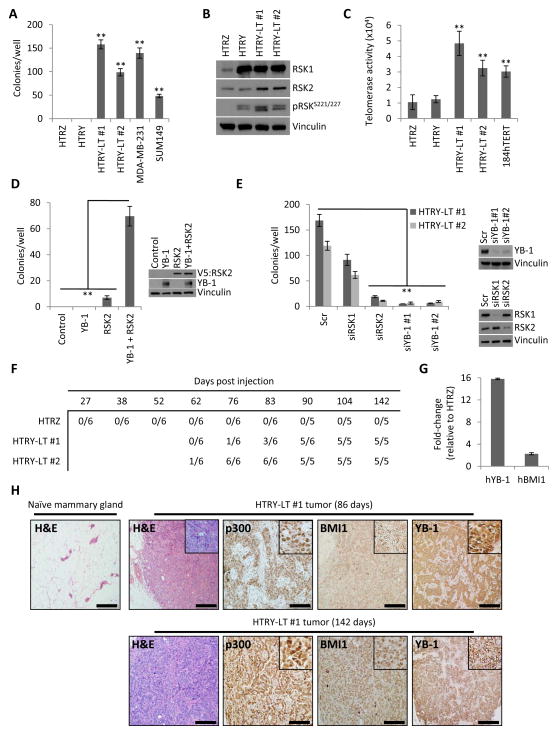
The ability of YB-1 to elicit phenotypes associated with malignant progression prompted us to investigate whether the oncogene could confer full transformation and ultimately tumor initiation. HTRY cells failed to form colonies in soft agar indicating that a single short-term pulse of YB-1 was insufficient to allow for anchorage-independent growth (Figure 5A). However, long-term expression of YB-1 in the HTRY-LT #1 and HTRY-LT #2 cell lines led to colony formation at a level comparable to SUM149 and MDA-MB-231 cells (Figure 5A). At the molecular level, HTRY-LT cells displayed elevated RSK1 and RSK2 protein expression relative to HTRZ cells (Figure 5B). This was particularly interesting as previous work from our group and others has established that BLBCs are dependent on RSK2 signaling for growth and survival [35, 36]. To understand why these cells formed colonies in soft agar we asked whether this involved telomerase (hTERT) activity given its established role in neoplastic transformation. Notably, hTERT activity was detected in the HTRY-LT cell lines, but less so in HTRZ and HTRY cells (Figure 5C).
Figure 5. Synergism between YB-1, RSK2, and hTERT conferred complete transformation.
(a) Quantification of HTRZ, HTRY, and HTRY-LT cell growth under anchorage-independent conditions. MDA-MB-231 and SUM149 cells acted as a positive control. (b) Immunoblot assessing RSK expression and activation. (c) Telomerase activity in HTRZ, HTRY, and HTRY-LT cell lysate. 184hTERT cells were used as a positive control. (d) Soft agar colony growth of HTRY cells expressing YB-1, RSK2, or YB-1 and RSK2. Uninduced HTRY cells served as the control. Immunoblotting confirmed transgene expression at 96 hours post-transfection. (e) Quantification of soft agar colony growth following treatment of HTRY-LT cells with scrambled (scr) or YB-1, RSK1, and RSK2 siRNA. Immunoblotting confirmed gene silencing. (f) The ability of HTRZ, HTRY-LT #1, and HTRY-LT #2 cells to form palpable tumors when transplanted into the mammary fat pad of NSG mice (6 mice/group). (g) qRT-PCR analysis of human YB-1 and BMI1 in tumors isolated from NSG mice inoculated with HTRZ and HTRY-LT #2 cells. Gene expression was normalized using eukaryotic 18S rRNA. (h) Hematoxylin-eosin (H&E) and immunoperoxidase staining with p300, BMI1, and YB-1 antibody in tumor tissue explanted 86 days and 142 days after implantation of HTRY-LT #1 cells into the mammary fat pad of NSG mice. The contralateral naïve gland served as a control. Scale bar represents 500 μm. Data represented as mean ± SEM. P values were determined using t test. **, P < 0.01.
We endeavored to uncover the minimal combination of genes necessary for the evolution of HTRY cells into fully transformed HTRY-LT cells. Uninduced HTRY cells acquired the ability to form soft agar colonies following YB-1/RSK2 double-transfection. Expression of YB-1 or RSK2 alone did not significantly enhance soft agar colony formation (Figure 5D). In a reciprocal experiment, we silenced YB-1 and RSK2 in the HTRY-LT cell lines using siRNA. This led to a repression in the number of soft agar colonies. Knockdown of RSK1 had only a moderate effect highlighting the unique importance of the RSK2 isoform (Figure 5E). Taken together, these results suggest that interplay between YB-1, RSK2, and hTERT is necessary for transformation. One cannot exclude that the H16N2 genetic background may have influenced the transformative potential of these cells; therefore, we introduced YB-1 into a second mammary epithelial cell line, 184hTERT. Following stable transfection, 184hTERT clones emerged that expressed YB-1, RSK1, and RSK2 to a similar level observed in MDA-MB-231 cells (Figure S9A). Moreover, compared to cells transfected with empty vector, 184hTERT YB-1-expressing cells exhibited increased soft agar colony formation (Figure S9B) and invasion through Matrigel-coated Transwell chambers (Figure S9C). Together these data suggest that the addition of RSK2 enabled YB-1-expressing cells to become fully transformed through collaboration with hTERT.
Given the observation that RSK1 and RSK2 levels were increased in the HTRY-LT cell lines we asked whether they would be sensitive to pan-RSK inhibition. Treating the HTRY-LT cell lines with BI-D1870 suppressed YB-1 activation (Figure S10A). This resulted in decreased growth; however, the drug had no effect on the HTRZ cells at low dose (≤2 μM) (Figure S10B). This growth inhibitory effect translated into a significant reduction in the ability of BI-D1870-treated HTRY-LT cells to form soft agar colonies (Figure S10C). Moreover, the capacity of these cells to grow as mammospheres was also abrogated (Figure S10D). This strongly implies that BI-D1870 is not only eradicating bulk tumor cells, but also the TIC subpopulation. Importantly, we demonstrate that treatment with BI-D1870 induced apoptosis in the HTRY-LT cell lines as measured by annexin V positivity (Figure S10E). Thus this model further demonstrates the potential for RSK inhibitors to control the growth of transformed cells that depend on YB-1.
To assess the tumorigenicity of HTRY-LT cells in vivo, we injected them into the mammary glands of NSG mice. HTRY-LT #1 and HTRY-LT #2 cells were characterized as expressing 7- and 4-fold higher level of YB-1 mRNA, respectively, compared to HTRZ cells immediately prior to in vivo transplantation (Figure S11A). RSK2 was also elevated (Figure S11B), while p16INK4a was depressed (Figure S11C), in the HTRY-LT cells relative to HTRZ cells. At 142 days post-transplantation, none of the HTRZ group grew tumors; however, all of the mice injected with HTRY-LT #1 and HTRY-LT #2 cells developed mammary tumors (Figure 5F and S11D) ranging in weight from 40 to 1480 milligrams (Figure S11E). The tumor expressed 16-fold higher human-YB-1 mRNA and 2.3-fold higher human-BMI1 mRNA relative to mammary glands injected with HTRZ cells (Figure 5G). In addition, human EP300 was readily detectable in the tumors by qRT-PCR (Ct=27) but undetectable in mammary glands from the HTRZ group (data not shown). Microscopic examination of mammary fat pads transplanted with HTRY-LT #1 cells showed the presence of solid masses of neoplastic cells (Figure 5H) with a high proliferative index based on human Ki67 staining (Figure S11F). The tumor cells also expressed high levels of p300 and BMI1 protein at 86 days post-injection and the levels were sustained out to 142 days (Figure 5H). YB-1 protein was also expressed at both time points (Figure 5H). To ascertain whether HTRY-LT cells could form tumors at limiting dilution, we injected 105, 104, and 102 HTRY-LT #1 and HTRZ cells bilaterally into the mammary gland of NOD/SCID mice. As few as 100 HTRY-LT cells was sufficient for tumor initiation (Figure S12A). DCIS-like lesions and small palpable tumor masses were present in mammary glands transplanted with HTRY-LT cells (Figure S12B and S12C). While tumors developed in both the NSG and NOD/SCID strains by 90 days post-injection, they were notably larger in the former. We suspect that this is a consequence of the lack of a functional immune system in NSG mice and the co-injection of cells with Matrigel and collagen I to recapitulate the human mammary gland microenvironment.
YB-1 transforms HMECs into cells with a BLBC subtype
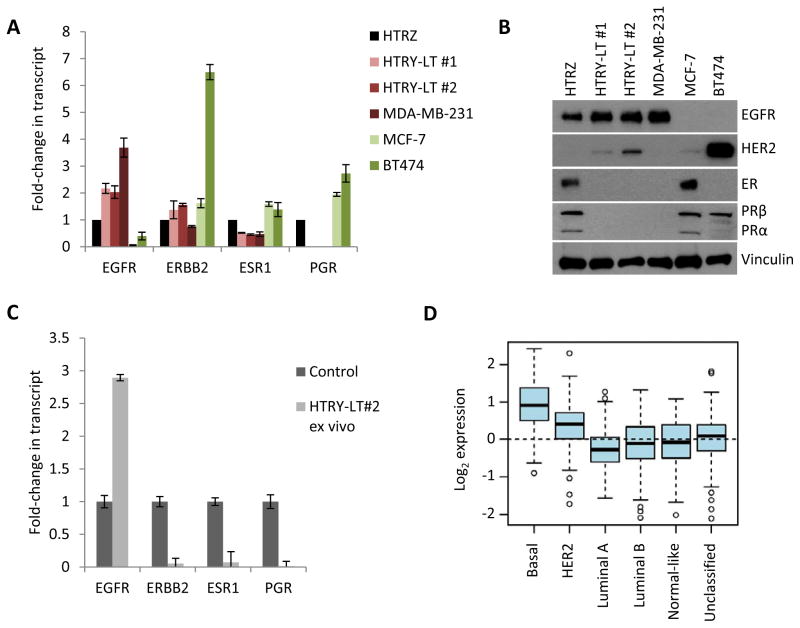
As breast cancer can be classified into distinct subtypes with differential aggressiveness and prognosis [37], we questioned which subtype the HTRY-LT cell line represented. During YB-1 mediated transformation, the HTRY-LT cells gained expression of EGFR and lost expression of ESR1 (encoding ER) and PGR (encoding PR) as measured by qRT-PCR based gene expression profiling (Figure 6A) and immunoblotting (Figure 6B). Negligible level of ERBB2 (HER2) was present in the HTRY-LT cell lines. Moreover, ex vivo analysis of the HTRY-LT tumors from NSG mice revealed that they expressed EGFR, but lacked ESR1, PGR, and ERBB2 (Figure 6C). Therefore, upon YB-1 induction HMECs were transformed from hormone receptor positive to triple-negative (lacking ER, PR, and HER2). More specifically, the transformed cells represent the BLBC subtype, which is clinically defined as ER, PR, HER2-negative and EGFR and/or CK5/6-positive [38]. The clinical relevance of our model can be appreciated by the fact that YB-1 transcript expression was significantly higher in high grade ER-negative tumors (p < 0.0001; Figure S13A and S13B), with the highest expression observed in the basal-like subtype (p < 0.0001; Figure 6D) when analyzed in a cohort of 1881 breast cancer patients using the Gene Expression Outcome for Breast Cancer Online algorithm (GOBO [39]).
Figure 6. HTRY-LT cells were molecularly classified as BLBC.
(a) qRT-PCR and (b) immunoblot analysis of subtype biomarkers. MDA-MB-231 (TNBC), MCF-7 (ER+), and BT474 (PR+/HER2+) cells were used as controls. (c) qRT-PCR analysis of subtype biomarkers in HTRY-LT #2-derived xenografts from NSG mice. Data are reported relative to the appropriate control for each gene: MDA-MB-231 (EGFR+), T47D (ER+), and BT474 (ERBB2+ and PR+). (d) Box plot analyses of YB-1 expression among HU subtypes, Basal (n = 194), HER2 (n = 78), Luminal A (n = 221), Luminal B (n = 122), Normal-like (n = 121), and unclassified (n = 191). Data obtained using Gene expression based Outcome for Breast cancer Online (GOBO).
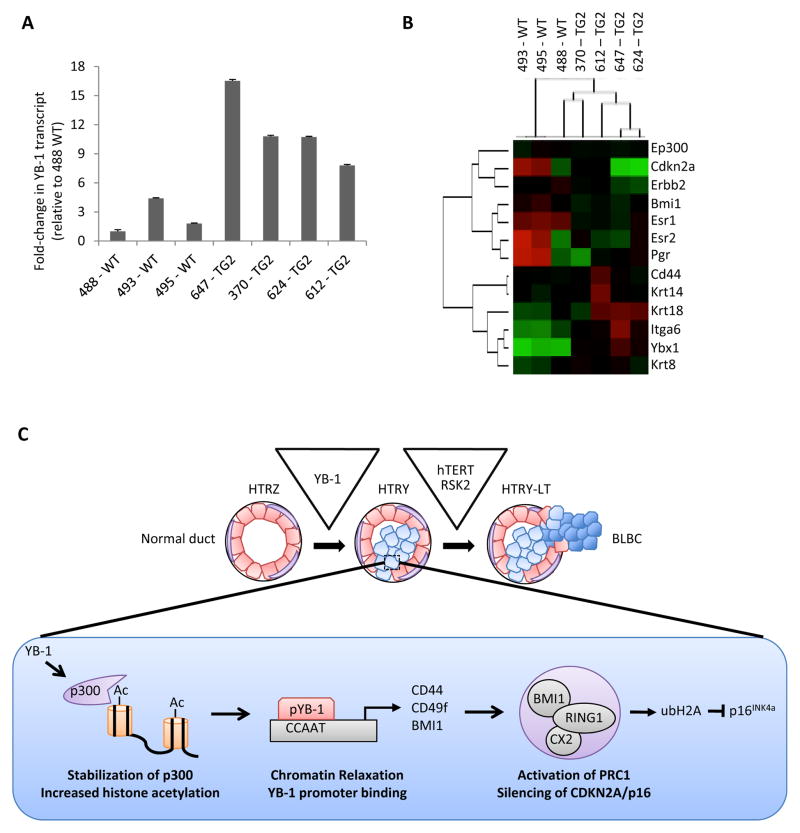
To further support the observation that YB-1 is inversely correlated with the hormone receptors we characterized YB-1 transgenic mouse tissues [26]. Tissues taken from 6–8 month old TG2 mice expressed high YB-1 as compared to age-matched wild-type (WT) mice (Figure 7A). Notably, we detected an increase in both human and murine YB-1 transcript in the TG2 tissue relative to WT (Figure S14A). This could be a consequence of human YB-1 inducing murine YB-1 via an autoregulatory mechanism [40]. It is important to note that the human YB-1 qRT-PCR probe exhibited slight cross-reactivity with murine YB-1, which rationalizes why human YB-1 was detected in WT mouse tissue (Figure S14B). NanoString gene expression profiling of the TG2 mammary glands showed a loss of ER1, ER2, PR and CDKN2A, while a gain of the luminal lineage marker KRT18 and the TIC genes CD44 and ITGA6 (encoding CD49f) (Figure 7B). These changes are consistent with the Tet-On expression model described above. However, in contrast to the human YB-1 driven system, EP300 and BMI1 levels were constant. This could be due, in part, to the timing in which the lesions were taken because by 8 months pre-neoplastic lesions were evident [4, 26].
Figure 7. YB-1 transgenic mice form hormone-receptor negative tumors.
(a) qRT-PCR analysis of hYB-1 mRNA transcript in wild-type (WT) and TG2 YB-1 transgenic mice normalized to 488 WT. (b) Gene expression analysis of YB-1 transgenic TG2 mice (n = 4) relative to the WT controls (n = 3) using the NanoString nCounter platform (red, high expression; green, low expression). (c) Depiction of the genetic and phenotypic features that define each step of YB-1-driven transformation of HMECs into a TNBC. The epigenetic alterations following YB-1 induction in the HTRY cells are shown in detail.
Using reverse phase protein arrays (RPPA) we validated the association between YB-1 and BLBC in a clinical cohort of 710 invasive breast cancers. By RPPA, YB-1 was inversely correlated with ER (r = -0.132, p = 0.00041), PR (r = -0.157, p = 2.7 × 10−5), and HER2 (r = -0.163, p = 1.3 × 10−5) (Table S1). Moreover, we addressed which signaling pathways are associated with YB-1 in primary breast cancers using this platform. We discovered several members of the MAPK signaling cascade to be correlated with YB-1 expression including MEK, pMEK, pS6 ribosomal protein, and pRSK (Table S1). This is a key finding because the MAPK pathway is linked to BLBC [41]. These clinical data support our finding that the induction of YB-1 in vitro converts hormone receptor-positive cells into a triple-negative cancer, which is consistent with its expression in primary BLBC.
In summary, our data convey that HMEC transformation by YB-1 occurs in a step-wise process. During an initial pre-malignant phase, YB-1-mediated activation of the HAT protein p300 alters the histone acetylation landscape. The relaxation of promoter-centered chromatin allows for YB-1 to bind and transcriptionally regulate BMI1 to instill stem/progenitor-like phenotypes, such as enhanced self-renewal and multipotent differentiation. Over time, pressures exerted by YB-1 leads to the emergence of tumorigenic cells with a BLBC subtype that express high levels of RSK2 and hTERT (Figure 7C).
Discussion
In the present study, we implicate YB-1 as a catalyst for the transformation of HMECs into BLBC. Up-regulation of YB-1 promoted p300-mediated chromatin remodeling that reeducated mature HMECs into stem/progenitor-like TICs via induction of BMI1, CD44, and CD49f. However, this was not sufficient for complete transformation. Over time, pressures exerted by sustained YB-1 expression promoted the emergence of cells with RSK2 and hTERT that were tumorigenic in vivo and were molecularly classified as BLBC. Notably, the strong clinical correlation between YB-1 expression and BLBC emphasizes the relevance of our in vitro studies in emulating the disease.
Royer first demonstrated that YB-1 was expressed in primary human breast cancers, but absent in normal breast tissue [18]. Following this, a transgenic mouse model was engineered that formed tumors with 100% penetrance thereby cementing YB-1 as an oncogene with a principal role in tumorigenesis [26]. We have now significantly furthered the field by dissecting the underlying mechanism of YB-1-mediated transformation. In this study, we found that YB-1 works in concert with p300 to facilitate the epigenetic reprogramming of HMECs into stem/progenitor-like TICs during the earliest stages of the neoplastic process. The de novo emergence of TICs is corroborated by the discovery of plasticity within both normal and malignant breast cells [42, 43]. We discovered that YB-1 expression skews the differentiation of stem/tumor-initiating cells along the luminal lineage. This is notable because BLBCs are believed to arise from luminal progenitors [8, 44, 45], suggesting that YB-1 primes cells to progress into the particularly aggressive BLBC subtype.
We have presented compelling evidence that epigenetic changes play a pivotal role in the earliest stages of breast cancer progression. In support of this paradigm shift, promoter methylation and inactivation of tumor suppressor genes such as CDKN2A [13, 46], HIC-1 [47], and RASSF1A [48] is frequently detected in pre-invasive breast lesions. The finding that YB-1 transcriptionally regulates BMI1 is especially exciting because, apart from miR-200c [49], it was previously unknown how its expression is controlled. The protein has a well-established role in altering histone ubiquitylation and promoter methylation to repress senescence, apoptotic, and differentiation pathways in stem/progenitor cells [50]. Our results suggest that these same pathways are silenced by YB-1 during early breast cancer development at least in part through BMI1.
In support of previous studies [1, 51], this work further solidifies the association between YB-1 and BLBC. Data from our RPPA analysis and corroborated by other groups [52] demonstrate that the MAPK signaling cascade is activated in triple-negative breast cancer. This is of particular interest because Ras/ERK/RSK regulates YB-1-dependent transcription of genes involved in executing malignant properties [53]. Moreover, YB-1 regulates MAPK signaling factors [54] suggesting that a self-reinforcing loop could potentiate breast cancer progression. Notably, aberrant MAPK activation has recently been implicated in the emergence of a tumor-initiating/stem cell-like phenotype in BLBC [55].
Conclusion
In summary, the expression of YB-1 in HMECs triggers a cascade of epigenetic events that promote self-renewal and cellular transformation. The finding that YB-1 specifically promotes the development of BLBC rationalizes why the oncogene is expressed in over 70% of clinical cases of the disease. However, the ubiquitous occurrence of YB-1 in cancer and the fact that it, as well as RSK2, have been shown to mediate hematopoietic transformation [56, 57] suggest that the implications of this study extend beyond breast cancer.
Supplementary Material
Acknowledgments
Financial Support: This work was largely supported by grants from the Canadian Institutes of Health Research (to S.E.D.), Child and Family Research Institute (to S.E.D.), Komen Foundation (to G.B.M.), and National Institutes of Health (to S.E.D. and G.B.M.), with additional support from a Frederick Banting and Charles Best Doctoral Award (to A.H.D.).
We thank Ms Lisa Xu (at the CFRI Flow Core Facility) for technical assistance.
Footnotes
Author Contributions:
Alastair H. Davies: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing
Kristen M. Reipas: Collection and/or assembly of data, Data analysis and interpretation
Mary Rose Pambid: Collection and/or assembly of data, Data analysis and interpretation
Rachel Berns: Collection and/or assembly of data, Data analysis and interpretation
Anna L. Stratford: Collection and/or assembly of data, Data analysis and interpretation
Abbas Fotovati: Collection and/or assembly of data
Natalie Firmino: Collection and/or assembly of data, Data analysis and interpretation
Arezoo Astanehe: Data analysis and interpretation
Kaiji Hu: Collection and/or assembly of data
Christopher Maxwell: Conception and design
Gordon B. Mills: Provision of study material or patients
Sandra E. Dunn: Conception and design, Manuscript writing, Final approval of manuscript
Conflicts of Interest
No potential conflicts of interest.
References
- 1.Habibi G, Leung S, Law JH, et al. Redefining prognostic factors for breast cancer: YB-1 is a stronger predictor of relapse and disease-specific survival than estrogen receptor or HER-2 across all tumor subtypes. Breast Cancer Res. 2008;10:R86. doi: 10.1186/bcr2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Lee C, Yokom D, et al. Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2. Cancer research. 2006;66:4872–4879. doi: 10.1158/0008-5472.CAN-05-3561. [DOI] [PubMed] [Google Scholar]
- 3.Stratford AL, Fry CJ, Desilets C, et al. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R99. doi: 10.1186/bcr2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To K, Fotovati A, Reipas KM, et al. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer research. 2010;70:2840–2851. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nature medicine. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 8.Molyneux G, Geyer FC, Magnay FA, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer research. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 10.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 11.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Holst CR, Nuovo GJ, Esteller M, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer research. 2003;63:1596–1601. [PubMed] [Google Scholar]
- 14.Dimri GP, Martinez JL, Jacobs JJ, et al. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer research. 2002;62:4736–4745. [PubMed] [Google Scholar]
- 15.Hoenerhoff MJ, Chu I, Barkan D, et al. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta S, Hoenerhoff MJ, Bommi P, et al. Bmi-1 cooperates with H-Ras to transform human mammary epithelial cells via dysregulation of multiple growth-regulatory pathways. Cancer research. 2007;67:10286–10295. doi: 10.1158/0008-5472.CAN-07-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romanov SR, Kozakiewicz BK, Holst CR, et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 18.Bargou RC, Jurchott K, Wagener C, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature medicine. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 19.Davies AH, Barrett I, Pambid MR, et al. YB-1 evokes susceptibility to cancer through cytokinesis failure, mitotic dysfunction and HER2 amplification. Oncogene. 2011;30:3649–3660. doi: 10.1038/onc.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berquin IM, Pang B, Dziubinski ML, et al. Y-box-binding protein 1 confers EGF independence to human mammary epithelial cells. Oncogene. 2005;24:3177–3186. doi: 10.1038/sj.onc.1208504. [DOI] [PubMed] [Google Scholar]
- 21.Astanehe A, Finkbeiner MR, Hojabrpour P, et al. The transcriptional induction of PIK3CA in tumor cells is dependent on the oncoprotein Y-box binding protein-1. Oncogene. 2009;28:2406–2418. doi: 10.1038/onc.2009.81. [DOI] [PubMed] [Google Scholar]
- 22.Muthuswamy SK, Li D, Lelievre S, et al. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cappella P, Gasparri F. Highly multiplexed phenotypic imaging for cell proliferation studies. Journal of biomolecular screening. 2014;19:145–157. doi: 10.1177/1087057113495712. [DOI] [PubMed] [Google Scholar]
- 24.Finkbeiner MR, Astanehe A, To K, et al. Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells. Oncogene. 2009;28:1421–1431. doi: 10.1038/onc.2008.485. [DOI] [PubMed] [Google Scholar]
- 25.Fotovati A, Abu-Ali S, Wang PS, et al. YB-1 bridges neural stem cells and brain tumor-initiating cells via its roles in differentiation and cell growth. Cancer research. 2011;71:5569–5578. doi: 10.1158/0008-5472.CAN-10-2805. [DOI] [PubMed] [Google Scholar]
- 26.Bergmann S, Royer-Pokora B, Fietze E, et al. YB-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer research. 2005;65:4078–4087. doi: 10.1158/0008-5472.CAN-04-4056. [DOI] [PubMed] [Google Scholar]
- 27.Zhong X, Jin Y. Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J Biol Chem. 2009;284:9168–9175. doi: 10.1074/jbc.M805562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 29.Bourguignon LY, Xia W, Wong G. Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates beta-catenin signaling and NFkappaB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J Biol Chem. 2009;284:2657–2671. doi: 10.1074/jbc.M806708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanyam K, Swaminathan V, Ranganathan A, et al. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 31.Poizat C, Sartorelli V, Chung G, et al. Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol Cell Biol. 2000;20:8643–8654. doi: 10.1128/mcb.20.23.8643-8654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland BW, Kucab J, Wu J, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 33.Stratford AL, Habibi G, Astanehe A, et al. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res. 2007;9:R61. doi: 10.1186/bcr1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 35.Stratford AL, Reipas K, Hu K, et al. Targeting p90 Ribosomal S6 Kinase (RSK) Eliminates Tumor-Initiating Cells by Inactivating Y-Box Binding Protein-1 (YB-1) in Triple-Negative Breast Cancers. Stem Cells. 2012 doi: 10.1002/stem.1128. [DOI] [PubMed] [Google Scholar]
- 36.Brough R, Frankum JR, Sims D, et al. Functional viability profiles of breast cancer. Cancer Discov. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 39.Ringner M, Fredlund E, Hakkinen J, et al. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda T, Ashizuka M, Nakamura T, et al. Characterization of the 5′-untranslated region of YB-1 mRNA and autoregulation of translation by YB-1 protein. Nucleic acids research. 2004;32:611–622. doi: 10.1093/nar/gkh223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan JS, Whittle MC, Nakamura K, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta PB, Fillmore CM, Jiang G, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 44.Gastaldi S, Sassi F, Accornero P, et al. Met signaling regulates growth, repopulating potential and basal cell-fate commitment of mammary luminal progenitors: implications for basal-like breast cancer. Oncogene. 2013;32:1428–1440. doi: 10.1038/onc.2012.154. [DOI] [PubMed] [Google Scholar]
- 45.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 46.Nuovo GJ, Plaia TW, Belinsky SA, et al. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci U S A. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujii H, Biel MA, Zhou W, et al. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16:2159–2164. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- 48.Dumont N, Crawford YG, Sigaroudinia M, et al. Human mammary cancer progression model recapitulates methylation events associated with breast premalignancy. Breast Cancer Res. 2009;11:R87. doi: 10.1186/bcr2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 51.Lasham A, Samuel W, Cao H, et al. YB-1, the E2F pathway, and regulation of tumor cell growth. J Natl Cancer Inst. 2012;104:133–146. doi: 10.1093/jnci/djr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balko JM, Cook RS, Vaught DB, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nature medicine. 2012;18:1052–1059. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurchott K, Kuban RJ, Krech T, et al. Identification of Y-box binding protein 1 as a core regulator of MEK/ERK pathway-dependent gene signatures in colorectal cancer cells. PLoS genetics. 2010;6:e1001231. doi: 10.1371/journal.pgen.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imada K, Shiota M, Kohashi K, et al. Mutual regulation between Raf/MEK/ERK signaling and Y-box-binding protein-1 promotes prostate cancer progression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:4638–4650. doi: 10.1158/1078-0432.CCR-12-3705. [DOI] [PubMed] [Google Scholar]
- 55.Balko JM, Schwarz LJ, Bhola NE, et al. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer research. 2013;73:6346–6358. doi: 10.1158/0008-5472.CAN-13-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang S, Dong S, Gu TL, et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhullar J, Sollars VE. YBX1 expression and function in early hematopoiesis and leukemic cells. Immunogenetics. 2011;63:337–350. doi: 10.1007/s00251-011-0517-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.