Abstract
Background
Anaerobic culture has been critical in our understanding of the oral microbiotas.
Highlight
Studies in advanced periodontitis in the 1970’s revealed microbial complexes that associated with different clinical presentations. Taxonomy studies identified species newly-observed in periodontitis as Aggregatibacter (Actinobacillus) actinomycetemcomitans, Campylobacter (Wolinella) rectus and other Campylobacter species, and Tannerella (Bacteroides) forsythia. Anaerobic culture of initial periodontitis showed overlap in the microbiota with gingivitis, and added Selenomonas noxia and Filifactor alocis as putative periodontal pathogens. Porphyromonas gingivalis and T. forsythia were found to be associated with initial periodontitis in adults. The dominant microbiota of dental caries differs from that of periodontitis. The major cariogenic species are acidogenic and acid tolerant species particularly Streptococcus mutans, and Lactobacillus and Bifidobacterium species. Anaerobic culture of severe early childhood caries revealed a widely diverse microbiota, comparable to that observed using cloning and sequencing. The PCR-based cloning approach, however, underestimated Actinobacteria compared with culture. Only a subset of the caries-associated microbiota was acid tolerant, with different segments of the microbiota cultured on blood agar compared to a low pH acid agar. While the major caries-associated species was S. mutans, a new species, Scardovia wiggsiae, was significantly associated with early childhood caries. Higher counts of S. wiggsiae were also observed in initial white spot carious lesions in adolescents.
Conclusion
In periodontitis and dental caries, anaerobic culture studies of advanced disease provided a comprehensive analysis of the microbiota of these infections. Anaerobic culture highlighted the limitation of PCR with standard primers that underestimate detection of Actinobacteria.
Keywords: Anaerobic culture, 16S rRNA, Microbiome, Caries, Periodontitis
1. Introduction Anaerobic culture of plaque biofilm samples
Dental plaque samples were first examined microscopically by van Leeuwenhoek, then by culture as methods and bacterial growth media were developed. The cultivability of dental plaque bacteria improved over time with the use of complex, blood-containing media, and with application of anaerobic methods to process and culture bacteria. Studies in the 1960’s were critical to demonstrate the value of anaerobic methods, which were successfully applied to both periodontal and caries samples in the 1970’s. Major investigators in anaerobic culture of plaque samples in the 1970’s and 1980’s were those at the Forsyth institute in the Socransky and co-workers laboratories, at the laboratories of Holdeman and Moore (Virginia Polytechnic Institute, USA), Walter Loesche (Michigan, USA), William Wade (UK), and Etsuro Hoshino (Nigata, Japan) among others.
Anaerobic cultural methods continue to expand our understanding of the oral microbiota of periodontitis and dental caries although approaches for strain identification have changed from biochemical tests to 16S RNA sequence based identifications. This review will focus on selected anaerobic culture studies that have provided the basis of our understanding of the oral microbiota. Non-cultural molecular analyses of plaque samples, mainly based on analysis of the 16S rRNA gene, have highlighted both strengths and limitations of culture to describe the complete oral microbiota. Nevertheless, when bacteria are detected by molecular methods, the focus then becomes to devise methods to cultivate them [1] which frequently involves use of anaerobic methods.
2. Periodontitis
2.1. Advanced Periodontitis
Clinically, periodontitis affects the tooth supporting structures and if left unchecked leads to tooth loosening and loss. Periodontitis is recognized by increased depth in the gingival sulcus leading to periodontal pockets, and by loss of periodontal attachment and the surrounding alveolar bone. Periodontitis is frequently associated with gingival inflammation, gingivitis, which may include spontaneous bleeding of gingival tissues.
Based on the observation that periodontal loss increased with age, in the 1960’s it had been assumed that this was a slowly progressing chronic infection that was difficult to arrest. One form of periodontitis variously recognized as periodontosis, juvenile periodontitis, and currently aggressive periodontitis, however, progressed very rapidly. Permanent teeth, principally central incisors and 1st permanent molars that erupted in childhood could be lost in adolescence. Anaerobic culture of bacteria from periodontal pockets associated with aggressive periodontitis in adolescents in Socransky’s laboratory in the 1970’s revealed a microbiota containing bacteria not previously recognized from periodontal samples of other patients [2, 3]. Further many isolates were difficult to maintain in culture. This form of rapidly destructive periodontitis in adolescents is now recognized as characterized by a microbiota frequently dominated by Aggregatibacter actinomycetemcomitans.
Advanced adult periodontitis sites were also found to have progressing disease as demonstrated by increasing alveolar bone loss observed from sequential radiographs. This contrasted from periodontal pockets that were not progressing and represented disease remission or repair. A second study of advanced periodontitis was undertaken in the 1970’s in Socransky’s laboratory. In this study adult periodontitis was defined as advanced disease with sites that either had a record of increasing alveolar bone loss on radiographs within the previous two years or young adults with very advanced periodontitis. Healthy, control sites in the same subjects (when present) had no attachment loss, and minimal if any gingival inflammation [4].
Subgingival samples were collected and processed using anaerobic methods, with prolonged incubation of samples since pilot sampling indicated that only a portion of the microbiota formed colonies with incubation times less than 10–14 days. Microbiological methods included use of prereduced anaerobically sterilized (PRAS) solutions in Hungate or roll tubes for sample processing and biochemical tests. Strain characterization relied on detection of acid end products of metabolism to define genera, and an array of fermentation and other tests for isolate speciation.
The major finding of clinical importance was that the microbiotas differed between subjects, and from that of the adolescent aggressive periodontitis. Furthermore, microbiotas of adult periodontitis could be grouped into distinct disease types. Hindsight allows renaming these disease groups from “young adult” to generalized aggressive periodontitis, “minimal inflammation” to post-antibiotics refractory periodontitis, and “moderate inflammation” to chronic periodontitis (Figure 1). Differentiation of clinical and microbial subgroups within periodontitis led to a profound change in the description of periodontitis with the recognition of distinct and different “periodontal diseases”.
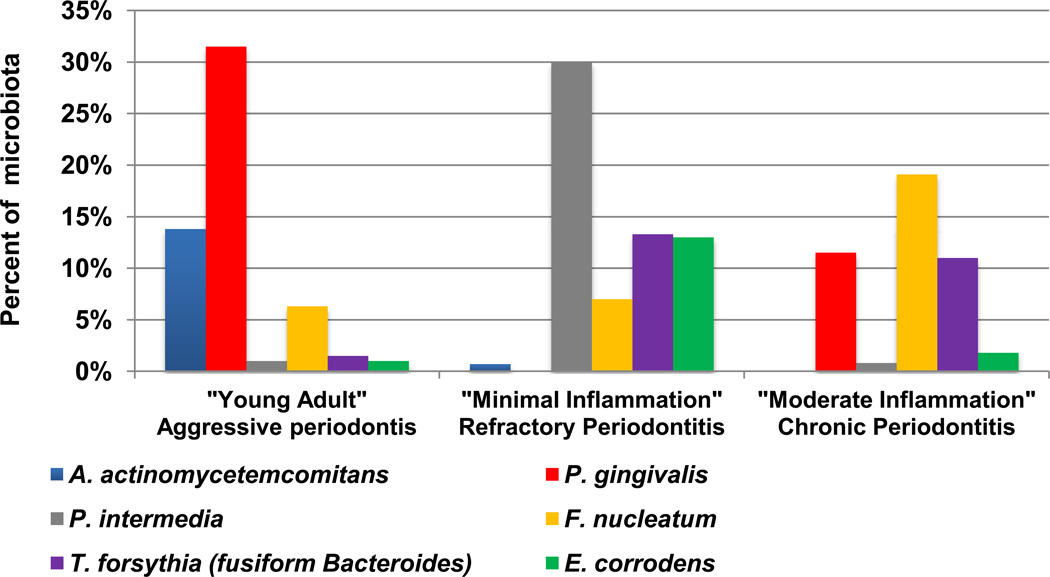
Figure 1. Gram negative species associated with progressing advanced periodontitis.
Samples from deep periodontal pockets were cultured anaerobically and species identified using biochemical tests. Three microbial complexes were observed in different clinical categories. Samples from “Young Adults” or aggressive periodontitis were dominated by A. actinomycetemcomitans and P. gingivalis; from “minimal inflammation” or refractory periodontitis by P. intermedia, T. forsythia and E. corrodens; and from “Moderate inflammation”, chronic periodontitis by P. gingivalis, F. nucleatum and T. forsythia. Data modified from Tanner et al. [4].
Healthy sulci microbes were dominated by gram positive species, mainly Streptococcus and Actinomyces species, although over 10% could not be identified. Gram negative anaerobic rods dominated the advanced periodontitis microbiotas (Figure 1). The “young adult”, aggressive periodontitis group was characterized with a species tentatively identified as A. actinomycetemcomitans with Bacteroides asaccharolyticus (now Porphyromonas gingivalis). The “minimal inflammation”, refractory periodontitis group harbored had higher proportions of Bacteroides melaninogenicus subsp. intermedius (now Prevotella intermedia), Eikenella corrodens and unidentified isolates grouped as “fusiform Bacteroides” (now Tannerella forsythia). The third “moderate inflammation”, chronic periodontitis group was dominated by a complex of P. gingivalis, F. nucleatum and “fusiform Bacteroides”. As in the previous Newman studies of periodontosis [3], many isolates did not fit species recognized from the earlier studies from periodontal pockets [5]. These unidentified isolates were grouped as A. actinomycetemcomitans-like, “vibrio-corroders” and “fusiform Bacteroides”.
2.2. Species new to periodontal pockets
Characterization of the A. actinomycetemcomitans-like isolates included determining guanine plus cytosine (G+C) content of cells and DNA-DNA hybridizations in addition to biochemical tests. Findings confirmed that some isolates were indeed A. actinomycetemcomitans [6]. The A. actinomycetemcomitans isolates included strain Y4 from adolescent aggressive periodontitis and this isolate has become a widely used reference strain. Isolates from advancing periodontitis included strains of the closely related Haemophilus aphrophilus [6].
The “vibrio-corroder group” presented a new challenge by showing minimal to no growth in conventional glucose broth, and when growth occurred, did not ferment carbohydrates so most routine biochemical tests were not useful for strain characterization. As a group, colonies were transparent and many pitted or corroded agar surfaces. Some isolates resembled Eikenella corrodens that uses amino acids as an energy source and nitrate as electron acceptor. Other isolates with identical colony morphologies were highly motile and resembled “Vibrio succinogens”, a cow rumen species that uses formate or hydrogen gas as energy sources, and fumarate (or other organic acids) as electron acceptors. A taxonomy study was performed and these strains were classified and identified as several new species, Wolinella recta (now Campylobacter rectus), Campylobacter concisus, and Bacteroides gracilis (now Campylobacter gracilis) [7]. Characterization of the stains used DNA hybridizations, as in the A. actinomycetemcomitans study [6], and sensitivity to a range of antibiotics, dyes and indicators. Companion reports classified the strains serologically [8] and described the cell ultrastructure of species [9].
The third group of unidentified isolates grew slowly as tiny colonies, composed of bacterial cells with tapered ends (fusiform) and formed succinate as an acid end product of metabolism, a characteristic of Bacteroides species, giving the group name of “fusiform Bacteroides”. Multiple additives were tested to enhance growth, without much success, although growth agar was enhanced by co-culturing with Fusobacterium nucleatum. To characterize the “fusiform Bacteroides” strains, SDS-PAGE (polyacrylamide gel electrophoresis for whole cell protein profiles) combined with inhibitor tests and DNA hybridizations were used. The strains did not resemble any reference named Bacteroides species and a new species Bacteroides forsythus was proposed [10]. Subsequently, 16S rRNA sequence data demonstrated that this species did not fit in Bacteroides, Prevotella, or Porphyromonas [11]. The species was reclassified to Tannerella forsythensis [12] with the final name of Tannerella forsythia proposed shortly afterwards [13].
Parallel microbiology studies on periodontal and gingival diseases using anaerobic methods were performed by Holdeman (Moore) and Moore at the Virginia polytechnic institute in Blacksburg, Virginia, USA. Their studies [14] led to description of several additional new species that are recognized as significant bacteria in the microbiology of periodontal infections. These new species included Selenomonas noxia, Selenomonas flueggei [15], Actinomyces georgiae, Actinomyces gerencseriae [16], Lactobacillus (now Olsenella) uli, Lactobacillus rimae [17], Oribaculum catoniae, Dialister pneumosintes [18], Prevotella tannerae, Prevotella enoeca [19]. Fusobacterium (now Filifactor) alocis and Fusobacterium (now Eubacterium) sulci [20]. Taxonomy studies from Wade’s laboratory further expands our understanding of the taxonomy of cultured oral species including for Shuttleworthia satelles [21], Dialister invisus [22], Prevotella marshii and Prevotella baroniae [23], and Propionibacterium acidifaciens [24]. Studies from Hishino’s group have further clarified the oral microbiology and taxonomy of oral anaerobes including for Mogibacterium diversum and Mogibacterium neglectum [25], Eubacterium saphenum [26], Eubacterium (Atopobium) minutum [27] and Eubacterium exiguum (now Slackia exigua) [28].
2.3. Initial periodontitis in adults
The early stages of adult periodontitis can be difficult to differentiate from gingivitis. Bacteria from periodontal pockets can spread around and infect gingivally healthy sites of the same individual [29], making healthy sites from subjects with gingivitis or periodontitis inappropriate as non-diseased control sites. Anaerobic cultural analysis of the microbiota of initial periodontitis in adults compared the microbiota of healthy sites from healthy subjects, a gingivitis site in subjects with gingivitis (but no evidence of recent periodontal attachment loss) and an active initial periodontitis site that showed attachment loss in the last few months [30]. Of particular interest was whether the species of advanced periodontitis would also be detected in initial periodontitis.
Microbiologically, differences were detected between health, gingivitis and active (progressing) periodontitis. Active periodontitis was characterized by T. forsythia, Selenomonas noxia, then a new Selenomonas species [15], C. rectus, Fusobacterium (now Filifactor) alocis [20] Eubacterium brachy (Table 1). Most of these species were also detected in gingivitis. Campylobacter gracilis and P. gingivalis were associated with gingivitis, whereas P. intermedia, P. nigrescens and E. corrodens were elevated in both initial periodontitis and gingivitis. Sites with progressing attachment loss were detected at buccal sites, typical of gingival recession. In contrast to the interproximal active sites, these sites harbored a microbiota similar to that of gingival health particularly Actinomyces naeslundii, Actinomyces israelii and Rothia dentocariosa (Table 1).
Table 1.
Microbiota of Initial Active Periodontitis compared with Health, Gingivitis and Recession (Mean % species ± SD) *
| Species | Healthy | Gingivitis | Active | Recession |
|---|---|---|---|---|
| N subjects (sites sampled) | 10 (14) | 10 (15) | 9 (10) | 5 (5) |
| Tannerella forsythia | 0.1±0.1ga | 5.3±2.9hr | 9.0±3.1hr | 0 |
| Selenomonas noxia [15] | 0.9±0.5a | 0.8±0.8a | 5.9±2.2hgr | 0a |
| Campylobacter rectus | 0.2±0.2a | 1.2±0.7 | 4.9±2.2hr | 0a |
| Selenomonas flueggeii [15] | 0 | 0.2±0.2 | 1.6±1.0 | 0 |
| Eubacterium lentum | 0 | 0 | 1.3±1.3 | 0 |
| F. nucleatum ss. fusiforme | 0.5±0.3 | 0.1±0.1 | 1.3±0.7 | 0.4±0.0 |
| Filifactor alocis [20] | 0a | 0.8±0.6 | 1.6±0.6h | 0 |
| Eubacterium brachy | 0.2±0.2ga | 2.3±0.7hr | 3.4±1.8h | 0g |
| Prevotella intermedia | 0 | 2.9±1.7 | 2.7±2.4 | 0.4±0.4 |
| Prevotella nigrescens | 2.2±2.0 | 2.2±1.3 | 2.2±1.8 | 0.8±0.8 |
| Streptococcus anginosus | 2.0±1.4 | 1.2±1.2 | 2.0±1.8 | 0.4±0.4 |
| Campylobacter gracilis | 1.9±0.8 | 4.5±1.9r | 1.9±0.5r | 0ga |
| Porphyromonas gingivalis | 0 | 2.8±2.2 | 0.7±0.7 | 0 |
| Capnocytophaga sputigena | 0.1±0.1 | 1.8±1.1 | 0.9±0.5 | 0 |
| Capnocytophaga gingivalis | 0.9±0.5 | 1.6±0.8 | 0.2±0.2 | 0 |
| Selenomonas sputigena | 0.7±0.6 | 1.5±0.6 | 0.6±0.4 | 0 |
| Olsenella uli [17] | 0 | 1.1±1.1 | 0 | 0 |
| Eikenella corrodens | 0.2±0.2 | 1.0±0.8 | 1.3±0.8 | 0.8±0.5 |
| Streptococcus oralis | 14.2±7.1ga | 0.8±0.3hr | 1.3±0.9h | 8.0±3.7g |
| Actinomyces naeslundii | 13.6±6.0 | 7.6±3.3r | 3.7±1.4r | 23.6±7.9ga |
| Actinomyces gerensceriae [16] | 10.8±5.9 | 2.6± 1.5 | 0.9±0.6 | 2.0±2.0 |
| Parvimonas micra | 4.9±2.3 | 3.4±1.6 | 3.2±1.0 | 0.8±0.5 |
| Streptococcus intermedius | 4.1±2.6 | 3.3±1.3 | 3.8±2.4 | 0.4±0.4 |
| Veillonella parvula | 2.5±1.1 | 0.8±0.5 | 2.7±2.2 | 0 |
| Streptococcus mutans | 2.4±1.8 | 0.1±0.1 | 1.8±1.2 | 0 |
| Eubacterium nodatum | 1.4±1.4 | 0.4±0.3 | 0 | 0 |
| Rothia dentocariousa | 1.3±1.3 | 0.3±0.2 | 0F | 14.4±12.0a |
| Gemella morbillorum | 1.5±1.0 | 0.3±0.2 | 0.2±0.2 | 0 |
| Streptococcus sanguinis | 0.7±0.6 | 0.4±0.4 | 0 | 8.4±7.9 |
| Actinomyces israelii | 0.2±0.2 | 2.3±1.6a | 0.0±0.0g | 10.0±10.0 |
| Actinomyces odontolyticus | 0.5±0.3 | 1.1±0.6 | 0.9±0.4 | 2.4±1.9 |
| Haemophilus aphropilus | 0.8±0.6 | 0.5±0.4 | 0.7±0.3 | 1.2±1.2 |
| Mean pocket depth (mm) | 2.5±0.5ar | 3.0±0.6ar | 4.0±1.0hgr | 1.5±0.4hga |
| Mean attachment level (mm) | 1.3±0.4ar | 1.9±0.8ar | 3.7±1.1hg | 3.3±0.5hg |
| Mean % of site with: | ||||
| Plaque | 10±32g | 70±42hr | 78±44r | 0ga |
| Redness | 10±32g | 80±42hr | 33±50g | 0ga |
| Bleeding on probing | 0 | 17±33 | 22±44 | 0 |
Superscripts values that differed between groups <0.05 Kriskal-Wallis rank test. h= health, g=gingivitis, a=interproximal active, r = buccal active/recession. 32 additional species were detected at less than 1% of the cultivable microbiota.
Data modified from Tanner et al. [30], with reference links to species newly described when paper published
Anaerobic culture of progressing initial periodontitis thus added Selenomonas noxia and Filifactor alocis as candidate periodontal pathogens, to those species associated advanced periodontitis. Other studies on the same patient population examined subgingival temperature and microbiota in initial periodontitis [31], Campylobacter species in health, gingivitis, and periodontitis [32], and Serum IgG reactivity to subgingival bacteria in initial periodontitis, gingivitis and healthy subjects [33].
While the strength of anaerobic culture is the ability to detect all species present cultured in a sample, culture is time consuming and labor intensive which limits the number of subjects that can be analyzed. Furthermore, culture can underestimate the presence of fastidious species. For example, we observed that more subjects were P. gingivalis positive when subgingival samples were assayed using an immunofluorescence assay compared with anaerobic culture [34], suggesting that culture was underestimating the P. gingivalis. Thus to test our culture findings of initial periodontitis, we designed a study with more subjects and assayed samples using molecular assays, 16S rRNA [35] and whole genomic probes [36], in reverse capture and conventional checkerboard assays respectively. To specifically target P. gingivalis and T. forsythia we used a multiplex PCR assay [37]. In this larger population of 190 subjects, P. gingivalis and T. forsythia were significantly associated with early periodontal loss in comparison with healthy and gingivitis sites, whereas P. gingivalis was detected more frequently in progressing compared with inactive sites, while the association of T. forsythia with active periodontitis did not reach significance (Figure 2). The gingival crevice fluid host in this population was also studied and a strong association between IL-1β and early periodontitis was observed [38].
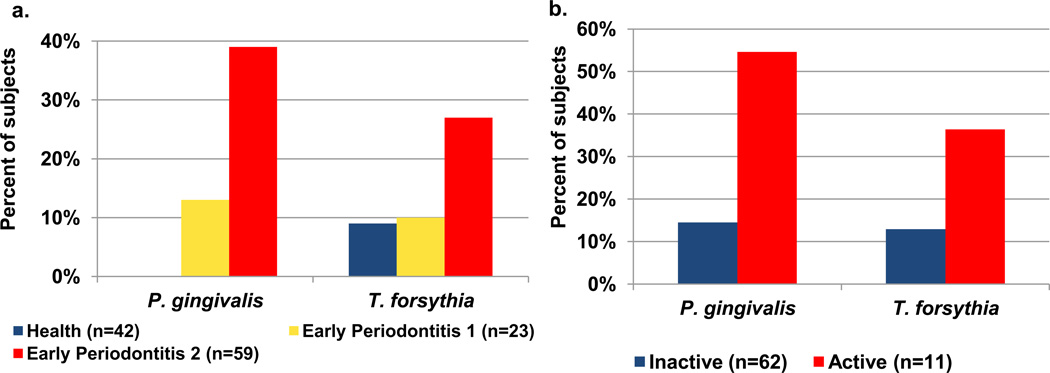
Figure 2. P. gingivalis and T. forsythia in (a) early periodontitis and (b) active initial periodontitis in adults.
Samples from periodontitis and healthy control sites were analyzed by PCR.
a. In the cross sectional analysis of early periodontitis, increased detection frequencies of P. gingivalis (p<0.001) and T. forsythia (p<0.03) were observed in subjects with over 1 site with at least 2 mm periodontal attachment loss.
b. In longitudinal analyses, detection frequency of P. gingivalis (p<0.01) was associated with progressing/active periodontitis whereas the association with T. forsythia did not reach significance (p=0.075). Data modified from Tanner et al. [35, 36].
2.4. Periodontitis by anaerobic culture
Findings from these anaerobic culture studies on advanced and initial adult periodontitis provided the species for DNA probes for checkerboard of Socransky and co-workers [39]. There is a vast literature in periodontal microbiology based on these species. Other studies using anaerobic microbiology to characterize periodontitis that improve our understanding of the cultivable microbiota, include those of Slots [40–42], Zambon [43], and most notably those of Holdeman and Moore [14]. Further culture of the related infections, pericoronitis [44], dentoalveolar abscesses [45] and endodontic infections [46] Wade’s group expand periodontal species and their role in oral infections. Additionally having cultured isolates provides strains for antimicrobial and periodontal pathogenicity testing.
In defining pathogens for human periodontitis, the associations of P. gingivalis and T. forsythia with advanced and initial periodontitis in adults, suggests a continuum of disease progression involving these species. They are not, however, the only periodontal pathogens, which likely vary between patients. Further, additional unnamed species that are better detected using molecular methods [47] need considering in the pathogenesis of periodontitis.
3. Dental Caries
Dental caries begins as decalcified enamel or white spot lesions which progresses into cavities in enamel and finally into dentin. If left untreated, bacteria in dentin invade the pulp then root canals. Dental caries is recognized to have three major risk factors; cariogenic bacteria, a susceptible host or compromised teeth and frequent ingestion of dietary carbohydrates which are used as substrates by bacteria to produce acid that will demineralize tooth surfaces. The rate of caries progression varies based on the levels of these risk factors. In young children dental caries can progress very rapidly and spread throughout the child’s dentition.
The major species most frequently associated with dental caries are the acid-producing and acid-tolerant Streptococcus mutans and Lactobacillus species [48]. Several other bacterial groups have been associated with carious lesions including non-mutans streptococci, Actinomyces and Bifidobacterium species [49–51]. A sequence of colonization of bacterial groups in the progression of dental caries was presented in the seminal paper of Takahashi and Nyvad [52]. Under this hypothesis an initial stage of stability, albeit associated with mild and infrequent acidification, was associated with non-mutans streptococci and Actinomyces. This could lead to second acidogenic stage characterized by low pH species. The last aciduric stage was characterized by species that were highly tolerant to acid, including Streptococcus mutans, Lactobacillus and Bifidobacterium species.
Many other species, however, have been detected in dental caries, particularly in studies performed in the last decade using 16S rRNA identifications of clones [53–55] or isolates [56]. The identification of a wide diversity of species, including uncultivated species, suggested that our knowledge of the cultural microbiota of early childhood caries is incomplete.
3.1. Early childhood caries
Study of dental caries using anaerobic culture examined a patient population of children with advanced, severe early childhood caries that were scheduled for treatment under general anesthesia for the extensive restorations and extractions needed [57]. This allowed clinical measurements and sampling procedures to be performed under anesthesia avoiding any behavioral or compliance issues that could interrupt clinical procedures in very young, and frequently frightened, children. A dietary and dental history survey was also obtained from the child’s parent or caregiver.
Samples for culture were taken and put in PRAS Ringers salt solution, processed and incubated anaerobically as for the previous studies in advanced and initial periodontitis. Samples from caries and caries-free control children were cultured on a complex blood agar to facilitate growth of fastidious anaerobes, including species without culture representatives, but that had been observed from cloning and sequencing analysis. Samples were also cultured on a pH 5 acidic agar medium to select for acid tolerant species, to enrich for novel caries pathogens [57]. Bacterial samples were co-processed by sequencing and cloning analysis [58] and by 16S rRNA (HOMIM) microarray [59].
There were several advantages to using an anaerobic culture method to study early childhood caries as has previously been observed for childhood caries [48] and carious dentin [60]. The cultural approach [57] detected a wider diversity of taxa than the parallel molecular cloning and sequencing [58] mainly because culture improved detection of Actinobacteria, including Actinomyces and Bifidobacterium/Scardovia species (Figure 3). Improved detection of Actinobacteria from dental caries by culture compared to cloning/sequencing analysis had been observed in samples from deep dentine [56] and was attributed to limitations in the 16S rRNA primers used to amplify the DNA in samples before cloning. Anaerobic culture on blood agar allowed cultivation of several unnamed and previously uncultured species, particularly from blood agar isolation and prolonged primary incubation of about 10 days. Newly cultured species were mainly in Streptococcus and Actinobacteria and these isolates are currently undergoing further characterization under the Human Oral Microbiome Database (HOMD) project [1, 61].
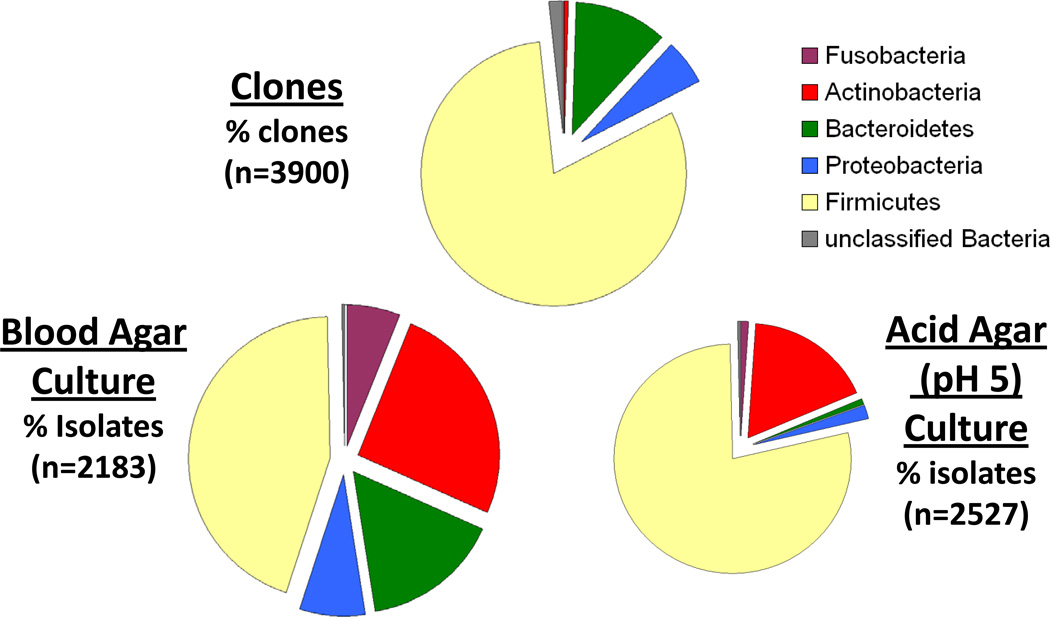
Figure 3. Bacterial phyla from clonal and cultural analyses of early childhood caries and caries-free children.
Samples from 80 children were processed by cloning and sequencing [58] and by anaerobic culture [57]. All clones and isolates were identified from 16S rRNA sequences in the Human Oral Microbiome Database (HOMD) [61]. Higher proportions of Actinobacteria were observed in cultural analysis on blood or acid agars compared with the cloning/sequencing analysis. Total bacterial counts on blood agar were 10 times higher than acid agar, and the greatest microbial diversity was observed from anaerobic blood agar isolation. Sequence based analyses by Natalia I Chalmers.
Several species were preferentially isolated on either blood (24 species) or the acidic (10 species) agar with marked differences within individual genera (Table 2) [57]. Species suppressed by the acidic agar included Streptococcus sanguinis, Actinomyces, Selenomonas, Capnocytophaga, Prevotella, Fusobacterium and Campylobacter species. This would suggest these species would not fit into species of the advanced aciduric phase of dental caries from the Takahashi/Nyvad model [52]. In contrast S. mutans, Streptococcus anginosus, Streptococcus salivarius, Lactobacillus gasseri, Scardovia wiggsiae, Parascardovia denticolens and Bifidobacterium dentium were detected more frequently from the acid agar and could be candidates for the advanced aciduric stage of caries progression.
Table 2.
Major phylotypes and species detected more frequently from either blood or acid (pH 5) agars from childhood caries and caries-free children
| Detected more frequently blood agar | Detected more frequently acid agar | ||
|---|---|---|---|
| Firmicutes | Streptococcus sanguinis (p<0.0001) | Firmicutes | Streptococcus thermophius (p< 0.0001) |
| Gemella morbillorum (p<0.0001) | Streptococcus mutans (p<0.0001) | ||
| Abiotrophia defectiva (p<0.0001) | Streptococcus anginosus (p<0.0001) | ||
| Actino- | Actinomyces naeslundii (p<0.0001) | Streptococcus salivarius (p= 0.001) | |
| bacteria | Actinomyces sp. HOT 171 (p<0.0001) | Lactobacillus gasseri (acid only) | |
| Actinomyces massiliensis (p<0.0001) | Actinobacteria | ||
| A. gerensceriae (p<0.0001) | Scardovia HOT195 (S. wiggsiae) (p<0.003) | ||
| Actinomyces sp. HOT 178 (p<0.0001) | Parascardovia denticolens (p=0.034) | ||
| Actinomyces sp. HOT 169 (p<0.0001) | Bifidobacterium dentium (acid only) (p=0.05) | ||
| Veillon- | Selenomonas noxia (p<0.0001) | Veillonellaceae | |
| ellaceae | Selenomonas infelix (p<0.0001) | Veillonella atypica (p<0.0001) | |
| Selenomonas artemidis (p<0.0001) | Veillonella dispar (p=0.003) | ||
| Selenomonas sputigena (p<0.0001) | |||
| Bacter- | Capnocytophaga granulosa (p<0.0001) | Bacteroides | NONE |
| oidetes | Capnocytophaga gingivalis (p<0.0001) | ||
| Capnocytophaga sp. HOT 326 (p<0.0001) | |||
| Capnocytophaga sp. HOT 336 (p<0.0001) | |||
| Prevotella melaninogenica (p<0.0001) | |||
| Prevotella nigrescens (p<0.0001) | |||
| Fusobacteria | F. nucleatum ss polymorphum (p<0.0001) | Fusobacteria | NONE |
| Leptotrichia bucallis (p<0.0001) | |||
| Proteo- | Campylobacter concisus (p<0.0001) | Proteobacteria | |
| bacteria | Campylobacter showae (p<0.0001) | NONE | |
| Eikenella corrodens (p<0.0001) | |||
Comparing the microbiota of severe early childhood caries with caries free children, the major caries-associated species from both blood and acid agar isolation were S. mutans and a new Bifidobacterium-group species, Scardovia wiggsiae [62] (Figure 4). Not all species isolated on the acid agar, however, were caries-associated including some unnamed Actinomyces species, Streptococcus gordonii, Streptococcus intermedius and Streptococcus thermophilus.
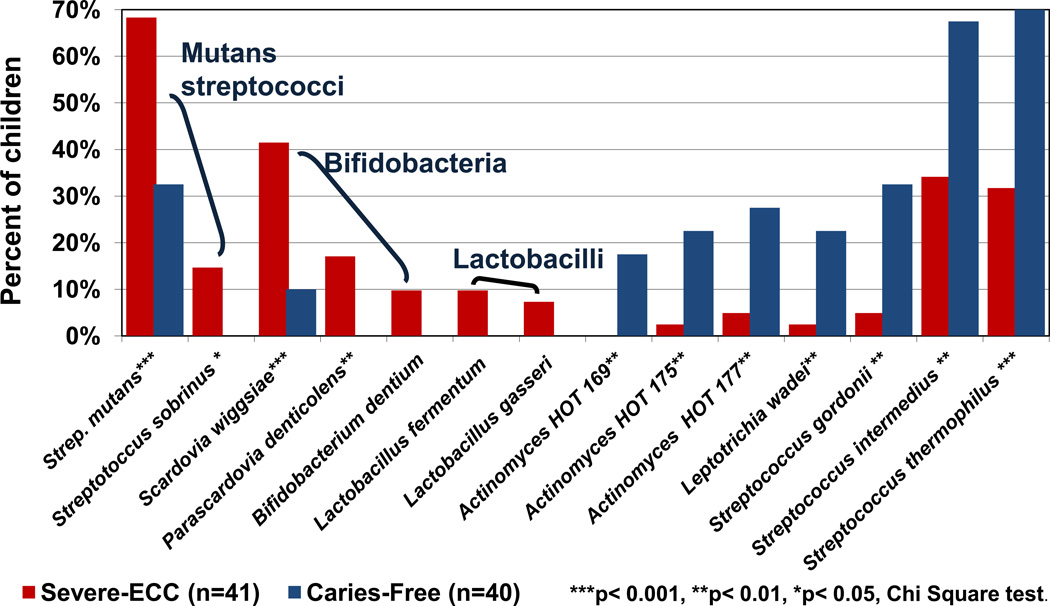
Figure 4. Major acid-tolerant species cultured from early childhood caries and caries-free children.
Samples were processed by anaerobic culture and isolates were identified from 16S rRNA sequences in HOMD. The detection frequencies of the major acid tolerant species associated with childhood caries were S. mutans, S. wiggsiae, Parascardovia denticolens and Streptococcus sobrinus. Other acid resistance species were detected more frequently from caries-free children including several non-mutans streptococci, and unnamed Actinomyces species as denoted by the Human Oral Taxon (HOT) number. Data modified from Tanner et al. [57].
The association of S. mutans with severe early childhood (nursing) caries from cultural analysis was also observed in previous culture studies with anaerobic incubation of samples [63, 64]. One study from David Beighton’s laboratory was a comprehensive analysis that incorporated selection of acid-tolerant bacteria from broth isolation. In all the anaerobic cultural analysis using non-selective blood agar for strain isolation, the dominant species detected were Streptococcus and Actinomyces species [57, 63, 64]. Cultural detection of several species in Bifidobacteriaceae was performed using a modified selective agar, and while S. wiggsiae was detected, the major species isolated childhood caries was Bifidobacterium dentium [65]. Bifidobacteriaceae including Bifidobacterium and Scardovia species, however, are major caries-associated species being highly associated with root caries and other lesions in older adults [49, 50] in addition to childhood caries [53, 66, 67]. A separate culture study of infected pulps in children identified high proportions of S. wiggsiae (as Bifidobacterium spp. 2) [68] suggesting that this species is a significant part of the microbial complex involved in the progression of deep dentinal caries.
Few Lactobacillus species were detected in the anaerobic cultivation of severe early childhood caries suggesting that they colonize at low levels of the microbiota in very young children [57]. Several Lactobacillus species were detected in the Beighton laboratory study of similar childhood caries infection, nursing caries, but only after use of selective isolation [64] which facilitates detection of species a low levels in samples. The major species detected, Lactobacillus fermentum was similar in both culture studies.
3.2. White spot initial carious lesions
The associations of S. mutans and S. wiggsiae with severe early childhood caries were observed from assay by species specific PCR [59] in addition to anaerobic culture. To evaluate whether S. mutans and S. wiggsiae were also involved in initial carious lesions, the PCR assay was modified to be quantitative (q-PCR) to examine the microbiota of initial white spot carious lesions [69]. The microbiota of white spot lesions in adolescents with fixed orthodontic bands was selected to study. White spots or zones parallel to the gingival margin representing enamel decalcification may develop within 6–12 months after orthodontic bands are placed, particularly when oral hygiene is poor [70]. White spot lesions are generally considered a cosmetic problem, although if enamel decalcification progresses to cavities, bands are generally removed to prevent further caries progression.
Several studies have examined the development of gingivitis associated with fixed orthodontic appliances [71, 72]. The microbiology of incipient white spot carious lesions was studied using anaerobic culture by George Bowden an co-workers [73]. The culture study focused on Streptococcus, Lactobacillus and Actinomyces species associated with the development of initial enamel decalcification below orthodontic bands and reported detection of S. mutans in only a few children.
Using qPCR, S. mutans and S. wiggsiae were associated with white spot lesions [69]. Both species were also associated with gingival inflammation, but, more importantly the caries associations remained significant after adjusting for presence of gingivitis. The caries-associations for S. mutans and S. wiggsiae were not significant using 16S rRNA probes in the HOMIM microarray that was performed in parallel with the qPCR assay [69]. Using the same HOMIM microarray caries-associations for S. mutans and S. wiggsiae were positive but were significant only for S. mutans [74]. Other white spot lesion-associated species varied between studies using the HOMIM microarray, possibly reflecting the longitudinal [74] compared to cross-sectional [69] study designs.
The detection of S. mutans and S. wiggsiae in initial caries, based on previous culture of these species using anaerobic methods of samples from advanced caries suggests that similar acidogenic and acid tolerant species can be involved in early and later stages of this disease, and could be good makers to detect aggressive disease.
4. Conclusions
Anaerobic culture of bacteria associated with advanced periodontitis and dental caries, compared to healthy, non-diseased, sites has proved extremely valuable in expanding our knowledge of the bacteria associated with these major clinical conditions of the oral cavity. By using rich non-selective media and anaerobic incubation long enough for the dominant microbiota to grow, a wide diversity of bacteria has been detected with improved detection of Actinobacteria compared to PCR-based methods using standard primers. Anaerobic culture studies have provided species for rapid detection using molecular methods which can be used on studies of larger populations of subjects. Anaerobic culture, however, is limited in the number of samples that can be processed as it remains a labor and time consuming approach. Furthermore, species in lower proportions of the overall microbiota, and species for which the nutritional requirements are as yet unknown remain undetected by anaerobic culture [1].
Of clinical importance, pathogens to advanced periodontal and carious lesions were detected in initial disease. This suggests that pathogens from advanced disease are candidates for disease risk assessment. Since dental pathogens may also colonize healthy sites assessment of periodontal and caries risk will require addition of other risk markers for example host factors in periodontitis [75] and diet in dental caries [76].
Acknowledgements
This work was conducted with support from USPHS grant DE- 016937 from the NICDR National Institutes of Health, and from the William Bingham 2nd Trust. I would like to thank Dr. Natalia Chalmers for the analysis in Figure 3 and Drs. Floyd Dewhirst and Christine Kressirer for suggestions to improve the manuscript.
Footnotes
Presented at the symposium in the annual meeting of Japanese Association for Oral Biology held in Okayama, Japan.
Ethical Approval
This review cites papers that have already been published and each had own ethical approval and thus was documented in the original publications.
Conflict of Interest
None.
References
- 1.Wade WG. Characterisation of the human oral microbiome. J Oral Biosci. 2013;55:143–148. [Google Scholar]
- 2.Newman MG, Socransky SS, Savitt ED, Propas DA, Crawford A. Studies of the microbiology of periodontosis. J Periodontol. 1976;47:373–379. doi: 10.1902/jop.1976.47.7.373. [DOI] [PubMed] [Google Scholar]
- 3.Newman MG, Socransky SS. Predominant cultivable microbiota in periodontosis. J Periodontal Res. 1977;12:120–128. doi: 10.1111/j.1600-0765.1977.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 4.Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 5.Socransky SS. Relationship of bacteria to the etiology of periodontal disease. J Dent Res. 1970;49:203–222. doi: 10.1177/00220345700490020401. [DOI] [PubMed] [Google Scholar]
- 6.Tanner AC, Visconti RA, Socransky SS, Holt SC. Classification and identification of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus by cluster analysis and deoxyribonucleic acid hybridizations. J Periodontal Res. 1982;17:585–596. doi: 10.1111/j.1600-0765.1982.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 7.Tanner ACR, Badger S, Lai CH, Listgarten MA, Visconti RA, Socransky SS. Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. Int J Syst Bacteriol. 1981;31:432–445. [Google Scholar]
- 8.Badger SJ, Tanner ACR. Serological studies of Bacteroides gracilis, Campylobacter concisus, Wolinella recta, and Eikenella corrodens, all from humans with periodontal disease. Int J Syst Bacteriol. 1981;31:446–451. [Google Scholar]
- 9.Lai CH, Listgarten MA, Tanner ACR, Socransky SS. Ultrastructures of Bacteroides gracilis, Campylobacter concisus, Wolinella recta, and Eikenella corrodens, all from humans with periodontal disease. Int J Syst Bacteriol. 1981;31:465–475. [Google Scholar]
- 10.Tanner ACR, Listgarten MA, Ebersole JL, Strzempko MN. Bacteroides forsythus sp. nov., a slow growing fusiform Bacteroides species from the human oral cavity. Int J Syst Bacteriol. 1986;36:213–221. [Google Scholar]
- 11.Paster BJ, Dewhirst FE, Olsen I, Fraser GJ. Phylogeny of Bacteroides, Prevotella and Porphyromonas spp. and related bacteria. J Bacteriol. 1994;176:725–732. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto M, Suzuki M, Umeda M, Ishikawa I, Benno Y. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52:841–849. doi: 10.1099/00207713-52-3-841. [DOI] [PubMed] [Google Scholar]
- 13.Maiden MF, Cohee P, Tanner AC. Proposal to conserve the adjectival form of the specific epithet in the reclassification of Bacteroides forsythus Tanner et al. 1986 to the genus Tannerella Sakamoto et al. 2002 as Tannerella forsythia corrig., gen. nov., comb. nov. Request for an Opinion. Int J Syst Evol Microbiol. 2003;53:2111–2112. doi: 10.1099/ijs.0.02641-0. [DOI] [PubMed] [Google Scholar]
- 14.Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 15.Moore LVH, Johnson JL, Moore WEC. Selenomonas noxia sp. nov., Selenomonas flueggei sp. nov., Selenomonas infelix sp. nov., Selenomonas dianae sp. nov., and Selenomonas artemidis sp. nov., from the human gingival crevice. Int J Syst Bacteriol. 1987;37:271–280. [Google Scholar]
- 16.Johnson JL, Moore LV, Kaneko B, Moore WE. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotype II in A.naeslundii genospecies 2. Int J Syst Bacteriol. 1990;40:273–286. doi: 10.1099/00207713-40-3-273. [DOI] [PubMed] [Google Scholar]
- 17.Olsen I, Johnson JL, Moore LV, Moore WE. Lactobacillus uli sp. nov., and Lactobacillus rimae sp. nov., from the human gingival crevice and emended descriptions of lactobacillus minutus and Streptococcus parvulus. Int J Syst Bacteriol. 1991;41:261–266. doi: 10.1099/00207713-41-2-261. [DOI] [PubMed] [Google Scholar]
- 18.Moore LV, Moore WE. Oribaculum catoniae gen. nov., sp. nov.; Catonella morbi gen. nov., sp. nov.; Hallella seregens gen. nov., sp. nov.; Johnsonella ignava gen. nov., sp. nov., and Dialister pneumosintes gen. nov., comb. nov., nom. rev., Anaerobic gram-negative bacilli from the human gingival crevice. Int J Syst Bacteriol. 1994;44:187–192. doi: 10.1099/00207713-44-2-187. [DOI] [PubMed] [Google Scholar]
- 19.Moore LV, Johnson JL, Moore WE. Descriptions of Prevotella tannerae sp. nov. and Prevotella enoeca sp. nov. from the human gingival crevice and emendation of the description of Prevotella zoogleoformans. Int J Syst Bacteriol. 1994;44:599–602. doi: 10.1099/00207713-44-4-599. [DOI] [PubMed] [Google Scholar]
- 20.Cato EP, Moore LVH, Moore WEC. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int J Syst Bacteriol. 1985;35:475–477. [Google Scholar]
- 21.Downes J, Munson MA, Radford DR, Spratt DA, Wade WG. Shuttleworthia satelles gen. nov., sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2002;52:1469–1475. doi: 10.1099/00207713-52-5-1469. [DOI] [PubMed] [Google Scholar]
- 22.Downes J, Munson M, Wade WG. Dialister invisus sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2003;53:1937–1940. doi: 10.1099/ijs.0.02640-0. [DOI] [PubMed] [Google Scholar]
- 23.Downes J, Sutcliffe I, Tanner AC, Wade WG. Prevotella marshii sp. nov., and Prevotella baroniae sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2005;55:1551–1555. doi: 10.1099/ijs.0.63634-0. [DOI] [PubMed] [Google Scholar]
- 24.Downes J, Wade WG. Propionibacterium acidifaciens sp. nov., isolated from the human mouth. Int J Syst Evol Microbiol. 2009;59:2778–2781. doi: 10.1099/ijs.0.010470-0. [DOI] [PubMed] [Google Scholar]
- 25.Nakazawa F, Poco SE, Jr, Sato M, Ikeda T, Kalfas S, Sundqvist G, Hoshino E. Taxonomic characterization of Mogibacterium diversum sp. nov. and Mogibacterium neglectum sp. nov., isolated from human oral cavities. Int J Syst Evol Microbiol. 2002;52:115–122. doi: 10.1099/00207713-52-1-115. [DOI] [PubMed] [Google Scholar]
- 26.Uematsu H, Nakazawa F, Ikeda T, Hoshino E. Eubacterium saphenum sp. nov., isolated from human periodontal pockets [corrected] Int J Syst Bacteriol. 1993;43:302–304. doi: 10.1099/00207713-43-2-302. [DOI] [PubMed] [Google Scholar]
- 27.Poco SE, Jr, Nakazawa F, Sato M, Hoshino E. Eubacterium minutum sp. nov., isolated from human periodontal pockets. Int J Syst Bacteriol. 1996;46:31–34. doi: 10.1099/00207713-46-1-31. [DOI] [PubMed] [Google Scholar]
- 28.Poco SE, Jr, Nakazawa F, Ikeda T, Sato M, Sato T, Hoshino E. Eubacterium exiguum sp. nov., isolated from human oral lesions. Int J Syst Bacteriol. 1996;46:1120–1124. doi: 10.1099/00207713-46-4-1120. [DOI] [PubMed] [Google Scholar]
- 29.Riviere GR, Smith KS, Tzagaroulaki E, Kay SL, Zhu X, DeRouen TA, Adams DF. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. J Periodontol. 1996;67:109–115. doi: 10.1902/jop.1996.67.2.109. [DOI] [PubMed] [Google Scholar]
- 30.Tanner ACR, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 31.Maiden MF, Tanner AC, Macuch PJ, Murray L, Kent RL., Jr Subgingival temperature and microbiota in initial periodontitis. J Clin Periodontol. 1998;25:786–793. doi: 10.1111/j.1600-051x.1998.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 32.Macuch PJ, Tanner AC. Campylobacter species in health, gingivitis, and periodontitis. J Dent Res. 2000;79:785–792. doi: 10.1177/00220345000790021301. [DOI] [PubMed] [Google Scholar]
- 33.Tanner AC, Kent RL, Jr, Maiden MF, Macuch PJ, Taubman MA. Serum IgG reactivity to subgingival bacteria in initial periodontitis, gingivitis and healthy subjects. J Clin Periodontol. 2000;27:473–480. doi: 10.1034/j.1600-051x.2000.027007473.x. [DOI] [PubMed] [Google Scholar]
- 34.Tanner AC, Maiden MF, Zambon JJ, Thoren GS, Kent RL., Jr Rapid chair-side DNA probe assay of Bacteroides forsythus and Porphyromonas gingivalis. J Periodontal Res. 1998;33:105–117. doi: 10.1111/j.1600-0765.1998.tb02299.x. [DOI] [PubMed] [Google Scholar]
- 35.Tanner AC, Paster BJ, Lu SC, Kanasi E, Kent R, Jr, Van Dyke TE, Sonis ST. Subgingival and tongue microbiota during early periodontitis. J Dent Res. 2006;85:318–323. doi: 10.1177/154405910608500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanner AC, Kent R, Jr, Kanasi E, Lu SC, Paster BJ, Sonis ST, Murray LA, Van Dyke TE. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol. 2007;34:917–930. doi: 10.1111/j.1600-051X.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- 37.Tran SD, Rudney JD. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus and Porphyromonas gingivalis. J Clin Microbiol. 1999;37:3504–3508. doi: 10.1128/jcm.37.11.3504-3508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stashenko P, Van Dyke T, Tully P, Kent R, Sonis S, Tanner AC. Inflammation and genetic risk indicators for early periodontitis in adults. J Periodontol. 2011;82:588–596. doi: 10.1902/jop.2010.100443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, Goodson JM. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 40.Slots J. The predominant cultivable microflora of advanced periodontitis. Scand J Dent Res. 1977;85:114–121. doi: 10.1111/j.1600-0722.1977.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 41.Slots J. Microflora in the healthy gingival sulcus in man. Scand J Dent Res. 1977;85:247–254. doi: 10.1111/j.1600-0722.1977.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 42.Slots J, Moenbo D, Langebaek J, Frandsen A. Microbiota of gingivitis in man. Scand J Dent Res. 1978;86:174–181. doi: 10.1111/j.1600-0722.1978.tb01929.x. [DOI] [PubMed] [Google Scholar]
- 43.Zambon JJ, Reynolds HS, Slots J. Black-pigmented Bacteroides spp. in the human oral cavity. Infect Immun. 1981;32:198–203. doi: 10.1128/iai.32.1.198-203.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wade WG, Gray AR, Absi EG, Barker GR. Predominant cultivable flora in pericoronitis. Oral Microbiol Immunol. 1991;6:310–312. doi: 10.1111/j.1399-302x.1991.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 45.Wade WG, Spratt DA, Dymock D, Weightman AJ. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin Infect Dis. 1997;25(Suppl 2):S235–S236. doi: 10.1086/516215. [DOI] [PubMed] [Google Scholar]
- 46.Munson MA, Pitt-Ford T, Chong B, Weightman A, Wade WG. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res. 2002;81:761–766. doi: 10.1177/0810761. [DOI] [PubMed] [Google Scholar]
- 47.Wade WG. The oral microbiome in health and disease. Pharmacol. Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Loesche WJ, Syed SA. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 1973;7:201–216. doi: 10.1159/000259844. [DOI] [PubMed] [Google Scholar]
- 49.Beighton D, Al-Haboubi M, Mantzourani M, Gilbert SC, Clark D, Zoitopoulos L, Gallagher JE. Oral Bifidobacteria: caries-associated bacteria in older adults. J Dent Res. 2010;89:970–974. doi: 10.1177/0022034510369319. [DOI] [PubMed] [Google Scholar]
- 50.Van Houte J, Lopman J, Kent R. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J Dent Res. 1996;75:1008–1014. doi: 10.1177/00220345960750040201. [DOI] [PubMed] [Google Scholar]
- 51.Beighton D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent Oral Epidemiol. 2005;33:248–255. doi: 10.1111/j.1600-0528.2005.00232.x. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 53.Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, Griffen AL. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 2012;7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanner AC, Mathney JM, Kent RL, Chalmers NI, Hughes CV, Loo CY, Pradhan N, Kanasi E, Hwang J, Dahlan MA, Papadopolou E, Dewhirst FE. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011;49:1464–1474. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanasi E, Dewhirst FE, Chalmers NI, Kent R, Jr, Moore A, Hughes CV, Pradhan N, Loo CY, Tanner AC. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanner AC, Kent RL, Jr, Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. Microbiota of severe early childhood caries before and after therapy. J Dent Res. 2011;90:1298–1305. doi: 10.1177/0022034511421201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoshino E. Predominant obligate anaerobes in human carious dentin. J Dent Res. 1985;64:1195–1198. doi: 10.1177/00220345850640100301. [DOI] [PubMed] [Google Scholar]
- 61.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Downes J, Mantzourani M, Beighton D, Hooper S, Wilson MJ, Nicholson A, Wade WG. Scardovia wiggsiae sp. nov., isolated from the human oral cavity and clinical material, and emended descriptions of the genus Scardovia and Scardovia inopinata. Int J Syst Evol Microbiol. 2011;61:25–29. doi: 10.1099/ijs.0.019752-0. [DOI] [PubMed] [Google Scholar]
- 63.Milnes AR, Bowden GH. The microflora associated with developing lesions of nursing caries. Caries Res. 1985;19:289–297. doi: 10.1159/000260858. [DOI] [PubMed] [Google Scholar]
- 64.Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. The predominant microflora of nursing caries lesions. Caries Res. 2001;35:397–406. doi: 10.1159/000047482. [DOI] [PubMed] [Google Scholar]
- 65.Mantzourani M, Gilbert SC, Sulong HN, Sheehy EC, Tank S, Fenlon M, Beighton D. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 2009;43:308–313. doi: 10.1159/000222659. [DOI] [PubMed] [Google Scholar]
- 66.Kaur R, Gilbert SC, Sheehy EC, Beighton D. Salivary levels of Bifidobacteria in caries-free and caries-active children. Int J Paediatr Dent. 2013;23:32–38. doi: 10.1111/j.1365-263X.2011.01220.x. [DOI] [PubMed] [Google Scholar]
- 67.Palmer CA, Kent R, Jr, Loo CY, Hughes CV, Stutius E, Pradhan N, Dahlan M, Kanasi E, Arevalo Vasquez SS, Tanner AC. Diet and caries-associated bacteria in severe early childhood caries. J Dent Res. 2010;89:1224–1229. doi: 10.1177/0022034510376543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ledezma-Rasillo G, Flores-Reyes H, Gonzalez-Amaro AM, Garrocho-Rangel A, Ruiz- Rodriguez Mdel S, Pozos-Guillen AJ. Identification of cultivable microorganisms from primary teeth with necrotic pulps. J Clin Pediatr Dent. 2010;34:329–333. doi: 10.17796/jcpd.34.4.20124lu111544377. [DOI] [PubMed] [Google Scholar]
- 69.Tanner AC, Sonis AL, Lif Holgerson P, Starr JR, Nunez Y, Kressirer CA, Paster BJ, Johansson I. White-spot lesions and gingivitis microbiotas in orthodontic patients. J Dent Res. 2012;91:853–858. doi: 10.1177/0022034512455031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lovrov S, Hertrich K, Hirschfelder U. Enamel demineralization during fixed orthodontic treatment - incidence and correlation to various oral-hygiene parameters. J Orofac Orthop. 2007;68:353–363. doi: 10.1007/s00056-007-0714-1. [DOI] [PubMed] [Google Scholar]
- 71.van Gastel J, Quirynen M, Teughels W, Coucke W, Carels C. Longitudinal changes in microbiology and clinical periodontal variables after placement of fixed orthodontic appliances. J Periodontol. 2008;79:2078–2086. doi: 10.1902/jop.2008.080153. [DOI] [PubMed] [Google Scholar]
- 72.Rego RO, Oliveira CA, dos Santos-Pinto A, Jordan SF, Zambon JJ, Cirelli JA, Haraszthy VI. Clinical and microbiological studies of children and adolescents receiving orthodontic treatment. Am J Dent. 2010;23:317–323. [PubMed] [Google Scholar]
- 73.Boyar RM, Thylstrup A, Holmen L, Bowden GH. The microflora associated with the development of initial enamel decalcification below orthodontic bands in vivo in children living in a fluoridated-water area. J Dent Res. 1989;68:1734–1738. doi: 10.1177/00220345890680120301. [DOI] [PubMed] [Google Scholar]
- 74.Torlakovic L, Klepac-Ceraj V, Ogaard B, Cotton SL, Paster BJ, Olsen I. Microbial community succession on developing lesions on human enamel. J Oral Microbiol. 2012;4:e16125. doi: 10.3402/jom.v4i0.16125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kinney JS, Morelli T, Braun T, Ramseier CA, Herr AE, Sugai JV, Shelburne CE, Rayburn LA, Singh AK, Giannobile WV. Saliva/pathogen biomarker signatures and periodontal disease progression. J Dent Res. 2011;90:752–758. doi: 10.1177/0022034511399908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bratthall D, Hansel Petersson G. Cariogram--a multifactorial risk assessment model for a multifactorial disease. Community Dent Oral Epidemiol. 2005;33:256–264. doi: 10.1111/j.1600-0528.2005.00233.x. [DOI] [PubMed] [Google Scholar]