Abstract
The C. elegans gene unc-89 encodes a set of mostly giant polypeptides (up to 900 kDa) that contain multiple Ig and Fn3, a triplet of SH3-DH-PH, and two protein kinase domains. The loss of function mutant phenotype and localization of antibodies to UNC-89 proteins, indicate that the function of UNC-89 is to help organize sarcomeric A-bands, especially M-lines. Recently, we reported that each of the protein kinase domains interact with SCPL-1, which contains a CTD type protein phosphatase domain. Here, we report that SCPL-1 interacts with LIM-9 (FHL), a protein that we first discovered as an interactor of UNC-97 (PINCH) and UNC-96, components of an M-line costamere in nematode muscle. We also show that LIM-9 can interact with UNC-89 through its first kinase domain and a portion of unique sequence lying between the two kinase domains. All the interactions were confirmed by biochemical methods. A yeast three-hybrid assay demonstrates a ternary complex between the two protein kinase regions and SCPL-1. Evidence that the UNC-89/SCPL-1 interaction occurs in vivo was provided by showing that overexpression of SCPL-1 results in disorganization of UNC-89 at M-lines. We suggest two structural models for the interactions of SCPL-1 and LIM-9 with UNC-89 at the M-line.
Keywords: C. elegans, obscurin, FHL, CTD phosphatase, muscle
INTRODUCTION
The fundamental unit of muscle contraction, the sarcomere, is an exquisitely organized assemblage of hundreds of different proteins. To gain insight into the functions of individual sarcomeric proteins, their interactions and the assembly and maintenance of the sarcomere, a number of laboratories profitably utilize the roundworm C. elegans 1, 2. C. elegans is an excellent model genetic organism due to its simple anatomy, sequenced genome and powerful forward and reverse genetics. Since sarcomere architecture and components are largely conserved, results from C. elegans can be extrapolated to other organisms, including man. Sarcomeres contain a number of unusually large polypeptides (0.7–4 MDa in molecular mass) related to mammalian titin 3. Members of the titin family consist primarily of multiple copies of immunoglobulin (Ig) and fibronectin type 3 (Fn3) domains, and one or even two protein kinase domains. C. elegans striated muscle of the body wall contains three such polypeptides 4: twitchin (754,000 Da) located in the A-band 5–7 , TTN-1 (2.2 MDa) located in the I-band 8, and UNC-89 (up to 900,000 Da) located at the M-line 9, 10. Loss of function unc-89 mutants display disorganized myofibrils, especially at the A-band, and usually lack M-lines 11, 12. unc-89 is a complex gene: through the use of three promoters and alternative splicing, at least 6 major polypeptides are generated, ranging in size from 156,000 to 900,000 Da 4, 10. The largest of these isoforms, UNC-89-B and UNC-89-F, which are each ~900,000 Da, consist of 52 Ig domains, 2 Fn3 domains, a triplet of SH3, DH and PH domains near their N-termini, and two protein kinase domains (called PK1 and PK2) near their C termini. Most recently, we have demonstrated that the DH/PH region of UNC-89 has guanine nucleotide exchange factor activity towards RHO-1 (RhoA in C. elegans) and this is important for organization of myosin thick filaments in nematode body wall muscle cells 13. Although neither kinase domain has thus far demonstrated catalytic activity, molecular modeling suggests that PK2 is active, while PK1 is inactive 10.
For several reasons, we are very interested in exploring the structures and functions of the kinase domains of the giant titin-like proteins. First, a mutation in the human titin protein kinase domain causes hereditary muscle disease 14 . Second, in C. elegans, two mutant alleles of unc-89 that lack the kinase domain region result in disorganization of myofibrils 4. Third, the normally autoinhibited protein kinase domains of these giant proteins may be activated by mechanical forces that occur during muscle contraction 15–17. One way to gain insight into the function of a protein is to identify its binding partners. We reported recently 18 that the two protein kinase domains of UNC-89 interact with SCPL-1 (small CTD phosphatase-like-1), which contains a CTD type phosphatase domain. By antibody staining SCPL-1 co-localizes with UNC-89 at the M-line. Knockdown of scpl-1 mRNA results in no detectable phenotype in body wall muscle, but it does result in a defect in the function of egg-laying muscles.
The human homolog of UNC-89 is called obscurin 19, 20. Although obscurin contains all the same domains as UNC-89, the SH3, DH and PH domains are located near the C terminus rather than near the N terminus as they are in UNC-89. Another difference is that although UNC-89 is located only at the M-line, various obscurin isoforms are located at either the M-line, the A/I junction or at the Z-disk and Z/I junction 21. Progress has also been made on finding binding partners for obscurin: Ig48 and Ig49 within the C-terminal third of obscurin interact with specific Ig domains within the Z-disk portion of titin 20 and with a C-terminal portion of the 700 kDa isoform of titin called “novex-3 titin” 22. A sequence of 25 residues located near the C terminus of obscurin interacts with a small isoform of ankyrin 1, and this is postulated to provide a link between myofibrils and the sarcoplasmic reticulum 23–25. Overexpression of part of the C terminus of obscurin in primary skeletal myotubes results in disorganization of the A-band, and co-immunoprecipitation experiments suggest that obscurin interacts with myosin 26. This indicates that obscurin, like UNC-89, has a role in the assembly or maintenance of A-bands. The DH domain of obscurin has been shown to interact with the scaffolding protein called Ran binding protein 9 (RanBP9), and both obscurin’s DH domain and RanBP9 interact with titin to regulate titin’s incorporation into Z-disks 27. Most recently, the N-terminal most Ig domains (1–3) of obscruin have been shown to interact with the C-terminal most Ig domain of titin, and also with the M-line protein myomesin. 28. Whether nematode UNC-89 and mammalian obscurin have the same or even overlapping interacting partners is unknown.
To understand how nematode SCPL-1 carries out its function, we attempted to find additional proteins that SCPL-1 interacts with, and we discovered that SCPL-1 interacts with LIM-9 (FHL in mammals). We also found that LIM-9 can interact with UNC-89 directly, via the first kinase domain region (Fn3-Ig-PK1) and a portion of the interkinase region. We present a model that at the M-line, both SCPL-1 and LIM-9 interact with two portions of UNC-89 and function as bridges to stabilize the interaction of two UNC-89 polypeptides of opposite polarity, and/or to stabilize a loop created by interaction of the two kinase regions within one UNC-89 polypeptide.
RESULTS
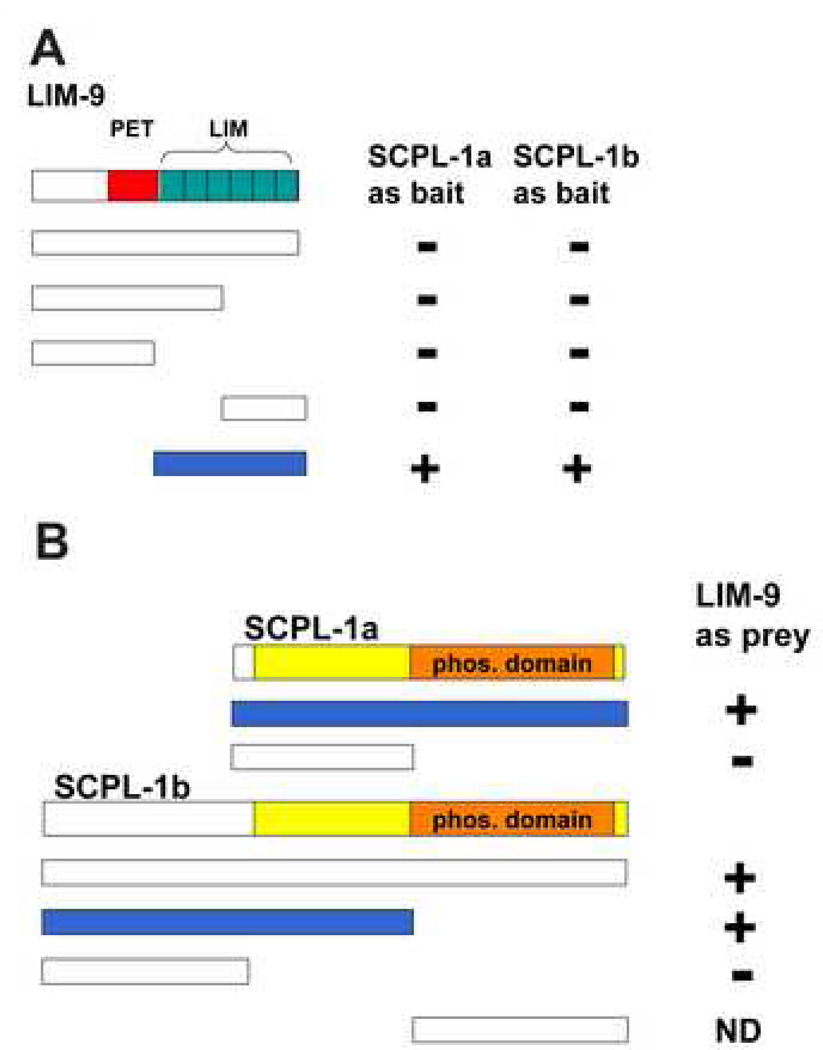
Given that we had determined that UNC-89 interacts with SCPL-1, we wished to determine which additional proteins SCPL-1 might interact with to carry out its function. Therefore, we used SCPL-1 to screen the 16-protein two-hybrid bookshelf of proteins known to reside at the nematode M-line 29. We discovered a 2-hybrid interaction between either SCPL-1a or −1b as bait and LIM-9 as prey (Figure 1A). LIM-9 contains one PET domain and six LIM domains, and is the closest worm homolog of mammalian FHL2. LIM-9 was characterized initially as an interactor of UNC-97 (PINCH) and UNC-96, and resides at least partially at M-lines 29. Domain mapping indicates that the minimum region of LIM-9 required for interaction with either SCPL-1a or −1b consists of the 6 LIM domains of LIM-9 (Figure 1A). Further, we can show that to obtain interaction with LIM-9, only the N-terminal, non-phosphatase region of SCPL-1b is sufficient, whereas the entire SCPL-1a protein is required (Figure 1B).
Figure 1.
Yeast two-hybrid assays demonstrate interaction between SCPL-1 and LIM-9. (A) LIM-9 contains a PET domain (red bar) and 6 LIM domains (green boxes). Full-length and deletion derivatives of LIM-9 as preys were tested for interaction with SCPL-1 or SCPL-1b as baits. The minimal interacting region of LIM-9 consists of the 6 LIM domains. (B) Both SCPL-1a and SCPL-1b have a protein phosphatase domain (orange bar) and share additional sequences (yellow). Full length SCPL-1a interacts with LIM-9. However, the N-terminal, non-phosphatase region of SCPL-1b is sufficient for interaction with LIM-9. + : growth on –Ade plates; −: no growth on –Ade plates. Blue bars represent the minimal portions required for interaction. ND: cannot be determined due to high background.
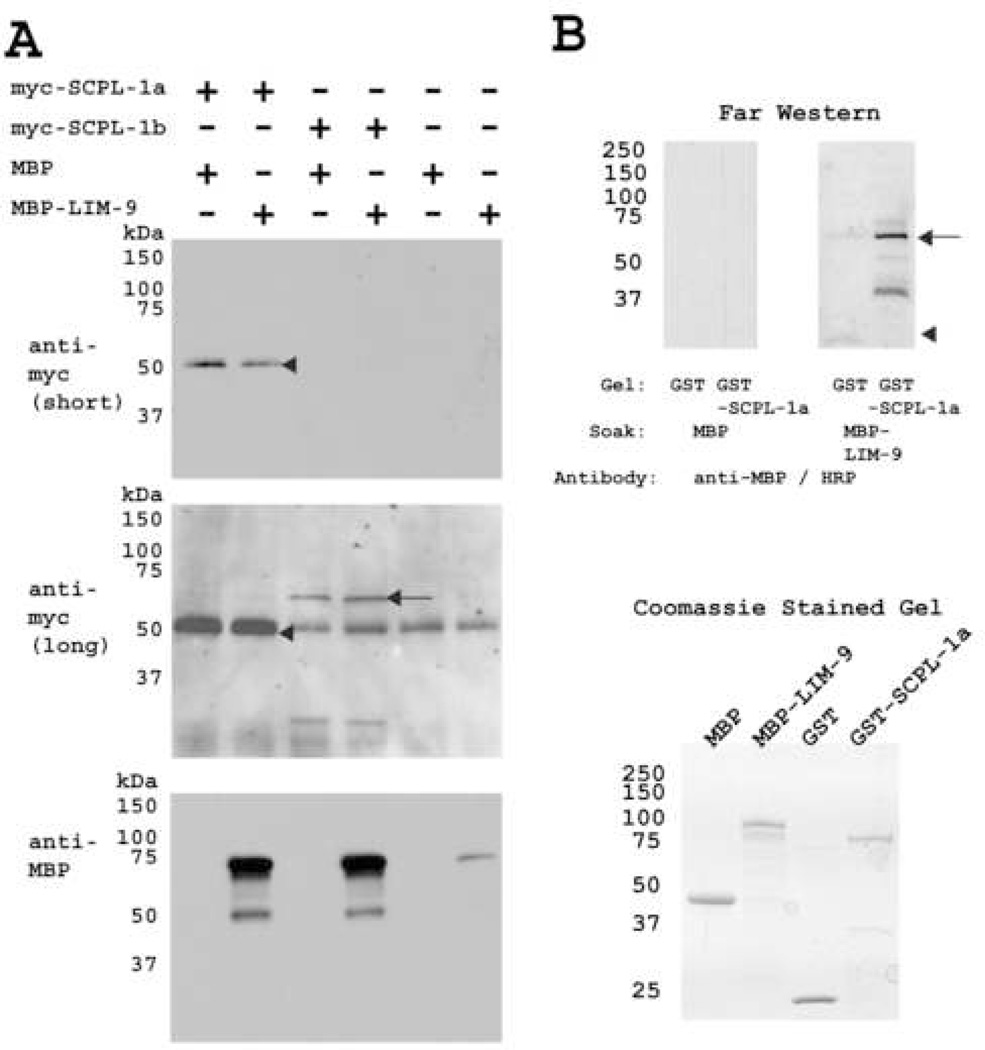
We confirmed the interaction between SCPL-1 and LIM-9 using two methods. First, we showed that yeast-expressed myc-tagged SCPL-1 can interact with a bacterially-expressed maltose binding protein (MBP) fusion of LIM-9 (LIM domains only). A total protein lysate from yeast expressing myc-SCPL-1a or –SCPL-1b was prepared and incubated with agarose beads coated with antibodies to myc, pelleted, washed, and incubated with MBP-LIM-9 or MBP. After pelleting and washing the beads, the proteins were eluted in Laemmli buffer and portions were separated on 3 gels, blotted and reacted with either antibodes to myc or antibodies to MBP. As shown in Figure 2A, as expected, both myc-SCPL-1a (top panel) and myc-SCPL-1b (middle panel) were precipitated. Reaction to anti-MBP (bottom panel) revealed that both myc-SCPL-1a and −1b co-precipitated MBP-LIM-9 but not MBP. Second, we showed that purified SCPL-1a interacts with purified LIM-9 (LIM domains only) by a far western assay (Figure 2B). GST and GST-SCPL-1a was separated on SDS-PAGE, transferred to membrane, and incubated with either MBP, or MBP fused to LIM-9 (LIM only). The binding reactions were visualized using antibodies to MBP followed by ECL. As shown in Figure 2B, MBP-LIM-9, but not MBP, interacts with SCPL-1a. Consistent with this direct interaction between SCPL-1 and LIM-9, we had found previously that each protein resides at M-lines 18, 29.
Figure 2.
Two methods confirm the interaction between SCPL-1 and LIM-9. (A) Yeast expressed myc tagged full length SCPL-1a or SCPL-1b were immunoprecipitated by anti-myc monoclonal antibody coated agarose beads, incubated with bacterially expressed MBP-LIM-9 (LIM only) or MBP, extensively washed, bound proteins were eluted and separated on SDS-PAGE gels, blotted and reacted with anti-myc and anti-MBP antibodies. myc-SCPL-1a (molecular weight ~50 kDa) was detected after short exposure, as shown by the arrow head. Long exposure revealed myc-SCPL-1b (molecular weight ~70 kDa), as shown by the arrow. (The 50 kDa band in the right four lanes of this central blot is likely to be rabbit immunoglobulin heavy chain resulting from cross reactivity with the anti-mouse secondary antibody used to detect the anti-myc monoclonal.) The bottom panel shows the results of reaction with anti-MBP and demonstrates that both SCPL-1a and SCPL-1b interact with MBP-LIM-9 (prominent bands at ~75 kDa) but not MBP. (B) SCPL-1a interacts with LIM-9 (LIM only) by far western assay. The bottom shows a Coomassie-stained SDS-PAGE of the starting materials: MBP, MBP fused to the 6 LIM domains of LIM-9, GST and GST fused to SCPL-1a. On the top is the far western: a gel similar to the one on the bottom was used to separate GST or GST-SCPL-1a, and the proteins transferred to a membrane. One half of the blot was incubated with MBP, the other with MBP-LIM-9 (LIM only), washed, and each half was incubated with antibodies to MBP conjugated to horseradish peroxidase. Reactions were visualized by ECL. Note that MBP-LIM-9 but not MBP interacts with SCPL-1a. The arrowhead indicates the position of GST; the arrow indicates the position of GST-SCPL-1a.
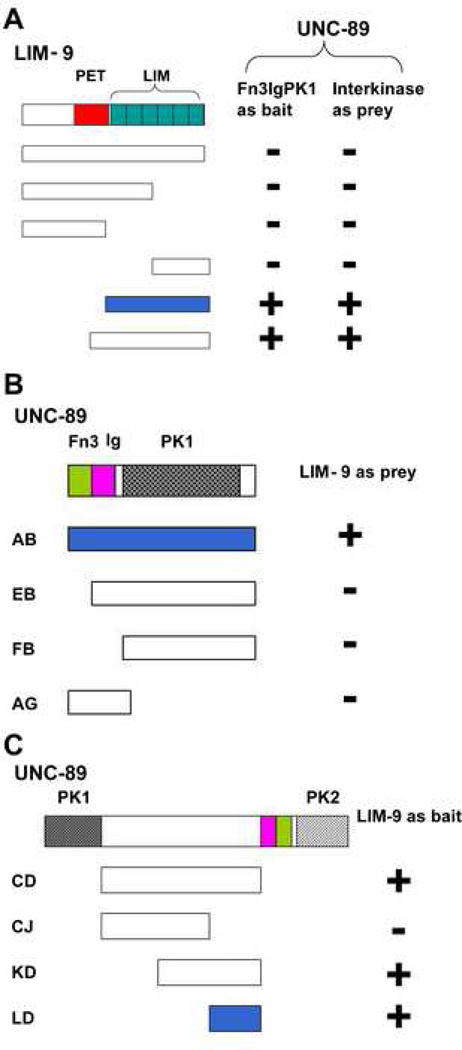
Given that LIM-9 is located at the M-line, we wondered if LIM-9 might interact directly with UNC-89. Consistent with this expectation, screening of the M-line bookshelf with Fn3-Ig-PK1 and interkinase regions of UNC-89 revealed that each interacts with LIM-9. We utilized 2-hybrid analysis to determine which portions of each protein that were involved in these interactions. As shown in Figure 3A, the smallest region of LIM-9 required to interact with either the Fn3-Ig-PK1 or interkinase regions of UNC-89 contains all 6 LIM domains, the identical region of LIM-9 required for interaction with SCPL-1 (Figure 1A). Note that full-length LIM-9 does not interact with SCPL-1, PK1 or interkinase regions. As shown in Figure 3B, in order to interact with LIM-9, the kinase domain of PK1 is not sufficient (segment FB): the upstream Fn3 and Ig domains are also required. This larger segment of UNC-89, Fn3-Ig-PK1 kinase domain, had also been shown 18 to be required for interaction with SCPL-1 (Figure 9A). As indicated in Figure 3C, a smaller segment of the interkinase region of UNC-89 is required for interaction with LIM-9: as little as the C-terminal 200 residues of interkinase are sufficient (segment LD).
Figure 3.
Yeast two-hybrid assays demonstrate interaction of LIM-9 with either the PK1 or interkinase regions of UNC-89. (A) The six LIM domains (green boxes) constitute the minimal portion of LIM-9 required for interaction with Fn3-Ig-PK1. The six LIM domains of LIM-9 are also sufficient for interaction with the interkinase region of UNC-89. (B) The entire PK1 region including the Fn3 and Ig domains N-terminal of the kinase domain (PK1) is required for interaction with LIM-9. (C) A C-terminal portion of the interkinase region of UNC-89 (about 200 amino acid residues) is sufficient for interaction with LIM-9. + : growth on –Ade plates; -: no growth on –Ade plates. Light green bars represent Fn3 domains and pink bars represent Ig domains. In UNC-89, the bar filled with heart dots denotes the PK1 protein kinase domain, and the bar filled in stripes denotes the PK2 protein kinase domain. Blue bars represent the minimal portions required for interaction.
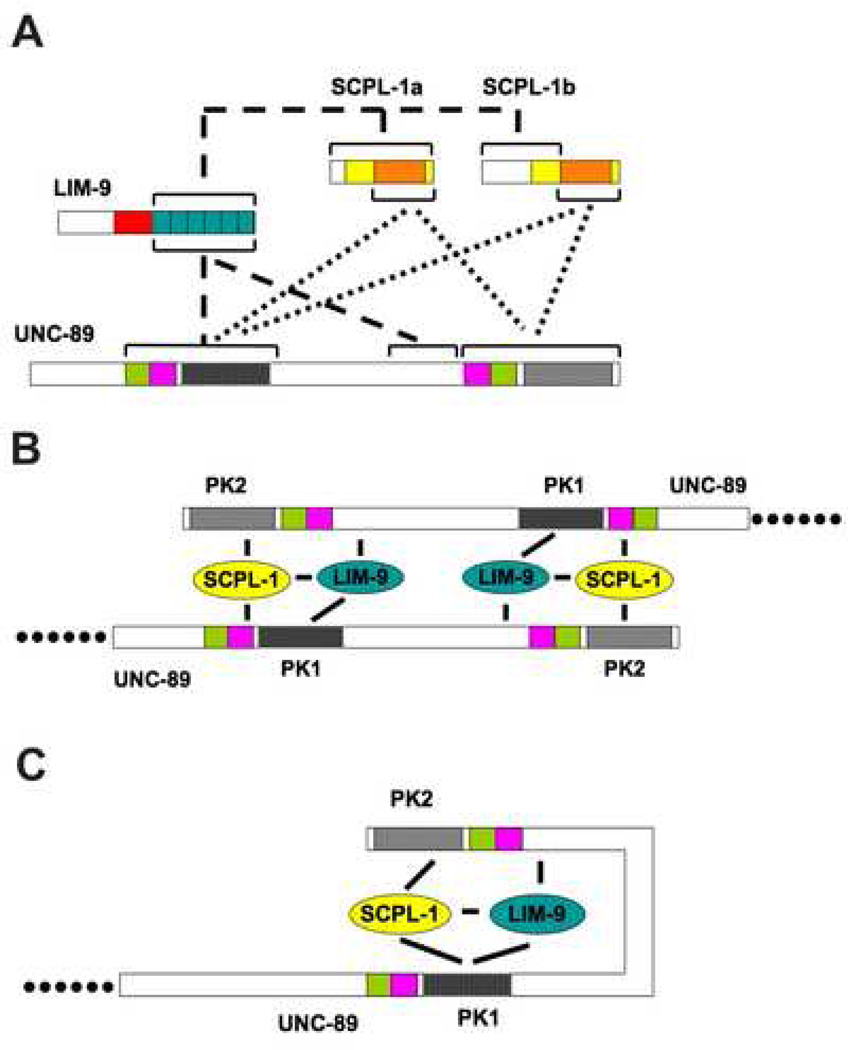
Figure 9.
Summary of interactions and two working models. (A) Summary of protein-protein interactions. The dotted lines show the interactions demonstrated previously 18. The phosphatase domain (orange bar) of either SCPL-1a or SCPL-1b interacts with Fn3-Ig-PK1 as well as Ig-Fn3-PK2 of UNC-89. The dashed lines show the interactions newly uncovered in this study. Only the 6 LIM domains of LIM-9 are required for interaction with SCPL-1a (or SCPL-1b), Fn3-Ig-PK1, or the interkinase region of UNC-89. (B) Model 1 involves two anti-parallel UNC-89 molecules linked at their C terminal regions through interactions with LIM-9 and SCPL-1. (C) In model 2, it is assumed that the non-domain interkinase region is flexible and forms a loop that is stabilized through interactions with LIM-9 and SCPL-1.
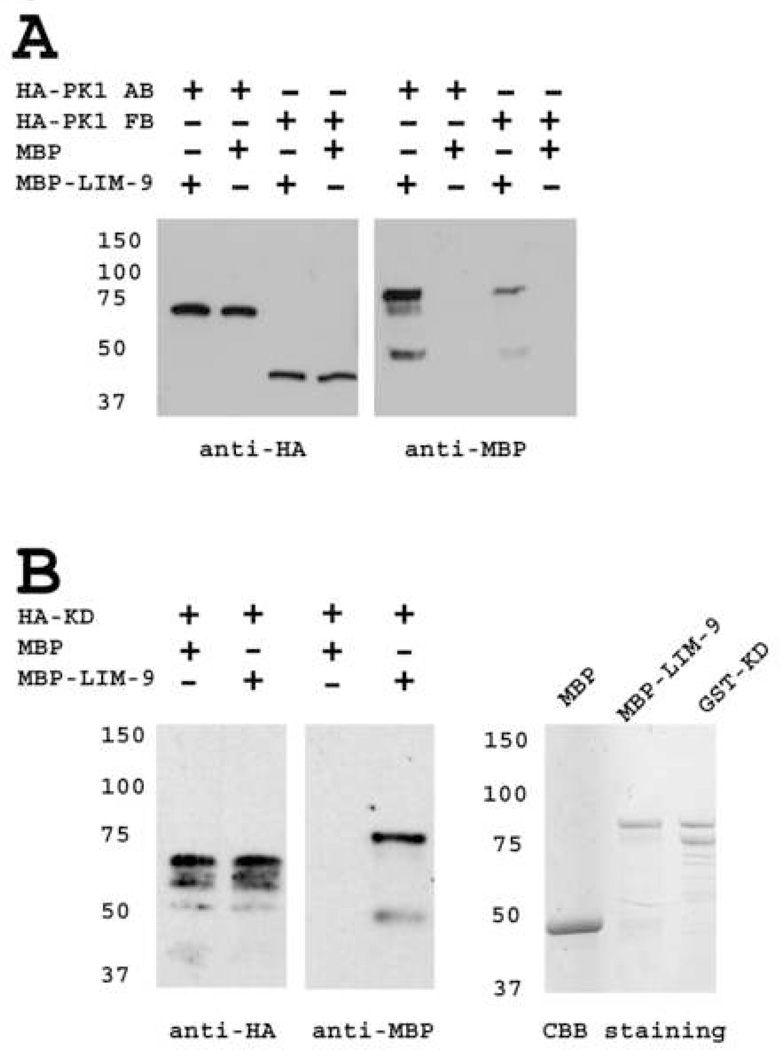
To confirm the 2-hybrid interactions of LIM-9 with the PK1 and interkinase regions of UNC-89, we used a similar assay as described above with yeast expressed PK1 or interkinase with bacterially expressed LIM-9. Yeast expressed PK1 AB or FB segments (described in Figure 3B) with HA tags were immunoprecipitated with agarose beads coated with antibodies to HA and tested for binding to either MBP-LIM-9 or MBP. As demonstrated in Figure 4A, consistent with the 2-hybrid results, AB fragment (Fn3-Ig-PK1 kinase) brought down much more MBP-LIM-9 than FB fragment (PK1 kinase alone), and neither segment brought down any detectable MBP. A similar strategy was used to verify interaction between LIM-9 and the interkinase region of UNC-89. In this case, yeast was used to express an HA tagged segment of the interkinase region (segment KD indicated in Figure 3C), which also interacts with LIM-9 by 2-hybrid. As shown in Figure 4B (left panels), the full-length KD fragment could be expressed and immunoprecipitated from yeast (upper band detected with anti-HA) although some apparent degradation bands were also present. Notably, this HA-KD protein was able to bring down MBP-LIM-9 but not MBP.
Figure 4.
Confirmation of interaction of LIM-9 with either the PK1 or interkinase regions of UNC-89. (A) Bacterially expressed LIM-9 interacts with yeast expressed HA-PK1. Yeast expressed HA tagged PK1 (AB or FB fragments, as shown in Figure 3) were immunoprecipitated by anti-HA monoclonal antibody-coated agarose beads, incubated with bacterially expressed MBP-LIM-9 or with MBP, extensively washed, the bound proteins were eluted and separated on SDS-PAGE gels, blotted and detected with anti-HA or anti-MBP antibodies. Consistent with the two-hybrid assay, Fn3-Ig-PK1 (AB fragment) demonstrates a stronger interaction with LIM-9, than does the kinase domain alone (FB fragment). No interaction was detected with MBP. (B) Bacterially expressed LIM-9 interacts with yeast expressed HA tagged interkinase region of UNC-89. Left, the C-terminal 2/3 of the interkinase region of UNC-89 (KD fragment, about 400 amino acid residues, as shown in Figure 3) interacts with MBP-LIM-9, but not MBP, using the same method described in (A). Right, coomassie brilliant blue (CBB) stained gel of MBP, MBP-LIM-9 and GST-KD. The top bands of MBP-LIM-9 and GST-KD are likely to be the full length fusion proteins, and the second bands may be degradation products. CBB staining demonstrates the molecular weight of MBP, MBP-LIM-9 and GST-KD. The bands of GST-KD on coomassie stained gel can be used to estimate the molecular weight of HA-KD.
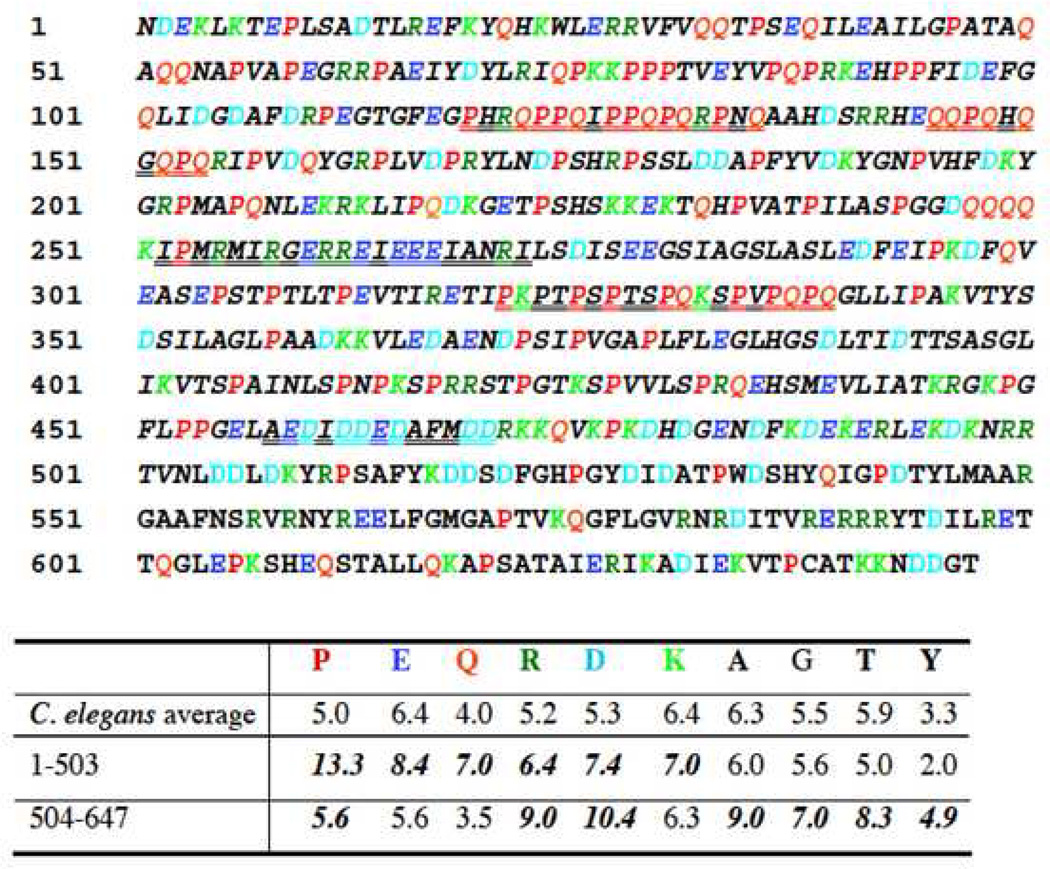
Given that the C-terminal 200 residues of the total 647 residues of the interkinase sequence were sufficient for binding to LIM-9, we wondered if there was anything special about the amino acid sequence or predicted structure of this binding region. As shown in Figure 5, the C-terminal 144 residues are different in several respects from the N-terminal 503 residues. In particular, residues 1–503 (italicized in Figure 5) is predicted to be a “NORS” region 30, that is, a sequence with little to no secondary structure and possibly structurally flexible 31. Residues 1–503 are also predicted to have high solvent accessibility (80%), and have a high probability for assigning neither α-helix, nor β-strand (85%). Residues 1–470 contain 5 segments ranging from 10 to 21 residues predicted 32 to be “SEG low-complexity regions” (double underlined in Figure 5). Moreover, the amino acid composition of residues 1–503 and 504–647 are quite different: in 1–503, there is especially increased frequency of P, E, Q and D; consecutive Ps (up to 3) or Qs (up to 4) are found only in the N-terminal 453 residues (especially in residues 1– 250). Therefore, sequence analysis suggests that the N-terminal 450–500 residues of the interkinase might be highly elastic, whereas the C terminal 150–200 residues are different, possibly forming a domain that is suited to interact with LIM-9.
Figure 5.
Sequence features of the 647 residue interkinase region of UNC-89. The sequence is devoid of recognizable domains. Residues 1–503 are italicized to denote that this is predicted to be a NORS region, a sequence with little to no secondary structure and possible structural flexibility. Double underlining represents the 5 segments within the first 470 residues that are predicted to be SEG low sequence complexity regions. Amino acids in color are those amino acids that are present at frequencies 0.5% above the average frequencies of a given amino acid in the C. elegans proteome. Note the high P,E,Q,R,D and K in residues 1–503. The binding site for LIM-9 has been mapped to residues 447–647.
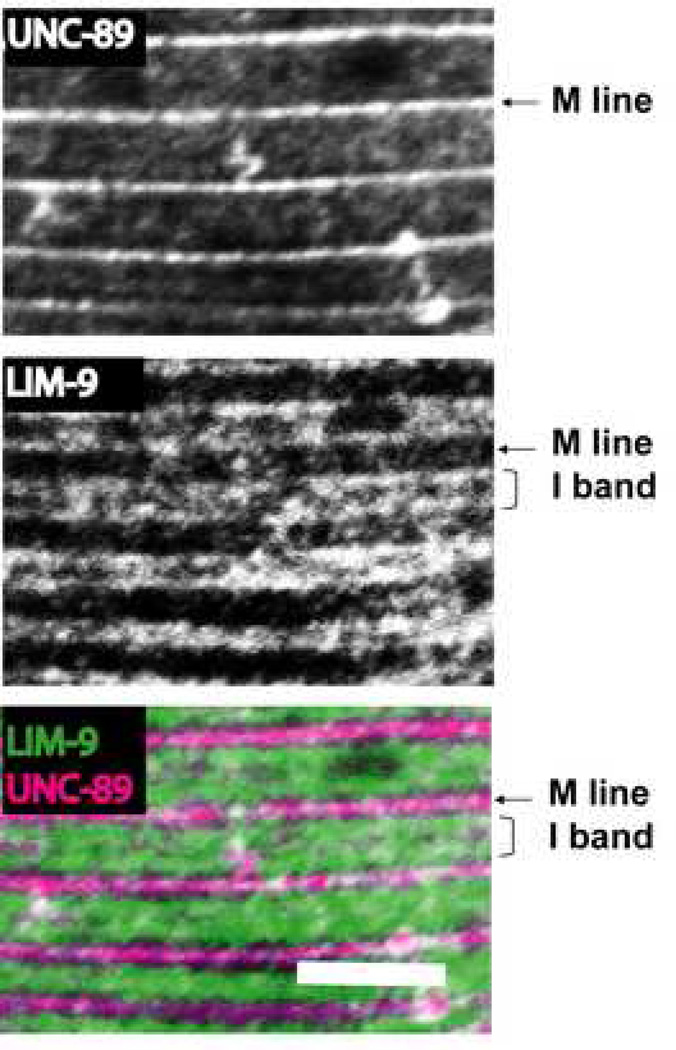
If LIM-9 interacts with UNC-89 in vivo, we would expect that the proteins co-localize in the sarcomere. Previously, we had shown that antibodies to LIM-9 localize to M-lines and to the I-band around and between dense bodies 29. We have also shown that antibodies to three different regions of UNC-89, including the interkinase region, localize to M-lines 10. Thus, we co-stained adult worms with anti-LIM-9 and anti-interkinase region of UNC-89. As demonstrated in Figure 6, there is partial co-localization between the interkinase region of UNC-89 and LIM-9 at the M-line (note the white dots at the bottom of the figure indicate co-localization).
Figure 6.
Co-localization of LIM-9 and the interkinase region of UNC-89 at the M lines in C. elegans body wall muscle. Anti-interkinase (of UNC-89) and anti-LIM-9 antibodies were co-incubated with wild type animals, and the body wall muscle was imaged by confocal microscopy. The images show a portion of one body wall muscle cell. Anti-UNC-89 is localized to M lines, anti-LIM-9 is localized to both M lines and I bands. Scale bar, 10µm.
Previously, we reported that SCPL-1 interacts with either Ig-Fn3-PK2 or Fn3-Ig-PK1 18. As described here, LIM-9 interacts with either Fn3-Ig-PK1, the C-terminus of the interkinase region, or SCPL-1 (Figure 9A). Additionally, we could not find direct interactions among Fn3-Ig-PK1, interkinase and Ig-Fn3-PK2 (data not shown). One hypothesis for the function of SCPL-1 binding to the two kinase domain regions is that in the sarcomere, SCPL-1 acts as a molecular bridge to bring together or stabilize two UNC-89 molecules or to stabilize a loop created by interaction of the two kinase regions within one UNC-89 molecule (Figure 9 B and C). This idea was first tested by a yeast 3-hybrid assay. As shown in Figure 7, when Fn-3-Ig-PK1 bait was presented to Ig-Fn3-PK2 prey, no growth of the yeast could be detected on adenine deficient media, indicating no detectable interaction (top row in Figure 7). However, when this bait and prey combination was expressed in the same yeast cell with myc-tagged SCPL-1 (either −1a or −1b isoforms), growth was detected on media missing adenine, indicating a three-way interaction (middle and bottom rows in Figure 7). Thus, the three hybrid results are consistent with SCPL-1 acting as a molecular bridge to bring together the PK1 and PK2 regions of UNC-89.
Figure 7.
Yeast three-hybrid assay demonstrates a ternary complex containing Fn3-Ig-PK1, SCPL-1 and Ig-Fn3-PK2. Fn3-Ig-PK1 as bait was coexpressed with myc tagged SCPL-1a or SCPL-1b (or empty myc-vector as control) and Ig-Fn3-PK2 as prey. +: growth on –Ade plates; −: no growth on –Ade plates. The right panel shows the yeast growth on –Ade plates from each experiment from three independent colonies.
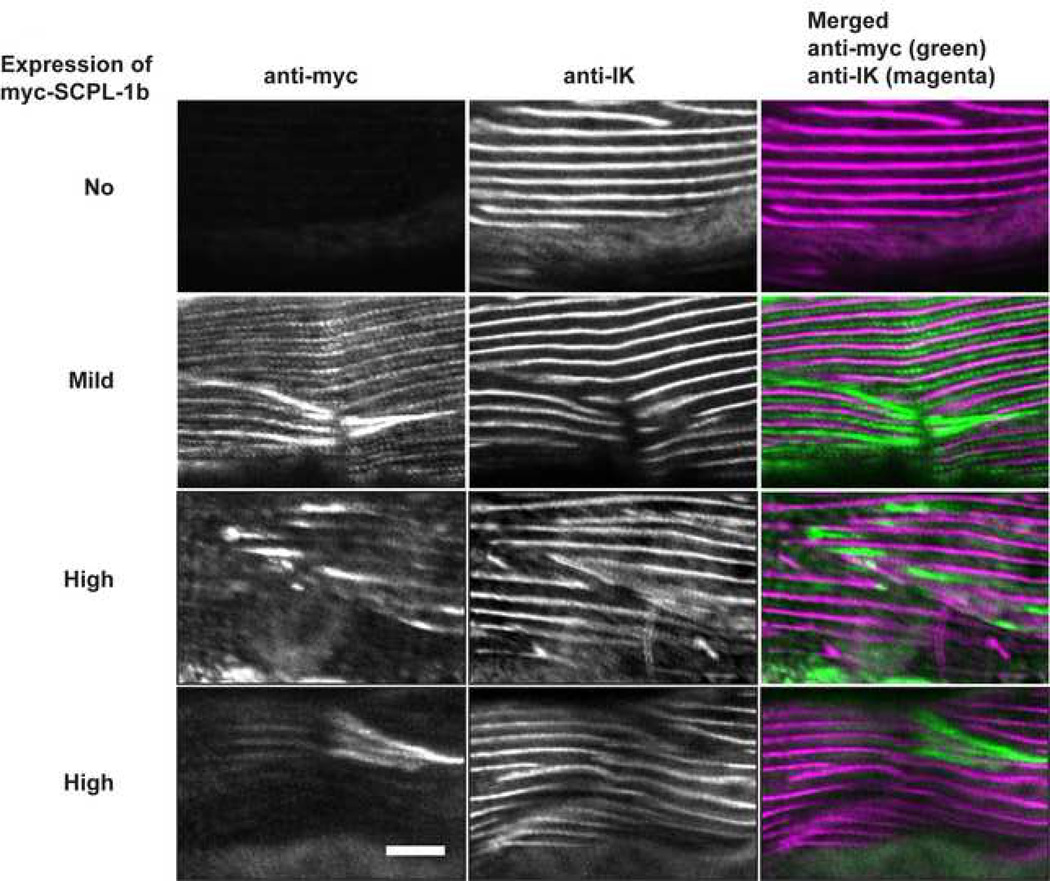
To obtain in vivo evidence for SCPL-1 functioning as a molecular bridge, we examined the muscle phenotype of worms overexpressing SCPL-1. We created several independent lines of wild type worms carrying extrachromosomal arrays containing a plasmid consisting of myc-tagged SCPL-1b full-length cDNA under the control of a heat shock promoter. After inducing expression by heat shock, worms were fixed and dually stained for anti-myc and anti-IK, the previously-described 10 antibody generated to the interkinase region of UNC-89. As shown in Figure 8, in body wall muscle cells not expressing the transgenic myc-SCPL-1b, the pattern of anti-IK staining is normal: M-lines in parallel. However, in body wall muscle cells in which SCPL-1b is mildly or highly overexpressed, there are accumulations of myc-SCPL-1b, and near these areas of the myofilament lattice, the M-lines detected by anti-IK are broken, diffuse or even missing. This result is consistent with our model that SCPL-1 acts as a linker or bridge between neighboring UNC-89 polypeptides at their kinase domain-containing C-terminal portions (Figure 9 B and C).
Figure 8.
Transgenic overexpression of SCPL-1b results in disorganization of UNC-89 in body wall muscle. Transgenic worms were generated that carry extrachromosomal arrays of a plasmid consisting of myc-tagged SCPL-1b full-length cDNA under the control of a heat-shock promoter. Each row shows images of body wall muscle stained with both anti-myc and antibodies to the interkinase region of UNC-89 (anti-IK). These arrays are sometimes lost during the somatic cell divisions that give rise to the mature body wall muscle cells. Moreover, of those muscle cells that retain the array, there is a variation in the level of expression from that array. The top row shows immunofluorescent staining of a muscle cell that has lost the array: anti-IK shows the normal pattern of localization of parallel M-lines 10. The second row shows a muscle cell which retained the array but the level of myc-SCPL-1b was mild: in this cell notice that by anti-myc staining there is both normal localization to I-bands and M-lines (similar to the anti-SCPL-1 antibody staining previously reported 18), and also in abnormal accumulations. Near the accumulations, anti-IK shows broken M-lines. The bottom 2 rows show muscle cells, which judging from the level of anti-myc staining have even higher expression of myc-SCPL-1b. In these cells, when stained with anti-IK, there are many broken M-lines and diffuse M-lines around accumulations of myc-SCPL-1b. Scale bar, 10 µm.
DISCUSSION
In this study, we provide evidence that a complex of two proteins, LIM-9 (FHL) and SCPL-1 (SCP) interact with the C-terminal protein kinase domain-containing portion of the largest isoforms of UNC-89 (obscurin). Interactions were identified by the two-hybrid method and then confirmed by showing interaction between purified components in vitro, and demonstrating co-localization in sarcomeres. Both LIM-9 and SCPL-1 were shown to interact with two portions of UNC-89 in ternary complexes. Moreover, support for the in vivo relevance of these interactions was provided by showing that overexpression of SCPL-1 results in abnormal localization of UNC-89, a key component of sarcomeric M-lines.
We previously reported that SCPL-1 interacts with the two protein kinase domains of UNC-89 18, and reported first identification of LIM-9 as a component of the integrin-associated protein complex at M-lines, and that the closest homolog for LIM-9 in mammals is “four and one-half LIM protein”, FHL 29. Here, we show that SCPL-1 interacts with LIM-9, and that LIM-9 interacts with UNC-89. In fact, our finding that UNC-89 interacts with LIM-9, now allows us to link the giant multi-domain protein UNC-89 to the integrin associated protein complex at the M-line (Figure 10)—a continuous linkage of proteins from the extracellular matrix to the thick filaments 29, 33–36. Moreover, although the largest isoform of UNC-89 (UNC-89B) is enormous and contains many domains, it is likely that LIM-9 and SCPL-1 only interact with the protein kinase domain containing regions of UNC-89: Using LIM-9, SCPL-1a or SCPL-1b as either 2-hybrid bait or prey to screen 16 fragments that comprise the largest UNC-89 isoform, we failed to identify interaction with any other portions of UNC-89 (results to be described elsewhere).
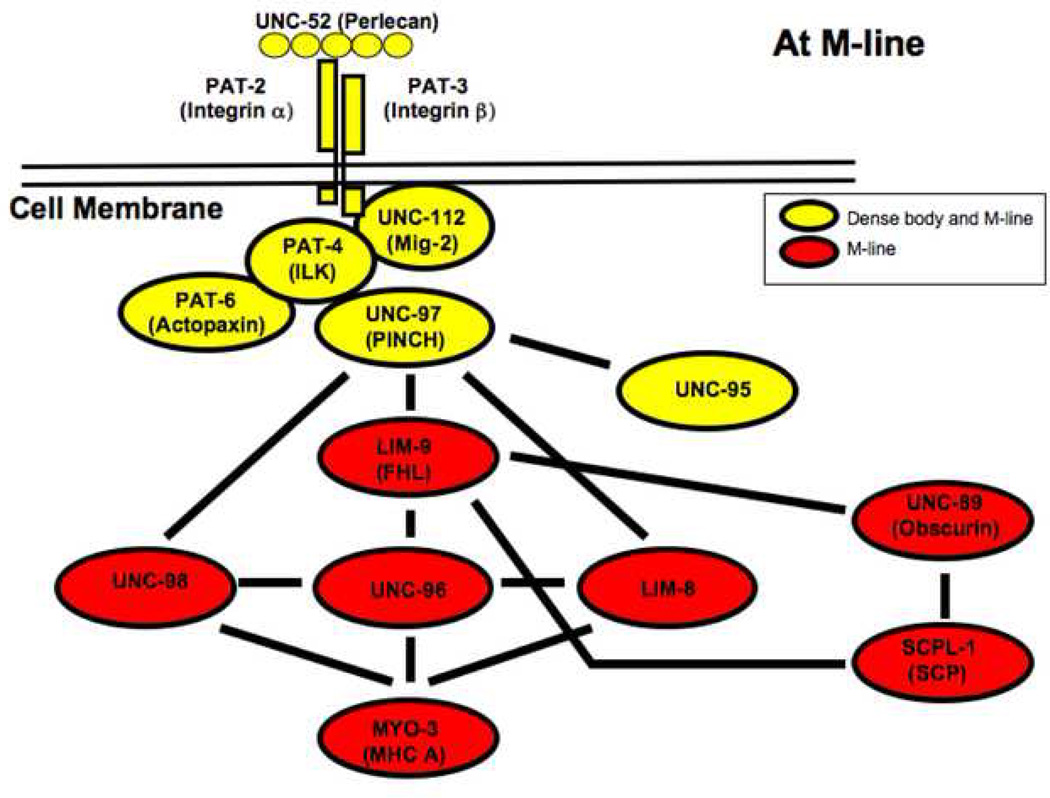
Figure 10.
Through their interactions with LIM-9, UNC-89 and SCPL-1 can now join the complex of proteins at the M-line costamere of C. elegans body wall muscle. This diagram is an updated version of the network of proteins described previously 29 that provide a continuous link from the ECM, through the muscle cell membrane to myosin heavy chain A (MHC A) in thick filaments at the M-line.
What is the function of LIM-9 in nematode muscle? Although two of the mammalian homologs of LIM-9 called FHL-2 and FHL-3 have been shown to interact with the cytoplasmic tails of several α- and β-integrins 37, 38, we have reported that LIM-9 failed to interact with the cytoplasmic tail of β-integrin 29. Nevertheless, LIM-9 was shown to interact with UNC-97, which is part of a 4 protein complex (including PAT-6 (actopaxin), PAT-4 (integrin linked kinase) and UNC-112(Mig-2)) that does associate with the tail of β-integrin. Thus, in both mammals and in nematodes, FHL proteins are linked to integrins, either directly or indirectly through integrin associated proteins.
Although knockout alleles of lim-9 have no obvious phenotype 29, mutations in one of the mammalian homologs of LIM-9 called FHL1 has been associated with three types of X-linked human myopathies 39, 40, 41. FHL1 is just one member of a family of four mammalian FHL proteins. Whereas FHL1 is ubiquitously expressed, FHL2 is primarily expressed in the heart 42 and localized to Z-disks and also to M-lines. FHL3 is localized to Z-disks in skeletal muscles 37. Recently, Cowling et al. have shown that overexpression of FHL1 in transgenic mice results in both skeletal muscle hypertrophy and enhancement of myoblast fusion in vitro; in addition, these authors showed that FHL1 interacts with NFATc1 and stimulates the transcription factor activity of NFATc1, probably leading to increased transcription of genes that result in increases in myofibrillar mass 43. Interestingly, our finding that LIM-9/FHL interacts with a giant protein kinase (in our case, UNC-89) is not unprecedented: FHL2 was found primarily to interact with 2 regions of mammalian titin—the N2B region, a heart-specific sequence of the I-band portion of titin, and the is2 region in the M-line portion of titin 44. These authors also showed that several muscle specific isoforms of metabolic enzymes such as creatine kinase, adenylate kinase and phosphofructokinase also interact with FHL2. Thus, our finding that LIM-9 interacts with both the first kinase domain of UNC-89 (PK1) and the SCPL-1 phosphatase adds two more enzymes to those that can associate with an FHL protein.
What is the function of the associations of LIM-9 and SCPL-1 with UNC-89? Previously, we reported that SCPL-1, under the conditions tested, was not a substrate for phosphorylation by the UNC-89 PK2 kinase domain 18. We note here that LIM-9 also could not be phosphorylated by UNC-89 PK2 (data not shown). Nevertheless, we propose two alternative models (Figure 9B and C) for the function of a LIM-9 / SCPL-1 complex in stabilizing interactions between two UNC-89 molecules, or between regions of one UNC-89 molecule, to form a loop at the C-terminal part of UNC-89. Support for the first model, that is, that an SCPL-1 / LIM-9 complex acts as a bridge between two adjacent UNC-89 polypeptides was provided by our finding that overexpression of SCPL-1 disrupts the organization of UNC-89 at the M-line (Figure 8). Perhaps excessive levels of SCPL-1 interfere with a ternary protein complex composed of SCPL-1 / LIM-9 / UNC-89, and this leads to disruption of the normal linkages of UNC-89 molecules through their kinase domain regions.
As shown in Supplementary Figure 1, RNAi knockdown for either lim-9, scpl-1, or both lim-9 and scpl-1 together, have no affect on the localization or organization of UNC-89. Given our model, we expected that such a “double mutant” might have a phenotype similar to unc-89 mutant alleles that lack the kinase domain-containing isoforms, UNC-89-B and UNC-89-F (alleles st79 and tm752 4). Because we observed normal localization of UNC-89 in the lim-9; scpl-1 double RNAi, it suggests yet an additional protein or proteins might link the kinase domain-containing portions of UNC-89. However, screening of the two-hybrid library with the interkinase domain of UNC-89 revealed no new interactors (data not shown). In the future we will attempt co-immunoprecipitation of UNC-89 with its binding partners followed by mass spectrometry to perhaps reveal these missing proteins.
The sequence of the interkinase region, in particular, the N-terminal 450–500 residues, suggests physical properties that are consistent with either model. This 450–500 amino acid region is predicted to have very little secondary structure, low sequence complexity and high proline content (Figure 5). These features suggest that this portion of the interkinase region is structurally flexible or even elastic. Flexibility would facilitate an intramolecular loop (the second model). It is also possible that elastic interkinase regions of two oppositely oriented UNC-89 molecules linked together by SCPL-1 and LIM-9 might allow a certain degree of structural expansion and contraction of the M-line. By two hybrid, the LIM-9 binding site has been mapped to the C-terminal 200 residues of the interkinase region (Figure 3). Again, sequence analysis is compatible with this result in that this sequence is different from the preceeding 450–500 residues, in that it is predicted to have some secondary structure, a lack of low sequence complexity regions and near normal proline content.
MATERIALS AND METHODS
Worm Strains and Culture
Bristol N2 was the wild-type strain, grown at 20°C on NGM plates with Escherichia coli strain OP50 as food source 45.
Yeast Two-hybrid and Three-bybrid Screens and Assays
The yeast two-hybrid method used to screen the nematode M-line bookshelf and to test for protein-protein interactions was performed as described 29, 34. The DNA sequences corresponding to fragments of a given protein were amplified by using synthesized primers and cDNA library template or plasmid templates isolated from the library during two-hybrid screening as described previously 29. The yeast three-hybrid assay was performed also as previously described 33.
Plasmid Construction
To construct yeast 2-hybrid plasmids for the interkinase region of UNC-89, we amplified the various portions of this region as depicted in Figure 3C using primers YIKC, YIKD, YIKJ, YIKK and YIKL as shown in Table 1. After cloning into pBluescript and verifying error-free sequences, the fragments were re-cloned into pGBDUC1 and pGADC1. The KD fragment from the pBluescript clone was also re-cloned into pGEXKK1 for expression of a GST-KD fusion protein. The method used to construct plasmids for yeast expression of myc-tagged SCPL-1a and b was described 18. To express hemagglutinin (HA)-tagged UNC-89 fragments in yeast, PCR was performed to amplify UNC-89 fragments by using the primers YIKK and YIKD for interkinase region (fragment KD in Figure 3C). The cDNA sequence was cloned into pKS-HA8 (Nhex2) (three HA-tagged vectors) by using SmaI and XhoI sites. HA-tagged UNC-89 interkinase region (KD fragment) was excised from this construct using NheI and inserted into pGAP-C-Nhe (yeast expression vector, TRP1 marker) by using the NheI site of the vector. Construction of the plasmid for expression of HA-tagged AB fragment of the PK1 region of UNC-89 was described 18. The plasmid used for expression of HA-tagged FB fragment of the PK1 region of UNC-89 was made by using primers U89-PK1-F and U89-PK1-B to PCR amplify and clone into pKS-HA8(Nhex2) (three HA-tagged vector) by using EcoRV and SalI sites.
Table 1.
PCR primers
| Primer name | Sequence |
|---|---|
| YIKC | GTACCCCGGGAACGATGAAAAGCTGAAAACTG |
| YIKD | CGTACTCGAGCTATCCATCATCATCATTCTTCTTTGT |
| YIKJ | CGTACTCGAGTCAACTCAACACAACTGGACTC |
| YIKK | GTACCCGGGCCACAAGACAAAGGAGAA |
| YIKL | GTACCCGGGTCTCCACGCCGTTCCACT |
| U89-PK1-B | CGTACTCGAGCTACGGACCAAGAATTGCTTCAAG |
| U89-PK1-F | GTACCCCGGGCGAGTGCGAGAGCTCTACGAA |
Purification of Bacterially expressed proteins
The procedures for purification of GST or MBP fusion proteins were described previously 46.
Assays to Confirm Interaction between Proteins
To confirm a protein-protein interaction detected by the yeast two-hybrid assay, yeast-expressed myc- or HA-tagged proteins were immunoprecipitated and tested for binding with bacterially expressed MBP-tagged proteins. To verify the interaction between SCPL-1 and LIM-9, myc-tagged SCPL-1a or −1b was expressed in yeast, and lysates were prepared as described 18. Myc-tagged SCPL-1a or −1b was immunoprecipitated from yeast lysates containing 300 mg of total protein in a volume of 500 µl for 1 h with shaking at 4°C in immunoprecipitation (IP) buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 0.25% gelatin, complete mini protease inhibitors, and 0.1% NP-40) using 50 µl of a 1:1 slurry of agarose beads conjugated to rabbit anti-myc antibodies (Sigma-Aldrich, cat. no. A7470). The beads were pelleted and washed 3X with IP buffer, and then incubated at 4° C for 1 h with shaking in a total volume of 500 µl with 10 µgs of either bacterially-expressed MBP-LIM-9 or MBP. The beads were washed 3X with IP buffer, transferred to clean Eppendorf tubes, pelleted, and the proteins eluted in 30 µl of Laemmli buffer. Two 10% SDS-PAGE were run and transferred to nitrocellulose membranes—one gel contained 5 µl of eluted protein per lane, and the other gel contained 25 µl per lane. The blot that had the 5 µl samples was reacted with mouse monoclonal to myc (Sigma-Aldrich cat. no. M4439) at 1:1000 followed by anti-mouse conjugated to horseradish peroxidase (HRP; Amersham) at 1:10,000, and detection by ECL. The blot containing 25 µl samples was incubated with HRP-conjugated anti-MBP (New England Biolabs) at 1:5000 and detected by ECL.
To confirm the interactions between the PK1 or interkinase regions of UNC-89 with LIM-9, we expressed in yeast HA-tagged PK1 AB or FB fragments, or HA-tagged interkinase region-KD, immunoprecipitated them with anti-HA bound agarose beads and conducted a binding assay with bacterially-expressed MBP or MBP-LIM-9, using methods described 18.
A far western assay was also used to confirm the interaction between SCPL-1a and LIM-9 (LIM domains only), performed essentially as previously described 29.
Immunostaining and Microscopy
Wild type adult worms were immunostained after fixation by the method described by Nonet et al. 47. Anti-interkinase region of UNC-89 (anti-IK 10) was used at 1:400 dilution, and anti-LIM-9 (Benian-9) was used at 1:100 dilution. Secondary antibodies and confocal microscopy were as described 29.
Amino acid sequence analysis
The interkinase sequence was analyzed using a combination of the Protean programs in the Lasergene package for tabulating amino acid composition and predicting secondary structure, and the website www.predictprotein.org for predicting SEG low-complexity and NORS regions. The average frequency of each amino acid in the C. elegans proteome was calculated by M. Borodovsky (pers. comm.).
RNAi for lim-9 and scpl-1
RNAi was performed by a feeding method 48. For feeding RNAi, the following plasmids were used: pPD129.36-lim-9 for lim-9, pPD129.36-scpl-1a for scpl-1, and pPD129.36-lim-9/scpl-1a for lim-9 and scpl-1 double RNAi. To construct pPD129.36-scpl-1a, the XhoI fragment of 25B (originally isolated from yeast 2-hybrid screening with UNC-89 PK2 as bait18 containing full length of scpl-1a) was cloned into pPD129.36 49 using its XhoI site. pPD129.36-lim-9 was made by cloning of a PCR-amplified LIM-9 N-terminal half fragment29 into pPD129.36 using BamHi and XhoI sites. For making pPD129.36-lim-9/scpl-1a double RNAi plasmid, the XhoI fragment of 25B (scpl-1a) was cloned into the XhoI site of pPD129.36-lim-9.
For immunostaining of RNAi worms, sufficient numbers of worms were obtained by the following protocol. Feeding bacteria carrying a specific plasmid were cultured in liquid 2xYT and induced to produce dsRNA by adding IPTG to a final concentration of 0.4 mM for 4 hrs. Induced bacteria cells were spotted onto NGM containing ampicillin (50 µg/ml) and tetracycline (15 µg/ml). Worms (the RNAi hypersensitive strain rrf-3 48) were picked onto five 6 cm RNAi plates (each containing 3 spots of induced bacteria) at 10 worms per plate and these plates were incubated at 20°C overnight to eliminate RNAi non-affected eggs. The next day, 10 worms were transferred from the 6 cm plates to 10 cm plates (11 spots of induced bacteria) and incubated at 20°C for 8 hrs. After 8 hrs, 10 worms were removed from the 10 cm plates. After 3 days incubation at 20°C, the F1 generation reached a good stage for immunostaining using the Nonet method 47.
Overexpression of myc-SCPL-1b
To construct a C. elegans expression plasmid of myc-SCPL-1b under the control of heat shock promoter, myc-tagged full length SCPL-1b fragment 18 was cloned into pPD49.78 and pPD49.83 using the NheI site. Mixtures of pPD49.78/83-myc-SPCL-1b(FL) with pTG96 (SUR-5::GFP, as a transformation marker) 50were injected into wild type (N2) for generating transgenic worms. Transgenic animals were identified by finding animals in which SUR-5::NLS::GFP was expressed in nearly all cells. Several independent lines were examined. Transgenic worms were placed at 30°C for 2 hours to induce expression of myc tagged full length SCPL-1b. After an additional 24 hours incubation at 20°C, worms were collected, and used for preparation of fixation by the Nonet method 47. Fixed worms were stained with anti-myc rabbit antibodies (Sigma Aldrich, 1:500 dilution) and anti-IK (1:500 dilution 10).
Supplementary Material
RNAi-mediated knockdown of either lim-9, scpl-1 or lim-9 and scpl-1 together, does not affect the localization or organization of UNC-89. The C. elegans RNAi hypersensitive strain rrf-3 were fed E. coli carrying the empty RNAi vector, or RNAi vectors with inserts for either lim-9, scpl-1, or both together in one plasmid. The resulting F1 animals were fixed and simultaneously stained with antibodies generated to two different regions of UNC-89—either to Ig 3–6 (near the N-terminus of the large UNC-89 isoforms), or to the interkinase region (near the C-terminus of the large UNC-89 isoforms). The images show one to several body wall muscle cells. Note that in all cases, there is normal localization of UNC-89 to parallel M-lines. Bar, 10 µm.
ACKNOWLEDGMENTS
We thank Kim Gernert for help with analysis of the interkinase sequence, we thank Mark Borodovsky (Georgia Institute of Techology) for providing us the average frequency of each amino acid in the C. elegans proteome, Andy Fire (Stanford University) for RNAi and transgenic vectors, and Tsuyoshi Kawano (Tottori University) for providing us the feeding method for 2-gene RNAi. Financial support was provided by National Institute of Arthritis & Musculoskeletal & Skin Diseases / National Institutes of Health grant AR-051466.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Moerman DG, Fire A. Muscle: structure, function and development. In: DL Riddle TB, Meyer BJ, JR, Priess, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- 2.Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. In: Commity TCeR., editor. WormBook. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat. Rev. Mol. Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara TM, Flaherty DB, Benian GM. Titin/connectin-related proteins in C. elegans: a review and new findings. J. Muscle Res. Cell Motil. 2005;26:435–447. doi: 10.1007/s10974-005-9027-4. [DOI] [PubMed] [Google Scholar]
- 5.Benian GM, Kiff JE, Neckelmann N, Moerman DG, Waterston RH. Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans . Nature. 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- 6.Benian GM, L’Hernault SW, Morris ME. Additional sequence complexity in the muscle gene, unc-22, and its encoded protein, twitchin, of Caenorhabditis elegans . Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moerman DG, Benian GM, Barstead RJ, Schriefer LA, Waterston RH. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans . Genes Dev. 1988;2:93–105. doi: 10.1101/gad.2.1.93. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty DB, Gernert KM, Shmeleva N, Tang X, Mercer KB, Borodovsky M, Benian GM. Titins in C.elegans with unusual features: coiled-coil domains, novel regulation of kinase activity and two new possible elastic regions. J. Mol. Biol. 2002;323:533–549. doi: 10.1016/s0022-2836(02)00970-1. [DOI] [PubMed] [Google Scholar]
- 9.Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J. Mol. Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Benian GM, Ayme-Southgate A, Tinley TL. The genetics and molecular biology of the titin/connectin-like proteins of invertebrates. Rev. Physiol. Biochem. Pharmacol. 1999;138:235–268. doi: 10.1007/BFb0119629. [DOI] [PubMed] [Google Scholar]
- 12.Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure of Caenorhabditis elegans . Dev. Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- 13.Qadota H, Blangy A, Xiong G, Benian GM. The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in Caenorhabditis elegans body wall muscle. J. Mol. Biol. 2008;383:747–752. doi: 10.1016/j.jmb.2008.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 15.Grater F, Shen J, Jiang H, Gautel M, Grubmuller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene DN, Garcia T, Sutton RB, Gernert KM, Benian GM, Oberhauser AF. Single-molecule force spectroscopy reveals a stepwise unfolding of Caenorhabditis elegans giant protein kinase domains. Biophys. J. 2008;95:1360–1370. doi: 10.1529/biophysj.108.130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schafer LV, Brandmeier B, Grater F, Grubmuller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci U S A. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM. A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (Obscurin) in Caenorhabditis elegans . Mol. Biol. Cell. 2008;19:2424–2432. doi: 10.1091/mbc.E08-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J. Muscle Res. Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- 20.Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzalez-Serratos H, Russell MW, Bloch RJ. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 23.Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, Russell MW, Bloch RJ. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- 25.Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am. J. Physiol. Cell. Physiol. 2004;287:C209–C217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- 27.Bowman AL, Catino DH, Strong JC, Randall WR, Kontrogianni-Konstantopoulos A, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Mol. Biol. Cell. 2008;19:3782–3792. doi: 10.1091/mbc.E08-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuzawa A, Lange S, Holt M, Vihola A, Carmignac V, Ferreiro A, Udd B, Gautel M. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J. Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- 29.Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Rost B. NORSp: Predictions of long regions without regular secondary structure. Nucleic Acids Res. 2003;31:3833–3835. doi: 10.1093/nar/gkg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Tan H, Rost B. Loopy proteins appear conserved in evolution. J. Mol. Biol. 2002;322:53–64. doi: 10.1016/s0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 32.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 33.Lin X, Qadota H, Moerman DG, Williams BD. C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr. Biol. 2003;13:922–932. doi: 10.1016/s0960-9822(03)00372-5. [DOI] [PubMed] [Google Scholar]
- 34.Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- 35.Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J. Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norman KR, Cordes S, Qadota H, Rahmani P, Moerman DG. UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Devolopmental Biology. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Samson T, Smyth N, Janetzky S, Wendler O, Müller JM, Schüle R, von der Mark H, von der Mark K, Wixler V. The LIM-only proteins FHL2 and FHL3 interact with alpha- and beta-subunits of the muscle alpha7 beta1 integrin receptor. J. Biol. Chem. 2004;279:28641–28652. doi: 10.1074/jbc.M312894200. [DOI] [PubMed] [Google Scholar]
- 38.Wixler V, Geerts D, Laplantine E, Westhoff D, Smyth N, Aumailley M, Sonnenberg A, Paulsson M. The LIM-only protein DRAL/FHL2 binds to the cytoplasmic domain of several alpha and beta integrin chains and is recruited to adhesion complexes. J. Biol. Chem. 2000;275:33669–33678. doi: 10.1074/jbc.M002519200. [DOI] [PubMed] [Google Scholar]
- 39.Quinzii CM, Vu TH, Min KC, Tanji K, Barral S, Grewal RP, Kattah A, Camaño P, Otaegui D, Kunimatsu T, Blake DM, Wilhelmsen KC, Rowland LP, Hays AP, Bonilla E, Hirano M. X-linked dominant scapuloperoneal myopathy is due to a mutation in the gene encoding four-and-a-half-LIM protein 1. Am. J. Hum. Genet. 2008;82:208–213. doi: 10.1016/j.ajhg.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Windpassinger C, Schoser B, Straub V, Hochmeister S, Noor A, Lohberger B, Farra N, Petek E, Schwarzbraun T, Ofner L, Löscher WN, Wagner K, Lochmüller H, Vincent JB, Quasthoff S. An X-linked myopathy with postural muscle atrophy and generalized hypertrophy,termed XMPMA, is caused by mutations in FHL1. Am. J. Hum. Genet. 2008;82:88–99. doi: 10.1016/j.ajhg.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schessl J, Zou Y, McGrath MJ, Cowling BS, Maiti B, Chin SS, Sewry C, Battini R, Hu Y, Cottle DL, Rosenblatt M, Spruce L, Ganguly A, Kirschner J, Judkins AR, Golden JA, Goebel HH, Muntoni F, Flanigan KM, Mitchell CA, Bonnemann CG. Proteomic identification of FHL1 as the protein mutated in human reducing body myopathy. J. Clin. Invest. 2008;118:904–912. doi: 10.1172/JCI34450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholl FA, McLoughlin P, Ehler E, de Giovanni C, Schäfer BW. DRAL is a p53-responsive gene whose four and a half LIM domain protein product induces apoptosis. J. Cell Biol. 2000;151:495–506. doi: 10.1083/jcb.151.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowling BS, McGrath MJ, Nguyen MA, Cottle DL, Kee AJ, Brown S, Schessl J, Zou Y, Joya J, Bonnemann CG, Hardeman EC, Mitchell CA. Identification of FHL1 as a regulator of skeletal muscle mass: implications for human myopathy. J. Cell Biol. 2008;183:1033–1048. doi: 10.1083/jcb.200804077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange S, Auerbach D, McLoughlin P, Perriard E, Schäfer BW, Perriard JC, Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J. Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- 45.Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol. Biol. Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- 48.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 49.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 50.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNAi-mediated knockdown of either lim-9, scpl-1 or lim-9 and scpl-1 together, does not affect the localization or organization of UNC-89. The C. elegans RNAi hypersensitive strain rrf-3 were fed E. coli carrying the empty RNAi vector, or RNAi vectors with inserts for either lim-9, scpl-1, or both together in one plasmid. The resulting F1 animals were fixed and simultaneously stained with antibodies generated to two different regions of UNC-89—either to Ig 3–6 (near the N-terminus of the large UNC-89 isoforms), or to the interkinase region (near the C-terminus of the large UNC-89 isoforms). The images show one to several body wall muscle cells. Note that in all cases, there is normal localization of UNC-89 to parallel M-lines. Bar, 10 µm.