Summary
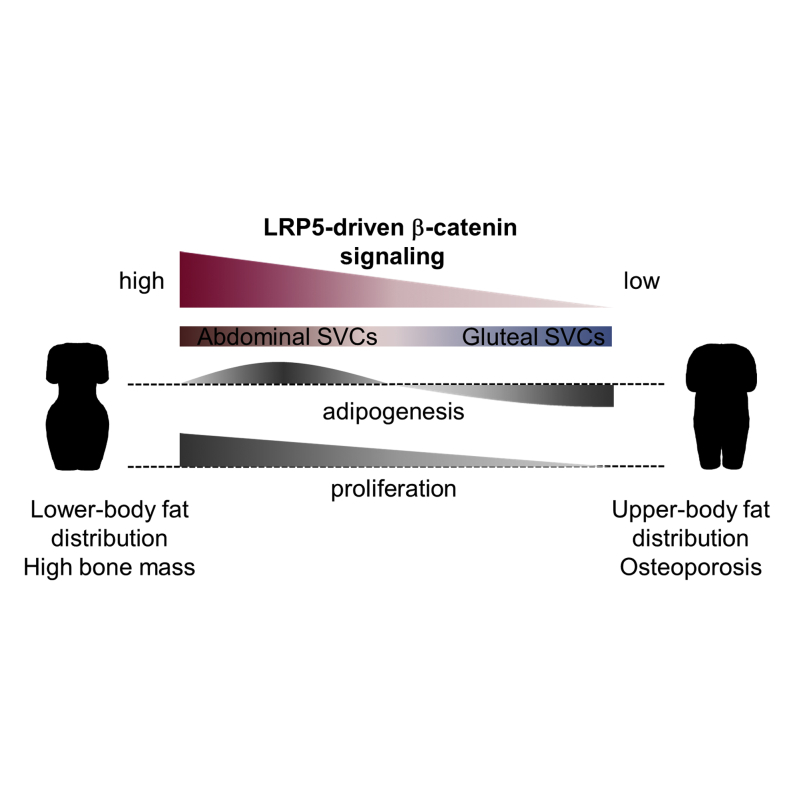
Common variants in WNT pathway genes have been associated with bone mass and fat distribution, the latter predicting diabetes and cardiovascular disease risk. Rare mutations in the WNT co-receptors LRP5 and LRP6 are similarly associated with bone and cardiometabolic disorders. We investigated the role of LRP5 in human adipose tissue. Subjects with gain-of-function LRP5 mutations and high bone mass had enhanced lower-body fat accumulation. Reciprocally, a low bone mineral density-associated common LRP5 allele correlated with increased abdominal adiposity. Ex vivo LRP5 expression was higher in abdominal versus gluteal adipocyte progenitors. Equivalent knockdown of LRP5 in both progenitor types dose-dependently impaired β-catenin signaling and led to distinct biological outcomes: diminished gluteal and enhanced abdominal adipogenesis. These data highlight how depot differences in WNT/β-catenin pathway activity modulate human fat distribution via effects on adipocyte progenitor biology. They also identify LRP5 as a potential pharmacologic target for the treatment of cardiometabolic disorders.
Graphical Abstract
Highlights
-
•
Carriers of LRP5 variants display altered body fat distribution
-
•
LRP5 is more highly expressed in abdominal versus gluteal fat progenitor cells
-
•
LRP5 knockdown in both progenitor types leads to different biological responses
-
•
LRP5 modulates fat progenitor biology by controlling β-catenin signaling dosage
Loh et al. identify the WNT co-receptor LRP5 as a regulator of human body fat distribution, an independent predictor of diabetes and cardiovascular disease risk. Studying LRP5 gene variant carriers and human fat progenitors, they show that LRP5 differentially modulates regional adipose progenitor biology by titrating WNT/β-catenin signaling dosage.
Introduction
Obesity is associated with the development of insulin resistance, linked to the pathogenesis of type 2 diabetes (T2D) and cardiovascular disease (CVD). Nonetheless, adverse metabolic sequelae are not uniformly observed in obese individuals. While subjects with abdominal obesity display an increased prevalence of CVD, similarly overweight individuals with gluteofemoral fat distribution are protected from cardiometabolic disorders (Yusuf et al., 2005). Consistent with epidemiologic findings, physiological studies have shown that the gluteofemoral white adipose tissue (WAT) depot displays differential fatty acid (FA) handling compared to the subcutaneous (SC) abdominal WAT depot (Jensen, 2008; Karpe and Pinnick, 2014). By favoring the long-term storage of FAs, gluteofemoral fat may protect skeletal muscle from ectopic lipid accumulation and lipotoxicity, which triggers insulin resistance (Schenk et al., 2008). Gluteofemoral fat may also contribute to improved metabolic risk by secreting a more beneficial adipocytokine profile than SC abdominal and visceral fat (Fontana et al., 2007; Turer et al., 2011). Adipose-derived hormones and cytokines directly modulate systemic insulin sensitivity (Qatanani and Lazar, 2007).
WAT expands by an increase in adipocyte number (hyperplasia) and size (hypertrophy) (Spalding et al., 2008; Tchoukalova et al., 2010). Adipocytes derive from mesenchymal stem cells (MSCs) and preadipocytes that reside in the stromovascular fraction of WAT. Several clinical and experimental studies indicate that discrete fat depots arise from distinct precursors with inherently different proliferative and adipogenic properties (Billon and Dani, 2012; Semple et al., 2011; Tchkonia et al., 2006). It is further postulated that developmental pathways play a key role in establishing the distinct identities of adipose progenitors from separate locations and thus in determining (1) the relative size of fat depots, by determining adipocyte number (and size) within each depot, and (2) the function of WAT depots, by modulating expression of adipogenic genes and their downstream targets. Consistent with this hypothesis, stromovascular cells (SVCs) isolated from discrete fat depots exhibit distinct developmental gene expression profiles (Gesta et al., 2006; Tchkonia et al., 2007). Furthermore, in a genome-wide association study (GWAS) meta-analysis, 4 of the 13 identified loci associated with body mass index (BMI)-adjusted waist-to-hip ratio (WHR), a measure of body fat distribution, mapped in or near developmental genes (Heid et al., 2010). Notably two of these, RSPO3 and ZNRF3, constitute WNT signaling modulators. A locus near RSPO3, distinct from the WHR-associated signal, was also shown to be associated with bone mineral density (BMD) at genome-wide significance (Duncan et al., 2011; Estrada et al., 2012).
WNTs are a family of 19 secreted glycoproteins acting locally via multiple pathways to regulate adult tissue homeostasis (Clevers and Nusse, 2012). In the β-catenin (“canonical”) cascade, WNT binding to Frizzled (FZD) receptors and low-density lipoprotein receptor-related protein (LRP) 5/6 co-receptors leads to nuclear accumulation of the transcriptional co-activator β-catenin, which, paired with LEF/TCF transcription factors, dose-dependently modulates (generally activates) WNT target gene expression. WNTs also signal through “non-canonical” pathways. In the planar cell polarity (PCP) pathway, WNT binding to FZD activates JNK and stimulates AP1-dependent transcription. In the WNT/Ca2+ pathway, WNT/FZD interaction triggers intracellular Ca2+ release and calmodulin-dependent protein kinase-2A (CAMK2A) activation. Canonical and non-canonical pathways are thought to be mutually antagonistic (Grumolato et al., 2010; Weidinger and Moon, 2003).
WNT signaling is a key regulator of MSC biology (Christodoulides et al., 2009; Krishnan et al., 2006). Canonical signaling, the best-studied pathway, which critically relies on LRP5 and LRP6 co-receptors for activation, has been shown to repress adipogenesis and stimulate osteoblastogenesis. Accordingly, patients carrying rare gain-of-function (GoF) LRP5 mutations exhibit high bone mass (HBM) (Boyden et al., 2002; Little et al., 2002). Reciprocally, rare loss-of-function (LoF) LRP5 mutations lead to osteoporosis (Ai et al., 2005; Gong et al., 2001), which, in a study of 12 affected probands from two families, was coupled with an increased prevalence of T2D (Saarinen et al., 2010). Finally, rare inactivating missense mutations in LRP6 result in autosomal dominant CVD, features of the metabolic syndrome, and osteoporosis (Mani et al., 2007; Singh et al., 2013). Prompted by these and the aforementioned GWAS findings (Heid et al., 2010), we sought to determine the role of LRP5 in human WAT biology and fat distribution. Our interest in LRP5 was also stimulated by preliminary analyses showing that it was differentially expressed between SC abdominal and gluteal SVCs. Furthermore, WAT LRP5 mRNA levels correlated with measures of regional adiposity and systemic insulin sensitivity. Herein we demonstrate that LRP5-driven β-catenin signaling regulates adipose progenitor proliferation and differentiation in a dose- and depot-specific manner, thereby modulating human body fat distribution.
Results
HBM-Causing LRP5 Mutations Are Associated with Lower-Body Fat Accumulation
We examined the adipose and metabolic phenotype of three pedigrees with extreme HBM secondary to rare heterozygous GoF LRP5 mutations. Compared to age-, gender-, and BMI-matched Oxford Biobank (OBB) controls, HBM LRP5 mutation carriers had an increased amount of lower-body fat as determined by whole-body dual energy X-ray absorptiometry (DXA). In particular, all overweight/obese (BMI ≥ 25) HBM subjects with LRP5 mutations (n = 5 of 6 individuals in total; S1–S3, S5, S6) had a higher tissue percent fat specifically in their legs (Table 1). Furthermore, all affected individuals displayed lower android/leg, android/total, and central/peripheral fat mass ratios (Table 1). This adipose phenotype was not driven by the HBM, as HBM LRP5 mutation carriers had a decreased upper-to-lower-body fat ratio even when compared with matched non-LRP5 HBM cases (n = 18) (Table 2). Similar results were obtained when comparing age-, gender-, and BMI-adjusted DXA data from LRP5 HBM cases versus the rest of the (non-LRP5) HBM cohort (n = 134) (Table S1). LRP5 HBM individuals also exhibited enhanced insulin sensitivity as determined by lower HOMA-IR and fasting insulin levels relative to OBB controls (Tables 1 and S2). One exception was subject S2 (68 years old), whom we were able to compare only with 49–50 year old gender- and BMI-matched controls. Finally, ex vivo gene expression analyses of fractionated SC adipose cells revealed lower inflammatory gene transcript levels in LRP5 HBM individuals (n = 4; subjects S1, S4–S6) versus OBB controls (n = 24–25) (Figure S1A). We conclude that rare, GoF LRP5 mutations are associated with enhanced lower-body fat accumulation, a favorable metabolic profile, and reduced WAT inflammation.
Table 1.
Anthropometry, Plasma Biochemistry, and DXA-Derived Measures of Body Fat Distribution of HBM Patients with LRP5 Gain-of-Function Mutations from Three Kindreds versus Those of Their Age-, Gender-, and BMI-Matched OBB Controls
| S1 | Controls for S1 | S2 | Controls for S2 | S3 | Controls for S3 | S4 | Controls for S4 | S5 | Controls for S5 | S6 | Controls for S6 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | – | 129 (77a) | – | 74 (42a) | – | 105 (65a) | – | 51 (33a) | – | 55 (33a) | – | 102 (59a) |
| Age (years) | 45 | 44.8 (44.3–45.2) | 68c | 49.1 (48.9–49.3) | 50c | 48.0 (47.7–48.4) | 28c | 31.9 (31.5–32.2) | 38 | 38.1 (36.9–39.3) | 34c | 34.6 (34.1–35.1) |
| Gender | F | F | F | F | F | F | M | M | F | F | M | M |
| BMI (kg/m2) | 27.2 | 27.2 (27.1–27.3) | 27.3 | 27.2 (27.0–27.3) | 26.0 | 26.0 (25.9–26.0) | 21.8 | 22.0 (21.8–22.1) | 35.1 | 35.0 (34.8–35.2) | 28.8c | 28.4 (28.2–28.5) |
| HOMA-IR | 0.6c | 1.7 (1.6–1.8) | 1.7 | 1.7 (1.6–1.9) | 1.6b,c | 1.7 (1.6–1.8) | 0.6c | 1.5 (1.3–1.6) | 1.2c | 2.4 (2.1–2.7) | 1.3c | 2.1 (2.0–2.3) |
| Tissue legs, % fat | 43.5c | 40.0 (38.9–41.1) | 44.2c | 39.6 (38.1–41.0) | 48.4c | 39.8 (38.7–40.8) | 19.7 | 20.5 (18.9–22.1) | 50.2c | 45.0 (43.1–46.8) | 31.1c | 26.4 (25.1–27.6) |
| Tissue android, % fat | 35.6c | 41.5 (40.0–42.9) | 43.7 | 42.4 (40.2–44.7) | 43.9c | 41.1 (39.4–42.8) | 11.6c | 21.0 (18.5–23.5) | 55.3 | 53.9 (52.3–55.5) | 35.5c | 39.6 (38.3–40.9) |
| Android:leg fat ratio (%) | 16.4c | 22.6 (21.1–24.1) | 21.5c | 24.7 (22.0–27.3) | 18.9c | 23.2 (21.4–25.0) | 11.0c | 22.0 (19.7–24.4) | 21.5c | 29.2 (26.3–32.1) | 24.8c | 35.9 (33.8–37.9) |
| Android:total fat ratio (%) | 6.7c | 7.8 (7.6–8.1) | 7.5c | 8.2 (7.7–8.6) | 7.0c | 7.9 (7.6–8.2) | 4.3c | 7.1 (6.6–7.5) | 8.6c | 9.3 (8.9–9.8) | 8.6c | 9.8 (9.6–10.1) |
| Central:peripheral fat mass ratio (g/g) | 12.9c | 17.0 (16.0–18.0) | 16.3c | 18.4 (16.6–20.1) | 14.1c | 17.4 (16.2–18.6) | 8.7c | 16.9 (15.2–18.6) | 18.6c | 21.7 (19.8–23.6) | 19.1c | 26.3 (25.0–27.6) |
Data for controls are means (95% CI). n, number of age-, gender-, and BMI-matched controls. Subjects from Kindred 1 (S1, S2) and Kindred 2 (S3, S4) carry the A242T mutation; those from Kindred 3 (S5, S6) carry the N198S mutation. NB: We were unable to match subjects S2 and S4 for age. HBM, high bone mass; CI, confidence interval; BMI, body mass index; HOMA-IR, homeostasis model assessment-estimated insulin resistance. See also Table S2 and Figure S1.
Number of controls with DXA measurements.
Actual value is below the 95% CI.
Values below/above the 95% CI.
Table 2.
Comparison of Anthropometry and DXA-Derived Measures of Body Fat Distribution of HBM Subjects With and Without LRP5 Gain-of-Function Mutations, Matched for Age, Gender, and BMI
| LRP5 HBM (n = 6) | Non-LRP5 HBM (n = 18) | |
|---|---|---|
| Mean ± SD | Mean (95% CI) | |
| Age (years) | 43.0 ± 14.5 | 41.9 (31.0, 52.7) |
| BMI (kg/m2) | 27.7 ± 4.3 | 27.7 (25.5, 29.9) |
| Tissue legs, % fat | 39.5 ± 11.8 | 36.3 (30.3, 42.2) |
| Tissue android, % fat | 37.6 ± 14.7 | 46.8 (40.3, 53.3)a |
| Android:leg fat ratio (%) | 19.0 ± 4.8 | 29.5 (26.5, 32.4)b |
| Android:total fat ratio (%) | 7.1 ± 1.6 | 9.1 (8.4, 9.7)b |
| Central:peripheral fat mass ratio (g/g) | 15.0 ± 3.9 | 22.0 (19.9, 24.1)b |
LRP5 HBM subjects (Tables 1 and S2) were matched with non-LRP5 HBM subjects on age (within 6 years), sex, and BMI (kg/m2), based on radius matching with a ratio 3:1, using a multi-level regression model clustering by match set. HBM, high bone mass; BMI, body mass index; CI, confidence interval. See also Table S1.
p < 0.05.
p ≤ 0.001.
A Low BMD-Associated Allele in LRP5 Correlates with Upper-Body Fat Accumulation
To further investigate the role of LRP5 in regulating regional adiposity, we explored the association between a common LRP5 single nucleotide polymorphism (SNP) and DXA-derived measures of fat distribution in 3,289 OBB volunteers. Rs599083 is an intronic SNP shown to be significantly associated with lumbar spine BMD in a GWAS meta-analysis (Rivadeneira et al., 2009). Of note, rs599083 showed modest evidence of association with fat distribution within the OBB cohort (Table 3). Specifically, the low BMD-associated minor allele at this locus correlated with increased age-, gender-, and BMI-adjusted android tissue percent fat and android/total and central/peripheral fat mass ratios. These associations were attenuated following adjustment for BMD, in keeping with rs599083 being an overlapping signal for bone and fat traits (Table 3). Based on the established association between LoF LRP5 mutations and osteoporosis (Ai et al., 2005; Gong et al., 2001), we presume that this allele is associated with reduced LRP5 function, which, according to gene expression analyses from 37 OBB subjects (Figure S2A) and eQTL data from the MuTHER consortium (http://www.muther.ac.uk), is not driven by changes in adipose LRP5 mRNA levels. We also undertook histological analyses of SC abdominal WAT from 18 overweight and obese individuals. Subject characteristics are shown in Table S3. These revealed that adipocyte numbers in android fat tended to be lower in carriers of the low BMD-associated allele at rs599083 (GG, GT) versus homozygous carriers of the common allele (TT) (p = 0.05). This effect was primarily due to a reduction in small adipocytes (Figures S2B and S2C). No associations between rs599083 and anthropometric measures of fat distribution were identified within the OBB (Table S4). Nonetheless, and consistent with our findings, this SNP was weakly associated with BMI-adjusted waist circumference (WC) in females (p = 0.001 for association, β = 0.016 for the minor allele) in the publicly available sex-stratified GIANT data set (Randall et al., 2013). It should be noted that associations between waist and hip circumferences and the respective regional fat masses (android and gynoid) within the OBB had rho values of ∼0.5 (Table S5). We conclude that, mirroring the effects of rare GoF LRP5 mutations, a common LRP5 allele that is presumably associated with reduced LRP5 function correlates with modestly increased upper-body fat accumulation.
Table 3.
Association Studies of the BMD-Associated LRP5 SNP Rs599083 and DXA-Derived Measures of Body Fat Distribution of Subjects from the Oxford Biobank
| Rs599083 |
||||||
|---|---|---|---|---|---|---|
| Adjusted for age, gender, and BMI | Adjusted for age, gender, BMI, and BMD | |||||
| Trait | p value | β | n | p value | β | n |
| Tissue leg, % fat | 0.4 | 0.007 | 3,289 | 1 | 0.002 | 3,289 |
| Tissue android, % fat | 0.004a | 0.033 | 3,289 | 0.02a | 0.027 | 3,289 |
| Android:leg fat ratio (%) | 0.008a | 0.028 | 3,289 | 0.02a | 0.025 | 3,289 |
| Android:total fat ratio (%) | 0.006a | 0.031 | 3,289 | 0.01a | 0.027 | 3,289 |
| Central:peripheral ratio (g/g) | 0.007 a | 0.028 | 3,289 | 0.02a | 0.024 | 3,289 |
| BMD (g/cm2) | 9 × 10−5a | −0.058 | 3,289 | – | – | – |
Effect allele: guanine nucleotide. Effect allele frequency: 0.34. BMD, bone mineral density; p value, empirical p value; β, standardized beta value; n, number of subjects. Data are from 1,438 men and 1,851 women. See also Tables S3–S5 and Figure S2.
Significant p values.
LRP5 Is More Highly Expressed in Abdominal Than Gluteal Adipose Progenitors
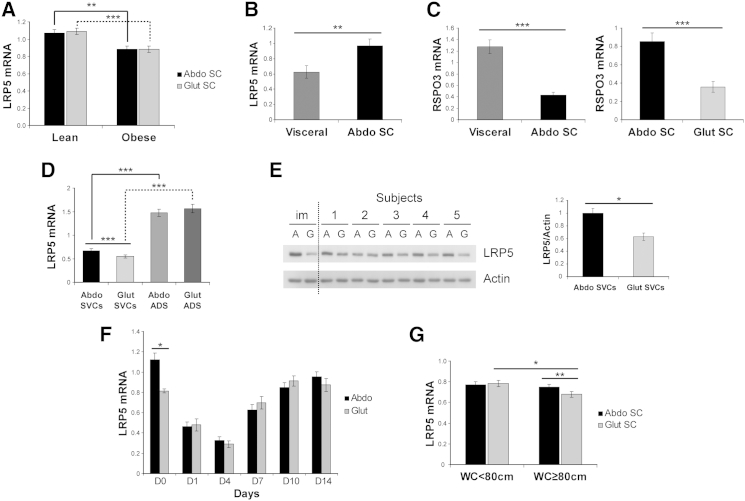
In order to gain mechanistic insights into the effects of LRP5 on fat distribution, we examined the LRP5 gene expression pattern in paired SC abdominal and gluteal fat from 20 lean and 20 obese OBB volunteers (Table S6). LRP5 expression was identical in the two depots, albeit lower in obese versus lean subjects (Figure 1A). We also compared LRP5 gene expression in paired SC abdominal and visceral fat from 16 subjects undergoing surgery. LRP5 mRNA levels were higher in SC abdominal WAT, in contrast to RSPO3 gene expression, which was higher in visceral versus SC fat (Figures 1B and 1C). We next determined the expression of LRP5 in fractionated SC abdominal and gluteal WAT (Figure 1D). LRP5 gene expression was higher in mature adipocytes compared with SVCs from both depots. No difference in LRP5 transcript levels was observed between abdominal and gluteal adipocytes. In contrast, LRP5 expression was significantly higher in abdominal versus gluteal SVCs (p = 4 × 10−6, with Bonferroni correction). We corroborated this finding by demonstrating higher abdominal LRP5 protein level in five independent pairs of primary and one pair of immortalized (see below) SVCs (Figure 1E). Finally, we examined the LRP5 gene expression profile in differentiating SVCs from a further five subjects. LRP5 mRNA levels were higher in undifferentiated abdominal versus gluteal SVCs but, following induction of adipogenesis, became indistinguishable between abdominal and gluteal adipocytes (Figure 1F). In summary, LRP5 is more highly expressed in SC versus visceral fat. Furthermore, when comparing SC depots, LRP5 mRNA and protein levels are specifically higher in abdominal compared with gluteal adipose progenitors.
Figure 1.
LRP5 Expression in Human SC Abdominal, Gluteal, and Visceral WAT
(A and B) LRP5 mRNA levels in (A) paired SC abdominal (Abdo) and gluteal (Glut) fat biopsies from lean and obese subjects (n = 20/group) and (B) paired SC abdominal and visceral fat biopsies from 16 individuals undergoing surgery. n = 7 women (age 43.6 ± 15.1 years [range 21.2–61]; BMI 30.0 ± 9.1 kg/m2 [range 19.1–42]) and 9 men (age 63.9 ± 9.3 years [range 48–76]; BMI 26.5 ± 5 kg/m2 [range 18.9–33.7]). Age and BMI are means ± SD.
(C) RSPO3 mRNA levels in paired visceral versus SC abdominal fat (n = 16) and SC abdominal versus gluteal fat (n = 20).
(D) LRP5 mRNA levels in cultured SVCs (n = 25) and mature adipocytes (ADS) (n = 24).
(E) Western blot and protein densitometry of LRP5 in immortalized (im) and primary (n = 5 pairs) SVCs from healthy subjects. A, abdominal; G, gluteal.
(F) LRP5 mRNA levels in differentiating primary abdominal and gluteal SVCs (n = 5 pairs).
(G) Comparisons of LRP5 mRNA levels in paired SC abdominal and gluteal WAT from women with gynoid (WC < 80 cm, n = 23) versus android (WC ≥ 80 cm, n = 24) fat distribution.
qRT-PCR data were normalized to PGK1 and PPIA (A–C and G) and to 18S (D and F). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, corrected for multiple testing. Histogram data are means ± SEM. See also Tables S6, S7, and S8.
To determine whether different patterns of fat distribution (upper versus lower) are associated with differences in WAT LRP5 gene expression, we analyzed LRP5 mRNA levels in paired SC abdominal and gluteal fat from 47 females recruited based on WC (Table S7). A WC ≥ 80 cm is a measure of central obesity, with increased CVD risk in Europid women as defined by the International Diabetes Federation (Alberti et al., 2005). LRP5 gene expression was selectively lower in the gluteal depot of women with android obesity (WC ≥ 80 cm) (Figure 1G). Furthermore, in partial correlation analyses, gluteal (but not abdominal) LRP5 expression correlated negatively with upper-body fat accumulation, systemic insulin resistance, and markers of inflammation after adjustment for age, BMI, and menopausal status (Table S8). We conclude that reduced gluteal WAT LRP5 gene expression correlates with upper-body fat accumulation and an adverse metabolic and inflammatory profile.
LRP5 Knockdown Has Distinct Biological Effects in Abdominal and Gluteal Progenitors
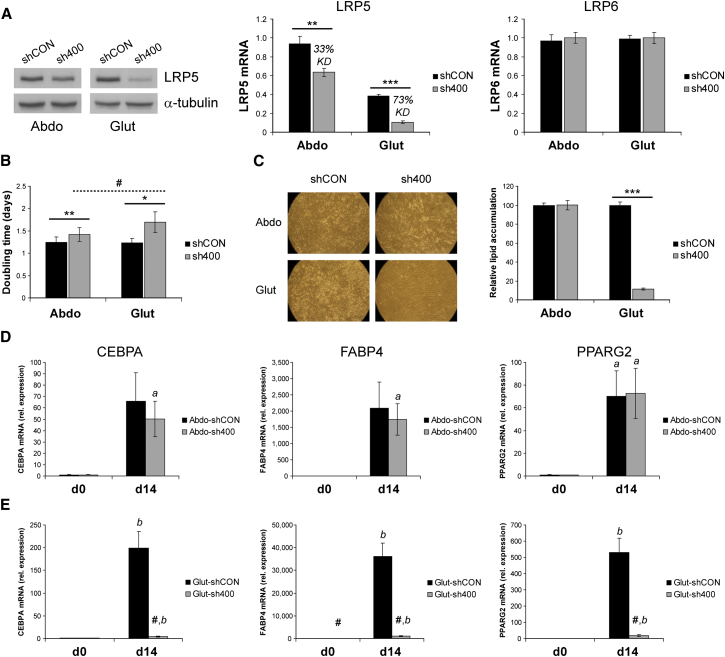
In light of our earlier findings, we investigated the role of LRP5 in depot-specific adipose progenitor biology using an immortalized pair of SC abdominal and gluteal SVCs. While displaying enhanced proliferation and adipogenesis (our unpublished data), these cells retain their depot-specific gene expression signatures (Pinnick et al., 2014). Stable LRP5 knockdown (KD) in these immortalized SVCs was achieved by lentiviral transduction with two independent shRNAs (Figures 2A and S3A). Both shRNAs targeted the two protein-coding LRP5 transcripts (http://www.ensembl.org) and were specific to LRP5 as no change in LRP6 mRNA levels was detected. The aim of these experiments was to compare the biological effects of equivalent LRP5 gene dosage reduction in abdominal and gluteal SVCs rather than to completely silence LRP5. Clone TRCN0000033400 (sh400) gave the more efficient KD. As intended, the KD magnitude was equivalent in abdominal and gluteal progenitors (see LRP5 gene expression panel in Figure 2A). Nonetheless, due to the higher LRP5 gene expression in abdominal SVCs, the percentage KD achieved was 33% in abdominal versus 73% in gluteal cells compared to scrambled shRNA-transduced SVCs. Less efficient LRP5 gene silencing (24% in abdominal versus 56% in gluteal SVCs) was achieved with shRNA clone TRCN0000033401 (sh401); again, in absolute terms LRP5 KD was near-identical in abdominal and gluteal cells (Figure S3A). LRP5 KD using either shRNA was associated with impaired proliferation in both abdominal and gluteal SVCs (Figure 2B and our unpublished data), a finding confirmed by LRP5 KD in primary SVCs derived from a female subject (Figure S4A). Decreased LRP5 expression in gluteal cells was also associated with a marked and dose-dependent inhibition of differentiation as ascertained by lipid accumulation and adipogenic gene expression (Figures 2C, 2E, S3B, and S3D). LRP5-KD gluteal adipocytes further exhibited heightened inflammation as determined by increased IL6 and MCP1 transcript levels (Figures S1B and S1C). In contrast, LRP5 KD in abdominal progenitors using sh400 was not associated with changes in adipogenesis or adipocyte inflammation (Figures 2C, 2D, and S1B); more strikingly, stable expression of sh401 in abdominal SVCs led to enhanced differentiation (Figures S3B and S3C). Similar effects on adipogenesis were observed following sh400-mediated LRP5 KD in female primary adipose SVCs (Figure S4B). In summary, equivalent absolute KD of LRP5 in abdominal and gluteal progenitors leads to distinct biological outcomes that may be driven by the differential LRP5 gene expression between the two progenitor populations.
Figure 2.
LRP5 KD in Immortalized Abdominal and Gluteal SVCs Alters Cell Proliferation and Adipogenesis
(A) LRP5 KD was confirmed by qRT-PCR and western blot analyses. LRP6 mRNA expression was not altered by LRP5 KD. shCON, control; sh400, LRP5-KD cells. ∗∗p < 0.01, ∗∗∗p < 0.001. α-tubulin was used as a western blot loading control.
(B) Doubling time of shCON and sh400 abdominal and gluteal SVCs. ∗p < 0.05, ∗∗p < 0.01, shCON versus sh400; #p < 0.05, Abdo-sh400 versus Glut-sh400.
(C) Representative micrographs of shCON and sh400 abdominal and gluteal SVCs at day 14 of adipogenic differentiation and histogram showing relative lipid accumulation, assessed by AdipoRed staining (n = 42 wells/group). ∗∗∗p < 0.001.
(D and E) Relative mRNA levels of adipogenic genes CEBPA, FABP4, and PPARG2 in (D) abdominal and (E) gluteal cells at baseline (d0) and day 14 (d14) of adipogenic differentiation. shCON versus sh400 cells: #p < 0.01; d0 versus d14 cells: ap < 0.05, bp < 0.01. Histogram data are means ± SEM. qRT-PCR data were normalized to 18S. n = 5–7 independent experiments. See also Figures S1, S3, and S4.
LRP5 KD Dose-Dependently Impairs Canonical WNT Signaling in Adipose Progenitors
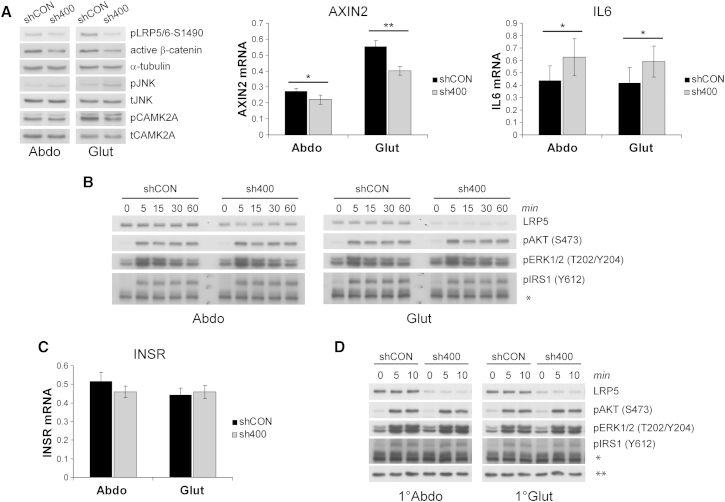
We examined which WNT pathway(s) were responsible for the biological actions of LRP5. LRP5 KD in both abdominal and gluteal cells led to impaired canonical WNT signaling, as determined by decreased active β-catenin and phosphorylated (active) LRP5/6 protein levels and reduced expression of AXIN2, a universal β-catenin target gene (Figures 3A and S3E). Given that β-catenin is generally thought to restrain adipogenesis, these findings are prima facie counterintuitive to the block in differentiation seen in gluteal SVCs. We next examined whether non-canonical WNT pathways were differentially regulated following LRP5 KD in abdominal and gluteal progenitors. However, both PCP and WNT/Ca2+ signaling were modulated in a directionally uniform manner in LRP5-KD cells. Specifically, LRP5 KD using sh400 (i.e., the more efficient shRNA) led to increased JNK phosphorylation and elevated IL6 expression in both abdominal and gluteal SVCs, consistent with PCP pathway activation (Figure 3A). Conversely, the phosphorylated to total CAMK2A ratio was uniformly decreased in abdominal and gluteal LRP5-KD cells (Figures 3A, S3E, and S3F). LRP5 was shown to promote insulin signaling and adipogenesis in 3T3-L1 preadipocytes (Palsgaard et al., 2012). Hence, we asked whether insulin/IGF1 signaling was driving the actions of LRP5 on adipose progenitor biology. As shown in Figures 3B and S3G, however, neither basal nor stimulated phosphorylation of IRS1, AKT, or ERK1/2 following treatment with 100 nM insulin (i.e., the same dose used to induce adipogenesis) were altered with LRP5 KD. Consistent with these data, no baseline change in insulin receptor (INSR) gene expression was detected in response to LRP5 KD in either abdominal or gluteal SVCs (Figures 3C and S3H). We confirmed and extended these findings in LRP5-KD primary SVCs treated with a more physiological insulin dose (10 nM) (Figure 3D).
Figure 3.
Effect of LRP5 KD on Canonical and Non-Canonical WNT and Insulin Signaling Pathways in Abdominal and Gluteal SVCs
(A) Western blots for pLRP5/6-Ser1490, active β-catenin, pJNK, and pCAMK2A and qRT-PCR analyses of AXIN2 and IL6, in control (shCON) and LRP5-KD (sh400) abdominal and gluteal immortalized SVCs. α-tubulin, total-JNK (tJNK), and total CAMK2A (tCAMK2A) were western blot loading controls. ∗p < 0.05, ∗∗p < 0.01.
(B) Representative western blots of shCON and sh400 abdominal and gluteal immortalized SVCs stimulated with 100 nM insulin for indicated duration. ∗non-specific band, used as loading control.
(C) INSR mRNA levels in shCON and sh400 abdominal and gluteal immortalized SVCs.
(D) Representative western blots of shCON and sh400 abdominal and gluteal primary (1°) SVCs stimulated with 10 nM insulin for indicated duration. ∗non-specific band detected with anti-pIRS1 (Y612) rabbit pAb, ∗∗non-specific band detected with anti-LRP5 rabbit mAb, used as loading controls. Histogram data are means ± SEM. n = 5–7 independent experiments. qRT-PCR data were normalized to 18S. See also Figure S3.
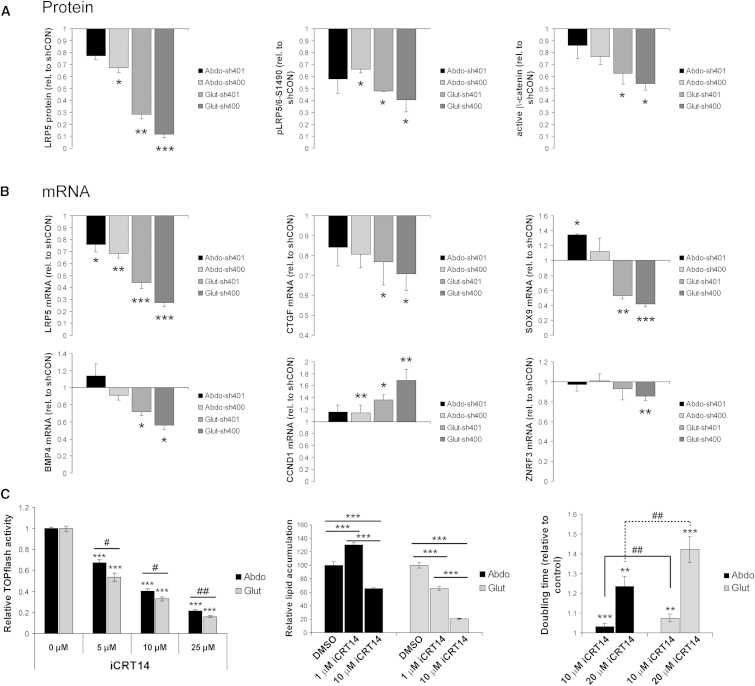
β-catenin has been shown to dose-dependently modulate target gene expression and stem/progenitor cell cycling and fate-determination (Hirata et al., 2013; Kielman et al., 2002; Luis et al., 2011). Given the graded effects of LRP5 KD on abdominal and gluteal SVC proliferation and differentiation, we examined whether active β-catenin levels and β-catenin target genes were regulated in a dose-like fashion. This was indeed the case (Figures 4A and 4B). Mirroring these findings, selectively attenuating β-catenin transcriptional activity with use of the small-molecule inhibitor iCRT14 (Gonsalves et al., 2011) dose-dependently impaired adipogenesis in immortalized gluteal SVCs (Figure 4C). In contrast, low-dose iCTR14 (1 μM) enhanced adipogenesis in abdominal progenitors while higher dose (10 μM) impaired differentiation, albeit to a lesser extent than the equivalent dose in gluteal cells. Accordingly, iCTR14-induced inhibition of β-catenin-dependent promoter activity was more pronounced in gluteal than abdominal SVCs. iCRT14 also dose-dependently impaired adipose progenitor proliferation. As expected, this effect was more marked in gluteal versus abdominal cells (Figure 4C). We conclude that the biological effects of LRP5 KD in adipose progenitors are driven by dose-dependent reductions in β-catenin transcriptional activity, which, for an equivalent decrease in LRP5 gene dosage, is more potently blocked in gluteal than abdominal SVCs.
Figure 4.
LRP5-KD in Abdominal and Gluteal SVCs Dose-Dependently Modulates β-Catenin Signaling
(A) Protein densitometry of LRP5, pLRP5/6-S1490, and active β-catenin in Abdo-sh401, Abdo-sh400, Glut-sh401, and Glut-sh400 SVCs. Densitometry data were normalized to α-tubulin and are shown relative to their respective shCON levels. n = 3–4 independent experiments.
(B) Gene expression profiling of β-catenin target genes in Abdo-sh401, Abdo-sh400, Glut-sh401, and Glut-sh400 SVCs. mRNA data were normalized to 18S and are shown relative to their respective shCON levels. n = 4–7 independent experiments.
(C) Treatment with the β-catenin small-molecule inhibitor iCRT14 dose-dependently modulates TOPflash promoter activity, adipogenesis, and proliferation in immortalized abdominal and gluteal SVCs (n = 6–7 replicates). Histogram data are means ± SEM. ∗,#p < 0.05, ∗∗,##p < 0.01, ∗∗∗p < 0.001. ∗,∗∗,∗∗∗ within group comparisons; #,## between group comparisons.
Discussion
Fat distribution is a heritable trait that strongly predicts diabetes and CVD risk independent of obesity. In this study we identify the WNT co-receptor LRP5 as a regulator of adipose progenitor biology and regional adiposity. By examining three HBM pedigrees, we show that rare GoF LRP5 mutations are associated with an increased amount of lower-body fat. Complimentary to these findings we also demonstrate, through the analysis of DXA data from > 3,000 individuals, that the minor allele of a common SNP in LRP5 associated with low BMD correlates with modestly increased upper-body fat accumulation. From the direction of its effect upon BMD, this allele is likely to be associated with reduced LRP5 function. Based on our in vitro KD studies, the increased central adiposity observed in rs599083 minor allele carriers is likely to be driven by enhanced cellularity of the SC abdominal relative to the gluteofemoral fat depot. Nonetheless, in these same experiments, LRP5-KD SC abdominal SVCs exhibited impaired proliferation and enhanced inflammation. We speculate that this is likely to impact negatively on the size and quality of the SVC pool and ultimately adipocyte number in this depot, too, a hypothesis consistent with the histological analysis of SC abdominal WAT from a limited number of rs599083 minor versus homozygous major allele carriers (Figures S2B and S2C).
LRP5 HBM individuals also exhibited enhanced insulin sensitivity versus age-, gender-, and BMI-matched controls. This may be at least partly due to their more favorable body fat distribution coupled with greater WAT cellularity (i.e., fat storage capacity, thought to protect against ectopic lipid deposition and lipotoxicity) due to enhanced SVC proliferation and, in the case of gluteal SVCs, potentially also differentiation. In contrast, rs599083 was not associated with T2D risk or measures of BMI-adjusted insulin sensitivity in publically available data sets from the DIAGRAM (Morris et al., 2012) and MAGIC (Manning et al., 2012) consortia, respectively. However, power to detect phenotypic associations between WHR- and BMI-associated variants and related metabolic parameters can be low. Accordingly, not all signals associated with fat distribution showed significant associations with surrogates of insulin resistance in the meta-analysis by Heid et al. (Heid et al., 2010). This is likely due to the limited power to detect downstream phenotypes, given the relatively low effect sizes at individual loci, even when the overall phenotypic associations are strong.
The lack of and weak association between rs599083 and anthropometric measures of fat distribution in the OBB (Table S4) and GIANT data sets, respectively, is notable. In this regard we detected only modest age-, gender-, and BMI-adjusted correlations between anthropometric and DXA-derived measures of fat distribution within the OBB (Table S5). Furthermore, alterations in body shape and/or skeletal geometry consequent to the actions of LRP5 on bone may mask the associations between LRP5 variants and anthropometric surrogates of adiposity. In keeping with this, rs3736228, a non-synonymous exonic SNP in LRP5 that is in linkage disequilibrium with rs599083, was shown to modulate femoral neck width, femoral shaft geometry, and vertebral body size (Boudin et al., 2013; van Meurs et al., 2006). Moreover, in a study of 258 individuals with unexplained HBM, HBM cases had significantly broader skeletal frames compared with controls (Gregson et al., 2012). Accordingly, we detected no consistent differences in anthropometric measures of fat distribution between HBM LRP5 mutation carriers and matched OBB controls (Table S2). Finally, distinct from the WAT expression pattern of LRP5, RSPO3 (Figure 1C) and ZNRF3 (Schleinitz et al., 2014) are more highly expressed in visceral versus SC fat. These contrasting gene expression profiles may also account for the more robust association between SNPs within these genes and body fat distribution in GWAS.
Much of the impetus for this work was based on an earlier search for WNT pathway genes that were differentially expressed between SC abdominal and gluteal SVCs. We had reasoned that these may underlie intrinsic differences in WNT pathway tone (signal strength) and/or specificity (canonical versus non-canonical pathway activation) between abdominal and gluteal progenitors. Such changes in turn might endow SVCs from these depots with distinct functional properties, thereby driving changes in fat distribution. As part of those analyses, LRP5 was found to be more highly expressed in abdominal versus gluteal progenitors. Herein we show that, consistent with our original hypothesis, equivalent magnitude of LRP5 KD in abdominal and gluteal SVCs leads to markedly different proportional reduction in LRP5 levels/activity and distinct biological outcomes; LRP5 KD impaired proliferation to a greater extent in immortalized gluteal than abdominal progenitors (Figure 2B). Furthermore, while LRP5 KD in gluteal SVCs led to impaired adipogenesis, differentiation in abdominal cells was unchanged and even enhanced following modest and low-level LRP5 KD, respectively. Finally, compared with controls, gluteal but not abdominal LRP5-KD adipocytes were inflamed. We speculate that by modulating regional WAT cellularity, these diverse biological responses underlie, at least partly, the effects of LRP5 on fat distribution. Moreover, they are rooted in the different levels of LRP5 expression between abdominal and gluteal progenitors coupled with dose-dependent effects of LRP5 on SVC function. As such, due to the lower LRP5 transcript and protein levels in gluteal versus abdominal SVCs (Figures 1D and 1E), gluteal progenitors are likely to be more sensitive than abdominal progenitors to changes in (1) LRP5 function, due to missense variants, and (2) LRP5 mRNA levels, e.g., decreased expression driven by obesity or intronic SNPs. In agreement with this hypothesis, we found that LRP5 gene expression in gluteal but not SC abdominal WAT correlated negatively with upper-body fat accumulation in females (Table S8). Adipocyte size (an inverse correlate of WAT cellularity) has also been reported to be larger in the gluteal versus abdominal depot of women (Votruba and Jensen, 2007). Complicating the interpretation of this latter finding, however, hormonal signals—e.g., sex-steroids—can interact with WNT signaling tone in vivo to modulate adipocyte size (Elbers et al., 1999). Additionally, in weight-gaining adults, abdominal WAT is thought to expand by hypertrophy (Spalding et al., 2008; Tchoukalova et al., 2010) and gluteofemoral WAT by hyperplasia (Tchoukalova et al., 2010).
Mechanistically, the effects of LRP5 KD on SVC proliferation arise from inhibition of canonical WNT signaling. This conclusion was confirmed by the dose-dependent decrease in both β-catenin-driven promoter activity and proliferation in adipose SVCs following treatment with iCTR14. Furthermore, the link between adipose cell inflammation and WNT/LRP5 signaling is likely to have its basis in the mutually antagonistic actions of the β-catenin and PCP pathways (Grumolato et al., 2010). Thus, suppressing canonical WNT signaling promotes JNK activation, thereby driving inflammation. Likewise, the enhanced differentiation seen in abdominal cells following low-level LRP5-KD is in keeping with the well-established anti-adipogenic action of WNT/β-catenin signaling. In contrast, further reduction in LRP5 levels progressively restrains adipocyte differentiation. In line with this, LRP5-KD gluteal SVCs, which exhibit the lowest LRP5 protein levels, display a “paradoxical” block in adipogenesis. The anti-adipogenic actions of LRP5 deficiency are not mediated via altered non-canonical WNT signaling, as neither the PCP nor the WNT/Ca2+ pathways were differentially modulated in abdominal versus gluteal LRP5-KD cells. Moreover, in contrast to Palsgaard et al. (Palsgaard et al., 2012), we were unable to detect any changes in insulin/IGF1 pathway activity in response to LRP5 KD (Figures 3B and S3G). Instead, utilizing iCRT14, we demonstrate that the anti-adipogenic effect of LRP5 deficiency is likely to be driven by impaired β-catenin transcriptional activity (Figure 4C). We note that we have not corroborated this conclusion by rescuing adipogenesis in LRP5-KD gluteal SVCs through augmenting β-catenin signaling. Such a rescue experiment, however, is technically challenging, since constitutively increasing β-catenin activity in adipose progenitors potently blocks adipogenesis (Christodoulides et al., 2009). Hence, rescuing differentiation in LRP5-KD gluteal cells would require precisely titrating the degree as well as the timing and duration of β-catenin signaling activation prior to and/or during differentiation. Our results suggest that, similar to its mode of action in stem/progenitor cells from other organs/tissues (Hirata et al., 2013; Kielman et al., 2002; Luis et al., 2011), canonical WNT signaling in human adipose SVCs is more complex than an on/off switch. Rather, it is associated with different biological outcomes dependent on a gradient of β-catenin transcriptional activity. Finally, in conjunction with earlier findings (Donati et al., 2014; Ross et al., 2000), our data also show that autonomous as well as non-autonomous WNT/β-catenin signals can both promote and inhibit adipogenesis.
In summary, we have established that LRP5 acts by titrating β-catenin signal strength to modulate adipose progenitor biology in a depot-specific manner, thus promoting lower-body fat accumulation. Rational manipulation of LRP5 expression/activity to achieve a more beneficial pattern of fat distribution may offer a potential approach to treat obesity-associated CVD. We note in this respect that treatment with humanized antibodies against SOST (a secreted LRP5 antagonist) is currently in phase III trials for osteoporosis management (McClung and Grauer, 2014). Once such treatments become the mainstay for osteoporosis, it will be important to determine their effects on fat distribution and metabolic health.
Experimental Procedures
Study Population and Sample Collection
Control study subjects were recruited from the OBB, a population-based collection of healthy subjects aged 30–50 years (http://www.oxfordbiobank.org.uk). HBM LRP5 mutation carriers were recruited from the UK-based HBM study cohort (Gregson et al., 2012) (see Supplemental Information). Paired SC abdominal and gluteal WAT specimens were obtained by needle biopsy as described (McQuaid et al., 2011). Paired SC abdominal and visceral fat samples were obtained from patients undergoing elective surgery as part of the MolSURG study. All studies were approved by the Oxfordshire Clinical Research Ethics Committee and the Bath multi-centre Research Ethics Committee, and all volunteers gave written, informed consent. Other aspects of the study in pre- and post-menopausal women have previously been reported (Hodson et al., 2014).
Dual Energy X-Ray Absorptiometry
Whole-body DXAs were performed using a Lunar iDXA (GE Healthcare). Acquired images were processed using enCORE v14.1 software. Central-to-peripheral fat ratio was calculated as android fat (g) ÷ (arms + legs) fat (g).
Isolation, Culture, and Differentiation of Human SVCs
SVCs were isolated from WAT biopsies, cultured, and differentiated as described (Collins et al., 2010) (see Supplemental Information). SVCs isolated from a male subject were immortalized by co-expressing human telomerase reverse transcriptase and human papillomavirus type-16 E7 oncoprotein (Pinnick et al., 2014). To study the effects of iCRT14 (Sigma-Aldrich) on adipogenesis, SVCs were seeded in type I collagen-coated 96-well plates. Culture media were replaced after 24 hr with growth media containing iCRT14 (1 μM or 10 μM) or DMSO. Confluent cells were differentiated 48 hr later in the same concentration of iCRT14 (or DMSO) throughout for 21 days. Media were changed every 2–3 days. Intracellular lipid levels were quantified using the AdipoRed assay reagent (Lonza) and a CytoFluor Multi-Well Plate Reader series 4000 (PerSeptive Biosystems).
Estimation of Cell Doubling Time
Equal numbers of control and LRP5-KD SVCs, i.e., 400,000 or 150,000, were seeded in T175 and T75 flasks, respectively. Cells were trypsinized and double counted every 72 hr. Doubling time was calculated using the formula Td = (t2 − t1) × [log(2) ÷ log(q2 ÷ q1)], where t = time (days) and q = cell number.
Lentiviral Constructs and Generation of Stable Cell Lines
MISSION LRP5 shRNA and control plasmid DNA vectors were obtained from Sigma-Aldrich. The 7TFP WNT reporter lentiviral vector (Fuerer and Nusse, 2010) was obtained from Addgene. Lentiviral particles were produced by transient co-transfection of HEK293 cells with the vector of interest and packaging vectors (MISSION [Sigma-Aldrich] and ViraPower [Invitrogen] packaging mixes). Stable cell lines were generated by transduction of SVCs with lentiviral particles followed by selection in growth media containing 2 μg/ml puromycin.
Insulin Stimulation Studies of SVCs
Stimulation experiments with 10 nM or 100 nM insulin (Sigma-Aldrich) were performed as described (Palsgaard et al., 2012).
Genotyping, Quantitative Real-Time PCR, Western Blot Analyses, and Measurement of Adipocyte Size and Number
Genotyping and qRT-PCR were performed using TaqMan assays. Western blot analyses were performed using standard protocols. Adipocyte sizing was performed using a histological method. Full details are found in Supplemental Information.
Luciferase Reporter Assay
Immortalized SVCs stably expressing 7TFP were grown to confluence in type I collagen-coated 24-well plates in complete growth media, then treated with indicated concentrations of iCRT14 (or DMSO) in serum-free media for 24 hr. Cell lysates were harvested in passive lysis buffer, and reporter activity was measured using the Luciferase Assay System (Promega) and a Veritas Microplate Luminometer (Turner Biosystems). Data were normalized to protein concentration.
Statistical Analysis
Statistical analyses for association studies between LRP5 SNPs and quantitative traits were performed using the PLINK program v.1.07 (http://pngu.mgh.harvard.edu/∼purcell/plink/) (Purcell et al., 2007). All quantitative traits were log transformed and analyzed with a linear regression model. Data were adjusted for age, gender, and BMI. Significance is presented as empirical p values as calculated by the additional implementation of permutation procedures (default under PLINK) to the linear regression model. For parametric data, statistical significance was determined by pairwise comparisons using a two-tailed paired or unpaired Student’s t test, as appropriate. For non-parametric data, group differences were determined using the Kruskal-Wallis one-way analysis of variance. Statistical significance was determined by pairwise comparisons using a two-tailed Mann-Whitney test. Statistical tests generating a p < 0.05 were considered significant. Partial correlations of non-parametric data were performed using the SPSS Statistics software package and adjusted for age, gender, and BMI. For studies of the HBM cohort, matched analyses used radius matching based on age, gender, and BMI with a 3:1 ratio, using a multi-level regression model clustering by match set. All data are presented as means ± SEM unless otherwise stated.
Author Contributions
N.Y.L. and C.C. designed and performed experiments, analyzed data, and co-wrote paper; M.J.N. analyzed genotyping data; K.M. undertook adipocyte sizing studies; S.A.H., J.H.T., and C.L.G. provided access to the HBM cohort; C.L.G. and E.L.D. undertook genotyping of the HBM cohort and analyzed data; B.A.F. and K.M. provided access to data and WAT from the pre- and post-menopausal women cohort; M.I.M. facilitated the generation of OBB exome chip array data and provided access to MolSURG WAT samples; F.K. provided access to the OBB, designed experiments, and co-wrote paper. All authors critically reviewed the manuscript.
Acknowledgments
This research is supported by a Clinical Research Grant Programme from the European Foundation for the Study of Diabetes and a Project Grant from the British Heart Foundation (BHF) (PG/12/78/29862). K.M. is supported by the European Commission (FP7-PEOPLE-2011-IEF). The menopause study was funded by the BHF (PG/09/003). We are grateful to all the human volunteers and the CRU staff (L. Dennis, M. Gilbert, and J. Cheeseman in particular). We thank L. Hodson, co-investigator of the menopause study. We thank A. Clark for help with adipocyte histology. We thank K. Addison, J. Harris, G. Clark, and L. Wheeler for their careful sequencing, performed in the laboratory of Prof. M. Brown, with thanks. We are grateful to A. Sayers for writing the radius-matching Stata program used and S.K. Thomsen for helpful discussions.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Fredrik Karpe, Email: fredrik.karpe@ocdem.ox.ac.uk.
Constantinos Christodoulides, Email: costas.christodoulides@ocdem.ox.ac.uk.
Supplemental Information
References
- Ai M., Heeger S., Bartels C.F., Schelling D.K., Osteoporosis-Pseudoglioma Collaborative Group Clinical and molecular findings in osteoporosis-pseudoglioma syndrome. Am. J. Hum. Genet. 2005;77:741–753. doi: 10.1086/497706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti K.G., Zimmet P., Shaw J., IDF Epidemiology Task Force Consensus Group The metabolic syndrome—a new worldwide definition. Lancet. 2005;366:1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- Billon N., Dani C. Developmental origins of the adipocyte lineage: new insights from genetics and genomics studies. Stem Cell Rev. 2012;8:55–66. doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- Boudin E., Steenackers E., de Freitas F., Nielsen T.L., Andersen M., Brixen K., Van Hul W., Piters E. A common LRP4 haplotype is associated with bone mineral density and hip geometry in men-data from the Odense Androgen Study (OAS) Bone. 2013;53:414–420. doi: 10.1016/j.bone.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Boyden L.M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M.A., Wu D., Insogna K., Lifton R.P. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Christodoulides C., Lagathu C., Sethi J.K., Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Collins J.M., Neville M.J., Hoppa M.B., Frayn K.N. De novo lipogenesis and stearoyl-CoA desaturase are coordinately regulated in the human adipocyte and protect against palmitate-induced cell injury. J. Biol. Chem. 2010;285:6044–6052. doi: 10.1074/jbc.M109.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G., Proserpio V., Lichtenberger B.M., Natsuga K., Sinclair R., Fujiwara H., Watt F.M. Epidermal Wnt/β-catenin signaling regulates adipocyte differentiation via secretion of adipogenic factors. Proc. Natl. Acad. Sci. USA. 2014;111:E1501–E1509. doi: 10.1073/pnas.1312880111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan E.L., Danoy P., Kemp J.P., Leo P.J., McCloskey E., Nicholson G.C., Eastell R., Prince R.L., Eisman J.A., Jones G. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers J.M., de Jong S., Teerlink T., Asscheman H., Seidell J.C., Gooren L.J. Changes in fat cell size and in vitro lipolytic activity of abdominal and gluteal adipocytes after a one-year cross-sex hormone administration in transsexuals. Metabolism. 1999;48:1371–1377. doi: 10.1016/s0026-0495(99)90146-4. [DOI] [PubMed] [Google Scholar]
- Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Oei L., Albagha O.M., Amin N., Kemp J.P. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L., Eagon J.C., Trujillo M.E., Scherer P.E., Klein S. Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes. 2007;56:1010–1013. doi: 10.2337/db06-1656. [DOI] [PubMed] [Google Scholar]
- Fuerer C., Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S., Blüher M., Yamamoto Y., Norris A.W., Berndt J., Kralisch S., Boucher J., Lewis C., Kahn C.R. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. USA. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D., Osteoporosis-Pseudoglioma Syndrome Collaborative Group LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Gonsalves F.C., Klein K., Carson B.B., Katz S., Ekas L.A., Evans S., Nagourney R., Cardozo T., Brown A.M., DasGupta R. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. USA. 2011;108:5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson C.L., Steel S.A., O’Rourke K.P., Allan K., Ayuk J., Bhalla A., Clunie G., Crabtree N., Fogelman I., Goodby A. ‘Sink or swim’: an evaluation of the clinical characteristics of individuals with high bone mass. Osteoporos. Int. 2012;23:643–654. doi: 10.1007/s00198-011-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumolato L., Liu G., Mong P., Mudbhary R., Biswas R., Arroyave R., Vijayakumar S., Economides A.N., Aaronson S.A. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Mägi R., MAGIC Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A., Utikal J., Yamashita S., Aoki H., Watanabe A., Yamamoto T., Okano H., Bardeesy N., Kunisada T., Ushijima T. Dose-dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development. 2013;140:66–75. doi: 10.1242/dev.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson L., Harnden K., Banerjee R., Real B., Marinou K., Karpe F., Fielding B.A. Lower resting and total energy expenditure in postmenopausal compared with premenopausal women matched for abdominal obesity. J Nutr Sci. 2014;3:e3. doi: 10.1017/jns.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.D. Role of body fat distribution and the metabolic complications of obesity. J. Clin. Endocrinol. Metab. 2008;93(1):S57–S63. doi: 10.1210/jc.2008-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe F., Pinnick K.E. Biology of upper-body and lower-body adipose tissue-link to whole-body phenotypes. Nat. Rev. Endocrinol. 2014 doi: 10.1038/nrendo.2014.185. Published online November 4, 2014. [DOI] [PubMed] [Google Scholar]
- Kielman M.F., Rindapää M., Gaspar C., van Poppel N., Breukel C., van Leeuwen S., Taketo M.M., Roberts S., Smits R., Fodde R. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat. Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- Krishnan V., Bryant H.U., Macdougald O.A. Regulation of bone mass by Wnt signaling. J. Clin. Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., Carulli J.P., Del Mastro R.G., Dupuis J., Osborne M., Folz C., Manning S.P., Swain P.M., Zhao S.C., Eustace B. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis T.C., Naber B.A., Roozen P.P., Brugman M.H., de Haas E.F., Ghazvini M., Fibbe W.E., van Dongen J.J., Fodde R., Staal F.J. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011;9:345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Mani A., Radhakrishnan J., Wang H., Mani A., Mani M.A., Nelson-Williams C., Carew K.S., Mane S., Najmabadi H., Wu D., Lifton R.P. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Multiple Tissue Human Expression Resource (MUTHER) Consortium A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung M.R., Grauer A. Romosozumab in postmenopausal women with osteopenia. N. Engl. J. Med. 2014;370:1664–1665. doi: 10.1056/NEJMc1402396. [DOI] [PubMed] [Google Scholar]
- McQuaid S.E., Hodson L., Neville M.J., Dennis A.L., Cheeseman J., Humphreys S.M., Ruge T., Gilbert M., Fielding B.A., Frayn K.N., Karpe F. Downregulation of adipose tissue fatty acid trafficking in obesity: a driver for ectopic fat deposition? Diabetes. 2011;60:47–55. doi: 10.2337/db10-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsgaard J., Emanuelli B., Winnay J.N., Sumara G., Karsenty G., Kahn C.R. Cross-talk between insulin and Wnt signaling in preadipocytes: role of Wnt co-receptor low density lipoprotein receptor-related protein-5 (LRP5) J. Biol. Chem. 2012;287:12016–12026. doi: 10.1074/jbc.M111.337048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnick K.E., Nicholson G., Manolopoulos K.N., McQuaid S.E., Valet P., Frayn K.N., Denton N., Min J.L., Zondervan K.T., Fleckner J., MolPAGE Consortium Distinct developmental profile of lower-body adipose tissue defines resistance against obesity-associated metabolic complications. Diabetes. 2014;63:3785–3797. doi: 10.2337/db14-0385. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qatanani M., Lazar M.A. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Genes Dev. 2007;21:1443–1455. doi: 10.1101/gad.1550907. [DOI] [PubMed] [Google Scholar]
- Randall J.C., Winkler T.W., Kutalik Z., Berndt S.I., Jackson A.U., Monda K.L., Kilpeläinen T.O., Esko T., Mägi R., Li S., DIAGRAM Consortium. MAGIC Investigators Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira F., Styrkársdottir U., Estrada K., Halldórsson B.V., Hsu Y.H., Richards J.B., Zillikens M.C., Kavvoura F.K., Amin N., Aulchenko Y.S., Genetic Factors for Osteoporosis (GEFOS) Consortium Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S.E., Hemati N., Longo K.A., Bennett C.N., Lucas P.C., Erickson R.L., MacDougald O.A. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Saarinen A., Saukkonen T., Kivelä T., Lahtinen U., Laine C., Somer M., Toiviainen-Salo S., Cole W.G., Lehesjoki A.E., Mäkitie O. Low density lipoprotein receptor-related protein 5 (LRP5) mutations and osteoporosis, impaired glucose metabolism and hypercholesterolaemia. Clin. Endocrinol. (Oxf.) 2010;72:481–488. doi: 10.1111/j.1365-2265.2009.03680.x. [DOI] [PubMed] [Google Scholar]
- Schenk S., Saberi M., Olefsky J.M. Insulin sensitivity: modulation by nutrients and inflammation. J. Clin. Invest. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleinitz D., Klöting N., Lindgren C.M., Breitfeld J., Dietrich A., Schön M.R., Lohmann T., Dreßler M., Stumvoll M., McCarthy M.I. Fat depot-specific mRNA expression of novel loci associated with waist-hip ratio. Int J Obes (Lond) 2014;38:120–125. doi: 10.1038/ijo.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple R.K., Savage D.B., Cochran E.K., Gorden P., O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocr. Rev. 2011;32:498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- Singh R., Smith E., Fathzadeh M., Liu W., Go G.W., Subrahmanyan L., Faramarzi S., McKenna W., Mani A. Rare nonconservative LRP6 mutations are associated with metabolic syndrome. Hum. Mutat. 2013;34:1221–1225. doi: 10.1002/humu.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding K.L., Arner E., Westermark P.O., Bernard S., Buchholz B.A., Bergmann O., Blomqvist L., Hoffstedt J., Näslund E., Britton T. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- Tchkonia T., Giorgadze N., Pirtskhalava T., Thomou T., DePonte M., Koo A., Forse R.A., Chinnappan D., Martin-Ruiz C., von Zglinicki T., Kirkland J.L. Fat depot-specific characteristics are retained in strains derived from single human preadipocytes. Diabetes. 2006;55:2571–2578. doi: 10.2337/db06-0540. [DOI] [PubMed] [Google Scholar]
- Tchkonia T., Lenburg M., Thomou T., Giorgadze N., Frampton G., Pirtskhalava T., Cartwright A., Cartwright M., Flanagan J., Karagiannides I. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am. J. Physiol. Endocrinol. Metab. 2007;292:E298–E307. doi: 10.1152/ajpendo.00202.2006. [DOI] [PubMed] [Google Scholar]
- Tchoukalova Y.D., Votruba S.B., Tchkonia T., Giorgadze N., Kirkland J.L., Jensen M.D. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proc. Natl. Acad. Sci. USA. 2010;107:18226–18231. doi: 10.1073/pnas.1005259107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turer A.T., Khera A., Ayers C.R., Turer C.B., Grundy S.M., Vega G.L., Scherer P.E. Adipose tissue mass and location affect circulating adiponectin levels. Diabetologia. 2011;54:2515–2524. doi: 10.1007/s00125-011-2252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meurs J.B., Rivadeneira F., Jhamai M., Hugens W., Hofman A., van Leeuwen J.P., Pols H.A., Uitterlinden A.G. Common genetic variation of the low-density lipoprotein receptor-related protein 5 and 6 genes determines fracture risk in elderly white men. J. Bone Miner. Res. 2006;21:141–150. doi: 10.1359/JBMR.050904. [DOI] [PubMed] [Google Scholar]
- Votruba S.B., Jensen M.D. Sex differences in abdominal, gluteal, and thigh LPL activity. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1823–E1828. doi: 10.1152/ajpendo.00601.2006. [DOI] [PubMed] [Google Scholar]
- Weidinger G., Moon R.T. When Wnts antagonize Wnts. J. Cell Biol. 2003;162:753–755. doi: 10.1083/jcb.200307181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf S., Hawken S., Ounpuu S., Bautista L., Franzosi M.G., Commerford P., Lang C.C., Rumboldt Z., Onen C.L., Lisheng L., INTERHEART Study Investigators Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet. 2005;366:1640–1649. doi: 10.1016/S0140-6736(05)67663-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.