Abstract
Familial cold autoinflammatory syndrome (FCAS, MIM 120100), commonly known as familial cold urticaria (FCU), is an autosomal- dominant systemic inflammatory disease characterized by intermittent episodes of rash, arthralgia, fever and conjunctivitis after generalized exposure to cold1–4. FCAS was previously mapped to a 10-cM region on chromosome 1q44 (refs. 5,6). Muckle–Wells syndrome (MWS; MIM 191900), which also maps to chromosome 1q44, is an autosomal-dominant periodic fever syndrome with a similar phenotype except that symptoms are not precipitated by cold exposure and that sensorineural hearing loss is frequently also present6–8. To identify the genes for FCAS and MWS, we screened exons in the 1q44 region for mutations by direct sequencing of genomic DNA from affected individuals and controls. This resulted in the identification of four distinct mutations in a gene that segregated with the disorder in three families with FCAS and one family with MWS. This gene, called CIAS1, is expressed in peripheral blood leukocytes and encodes a protein with a pyrin domain9–11, a nucleotide-binding site (NBS, NACHT subfamily12) domain and a leucine-rich repeat (LRR) motif region13, suggesting a role in the regulation of inflammation and apoptosis.
We previously identified a locus for FCAS on chromosome 1q44 between markers D1S423 and D1S2682 (ref. 5). We developed a physical contig of bacterial artificial chromosomes (BACs) and P1-derived artificial chromosomes and narrowed the critical region to less than 1 Mb using haplotype analysis (data not shown). We used ESTs mapping to this region and exon-prediction programs to identify potential exons in the publicly available genome sequence. We sequenced more than 80 exons with the flanking intron sequence from the genomic DNA of affected individuals and controls after amplification by PCR using flanking intronic primers. This analysis revealed four different missense mutations in a 1,753–base pair (bp) exon (exon 3) of CIAS1 that segregated with the disease in each family (Table 1 and Figs. 1 and 2).
Table 1.
Mutations found in families with FCAS and MWS
| Family | Exon | Nucleic-acid change | Amino-acid change | Normal control frequency |
|---|---|---|---|---|
| 1 | 3 | C1316T | A439V | 0/113 |
| 2 | 3 | G592A | V198M | 0/109 |
| 3 | 3 | A1880G | E627G | 0/114 |
| 4 | 3 | C1055T | A352V | 0/108 |
Nucleotide and amino acid coordinates are given assuming that the A of the second ATG codon is nucleotide 1. The normal control frequency refers to the number of times an indicated nucleotide change was found in the indicated number of control DNA samples analyzed.
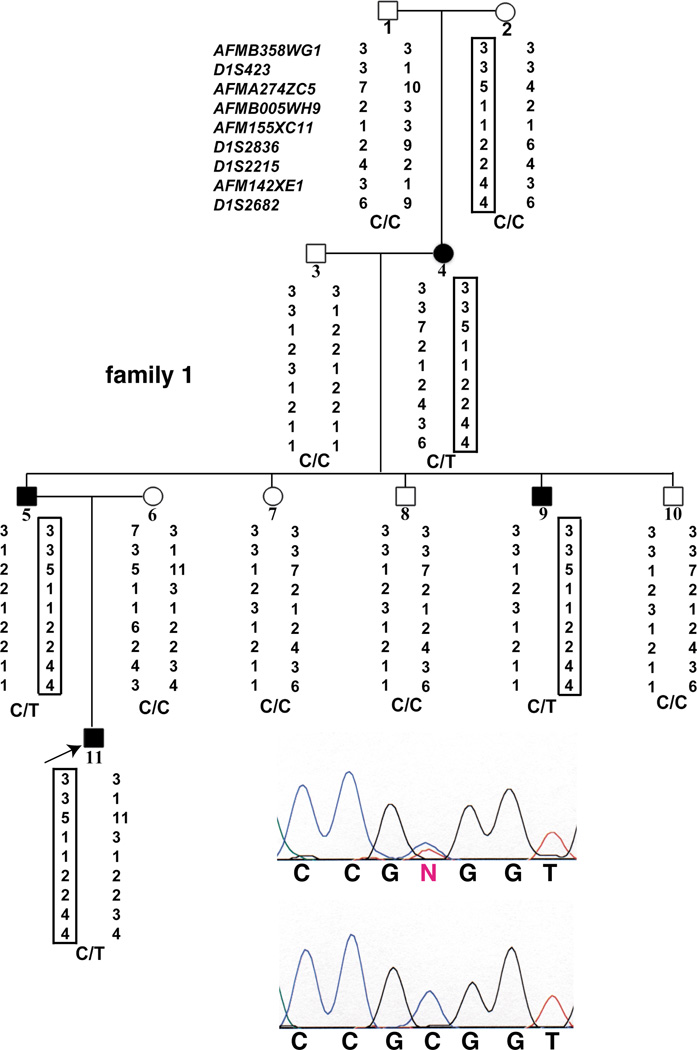
Fig. 1.
Pedigree, linkage and mutational analysis of a family with FCAS with a de novo mutation. Illustrated is the pedigree of family 1, with filled squares (male) and circles (female) indicating individuals with FCAS and open squares and circles indicating unaffected individuals; the subject number is indicated below each symbol. Listed to the left of subject 1 are the microsatellite markers used to genotype the family. Below each subject are indicated the allele numbers for each microsatellite marker observed for the two chromosome 1q44 haplotypes inherited by each individual; the disease haplotype is boxed. Indicated below each pair of haplotypes is the genotype for each subject, C being the wild type allele (C1317) and T the mutant allele (T1317). The upper sequencing chromatogram is a representative chromatogram from a mutation carrier (the subject being indicated by the arrow) showing the heterozygous genotype (C/T), and the lower sequencing chromatogram is a representative chromatogram showing the same region from an unaffected individual.
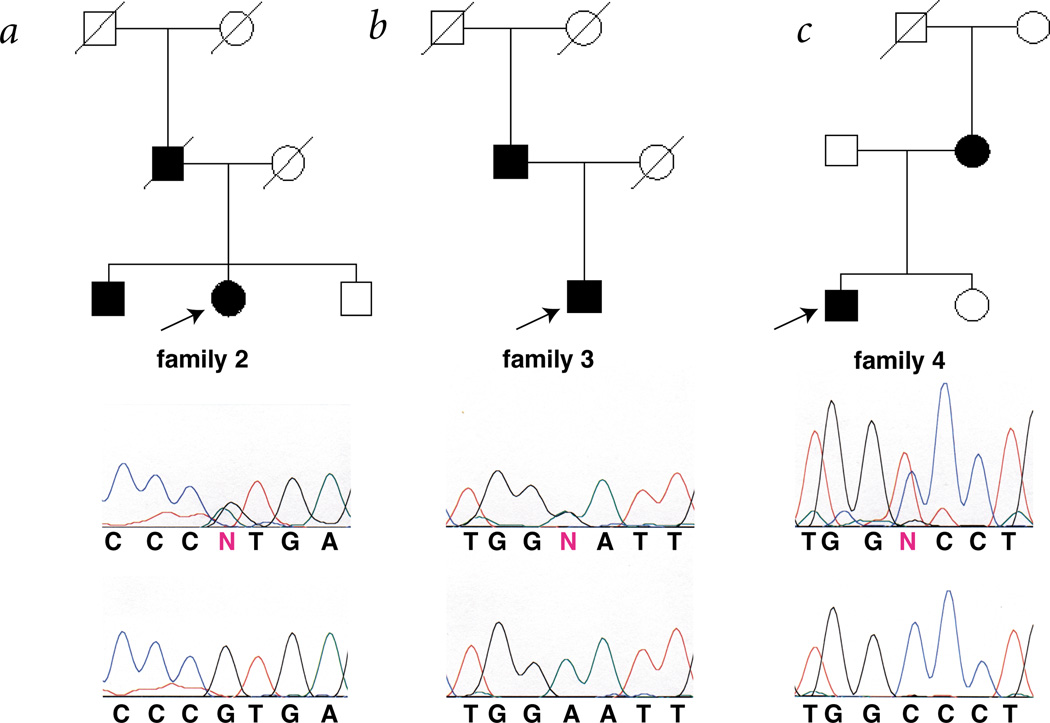
Fig. 2.
Pedigree and mutational analysis of two families with FCAS and one with MWS. The key to the pedigrees and sequence analysis is as described in Fig. 1. a,b, Families with FCAS possessing the G592A and A1880G mutations, respectively. c, A family with MWS possessing the C1055T mutation. All living family members shown were studied. All of the affected individuals shown were found to contain the indicated mutation; arrows indicate the individual whose DNA was sequenced to generate the mutation-containing chromatogram presented below each pedigree.
The four-generation family with FCAS (family 1; Fig. 1) has an apparent de novo mutation that first appeared in subject 4 on a chromosome inherited from her mother. Only the affected members of subsequent generations of the family (subjects 5, 9 and 11) inherited this chromosome. We found a missense mutation in subject 4 that segregated with the disease haplotype in subsequent generations (subjects 5, 9 and 11). This mutation was not present in either of her parents (subjects 1 and 2), even though the unaffected mother (subject 2) possessed the haplotype that segregates with disease in this family.
The other two families with FCAS (families 2 and 3) also had different missense mutations in the same exon; these were present in all affected family members (Fig. 2). In the one family studied with affected members with a diagnosis of MWS (family 4), both affected individuals had sensorineural hearing loss (Fig. 2). This family also had a missense mutation in the same exon as the families with FCAS. As in family 1, the phenotype of family 4 resulted from a de novo mutation. The unaffected mother (DNA was not available from the father) in family 4 possessed the haplotype that segregates with the disease in subsequent generations (data not shown), but she did not have the mutation that was found in all of the affected family members. These missense mutations were not found in any unaffected individuals in these families or in random North American blood-bank samples, indicating that these missense mutations are not common polymorphisms (Table 1; over 100 controls were analyzed for each missense mutation). The observation of four different mutations including two de novo mutations, and the absence of all of the missense mutations in normal control samples, provide strong evidence that these mutations cause FCAS and MWS.
To further define CIAS1, we identified two ESTs containing all or part of exon 3 through BLAST searches of the public databases (AK027194 and AF054176). We identified seven exons by homology to the publicly available genome sequence or sequence from BACs containing this region. Subsequent sequencing of RT–PCR products revealed two additional exons and extensive alternative splicing in the carboxy-terminal region of the gene (data not shown).
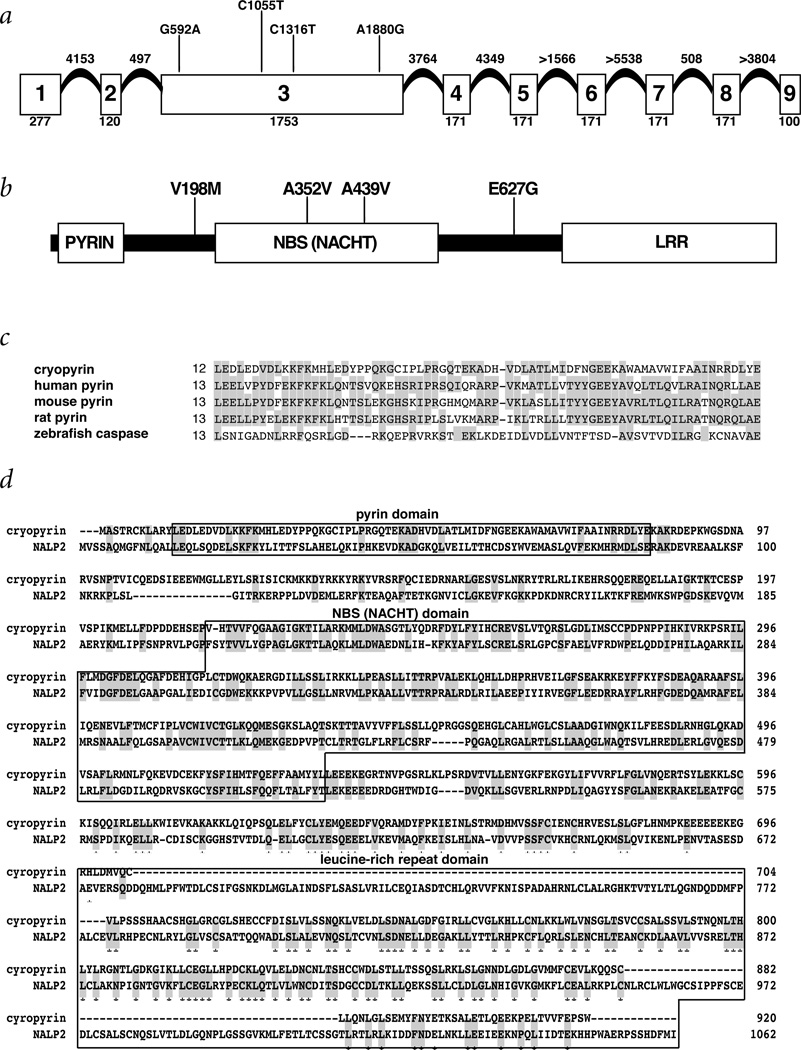
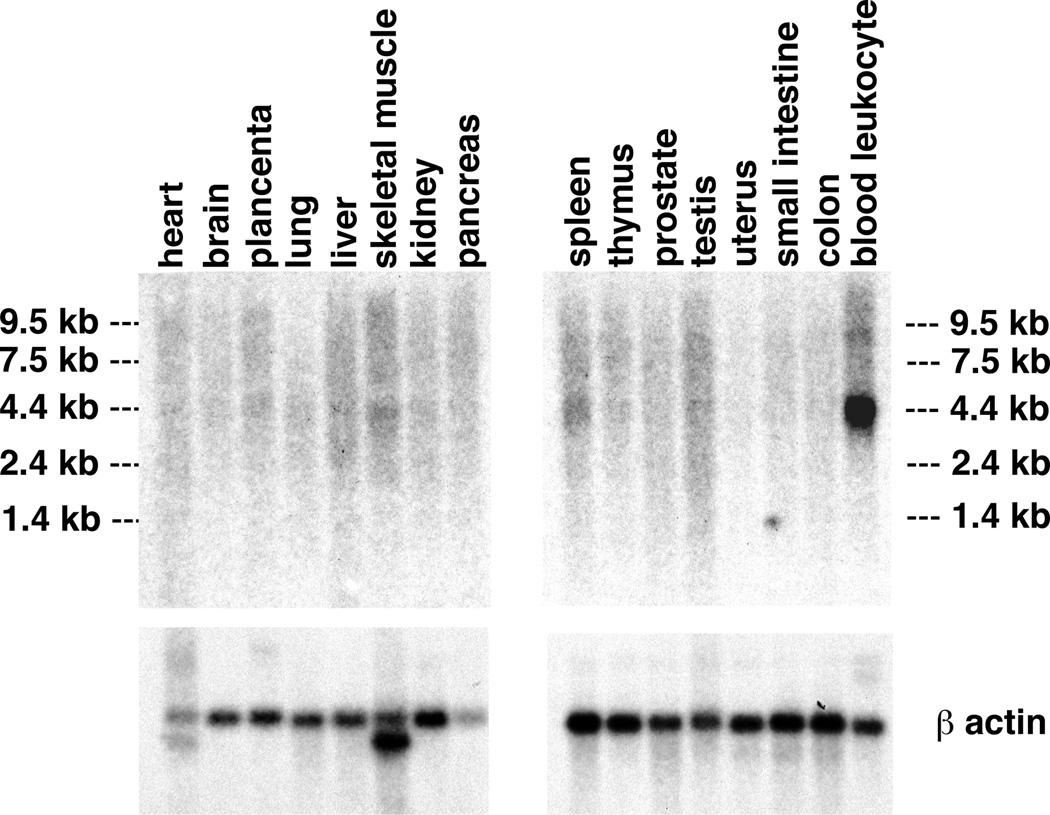
We characterized each exon and its flanking intron sequence by direct sequencing of PCR products amplified from human genomic DNA and from RPI11 BAC clones (433K2, 978I15 and 482N10) from chromosome 1q44 (Table 2). The predicted structure of the gene and the position of identified mutations are shown (Fig. 3a). The nine-exon gene is predicted to encode an ORF of 3,105 bp with two potential start codons in exon 1, the second start codon meeting more Kozak criteria14, and a stop codon in exon 9. Northern-blot hybridization using probes derived from the coding sequence contained in exons 1, 3 and 7–9 each identified an approximately 4-kb mRNA, migrating as a broad band, expressed at a low level in peripheral blood leukocytes; little or no expression was detectable in other tissues (Fig. 4). This is identical to the expression pattern seen with MEFV, the gene involved in familial Mediterranean fever15.
Table 2.
Intron–exon junction sequences of CAIS1
| exon 1 | ||
| GCAGATGAAGatggcaagca | (–752–277) | ccgaagtgggGTGAGTGGAA |
| exon 2 | ||
| TCCCTTTTAGgttcagataa | (278–397) | atgaagaaagGTAAGCGACT |
| exon 3 | ||
| TTTGCCGTAGattaccgtaa | (398–2150) | gttctcatggGTAAGGAAAC |
| exon 4 | ||
| TAATTCCTAGattggtgaac | (2151–2321) | ggagattgtgGTGAGTCCCC |
| exon 5 | ||
| TGGCATGAAGgttggggcgc | (2322–2492) | agaagctctgGTGAGTCGAG |
| exon 6 | ||
| TCTATGGAAGgttggtcagc | (2493–2663) | agaaactgggGTAAGTCTTC |
| exon 7 | ||
| CCTTCTACAGgttggtgaat | (2664–2834) | aggtgttggaGTAAGTCCTT |
| exon 8 | ||
| CTTTTTGCAGattagacaac | (2835–3005) | agaacctgggGTGAGTGTGC |
| exon 9 | ||
| GCTTTTACAGgttgtctgaa | (3006–3105) | ttcttggtagGAGTGGAAAC |
The coding sequence is in lower case and 5′ untranslated, 3′ untranslated and intron sequences are in upper case. Nucleotide coordinates are given assuming that the A of the ATG is nucleotide 1. The numbers in parentheses refer to: for exon 1, nucleotide coordinates starting at the 5′ end of the largest 5′ RACE product and ending at the last nucleotide before exon 1; for exons 2–8, nucleotide coordinates of the mRNA sequence encoded by that exon; for exon 9, nucleotide coordinates of the mRNA sequence encoded by that exon through the end of the longest 3′ RACE product obtained. The stop codon starts at nucleotide 3103 of the nine-exon predicted ORF.
Fig. 3.
Structural features of CIAS1. a, Illustration of the intron–exon structure of CIAS1. The coding exons are illustrated by the boxes, the number of coding nucleotides in each exon (including the stop codon) is indicated below the box and the relative positions of each mutation are identified above the exon 3 box. The exact and estimated sizes of the introns are indicated above each intervening segment. Exons 4–8 each encode two LRRs and exon 9 encodes one LRR. b, Schematic diagram of the structural features of the CIAS1 protein (cryopyrin), which are indicated by the boxes. The pyrin domain comprises aa 13–83, the NBS domain aa 217–533 and the LRR domain aa 697–920 (of the initial isoform). The positions of the amino-acid substitutions caused by the mutations identified are indicated above the illustration of the protein. c, Alignment of the amino-acid sequence of the pyrin domain of cryopyrin with the pyrin domain of human, mouse and rat pyrin and zebrafish caspase (GenBank accession numbers AAB70557, AAF03766, AAF03767 and AAF66964). d, Alignment of the amino-acid sequence of the seven exon initial isoform (exons 1–3, 5 and 7–9) with the amino-acid sequence of NALP2 (GenBank accession number AAG30289). The observed identity and similarity were 28% and 42%, respectively, the identities being indicated by gray shading. The positions within cryopyrin containing the pyrin, NBS and LRR domains are indicated by the boxes; these domains show significant homology between cryopyrin and NALP2. The large gaps in the alignment all lie in the LRR domain and result simply from the fact that NALP2 contains six more LRRs than this isoform of cryopyrin.
Fig. 4.
Northern-blot analysis of FCAS expression in different tissues. Northernblot analysis was carried out with an exon 1–specific probe. The upper panel illustrates hybridization with the exon 1 probe and the lower panel hybridization with the β-actin control probe. The positions of the molecular-mass markers are indicated to the left and right of the upper panels.
To verify the size of this mRNA, we amplified the predicted DNA segments by RT–PCR from peripheral blood leukocyte mRNA using primers designed from the genomic coding sequence, and also amplified the ends of the mRNA by 5′ and 3′ RACE. We cloned and sequenced these PCR products (data not shown). Analysis of these sequences and genomic exon sequence revealed extensive alternative splicing of exons 4–8 that resulted in mRNAs ranging in size from 3,315 bp to 4,170 bp, consistent with the northern-blot analysis.
The predicted protein encoded by the first splice form of CIAS1 identified (exons 1–3, 5 and 7–9), which we call cryopyrin, consists of 920 amino acids (aa) with a size of 105.7 kD and a pI of 6.16. The protein sequence contains several distinct motifs (Fig. 3b), including a pyrin domain (Fig. 3c) in the amino terminus (aa 13–83)9–11, a central NBS (NACHT subfamily12) domain in exon 3 (aa 217–533) and a c-terminal LRR domain containing seven LRRs (aa 697–920)13. No nuclear localization signals were identified and no clear transmembrane regions were found. The largest protein potentially encoded by the nine exons of CIAS1 consists of 1,034 aa with a size of 117.9 kD and 11 C-terminal LRRs. All of these features (Fig. 3b) suggest that cryopyrin is a signaling protein involved in the regulation of inflammation and apoptosis, although this remains to be verified experimentally.
Mutations in CIAS1 result in the phenotypically similar, yet distinct, periodic fever disorders of FCAS and MWS. They are distinguished clinically by the presence of cold sensitivity and a higher incidence of episodic conjunctivitis in FCAS, and the development of sensorineural deafness and a higher incidence of the development of amyloidosis in MWS16. The phenotypic variation between these two conditions with the same apparent genetic basis may reflect subtle differences in the effects of different mutations or may be the result of differing genetic backgrounds or environmental factors. Further analysis is necessary to determine the mechanisms underlying cold sensitivity in FCAS and deafness in MWS. Further phenotypic variability may be seen if CIAS1 mutations are also found in a family with another clinically distinct autosomal dominant periodic fever disorder that was recently mapped to chromosome 1q44 (ref. 17). Identification of the gene and predicted protein structure further supports the inclusion of FCAS in the group of autoinflammatory syndromes. Because of the genetic evidence and phenotypic similarity to other hereditary periodic fevers, we have proposed ‘familial cold autoinflammatory syndrome’ as a more accurate and preferable name for the disorder previously referred to as ‘familial cold urticaria’16.
The discovery of a gene for FCAS and MWS follows the determination of the genetic basis of several other autoinflammatory syndromes, including the involvement of MVK in hyper-IgD syndrome18,19 and of tumor necrosis factor receptor (TNFR1SF1A) in familial Hibernian fever20. Familial Mediterranean fever is caused by mutations in MEFV15,21, which encodes a protein called pyrin (also known as marenostirin) that is expressed predominantly in mature neutrophils and is localized to the cytoplasm bound to bundle microtubules22–24. Recently, the N-terminal domain of pyrin has been included in a family of protein modules involved in apoptosis and inflammation, including the death domain, the death effector domain and the caspase activation and recruitment domain (CARD)9–11. These proteins seem to mediate protein–protein interactions involved in signal transduction from death receptors such as Fas and TNFR1 and often activate nuclear factor-κB25.
The cryopyrin protein contains an N-terminal pyrin domain, a central NACHT domain and a LRR motif-containing region, which could be involved in innate immune responses as well as in apoptosis and inflammation. Two proteins, NALP1 (CARD7 or DEFCAP) and NALP2 (NBS1), were recently identified with the same three domains (GenBank accession numbers AAG30288 and AAG30289)9. NALP2 shares the most homology with cryopyrin (Fig. 3d), but there are several additional proteins with apoptosis or innate immune function that have a similar modular structure and also share homology with cryopyrin, including Nod1 (CARD4), Nod2 (CARD15), ASC and plant resistance genes such as RPS2 (GenBank accession numbers AAD43922, AAG33677, XP_050168 and AAK38117; refs 9–11). Mutations of the gene encoding Nod2 have recently been implicated in two inflammatory diseases, Blau syndrome and Crohn disease26–28. Functional analysis of cryopyrin will help to elucidate the mechanisms regulating apoptosis and inflammation as well as the biochemical basis for cold-sensitive phenotypes.
Methods
Patients
We studied three families with FCAS and one family with MWS. Informed consent was obtained with a protocol approved by the University of California at San Diego institutional review board. Diagnosis was based on a characteristic history of recurrent episodes of rash, arthralgia and fever. Individuals with FCAS reported symptoms primarily after generalized cold exposure, whereas those with MWS did not associate episodes with cold exposure. The two individuals with MWS also had sensorineural hearing loss, a common feature of MWS. Families 1–3 were previously reported in our linkage study5.
Genotyping
We isolated genomic DNA from peripheral blood using kits from PureGene. We carried out PCR and automated fluorescent genotyping using standard procedures as previously described5. All microsatellite markers used were derived from the public databases. Marker order was derived from multiple databases and confirmed by radiation-hybrid mapping.
Analysis of cDNA and identification of coding region
We identified ESTs from the public databases, sequenced these cDNA clones and assembled, extended and confirmed their sequence using SEQUENCHER 3.1 (Gene Codes). We analyzed the genomic sequence from the chromosome 1q44 region in the public database for coding regions using BLAST homology searching with the EST/cDNA sequences and the GENSCAN gene prediction program. We carried out 5′ and 3′ RACE with a cDNA from peripheral blood leukocytes using a Marathon ready-cDNA kit (Clontech) and the primers 5′–TTGTGACACAGAGGAGCCTG–3′ and 5′–CCTCGTTCTCCTGAATCAGAC–3′ according to the manufacturer’s instructions. We cloned the resulting PCR products using a TOPO TA cloning kit (Invitrogen) and sequenced them using the primers derived from CIAS1 coding sequence.
Genomic DNA amplification
We designed primers for PCR amplification with PRIMER III (Whitehead Institute), using exons with flanking intronic sequence. We amplified exons from genomic DNA in 20-µl PCR reactions using AmpliTaq DNA polymerase (0.8 U), dNTPs (250 µM), MgCl (2.5 mM), PE Buffer II (1×), primers (500 nM) and DNA (20–80 ng) using reagents from PE Applied Biosystems. PCR conditions were as follows: initial denaturation at 94 °C for 4 min followed by 10 cycles of 94 °C for 30 s, touchdown annealing (1 °C decrease per cycle) between 65 °C and 55 °C for 30 s, and extension at 74 °C for 1 min. For the remaining 25 cycles, we used an annealing temperature of 55 °C and finished with a final extension cycle at 72 °C for 7 min. The CIAS1 exon amplification primers are listed in Web Table A.
DNA sequencing and mutation detection
We amplified 90 genomic fragments containing more than 80 exons from the chromosome 1q44 region in four affected individuals from different families and one control individual. We purified PCR products and carried out automated sequencing as previously described29. We sequenced both the forward and reverse strands, and carried out data assembly and analysis using SEQUENCHER 3.1. For analysis of normal controls, we sequenced exon 3 of the cryopyrin gene from unaffected spouses from the families and over 100 normal blood-bank control DNA samples. We sequenced BACs and cDNA clones using standard protocols.
Northern-blot analysis
We radioactively labeled 363-bp, 1058-bp and 618-bp PCR fragments of the cryopyrin gene corresponding to nucleotides –87–267, 1093–2150 and 2353–2970 of the initial mRNA splice variant, respectively, and a β-actin control. We used these as hybridization probes for the analysis of two multiple-tissue northern blots (Blot H and H4, Clontech). We labeled the probes with [α32P]dCTP using the PrimeIt Kit (Stratagene) and purified them with a G-50 Sephadex column (Boehringer Mannheim). We then hybridized the filters for 1 h at 68 °C in ExpressHyb solution (Clontech), washed them at 50–65 °C according to the manufacturer’s instructions and analyzed them by autoradiography.
Protein prediction programs
We used InterPro (www.ebi.ac.uk/interpro), ProDom (prodes.toulouse.inra.fr/prodom/doc/prodom.html) and Predict-Protein (dodo.bioc.columbia.edu/predictprotein/predictprotein.html) to analyze protein structure and domains.
GenBank accession numbers
The sequences determined as part of this study have been submitted to GenBank and are available under accession numbers AF410477, AY051112, AY051113, AY051114, AY051115, AY051116, AY051117, AY056059, AY056060 and AF427617.
Supplementary Material
Acknowledgments
We are extremely grateful to all of the family members who participated in this research effort. We are indebted to R. Townley, R. Simon and F. Shih for their referral of patients. We thank K. Woods, M. Tressierras, D. Lightfoot and M. Kane for technical assistance, J. Weger and J. Sansone for technical assistance with genotyping and sequencing, B. Stevenson and V. Jongeneel for database sequence analysis, B. Riegle for computer support, and S. Soefje for editorial support. We also thank M. McDermott, B. Ross, W. Cavenee, R. White, R. Das Gupta, W. Biggs, C. Putnam, F. Wright and G. Hampton for their helpful advice. The research was carried out with the support of National Institute of Health (NIAID 5-K08 AI01508), the American Academy of Allergy, Asthma, and Immunology Education and Research Trust Faculty Development Award, the Arthritis Foundation San Diego Chapter Research Award and the Ludwig Institute of Cancer Research.
Footnotes
Note: Supplementary information is available on the Nature Genetics web site (http://genetics.nature.com/supplementary_info/).
References
- 1.Kile RL, Rusk HA. A case of cold urticaria with an unusual family history. JAMA. 1940;114:1067–1068. [Google Scholar]
- 2.Tindall JP, Beeker SK, Rosse WF. Familial cold urticaria. A generalized reaction involving leukocytosis. Arch. Intern. Med. 1969;124:129–134. [PubMed] [Google Scholar]
- 3.Zip CM, et al. Familial cold urticaria. Clin. Exp. Dermatol. 1993;18:338–341. doi: 10.1111/j.1365-2230.1993.tb02212.x. [DOI] [PubMed] [Google Scholar]
- 4.Ormerod AD, Smart L, Reid TM, Milford-Ward A. Familial cold urticaria. Investigation of a family and response to stanozolol. Arch. Dermatol. 1993;129:343–346. doi: 10.1001/archderm.129.3.343. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman HM, Wright FA, Broide DH, Wanderer AA, Kolodner RD. Identification of a locus on chromosome 1q44 for familial cold urticaria. Am. J. Hum. Genet. 2000;66:1693–1698. doi: 10.1086/302874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung M, et al. A locus for familial cold urticaria maps to distal chromosome1q: familial cold urticaria and Muckle–Wells syndrome are probably allelic. Am. J. Hum. Genet. 1996;59:A223. [Google Scholar]
- 7.Muckle TJ. The ‘Muckle–Wells’ syndrome. Br. J. Dermatol. 1979;100:87–92. doi: 10.1111/j.1365-2133.1979.tb03572.x. [DOI] [PubMed] [Google Scholar]
- 8.Cuisset L, et al. Genetic linkage of the Muckle–Wells syndrome to chromosome 1q44. Am. J. Hum. Genet. 1999;65:1054–1059. doi: 10.1086/302589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinon F, Hofmann K, Tschopp J. The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 2001;11:R118–R120. doi: 10.1016/s0960-9822(01)00056-2. [DOI] [PubMed] [Google Scholar]
- 10.Bertin J, DiStefano PS. The PYRIN domain: a novel motif found in apoptosis and inflammation proteins. Cell Death Differ. 2000;7:1273–1274. doi: 10.1038/sj.cdd.4400774. [DOI] [PubMed] [Google Scholar]
- 11.Pawlowski K, Pio F, Chu Z, Reed JC, Godzik A. PAAD—a new protein domain associated with apoptosis, cancer and autoimmune diseases. Trends Biochem. Sci. 2001;26:85–87. doi: 10.1016/s0968-0004(00)01729-1. [DOI] [PubMed] [Google Scholar]
- 12.Koonin EV, Aravind L. The NACHT family—a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 13.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 14.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.International FMF Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman HM, Wanderer AA, Broide DH. Familial cold autoinflammatory syndrome: phenotype and genotype of an autosomal dominant periodic fever. J. Allergy Clin. Immunol. doi: 10.1067/mai.2001.118790. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDermott MF, et al. An autosomal dominant periodic fever associated with AA amyloidosis in a north Indian family maps to distal chromosome 1q. Arthritis Rheum. 2000;43:2034–2040. doi: 10.1002/1529-0131(200009)43:9<2034::AID-ANR14>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 18.Houten SM, et al. Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nature Genet. 1999;22:175–177. doi: 10.1038/9691. [DOI] [PubMed] [Google Scholar]
- 19.Drenth JP, et al. Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome. International Hyper-IgD Study Group. Nature Genet. 1999;22:178–181. doi: 10.1038/9696. [DOI] [PubMed] [Google Scholar]
- 20.McDermott MF, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97:133–144. doi: 10.1016/s0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 21.French FMF Consortium. A candidate gene for familial Mediterranean fever. Nature Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 22.Centola M, et al. The gene for familial Mediterranean fever MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood. 2000;95:3223–3231. [PubMed] [Google Scholar]
- 23.Mansfield E, et al. The familial Mediterranean fever protein, pyrin, associates with microtubules and co-localizes with actin filaments. Blood. 2001;98:851–859. doi: 10.1182/blood.v98.3.851. [DOI] [PubMed] [Google Scholar]
- 24.Tidow N, et al. Hematopoietic-specific expression of MEFV, the gene mutated in familial Mediterranean fever, and subcellular localization of its corresponding protein, pyrin. Blood. 2000;95:1451–1455. [PubMed] [Google Scholar]
- 25.Martin SJ. Dealing the CARDs between life and death. Trends Cell Biol. 2001;11:188–189. doi: 10.1016/s0962-8924(01)01971-7. [DOI] [PubMed] [Google Scholar]
- 26.Miceli-Richard C, et al. CARD15 mutations in Blau syndrome. Nature Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 28.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 29.Kolodner RD, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.