Yang et al. provide new mechanistic insight into the role of kinase PDK1 in early NK cell development. Genetic deletion of PDK1 in mice led to a severe loss of NK cells and impaired antitumor activity in vivo. Their results indicate that PDK1 acts as a regulator of IL-15 signaling to induce expression of the circadian protein E4BP4, also modulating CD122 upregulation.
Abstract
E4BP4, a circadian protein, is indispensable for NK cell development. It remains largely unknown which signal is required to induce E4BP4 expression and what effects it has during NK cell differentiation. Here, we reveal that PDK1, a kinase upstream of mTOR, connects IL-15 signaling to E4BP4. Early deletion of PDK1 caused a severe loss of NK cells and compromised antitumor activity in vivo. PDK1-deficient NK cells displayed much weaker IL-15–induced mTOR activation and E4BP4 induction, as well as remarkable reduction in CD122, a receptor subunit specifying NK cell responsiveness to IL-15. The phenotypes were partially reversible by ectopic expression of E4BP4 or bypassed activation of mTOR. We also determined that PDK1-mediated metabolic signaling was dispensable for NK cell terminal maturation and survival. Thus, we identify a role for PDK1 signaling as a key mediator in regulating E4BP4 expression during early NK cell development. Our findings underscore the importance of IL-15 self-responsiveness through a positive feedback loop that involves PDK1–mTOR–E4BP4–CD122 signaling.
IL-15–IL-15 receptor signaling is considered a critical rate-limiting step for NK cell development (DiSanto et al., 1995; Suzuki et al., 1997; Vosshenrich et al., 2005). NK cell commitment is characterized by the expression of CD122, the receptor subunit that confers IL-15 responsiveness. Once they are committed, NK cells require sustained IL-15 signaling for subsequent early differentiation. Although the basal level of CD122 is sufficient for IL-2 signaling in T cells, NK cells require enhanced CD122 expression for responsiveness to IL-15 (Intlekofer et al., 2005). Mice lacking IL-15 or IL-15Rα selectively lose CD122high lineage cells, including NK cells, NK-T cells, and memory-phenotype CD8+ T cells. Significant advances have been made in deciphering the mechanisms by which NK cells preserve elevated levels of CD122. Unique roles have been identified for T-bet and Eomes, two transcription factors critical for NK cell development, in binding the promoter of Il2rb, the gene encoding CD122, and inducing enhanced expression of CD122 on NK cells (Intlekofer et al., 2005). Recently, nuclear factor, interleukin 3, regulated (NFIL3, also known as E4BP4) was demonstrated to bind to the Eomes promoter and to regulate the earliest stages of NK cell development (Male et al., 2014). Mice lacking E4BP4 exhibit a severe defect in early NK cell development (Gascoyne et al., 2009; Kamizono et al., 2009). Nevertheless, how E4BP4 regulates NK cell development is controversial. An earlier study from the same group revealed that E4BP4 plays a role in IL-15 signaling as well (Gascoyne et al., 2009). Despite this, it remains largely unknown which signal is required to induce E4BP4 expression in NK cells and what effects IL-15–induced E4BP4 has during NK cell differentiation.
As a circadian clock gene, E4BP4 expression is dynamic (Doi et al., 2004; Male et al., 2012). In mice, feeding can quickly induce the up-regulation of E4BP4 expression, whereas inhibition of insulin signaling can abolish this activity (Tong et al., 2010). These data raise the possibility that E4BP4 induction in NK cells relies on metabolic signaling, which may be required for NK cell development. The mammalian target of rapamycin (mTOR) is the central checkpoint molecule in the regulation of cell metabolism. mTOR senses and integrates diverse environmental cues, including nutrients and growth factors (Powell et al., 2012; Waickman and Powell, 2012), and exists in two complexes: mTOR complex 1 (mTORC1) and mTORC2. The well-established molecular function of mTORC1 is the initiation of protein translation by phosphorylating p70 S6 kinase (S6K) and the translation-initiating, eIF4E-binding protein (4EBP1). The intimate interaction between metabolism and immunity has attracted much attention (Chi, 2012; Powell et al., 2012; Waickman and Powell, 2012). Most of the metabolic control over cell fate is focused on the activation of adaptive immune cells, such as T cells (Kim et al., 2013; Zeng et al., 2013; Wu et al., 2014). In contrast, the function of mTOR signaling in the development of lymphocytes, particularly NK cells, is rarely reported. Recently, NK cell–specific deletion of mTOR revealed its critical, nonredundant role in the regulation of two key checkpoints in NK cell biology, proliferation in the bone marrow, and activation in the periphery (Marçais et al., 2014).
The PI3K pathway is a major upstream regulator of mTOR-dependent metabolic activation and plays a critical role in cell proliferation and differentiation. Mice simultaneously lacking the PI3K subunits P110 γ and δ exhibit a severe defect in early NK cell development (Tassi et al., 2007; Guo et al., 2008). Similarly, NK cell differentiation is also retarded in mice lacking the PI3K subunit p85 (Awasthi et al., 2008). 3′-phosphoinositide–dependent kinase 1 (PDK1) has been considered a critical metabolic regulator connecting PI3K and downstream mTOR activation (Finlay et al., 2012). An important role for PDK1 is to phosphorylate the T308 site of AKT and synergize with mTORC2 to fully activate downstream AKT. In the immune system, PDK1 has been shown to be critical for the development of T and B cells (Hinton et al., 2004; Park et al., 2013; Venigalla et al., 2013; Baracho et al., 2014). However, the role of PDK1 in NK cell development has not been directly addressed. Loss of NK cells in mice lacking PI3K activity could imply a role for PDK1 in NK cell development. However, PI3K signaling can also result in the activation of phospholipase C γ and the Vav family independent of the PDK1–Akt–mTOR pathway (Tassi et al., 2005, 2008; Graham et al., 2006).
To address whether induction of E4BP4 in NK cells is regulated by metabolic signaling and a potential role for PDK1 in NK cell development, we generated conditional PDK1-deficient mice to block the connection between PI3K and mTOR signaling. We found that PDK1-dependent metabolic signaling functions as a key regulator for IL-15 signaling to stimulate E4BP4 expression, which in turn regulates CD122 expression and confers IL-15 responsiveness.
RESULTS
PDK1–mTOR signaling is required for IL-15–induced E4BP4 expression in vitro
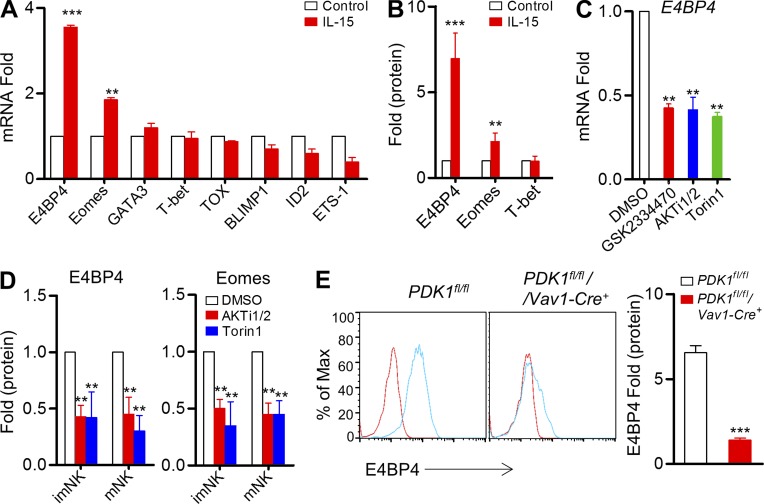
E4BP4 was reported to be the most specific transcription factor required for the NK cell lineage development (Gascoyne et al., 2009; Kamizono et al., 2009), but how it is regulated remains unknown. A real-time PCR assay revealed that IL-15 could preferentially and significantly up-regulate mRNAs encoding E4BP4 and Eomes (Fig. 1 A). To evaluate the E4BP4 protein level, we developed an intracellular staining assay to quantify its dynamic changes before and after IL-15 stimulation. To our surprise, E4BP4 was rarely detectable in resting NK cells. Upon IL-15 triggering, E4BP4 exhibited a more than fivefold increase in NK cells, and Eomes, which is considered to be involved downstream of E4BP4 (Male et al., 2014), exhibited slight up-regulation (Fig. 1 B). However, T-bet expression was not enhanced by IL-15 stimulation (Fig. 1 B).
Figure 1.
PDK1-mTOR signaling regulates IL-15–induced E4BP4 expression in vitro. (A) Quantitative RT-PCR analysis of NK cell–related genes in sorted wild-type CD3−NK1.1+ cells before and after stimulation with IL-15–IL-15R complexes; the results were normalized to those of β-actin and are presented relative to those of untreated cells, set as 1. Data were pooled from three independent experiments. **, P < 0.005; ***, P < 0.0005 versus control. (B) Intracellular staining was used to analyze the expression of E4BP4, Eomes, and T-bet on gated CD3−NK1.1+ cells by flow cytometry before and after stimulation with IL-15–IL-15R complexes; the results were normalized and are presented relative to MFI of untreated cells, set as 1. Data were pooled from four independent experiments. **, P < 0.005; ***, P < 0.0005 versus control. (C) Quantitative RT-PCR analysis of E4bp4 in sorted wild-type CD3−NK1.1+ cells after stimulation with IL-15–IL-15R complexes in the presence of the indicated inhibitors or of DMSO (control); the results were normalized to those of β-actin and are presented relative to those of DMSO-treated cells, set as 1. Data represent the mean ± SEM (3 repeats). **, P < 0.005 versus DMSO. (D) Intracellular staining was used to analyze the expression of E4BP4 and Eomes on gated CD3−NK1.1+ cells by flow cytometry after stimulation with IL-15–IL-15R complexes plus indicated inhibitors; the results were normalized and are presented relative to those of DMSO-treated cells, set as 1. Data represent the mean ± SEM (3 repeats). **, P < 0.005 versus DMSO. (E) Intracellular staining was used to analyze the expression of E4BP4 in CD3−CD122high NK1.1+ NK cells by flow cytometry from indicated mice before and after stimulated with IL-15–IL-15R complexes. Representative overlaid histogram is shown (left). Red lines, unstimulated; Blue lines, stimulated. Results are presented relative to MFI values before treatment. Data were pooled from two independent experiments (right, n = 4). ***, P < 0.0005.
To examine whether the induction of E4BP4 by IL-15 requires metabolic signaling, several pharmacological inhibitors of varying specificity, including GSK2334470, AKTi1/2, and Torin1, were chosen to suppress PDK1, AKT, and mTOR, respectively. As expected, pharmacological inhibition of their activities led to a twofold decrease in IL-15–triggered up-regulation of E4BP4 at the transcriptional and translational levels, (Fig. 1, C and D). To genetically confirm this finding, we generated hematopoietic cell–specific PDK1-deficient mice, PDK1fl/fl/Vav1-Cre+ (hereafter, referred to as PDK1−/−), and then detected E4BP4 expression in the splenic NK cells. As expected, PDK1−/− NK cells failed to significantly up-regulate E4BP4 expression upon IL-15 stimulation. These data indicate that PDK1-mediated metabolic signaling is indispensable for E4BP4 induction by IL-15.
PDK1 is intrinsically required for early NK cell development in vivo
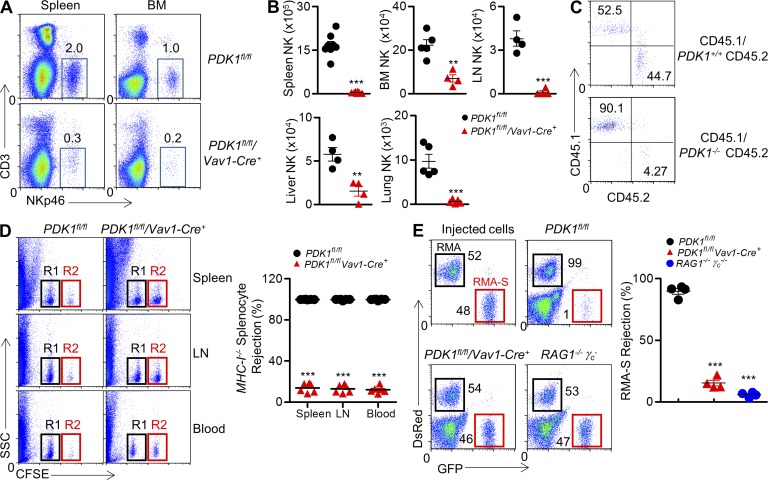
To identify a potential role for PDK1-medated signaling in NK cell physiology, we extensively analyzed NK cell development and function in PDK1−/− mice. Compared with PDK1-sufficient mice, PDK1−/− mice exhibited a nearly 95% reduction in the number of NK cells in the spleen and bone marrow (Fig. 2, A and B). The remarkable reduction in NK cell pool could also be found in other lymphoid organs, including the lymph nodes, liver, and lungs (Fig. 2 B). To investigate the cell-intrinsic effect of PDK1 for NK cell development, bone marrow mixtures were adoptively transferred into sublethally irradiated immunodeficient RAG1−/−γc− mice. In contrast to CD45.2+ WT, PDK1−/− bone marrow cells failed to reconstitute NK cell pool efficiently (Fig. 2 C), suggesting the critical requirement for PDK1 in NK cell development is cell intrinsic.
Figure 2.
Severely reduced NK cellularity and loss of NK cell activity in PDK1-deficient mice. (A) CD3−NKp46+ NK cells were isolated from spleen and bone marrow, and percentages are indicated in representative flow cytometric plots. (B) Absolute numbers of CD3−NKp46+ NK cells are also indicated in tissues and organs from PDK1fl/fl and PDK1fl/fl/Vav1-Cre+ mice. Each symbol represents an individual mouse; small horizontal lines indicate the mean. Data were pooled from two individual experiments (n = 4 or more). **, P < 0.005; ***, P < 0.0005. (C) Mice were treated with 5-FU before bone marrow was isolated and depleted of B220+ and CD3+ and NK1.1+ cells. Depleted bone marrow from CD45.1 WT mice was mixed with either WT or PDK1−/− bone marrow cells expressing CD45.2 at a 1:1 ratio. Cells were then injected into sublethally irradiated RAG1−/−γc− recipient mice. CD45.1 versus CD45.2 expression on gated CD3−CD122+NKp46+ was detected by flow cytometry. The numbers show the percentages in relevant quadrant. (D, left) Representative flow cytometry of CFSE+ cells obtained from spleen, LN, or blood of the indicated recipient mice 18 h after injection with an equal number of WT or β2-microglobulin (β2M)–deficient splenocytes labeled with various concentrations of the cytosolic dye CFSE. R1, CFSElow splenocytes from WT mice; R2, CFSEhigh splenocytes from β2M-deficient mice. (D, right) Quantification of the percent rejection of β2M-deficient splenocytes. Each symbol represents an individual mouse; small horizontal lines indicate the mean (n = 6 mice per group). (E) Representative flow cytometry plot (left) and quantification of the percentages (right, percent rejection) of RMA-S cells in the peritoneal cavity on 18 h after intraperitoneal injection in the indicated mice. A mixture of NK cell–sensitive RMA-S cells expressing green fluorescent protein (GFP; outlined in red) together with NK cell-resistant RMA cells expressing the fluorescent protein DsRed (outlined in black) were injected. The numbers near square boxes represent percentages of RMA-S or RMA relative to the total number of injected cells. Immunocompromised RAG1−/−γc− mice were used as a control. Injected cells are shown. Each symbol represents an individual mouse; small horizontal lines indicate the mean (n = 4 mice per group). Data are representative of two independent experiments. ***, P < 0.0005.
To confirm the diminished NK cell population in PDK1−/− mice, the ability to eliminate “missing-self” MHC-I−/− splenocytes was measured in vivo as previously described (Dong et al., 2009, 2012). In the model, mismatched MHC-I−/− splenocytes were preferentially killed by control mice due to NK cell activity. In contrast, minimal NK cell activity was detectable in PDK1−/− mice (Fig. 2 D). We also observed that in vivo NK cytotoxicity against hematopoietic tumor RMA-S cells was minimal in PDK1−/− mice as well, nearly comparable with NK cell–lacking RAG1−/−γc− mice (Fig. 2 E). Therefore, PDK1−/− mice effectively lack peripheral NK cells and innate cytotoxic activity.
PDK1 maintains CD122 expression and regulates NK-cell responsiveness to IL-15
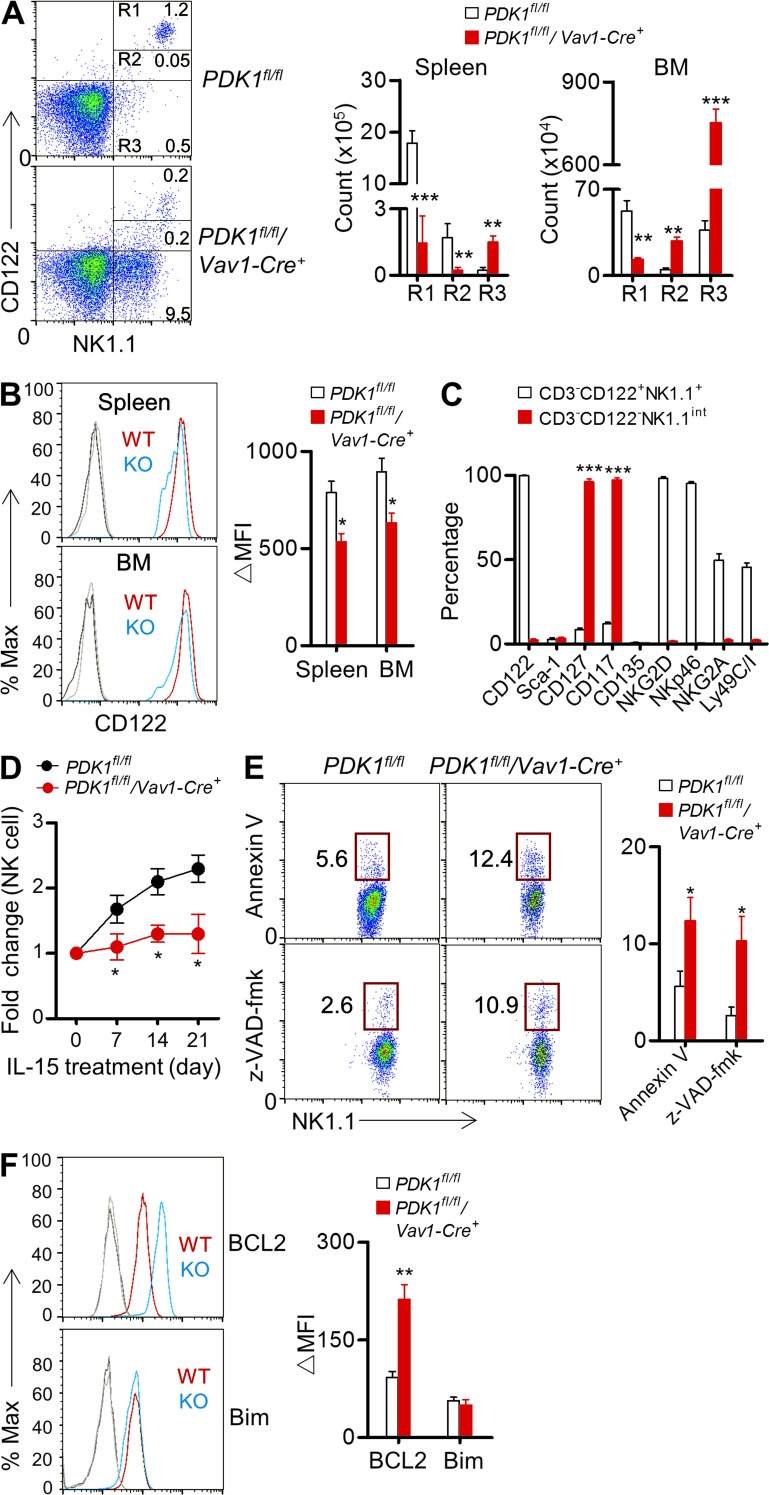
Because PDK1-deficient NK cells were resistant to IL-15–induced E4BP4 up-regulation, the impaired NK cell development probably resulted from the compromised IL-15 signaling. We initially detected IL-15–dependent lineage cells in PDK1−/− mice. The absolute numbers of NK cells (CD3−NK1.1+), NK-T cells (CD3+NK1.1int), and memory CD8+ T cells (CD8+CD122+) were remarkably diminished in PDK1−/− mice (unpublished data). To further clarify this effect, we analyzed CD122 expression on CD3−NK1.1+ cells. PDK1−/− mice exhibited dramatically fewer CD3− CD122 high NK1.1+ cells (R1) in the detected lymphoid organs, like bone marrow, spleen, lung, and liver (Fig. 3 A and not depicted). Additionally, the CD122 intensity on PDK1−/− CD3−NK1.1+ cells was significantly lower and had a tendency to be gradually attenuated (Fig. 3, A and B). Surprisingly, there was a noticeable population of CD122-negative NK cells, which exhibited the distinct phenotype of intermediate expression of NK1.1, called NK1.1int. These cells accumulated mainly in the bone marrow rather than other organs (data not shown), of PDK1−/− mice, whereas the population was rarely seen in WT mice (Fig. 3 A). To further characterize this population, we confirmed that the cells expressed high levels of CD117 and CD127, the two earliest markers for NK cells (Fig. 3 C), and were not reactive for CD1d tetramer binding (unpublished data); this excluded the possibility of the cells being NK-T cells, which also exhibit NK1.1int. The population was negative for NKp46, a marker for type I group of innate lymphoid cell (ILC1; Fig. 3 C). Collectively, these data indicate that PDK1 is essential for preserving a CD122high state during early NK cell development.
Figure 3.
PDK1 regulates CD122 expression and IL-15 responsiveness in vivo. (A) Representative flow cytometry profile of CD122 versus NK1.1 expression on CD3− bone marrow cells. Three populations, CD122highNK1.1+ (R1), CD122intNK1.1int (R2), and CD122−NK1.1int (R3), are outlined (left). Absolute numbers of indicated NK cell populations from spleen and bone marrow are also shown (right). Data were pooled from two independent experiments (n = 6). **, P < 0.005, ***, P < 0.0005. (B). Representative overlaid histograms demonstrating CD122 expression on gated R1 NK cells in the spleens and BM of PDK1fl/fl (WT) and PDK1fl/fl/Vav1-Cre+ (KO) mice (left); the absolute MFI (ΔMFI) were quantified (right). Data were pooled from two independent experiments (n = 6). *, P < 0.05. (C). Profiling developmental markers on BM CD3−CD122+NK1.1+ cells from PDK1fl/fl mice and BM CD3−CD122−NK1.1int NK cells in PDK1fl/fl/ Vav1-Cre+ mice. n = 5 per group. ***, P < 0.0005. (D). PDK1fl/fl or PDK1fl/fl/Vav1-Cre+ mice were injected with IL-15–IL-15R complexes every 3 d. The absolute number of peripheral blood CD3−NKp46+NK cells was monitored on the indicated days. Fold change was calculated simply as the ratio of the NK cell number at each time point after IL-15–IL-15R treatment to the initial NK cell number in untreated mice (day 0). Data represent the mean ± SEM of 3 mice per time point and are representative of two independent experiments. *, P < 0.05. (E) Representative flow cytometry plot showing Annexin V staining and Caspase activity in naive NK cells from the indicated mice (left). Numbers adjacent to the outlined areas (left) indicate the percentage of Annexin V– or caspase-positive cells. Quantifications were performed from two independent experiments (right, n = 5). *, P < 0.05. (F). Intracellular staining of Bcl2 and Bim in naive NK cells from the indicated mice (left). Quantifications were performed from two independent experiments (right, n = 5). **, P < 0.005.
NK cells need high status of CD122 to ensure IL-15 responsiveness. To detect the in vivo IL-15 responsiveness of PDK1−/− NK cells, mice were intraperitoneally injected with recombinant IL-15–IL-15R complexes. Peripheral blood NK cells underwent a greater than twofold expansion in PDK1-sufficient mice but not in PDK1−/− mice (Fig. 3 D). Therefore, PDK1−/− mice exhibit hyporesponsiveness to IL-15 stimulation in vivo.
Reduced or failed expression of CD122 likely causes NK cell death. We found that PDK1−/− mice indeed exhibited about twofold more Annexin V+ and Caspase3+ NK cells in the spleen (Fig. 3 E). Thus, increased NK cell apoptosis likely contributes to the compromised NK cell development in PDK1−/− mice. In parallel, we detected survival-related intracellular molecules, including antiapoptotic Bcl-2 and proapoptotic Bim, both of them were reported to be involved in NK cell survival or death (Huntington et al., 2007). Intriguingly, NK cells from PDK1−/− mice did not exhibit more Bim and less Bcl-2 expression as we expected; in contrast, Bcl2 were really up-regulated in PDK1−/− NK cells (Fig. 3 F), suggesting that the PDK1-deficient NK cells maybe undergo cell cycle arrest like B cells to a great extent, rather than apoptosis (Venigalla et al., 2013).
Ectopic expression of E4BP4 rescues NK cell development in PDK1−/− mice
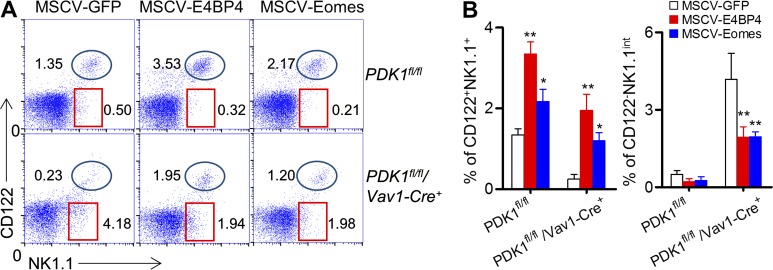
To investigate whether the compromised NK cell development and low expression level of CD122 correlated with less E4BP4 induction by IL-15 in PDK1−/− mice, actively dividing PDK1−/− bone marrow cells with enriched HSCs were infected by retroviruses encoding E4BP4 or Eomes. 6 wk after in vivo differentiation, exogenous expression of E4BP4 gave rise to an increase in the NK cell population in PDK1-intact bone marrow group, as reported elsewhere (Fig. 4, A and B; Gascoyne et al., 2009). Importantly, ectopic expression of E4BP4 in PDK1−/− bone marrow cells could recover the CD122+NK1.1+ NK cell population to a great extent (Fig. 4 A, B). Likewise, exogenous E4BP4 expression heightened the expression level of CD122 on PDK1−/− NK cells and prevented the accumulation of the arrested NK1.1int CD122− NK cells in the PDK1−/− bone marrow. Similar results were also obtained when Eomes was ectopically expressed (Fig. 4, A and B). Therefore, the impaired NK cell development in PDK1-deficient mice is largely caused by the defect in E4BP4 induction by IL-15.
Figure 4.
Ectopic expression of E4BP4 or Eomes rescues PDK1-deficient NK cell development. (A) PDK1fl/fl and PDK1fl/fl/Vav1-Cre+ mice were treated with 5-FU for 4 d, and bone marrow cells were collected for spin infection with MSCV retrovirus encoding control GFP, E4BP4, or Eomes, respectively. The infected BM cells were then transferred into RAG1−/−γc− mice. After 6 wk, CD122 versus NK1.1 expression on gated CD3− cells was analyzed by flow cytometry. The numbers in the outlined areas indicate the percent of CD3− cells. (B) Percentages of CD3−CD122+NK1.1+ and CD3−CD122−NK1.1int were enumerated as shown. Data were pooled from three independent experiments. n = 5–7. *, P < 0.05; **, P < 0.005.
PDK1 regulates IL-15–stimulated metabolic NK cell activation via activating mTOR
PDK1 is a master molecule linking the PI3K pathway with mTOR activation via Akt. To explore whether mTOR-dependent PDK1 signaling is involved in the early NK cell development promoted by IL-15, we stimulated PDK1−/− splenocyte with recombinant IL-15–IL-15R complexes and detected the expression of nutritional receptors and mTOR signaling in CD3−CD122highNK1.1+ cells, which could eliminate the developmental discrepancy in CD122 levels between the two genotypes. We found that after overnight IL-15 stimulation, PDK1-sufficient NK cells displayed a two-to fivefold increase in CD71 and CD98, two nutritional receptors, whereas PDK1−/− NK cells nearly lost the ability to up-regulate these receptors (Fig. 5 A), and these cells consistently displayed impaired activation of AKT-mTOR1 signaling upon IL-15 exposure (Fig. 5 B). To further exclude the possibility that the impaired metabolic activation by IL-15 is caused by the variation in CD122 levels between the genotypes, several pharmacological inhibitors were chosen. Blockage of PI3K–PDK1–mTOR activation largely prevented the IL-15–triggered up-regulation of CD71 and CD98 on bone marrow and splenic NK cells (Fig. 5 C and not depicted), further demonstrating that IL-15 is able to trigger NK cell metabolic activation, which requires mTOR-dependent PI3K signaling. To directly test mTOR signaling in NK cell development, wild-type mice were injected with Torin1 to suppress mTOR activity. As expected, the proproliferative role of IL-15 was notably diminished by mTOR inhibition (Fig. 5 D). Together, these data demonstrate that PDK1 signaling is required for NK cell metabolic activation and proliferation downstream of the IL-15 receptor via activating mTOR.
Figure 5.
IL-15 augments NK cell metabolic activation and proliferation via PI3K–PDK1–mTOR signaling. Splenocytes from PDK1fl/fl or PDK1fl/fl/Vav1-Cre+ mice were stimulated with recombinant IL-15–IL-15R complexes overnight. (A) Expression of CD71 and CD98 was detected by flow cytometry (left) and was quantified as mean fluorescence intensity. The results are presented relative to unstimulated cells, set as 1 (right). Data represent the mean ± SEM of 5 mice per group. **, P < 0.005. (B). Intracellular phosphorylated S6, AKT T308, and AKT S473 were detected by flow cytometry (left), and the MFI was calculated. The protein expression levels in PDK1 fl/fl NK cells are presented relative to PDK1fl/fl/Vav1-Cre+ mice, set as 1 (right). Data represent the mean ± SEM of 3 mice per group. Data are representative of two independent experiments. **, P < 0.005. (C) WT bone marrow cells were stimulated with recombinant IL-15–IL-15R complexes overnight in the presence of the indicated pharmacological inhibitors or DMSO, as a negative control. Flow cytometry was used to detect CD71 and CD98, and quantification is presented as the MFI. Data represent the mean ± SEM of 3 independent experiments. **, P < 0.005, ***, P < 0.0005. (D) WT mice were injected with IL-15–IL-15R complexes every 3 d together with the mTOR inhibitor Torin1 or DMSO. The absolute number of peripheral blood CD3−NKp46+NK cells was quantified on the indicated days. Fold change was calculated as describe in Fig. 3 D. Data represent the mean ± SEM of 3 mice per time point and are representative of two independent experiments. *, P < 0.05.
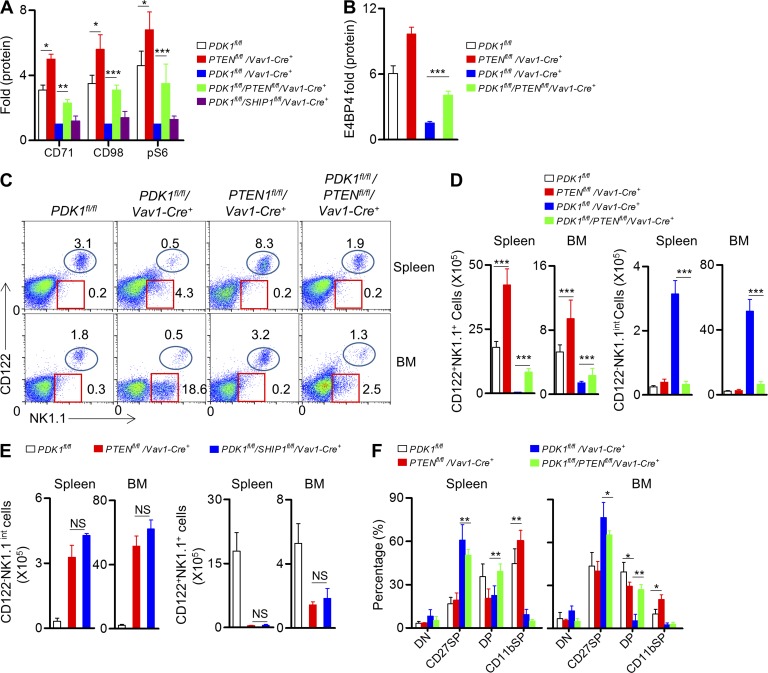
PTEN deletion partially rescues E4BP4 expression and NK cell development in the absence of PDK1
Previous studies have revealed that the deletion of PTEN resulted in enhanced mTOR activation, which consequently promoted more dendritic cells generation (Sathaliyawala et al., 2010). To ascertain whether PTEN deletion could increase NK cell number and rescue the impaired NK cell development in absence of PDK1, PDK1−/− mice were further bred with PTENfl/fl mice to yield PTENfl/fl/PDK1fl/fl/Vav1-Cre+ mice. Similarly, PDK1−/− mice were also bred onto a SHIP1fl/fl background. We first found that deletion of PTEN, but not SHIP1, could notably enhance metabolic activation of NK cell via IL-15 stimulation, even in the absence of PDK1 (Fig. 6 A). Most importantly, PTEN inactivation significantly restored IL-15–triggered E4BP4 expression by PDK1−/− NK cells (Fig. 6 B). To test whether the rescued mTOR activation and increased E4BP4 expression could recover NK cell generation in PTENfl/fl/PDK1fl/fl/Vav1-Cre+ mice, NK cell development was carefully examined (Fig. 3 A). In brief, PTEN deletion could significantly increase the absolute number of NK cells in the spleen and bone marrow (Fig. 6, C and D). Most importantly, the population of CD3−CD122−NK1.1int cells previously observed in PDK1−/− mice was also dramatically reduced (Fig. 6, C and D). In contrast, SHIP1 deletion had little effect on PDK1−/− NK cell development, likely due to its inability to up-regulate mTOR-dependent metabolic signaling (Fig. 6 E).We further evaluated the developmental process of the rescued NK cells in the PTENfl/fl/PDK1fl/fl/Vav1-Cre+mice and found that bypassed activation of mTOR could restore the transitional process through the NK progenitor to CD27+CD11b+ NK cell stage in the absence of PDK1. Nevertheless, this activation failed to complete the terminal maturation of NK cells (Fig. 6 F). Collectively, PDK1 plays a critical role in early NK cell differentiation mainly through activating mTOR signaling, which is critical for E4BP4 induction downstream IL-15 signaling.
Figure 6.
Deletion of PTEN, but not SHIP1, alternatively activates mTOR signaling and rescues the early development of PDK1-deficient NK cells. (A) Splenocytes from the indicated mice were stimulated with recombinant IL-15–IL-15R complexes overnight. Flow cytometry was used to detect CD71, CD98, and intracellular phosphorylated S6 (pS6). The results are presented relative to the expression in unstimulated CD3−CD122high NK1.1+ cells in the respective genotype mice. Data represent the mean ± SEM of 3 mice and are representative of two independent experiments. (B) Intracellular staining was used to analyze the expression of E4BP4 in CD3−CD122high NK1.1+ NK cells from the indicated mice before and after IL-15–IL-15R complexes. Results are presented relative to MFI value before treatment. Data were pooled from two independent experiments (n = 4). ***, P < 0.0005. (C) Expression of CD122 versus NK1.1 on gated CD3− splenocytes and BM cells from the indicated mice was examined by flow cytometry. The numbers outlined indicate the percent of cells in each mouse. (D) The quantification of absolute numbers of CD3−CD122+NK1.1+ (left) and CD3−CD122−NK1.1int NK cells (right) is also presented. Data represent the mean ± SEM of 3 mice and are representative of two independent experiments. ***, P < 0.0005. (E). Expression of CD122 versus NK1.1 on gated CD3−splenocytes and BM cells from the indicated mice was examined by flow cytometry. Absolute numbers of CD3−CD122−NK1.1int (left) and CD3−CD122+NK1.1+NK (right) cells are presented. Data were pooled from two independent experiments (n = 6). NS, not significant. (F) Expression of CD27 or CD11b on CD3−NKp46+ splenocytes and BM cells from the indicated mice was analyzed by flow cytometry. The representative profiles represent the four-stage development of NK cells, including CD27−CD11b−(DN), CD27+CD11b−(CD27SP), CD27+CD11b+(DP), and CD27−CD11b+(CD11b SP). The results are presented as the percentage of cells. Data represent the mean ± SEM of 3 mice and are representative of two independent experiments. *, P < 0.05, **, P < 0.005.
PDK1 signaling is not required for terminal NK cell maturation and survival
To accurately understand at which stage PDK1 signaling is required for NK cell development. We extensively analyzed NK cell receptor profile, and revealed that a population of NK1.1+NKp46− cells accumulated in the bone marrow of PDK1-deficient mice, indicating the critical role of PDK1 for relatively early NK cell development (Fig. 7 A). Then, we further investigated whether deletion of PDK1 at a late stage of NK cell differentiation could affect NK cell terminal maturation or survival. Ncr1-Cre mice have previously been used for NK cell evaluation by two other groups (Walzer et al., 2007; Eckelhart et al., 2011). We regenerated the mice with Cre expression under the control of the mouse Ncr1 promoter and established a stable line with Cre expression. To characterize this new line, Ncr1-Cre mice were intercrossed with Rosa26 DTASTOP mice. The splenic NK cell number was 75% reduced in Rosa26 DTASTOP/Ncr1-Cre+ mice (Fig. 7, B and C), and the remaining NK cells were mostly immature CD3−NK1.1+CD27−CD11b− (Fig. 7, B and C). Therefore, the newly generated Ncr1-Cre mice will be a useful tool for gene deletion at a late stage of NK cell differentiation.
Figure 7.
PDK1 signaling is dispensable for NK cell terminal maturation and survival. (A) Representative flow cytometric profiles of NK1.1 versus NKp46 expression in splenic and BM CD3− cells from the indicated mice. Numbers show the percentages in square boxes among gated CD3− cells. (B and C) Flow cytometry analysis of NK1.1 versus CD122 expression in splenic CD3− cells in Rosa26 DTASTOP and Rosa26 DTASTOP/Ncr1-Cre+mice (left). Expression of CD27 versus CD11b on CD3−NK1.1+CD122+ NK cells was further analyzed (right). The numbers indicate the percentages of cells in each quadrant. (C) Absolute number of splenic CD3−CD122+NK1.1+ cells (left) and four-stage differentiating NK cell subsets (left), including DN, CD27SP, DP, and CD11bSP, were quantified in Rosa26 DTASTOP and Rosa26 DTASTOP/Ncr1-Cre+ mice. n = 4. ***, P < 0.0005. (D) Flow cytometry analysis of NK1.1 versus CD122 expression in CD3-negative splenic and BM cells of PDK1fl/fl and PDK1fl/fl/Ncr1-Cre+ mice. Numbers show the percentages in indicated circles among gated CD3− cells. (E) The absolute numbers of NK cells (CD3−NKp46+) in the spleens and BM of PDK1fl/fl and PDK1fl/fl/Ncr1-Cre+ mice were also quantified. Data were pooled from two independent experiments (n = 5–7). NS, not significant. (F) Representative flow cytometric profiles of the four-stage NK cell development, including CD27−CD11b−(DN), CD27+CD11b−(CD27SP), CD27+CD11b+(DP), and CD27−CD11b+(CD11b SP), from gated CD3−NK1.1+ NK cells in the spleens and BM of PDK1fl/fl and PDK1fl/fl/Ncr1-Cre+ mice are shown. The numbers indicate the percentages of cells in each quadrant.
Next, our Ncr1-Cre mice were bred with PDK1fl/fl mice. Terminal deletion of PDK1 did not obviously impair the NK cell percentage or number either in the spleen or the bone marrow (Fig. 7, D and E). The NK cells in the PDK1fl/fl/Ncr1-Cre+ mice exhibited a normal profile of NK cell receptor expression (unpublished data), and the terminal transition from iNK to mNK was unchanged (Fig. 7 F). These data apparently suggest that PDK1 signaling is not required for terminal NK cell differentiation and maintenance of NK cell survival.
DISCUSSION
In this study, we used two different genetic approaches to disconnect PI3Ks with mTOR signaling at the early (Vav1-Cre) or late (Ncr1-Cre) stage of NK cell development. We revealed that the PDK1-mediated metabolic signaling works as a fundamental switch downstream of IL-15R signaling on early NK cells to induce the expression of E4BP4 that maintain IL-15 responsiveness via preserving the high level of CD122. Our study identifies a positive feedback loop via PDK1–mTOR–E4BP4–Eomes–CD122, a mechanism involving NK cell development.
NK cells develop from hematopoietic stem cells through multiple stages. The central issues are how NK cells become committed to be responsive to IL-15, how NK cells maintain IL-15 responsiveness during their differentiation. Previous study has reported that Eomes could bind CD122 promoter region, and Eomes deficiency caused significantly low CD122 expression on IL-15–dependent cell lineages, like NK cells and memory CD8+ T cells (Intlekofer et al., 2005). Recent studies highlighted the critical role of E4BP4 as upstream of Eomes, so that E4BP4 deficiency caused severe defects in NK cell development (Kamizono et al., 2009; Male et al., 2014). We reported here a novel role for IL-15 in regulating self-responsiveness via the induction of CD122 in a PDK1-dependent manner. PDK1-deficient NK cells exhibited reduced CD122 expression and were hyporesponsive to IL-15 stimulation in vivo. Ectopic expression of E4BP4 or Eomes could largely rescue CD122 expression and NK cell generation. Therefore, PDK1 signaling is critical for NK cell development via induction of E4BP4 and Eomes. However, we also observed distinct phenotypes in mice lacking either E4BP4 or PDK1. E4BP4 deficiency displayed more severe phenotype and did not result in a population of NK cells without CD122. This discrepancy between the two genotypes may be explained by recent findings that E4BP4 may also be involved in the earliest NK cell commitment at the preNK stage (Male et al., 2014). Thus, E4BP4 may have dual activity, establishing NK cell commitment by inducing CD122 expression and maintaining IL-15 responsiveness by preserving the CD122high status during early NK cell development. The PDK1–mTOR–E4BP4 axis is only required for early differentiation after NK cell commitment through maintaining IL-15 responsiveness, whereas E4BP4 has an additional role in establishing NK cell commitment in a PDK1–mTOR-independent manner. Additionally, Firth et al. (2013) showed that late-stage deletion of E4BP4 did not affect NK cell differentiation. Here, we also observed normal NK cell development in the absence of PDK1 at the terminal stages, indicative of the dispensable role of PDK1-mediated E4BP4 induction for NK cell terminal maturation and survival.
In addition to the essential role of E4BP4 in conventional NK cell development, more recent studies have revealed that innate lymphoid cells (ILCs) also need E4BP4 for their generation (Geiger et al., 2014; Seillet et al., 2014), and nonclassical tissue-resident NK cells in the liver, thymus, and salivary glands are developmentally E4BP4-independent (Cortez et al., 2014; Crotta et al., 2014; Sojka et al., 2014). In PDK1-deficient mice, there is a noticeable population with distinct phenotype, CD3−CD122−CD117+CD127+NK1.1int, which only accumulates in the bone marrow. We assume that the population belongs to NK cell lineage, but wonder how these cells maintain survival in the absence of IL-15 signaling. Although we ruled out the possibility of these cells being NKT cell and ILC1, it remains to be determined whether this unusual population is ILC progenitor or other ILCs, and whether PDK1-mediated metabolic signaling is required for the development of E4BP4-dependent ILCs.
The prevailing working mechanism of IL-15 is to activate the JAK-STAT family downstream of IL-15 β and γ, accounting for the fact that humans or mice that lack STAT5 have impaired NK cell development (Imada et al., 1998; Eckelhart et al., 2011). IL-15 also has the ability to biochemically activate PI3K pathways (Nandagopal et al., 2014). In this study, pharmacological or genetic inactivation of PDK1 activity could not completely prevent E4BP4 induction in NK cells by IL-15 triggering, suggesting that other signaling, particularly JAK-STAT pathway, is probably also involved in the induction of E4BP4 by IL-15, though PI3K-mediated metabolic signaling is the major regulator of E4BP4 expression.
Mice simultaneously lacking PI3K subunit P110 γ and δ had severe defect in early NK cell development (Tassi et al., 2007; Guo et al., 2008). We observed that PDK1 deficiency caused an almost 95% reduction in NK cells, comparable to p110 γ and δ doubly-deficient mice (Tassi et al., 2005), strongly suggesting that PI3K regulates NK cell development largely through the recruitment of PDK1, which may further activate AKT phosphorylation. Pharmacological inhibition of total AKT activity using AKTi, which inhibits all three isoforms, resulted in decreased metabolic activation of NK cells. Importantly, AKT inhibition recapitulated the phenotype that less E4BP4 induction by IL-15 was found in the absence of PDK1. Thus, the PI3K–PDK1–AKT axis is critical to NK cell development via regulating E4BP4 expressing downstream IL-15 signaling.
AKT activates mTOR by phosphorylating tuberous sclerosis complex 1, a negative regulator of mTOR signaling. As hematopoietic-specific deletion of mTOR is embryonic lethal to mice in our hands, we were hindered from extensively analyzing its role in early NK cell development directly. Fortunately, Marçais et al. (2014) revealed the critical role of mTOR kinase as key metabolic checkpoint for NK cell proliferation and activation during the manuscript preparation. In this study, we also revealed that pharmacological inhibition of mTOR via the chemical Torin1, which is an inhibitor specific for mTOR enzymatic activity as part of both mTORC1 and mTORC2, could suppress NK cell differentiation and abolish the proproliferative role of IL-15 in vivo. We confirmed these results when rapamycin, a widely used mTORC1 inhibitor, was used, suggesting the central role of mTORC1 in the PDK1-mediated regulation of early NK cell development, but we could not exclude the possibility that mTORC2 is also involved, as long-term rapamycin treatment likely suppresses mTORC2 activity via a feedback mechanism. Genetic disruption of the major components of mTORC1 and mTORC2, including RAPTOR and RICTOR, respectively, will be useful for further studies.
Tumor cells always exhibit altered, usually accelerated, energy consumption. Targeting tumor metabolism represents an alternative way to suppress tumor growth. PDK1 is considered as a potential target for tumor therapy (Raimondi and Falasca, 2011). Our current study has implications for the long-term therapeutic use of PDK1 inhibitors or mTOR inhibitors, such as rapamycin, for tumor patients, which may exhibit a risk of affecting NK cell development in human patients.
MATERIALS AND METHODS
Mice.
The Ncr1-Cre transgene was designed with the mouse ncr1 (gene encoding NKp46) promoter region containing 5-UTR 3,000 bp, an optimized variant of Cre recombinase and an SV40 polyA signal. The transgene was microinjected into the pronuclei of fertilized oocytes from C57BL/6 mice. One founder stably expressing Cre was established and used to generate mutant mice.
Hematopoietic or NK cell–specific PDK1-deficient mice were generated by crossing PDK1fl/fl (from D. Alessi, University of Dundee, Dundee, Scotland, UK) with Vav1-Cre (B6.Cg-Tg(Vav1-cre)A2Kio/J; The Jackson Laboratory) or newly generated Ncr1-Cre mice in our laboratory. RAG1−/−γc− mice were obtained by intercrossing B6.129S7-Rag1tm1Mom/J (deficient in recombination-activating gene 1) with NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; The Jackson Laboratory). PCR was used to identify the genotype. The mice were then backcrossed to pure B6 background at least 8 generations. β2m-deficient mice, C57BL/6 mice, and CD45.1 mice were purchased from The Jackson Laboratory. All mice were bred and maintained in specific pathogen-free animal facilities of Tsinghua University. All procedures involving animals were approved by the Animal Ethics Committee of Tsinghua University.
Reagent.
Antibodies recognizing mouse CD3 (mAb 145-2C11), NKp46 (mAb 29A1.4), NK1.1 (PK136), CD117 (2B8), CD127 (A7R37), Ly49A (A1), Ly49C/I (5E6), CD135 (A2F10), SCA-1 (D7), NKG2D (A10), NKG2A/C/E (20D5), CD11b (mAb M1/70), CD27 (LG.7F9), and isotype controls were purchased from eBioscience or BD. CD1d-PBS157 tetramer was provided by the National Institutes of Health tetramer facility. To inhibit the activities of PI3K–mTOR pathway, NK cells were treated with various inhibitors: 1 µM Torin1 (Tocris), 1 µM GSK2334470 (Sigma-Aldrich), 1 µM AKTi1/2 (EMD Millipore), or 10 µM Ly294002 (EMD Millipore).
Real-time PCR.
Splenic NK cells were sorted by fluorescence-activated cell sorting, and stimulated with IL-15–IL-15R complex overnight. Total RNA was extracted using TRIzol kit (Invitrogen), and reverse-transcribed using reverse transcription system (Promega). qPCR was performed using SYBR Green-based detection. The expression level of the genes of interest was determined relative to the expression of β-actin. The results are presented as relative to unstimulated, set as 1.
Flow cytometry.
For analysis of surface markers CD71, CD98, and CD122, cells were stained in PBS containing 2% (wt/vol) FBS with antibodies from eBioscience or BD. Their expression level was presented as net mean fluorescence intensity (ΔMFI), which was determined by subtracting MFI of isotype control, or as fold relative to unstimulated, set as 1. Intracellular staining was used for NK cell transcription factor and phosphorylated proteins. NK cell transcription factors E4BP4, Eomes, and T-bet were stained with anti–mouse E4BP4 antibody (S2M-E19; eBioscience), anti-Eomes (Dan11mag; eBioscience), and anti–T-bet (eBio4B10; eBioscience) before and after overnight stimulation with IL-15–IL-15R complex according to the manufacturer’s instruction (eBioscience), similar to Foxp3 staining, respectively. For detection of phosphorylated signaling proteins, NK cells were fixed with Phosflow Lyse/Fix buffer, followed by permeabilization with Phosflow Perm buffer III (BD) and staining with antibodies to S6 phosphorylated at Serc235 and Serc236 (D57.2.2E; Cell Signaling Technology), Akt phosphorylated at Serc473 (M89-61; BD), and Thrc308 (J1-223.371; BD). Flow cytometry data were acquired on LSRII or LSR Fortessa (BD) and analyzed using FlowJo software (Tree Star). Net mean fluorescence intensity (ΔMFI) was calculated. Expression levels were presented as fold relative to unstimulated, set as 1.
Detection of NK cell apoptosis.
Splenic NK cells were staining with Annexin V (BD), and Caspase activity was measured with FITC-conjugated z-VAD-fmk according to the manufacturer’s instruction (eBioscience).
Adoptive cell transfer.
Donor mice were treated with 5-fluorouracil (5-FU) for 4 d before bone marrow cells were harvested. A mixture of 2 × 105 CD3+CD19+NK1.1+-depleted bone marrow cells from either WT CD45.1 B6 mice with same number of WT or PDKfl/fl/Vav-Cre mice expressing CD45.2 were intravenously transferred into sublethally irradiated RAG1−/−γc− mice. For in vivo rescue, recipient RAG1−/−γc− mice were sublethally irradiated and then transplanted with 2 × 106 retrovirally transduced bone marrow cells from 5-FU–treated PDK1-deficient mice. Reconstitution of recipients was assessed by flow cytometry of bone marrow and spleen at 6 wk after transplantation.
In vivo NK cell functions.
In vivo splenocyte rejection assay has been previously described (Dong et al., 2009). For RMA-S clearance assay, mice treated with 200 µg Poly I:C for 18 h were intraperitoneally injected with a mixture of target cells, NK-sensitive RMA-S cells expressing GFP (106), and NK-nonsensitive RMA expressing DsRed. 18 h after the tumor injection, mice were killed, and cells in peritoneal cavity were collected by repeated washing with PBS containing 2 µM EDTA, and after centrifugation, cells were finally suspended in 1 ml PBS. The relative percentages of RMA-S and RMA cells were monitored with flow cytometry. The percentage of RMA-S cell rejection was calculated as followed formula: 100 × (1 − [percentage of residual GFP+ population in total GFP+ and DsRed+ of experimental group/percentage of residual GFP+ population in total GFP+ and DsRed+ cells of RAG1−/−γc− mice group]).
Detection of in vivo IL-15 responsiveness.
Mice were intravenously injected with 500 ng IL-15/IL-15 complexes every 3 d, and NK cell numbers were dynamically monitored by flow cytometric analysis of peripheral blood CD3−NKp46+ cells at indicated time-points. The data were shown as the increase fold relative to untreated mice.
Statistical analyses.
Unpaired Student’s t tests (two-tailed) were performed using Prism software.
Acknowledgments
We thank the senior members of Institute for Immunology, Tsinghua University, for critical reading of the manuscript. We also acknowledge Dr. André Veillette (Institute of Clinical Research of Montreal, Canada) for his provision of valuable comments and Dario R. Alessi (University of Dundee) for sharing the PDK1fl/fl mice.
This work was supported by grants to Dr. Zhongjun Dong’ laboratory from the Ministry of Science and Technology of China (2013CB944901), the Natural Science Foundation of China (81322041, 81273198, 81361128016, and 81471523), and the Beijing Natural Science Foundation (5132018).
The authors declare no competing financial interest.
M. Yang, D. Li, and Z. Dong performed and analyzed experiments. Z. Chang, Z. Yang, and Z. Tian provided critical reagents and advice. M. Yang and Z. Dong designed experiments, analyzed data, and wrote the manuscript.
Footnotes
Abbreviations used:
- Bim
- B-cell lymphoma 2 interacting mediator of cell death
- E4BP4
- E4 promoter-binding protein 4
- Eomes
- Eomesodermin
- mTOR
- mammalian target of rapamycin
- PDK1
- 3′-phosphoinositide-dependent kinase 1
- PTEN
- phosphatase and tensin homolog
- SHIP1
- Src homology-2 domain-containing inositol 5-phosphatase 1
- T-bet
- T-box expressed in T cells
References
- Awasthi A., Samarakoon A., Dai X., Wen R., Wang D., and Malarkannan S.. 2008. Deletion of PI3K-p85alpha gene impairs lineage commitment, terminal maturation, cytokine generation and cytotoxicity of NK cells. Genes Immun. 9:522–535 10.1038/gene.2008.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracho G.V., Cato M.H., Zhu Z., Jaren O.R., Hobeika E., Reth M., and Rickert R.C.. 2014. PDK1 regulates B cell differentiation and homeostasis. Proc. Natl. Acad. Sci. USA. 111:9573–9578 10.1073/pnas.1314562111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H.2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12:325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez V.S., Fuchs A., Cella M., Gilfillan S., and Colonna M.. 2014. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 192:4487–4491 10.4049/jimmunol.1303469 [DOI] [PubMed] [Google Scholar]
- Crotta S., Gkioka A., Male V., Duarte J.H., Davidson S., Nisoli I., Brady H.J., and Wack A.. 2014. The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development. J. Immunol. 192:2677–2688 10.4049/jimmunol.1302765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSanto J.P., Müller W., Guy-Grand D., Fischer A., and Rajewsky K.. 1995. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. USA. 92:377–381 10.1073/pnas.92.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M., Okano T., Yujnovsky I., Sassone-Corsi P., and Fukada Y.. 2004. Negative control of circadian clock regulator E4BP4 by casein kinase Iepsilon-mediated phosphorylation. Curr. Biol. 14:975–980 10.1016/j.cub.2004.05.043 [DOI] [PubMed] [Google Scholar]
- Dong Z., Cruz-Munoz M.E., Zhong M.C., Chen R., Latour S., and Veillette A.. 2009. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat. Immunol. 10:973–980 10.1038/ni.1763 [DOI] [PubMed] [Google Scholar]
- Dong Z., Davidson D., Pérez-Quintero L.A., Kurosaki T., Swat W., and Veillette A.. 2012. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 36:974–985 10.1016/j.immuni.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Eckelhart E., Warsch W., Zebedin E., Simma O., Stoiber D., Kolbe T., Rülicke T., Mueller M., Casanova E., and Sexl V.. 2011. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 117:1565–1573 10.1182/blood-2010-06-291633 [DOI] [PubMed] [Google Scholar]
- Finlay D.K., Rosenzweig E., Sinclair L.V., Feijoo-Carnero C., Hukelmann J.L., Rolf J., Panteleyev A.A., Okkenhaug K., and Cantrell D.A.. 2012. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209:2441–2453 10.1084/jem.20112607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth M.A., Madera S., Beaulieu A.M., Gasteiger G., Castillo E.F., Schluns K.S., Kubo M., Rothman P.B., Vivier E., and Sun J.C.. 2013. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med. 210:2981–2990 10.1084/jem.20130417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., and Brady H.J.. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Geiger T.L., Abt M.C., Gasteiger G., Firth M.A., O’Connor M.H., Geary C.D., O’Sullivan T.E., van den Brink M.R., Pamer E.G., Hanash A.M., and Sun J.C.. 2014. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J. Exp. Med. 211:1723–1731 10.1084/jem.20140212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D.B., Cella M., Giurisato E., Fujikawa K., Miletic A.V., Kloeppel T., Brim K., Takai T., Shaw A.S., Colonna M., and Swat W.. 2006. Vav1 controls DAP10-mediated natural cytotoxicity by regulating actin and microtubule dynamics. J. Immunol. 177:2349–2355 10.4049/jimmunol.177.4.2349 [DOI] [PubMed] [Google Scholar]
- Guo H., Samarakoon A., Vanhaesebroeck B., and Malarkannan S.. 2008. The p110 delta of PI3K plays a critical role in NK cell terminal maturation and cytokine/chemokine generation. J. Exp. Med. 205:2419–2435 10.1084/jem.20072327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton H.J., Alessi D.R., and Cantrell D.A.. 2004. The serine kinase phosphoinositide-dependent kinase 1 (PDK1) regulates T cell development. Nat. Immunol. 5:539–545 10.1038/ni1062 [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Puthalakath H., Gunn P., Naik E., Michalak E.M., Smyth M.J., Tabarias H., Degli-Esposti M.A., Dewson G., Willis S.N., et al. . 2007. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat. Immunol. 8:856–863 10.1038/ni1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K., Bloom E.T., Nakajima H., Horvath-Arcidiacono J.A., Udy G.B., Davey H.W., and Leonard W.J.. 1998. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 188:2067–2074 10.1084/jem.188.11.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. . 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Kamizono S., Duncan G.S., Seidel M.G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E.F., Haight J.P., Ohashi P.S., et al. . 2009. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206:2977–2986 10.1084/jem.20092176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Sklarz T., Banks L.B., Gohil M., Waickman A.T., Skuli N., Krock B.L., Luo C.T., Hu W., Pollizzi K.N., et al. . 2013. Natural and inducible TH17 cells are regulated differently by Akt and mTOR pathways. Nat. Immunol. 14:611–618 10.1038/ni.2607 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Male V., Nisoli I., Gascoyne D.M., and Brady H.J.. 2012. E4BP4: an unexpected player in the immune response. Trends Immunol. 33:98–102 10.1016/j.it.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Male V., Nisoli I., Kostrzewski T., Allan D.S., Carlyle J.R., Lord G.M., Wack A., and Brady H.J.. 2014. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J. Exp. Med. 211:635–642 10.1084/jem.20132398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais A., Cherfils-Vicini J., Viant C., Degouve S., Viel S., Fenis A., Rabilloud J., Mayol K., Tavares A., Bienvenu J., et al. . 2014. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 15:749–757 10.1038/ni.2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal N., Ali A.K., Komal A.K., and Lee S.H.. 2014. The Critical Role of IL-15-PI3K-mTOR Pathway in Natural Killer Cell Effector Functions. Front. Immunol. 5:187 10.3389/fimmu.2014.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.G., Long M., Kang J.A., Kim W.S., Lee C.R., Im S.H., Strickland I., Schulze-Luehrmann J., Hayden M.S., and Ghosh S.. 2013. The kinase PDK1 is essential for B-cell receptor mediated survival signaling. PLoS ONE. 8:e55378 10.1371/journal.pone.0055378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J.D., Pollizzi K.N., Heikamp E.B., and Horton M.R.. 2012. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 30:39–68 10.1146/annurev-immunol-020711-075024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi C., and Falasca M.. 2011. Targeting PDK1 in cancer. Curr. Med. Chem. 18:2763–2769 10.2174/092986711796011238 [DOI] [PubMed] [Google Scholar]
- Sathaliyawala T., O’Gorman W.E., Greter M., Bogunovic M., Konjufca V., Hou Z.E., Nolan G.P., Miller M.J., Merad M., and Reizis B.. 2010. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity. 33:597–606 10.1016/j.immuni.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Rankin L.C., Groom J.R., Mielke L.A., Tellier J., Chopin M., Huntington N.D., Belz G.T., and Carotta S.. 2014. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 211:1733–1740 10.1084/jem.20140145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka D.K., Plougastel-Douglas B., Yang L., Pak-Wittel M.A., Artyomov M.N., Ivanova Y., Zhong C., Chase J.M., Rothman P.B., Yu J., et al. . 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 3:e01659 10.7554/eLife.01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Duncan G.S., Takimoto H., and Mak T.W.. 1997. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. J. Exp. Med. 185:499–505 10.1084/jem.185.3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi I., Presti R., Kim S., Yokoyama W.M., Gilfillan S., and Colonna M.. 2005. Phospholipase C-gamma 2 is a critical signaling mediator for murine NK cell activating receptors. J. Immunol. 175:749–754 10.4049/jimmunol.175.2.749 [DOI] [PubMed] [Google Scholar]
- Tassi I., Cella M., Gilfillan S., Turnbull I., Diacovo T.G., Penninger J.M., and Colonna M.. 2007. p110gamma and p110delta phosphoinositide 3-kinase signaling pathways synergize to control development and functions of murine NK cells. Immunity. 27:214–227 10.1016/j.immuni.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Tassi I., Cella M., Presti R., Colucci A., Gilfillan S., Littman D.R., and Colonna M.. 2008. NK cell-activating receptors require PKC-theta for sustained signaling, transcriptional activation, and IFN-gamma secretion. Blood. 112:4109–4116 10.1182/blood-2008-02-139527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Muchnik M., Chen Z., Patel M., Wu N., Joshi S., Rui L., Lazar M.A., and Yin L.. 2010. Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. J. Biol. Chem. 285:36401–36409 10.1074/jbc.M110.172866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venigalla R.K., McGuire V.A., Clarke R., Patterson-Kane J.C., Najafov A., Toth R., McCarthy P.C., Simeons F., Stojanovski L., and Arthur J.S.. 2013. PDK1 regulates VDJ recombination, cell-cycle exit and survival during B-cell development. EMBO J. 32:1008–1022 10.1038/emboj.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshenrich C.A., Ranson T., Samson S.I., Corcuff E., Colucci F., Rosmaraki E.E., and Di Santo J.P.. 2005. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 174:1213–1221 10.4049/jimmunol.174.3.1213 [DOI] [PubMed] [Google Scholar]
- Waickman A.T., and Powell J.D.. 2012. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 249:43–58 10.1111/j.1600-065X.2012.01152.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T., Bléry M., Chaix J., Fuseri N., Chasson L., Robbins S.H., Jaeger S., André P., Gauthier L., Daniel L., et al. . 2007. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. USA. 104:3384–3389 10.1073/pnas.0609692104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Yang J., Yang K., Wang H., Gorentla B., Shin J., Qiu Y., Que L.G., Foster W.M., Xia Z., et al. . 2014. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J. Clin. Invest. 124:1685–1698 10.1172/JCI69780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., Neale G., Vogel P., and Chi H.. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 499:485–490 10.1038/nature12297 [DOI] [PMC free article] [PubMed] [Google Scholar]