Abstract
Objective(s):
Psoriasis is an autoimmune disease that appears on the skin. Although psoriasis is clinically and histologically well characterized, its pathogenesis is unknown in detail. The aims of this study were to evaluate the proteome of psoriatic patients' sera and to compare them with those of normal healthy human to find valuable biomarkers.
Materials and Methods:
In a case-control study, twenty cases of white patients with psoriasis vulgaris, 10 males and 10 females and sixteen healthy controls, 8 males and 8 females were enrolled in the study. The serum protein expression patterns obtained after depletion of albumin were compared by using two dimensional gel electrophoresis (2-DE) coupled to MALDI/TOF-TOF to identify disease associated proteins.
Results:
Differential expression of nine protein spots representing four unique proteins including alpha-1 antitrypsin, retinol binding protein, keratin 10 and an unknown protein (with pI 6.47 and molecular weight of 19941 Da), between psoriatic and healthy human serum were found. Furthermore, expression of four new alpha-1 antitrypsin isoforms with different molecular weight and isoelectric point were observed in psoriatic serums in this research for the first time.
Conclusion:
A unique proteomic profiling with abnormal expression of alpha-1 antitrypsin and presence of keratin 10 in sera of psoriasis patients were observed that may constitute new and useful findings of psoriasis and offer a clue to a better understanding of the inflammatory pathway.
Keywords: Alpha-1 antitrypsin, Keratin 10, Proteomics, Psoriasis, Retinol binding protein
Introduction
Psoriasis is an autoimmune disease that appears on the skin, transmits genetically, and affects approximately 2% of the world's population, with men and women being equally affected (1). It is a complex genetic disease with environmental and genetic components and in fact, the term represents a clinically heterogeneous group of diseases including psoriasis vulgaris, pustular psoriasis, psoriasis arthritis, erythrodermic psoriasis, nail psoriasis and psoriasis inversa (2). The most common phenotype is chronic plaque psoriasis with well-demarcated lesions typically located in the scalp and on the extensor surfaces (3). Although psoriasis is clinically and histologically well characterized, its pathogenesis is unknown in detail, and unlike other autoimmune diseases, does not have a generally accepted animal model. Our knowledge about its pathogenesis is derived exclusively from clinical studies and translational science performed in psoriatic patients.
Pathological conditions can induce some quality and quantity changes in sera and these characteristic changes provide a favorable biomarker to distinguish between healthy and sick states. Global gene expression (4-6) and protein analysis (7) in psoriasis have been investigated previously. Also, other aspects such as role of vitamins (8), immnunoglubolins (9), microRNAs (10), adipokines and cytokines (11) and methylglyoxal (12) in the pathophysiology of this autoimmune disease have been studied. Proteomic studies also are used in the evaluation of this disorder, but there are few published data about serum proteom alteration in psoriatic patients (13, 14) and more studies on this topic are needed. Proteome analysis can be regarded as a peptide screening approach aimed at documenting the overall distribution of proteins in cells, organs, or other samples (15). Therefore, the aim of this study was to explore serum proteomics patterns of psoriatic patients by using two dimensional gel electrophoresis (2-DE) followed by MALDI/TOF-TOF.
Materials and Methods
A total of 36 patients (18 males and 18 females) were selected. Twenty cases of white patients with psoriasis vulgaris, 10 males and 10 females (mean ± SD age 41.42±12.7 years), and sixteen healthy control peoples, 8 males and 8 females (mean ± SD age 46.90±16.7 years) were enrolled in the study. All patients were diagnosed as having psoriasis vulgaris and had no prior history of other diseases or any medications which may complicate the analysis since two months ago. The study was approved by the local Ethics Committee. Fasting blood samples were taken from the cubical vein into sterile vacutainers without anticoagulant and after centrifugation (2000g, 10min), serum stored at -20°C until used. Protein concentration was determined according to the method proposed by Bradford, using bovine serum albumin (BSA) as a standard protein (16). Albumin concentration was assayed by bromocresol green (BCG) method. TCA/acetone precipitation (17) and Aurum serum protein mini kit (18) were used to remove high abundant proteins. Processed serum samples were analyzed by SDS-PAGE in the presence of 2-mercaptoethanol (2-ME) according to the Laemmli method (19) using the running and stacking gel of 12% and 3.0% acrylamide (w/v), respectively. Gels were stained with Coomassie Brilliant blue (CBB) R-250. Two-dimensional gel electrophoresis was performed according to the previously reported methods and resolved proteins were detected by staining with either CBB R-250 or silver nitrate, but omitting the glutaraldehyde fixation (20). For each serum sample analysis, 2-D gels have been performed at least in triplicate, with independent protein preparations. 2-D gels were scanned and analyzed by Melanie software, version 6.0.2.0. For MALDI/TOF-TOF analysis, the protein spots of interest were excised carefully and analyzed by MALDI/TOF- TOF by the “The Technology Facility Proteomics & Analytical Biochemistry Laboratory” (The University of York, UK). Database interrogation was performed using the National Center for Biotechnology Information (NCBI) databases on a GPS workstation. In order to highlight the serum proteome changes in psoriasis patients, image analysis has been performed on three replicates, and variations on spots' area were confirmed by statistical analysis.
Statistical analysis
Values were expressed as the mean±SD. Differences between the healthy and disease sera were determined by two independent sample T test. Using SPSS version 16, a P-value equal to 0.05 was considered statistically significant.
Results
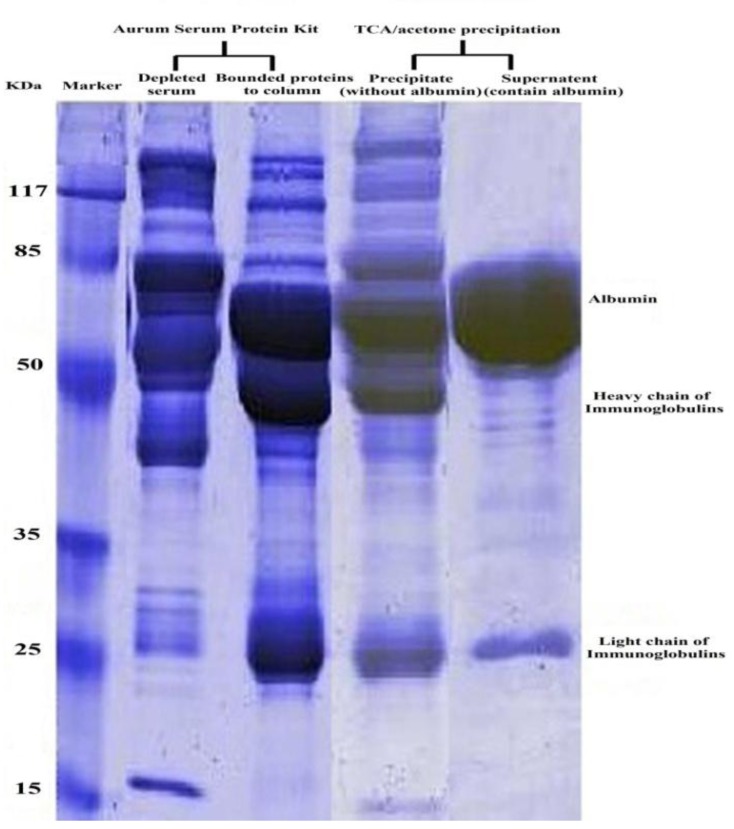
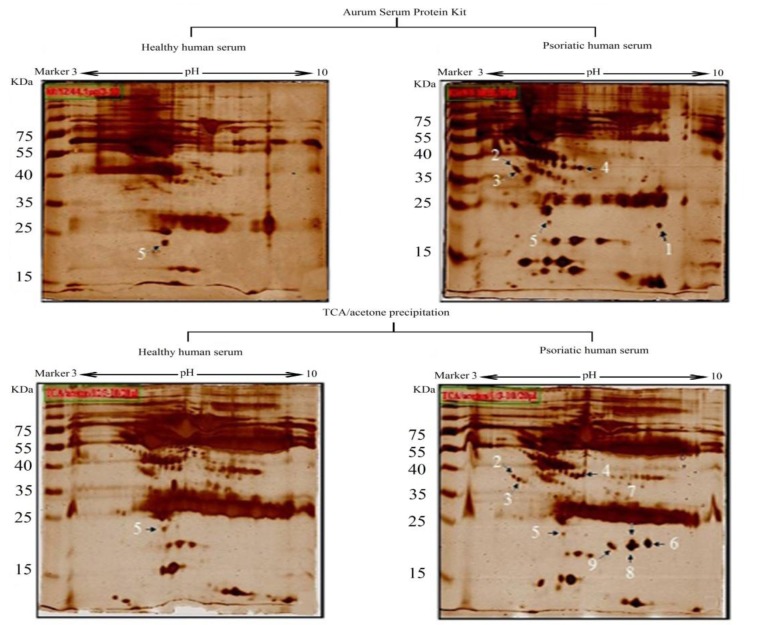
The serum protein yield in untreated, TCA/acetone or Aurum kit treated serums are summarized in Table 1. Figure 1 depicts typical SDS-PAGE results of serum proteins obtained by different treatments, and in each sample protein in 2 µl of serum were loaded to the gel. A comparison of the proteomics patterns of health and patient serum gels, as well as both high protein depletion methods are shown in Figure2.
Table 1.
Mean±SD of patients serum protein concentration and percentage of protein recovery obtained before and after treatment with TCA/acetone or Aurum kit
| Protein concentration (mg/ml) | Protein recovery (%) | |
|---|---|---|
| Untreated | 67.72±8.82 | 100 |
| Aurum kit treated | 29.25±7.15 | 43.1 |
| TCA/acetone treated | 8.27±3.14 | 12.2 |
Figure 1.
Human serum proteins stained with Commassie brilliant blue R-250 after depletion of high-abundant proteins using different depletion treatment on 12 % SDS-PAGE
Figure 2.
2-Dimentioanl gel electrophoresis profile of different treated of human serum which were stained by silver staining (Spot number according to Table 2)
Patient serum treated with TCA/acetone has four spots more than patient serum depleted with Aurum serum protein mini kit (Figure 2B and 2D, spot No. 6-9). As shown in Figure 2, nine protein spots were subjected to MALDI/TOF-TOF analysis. Table 2 lists the identities of the proteins which were analyzed in this experiment using MALDI/TOF-TOF and summarizes the differences in existence, absence or alteration in expression of spots between the two depletion methods and between patient and health control serums.
Table 2.
MALDI/TOF-TOF analysis of selected protein spots from 2-Dimentional gel electrophoresis that differently expressed in sera from psoriatic and healthy human between two depletion methods
| Spot no*. | Protein name | MW (Da) | P-value | NCBI ID | TCA/aceton† | Aurum kit | ||
|---|---|---|---|---|---|---|---|---|
| H | P | H | P | |||||
| 1 | Unknown protein | 19941 | 6.47 | gi|119569581 | - | + | - | - |
| 2 | Keratin 10 | 39832 | 4.72 | gi|186629 | - | + | - | + |
| 3 | Chain A, The S Variant of human α1-Antitrypsin | 39099 | 5.27 | gi|231240 | - | + | - | + |
| 4 | Chain A, Crystal structure of cleaved antitrypsin | 37622 | 5.43 | gi|7546268 | - | + | - | + |
| 5 | Retinol binding protein 4 | 23371 | 5.76 | gi|18088326 | ++ | + | ++ | + |
| 6 | α1-AT (isoform 1) | 22871 | 6.11 | gi/28637 | - | - | - | + |
| 7 | α1-AT (isoform 2) | 22871 | 6.11 | gi/28637 | - | - | - | + |
| 8 | α1-AT (isoform 3) | 22871 | 6.11 | gi/28637 | - | - | - | + |
| 9 | α1-AT (isoform 4) | 22871 | 6.11 | gi/28637 | - | - | - | + |
α1-AT: Alpha1- antitrypsin; H: healthy; P: patients.
Spot no. related to the annotation in Figure 2.
+ existence; - absence; ++ increased concentration
Discussion
It was found that three distinct proteins including different isoforms of α1-AT (according to a previous unpublished report by Wieland et al), retinol binding protein (RBP) and keratin 10 suffer changes in psoriatic serum against control healthy human serum. In contrast with our results, in a previous study conducted by Williamson et al (2013) it has been reported that abundance of more than 50 proteins was altered in psoriatic patent versus health ones. Between these proteins, profilin 1 is described as a candidate plasma biomarker of psoriasis (13). There are several reasons for this dissimilation such as differences in sample preparation, protein enrichment and detection.
Several investigators have emphasized the role of proteolytic enzymes, especially serine proteases in the pathogenesis of psoriasis (2, 21, 23). In the psoriatic lesion, MCTC cells (mast cells that contain several serine protease) are predominantly responsible for the mast cell infiltration into the papillary dermis (21). Alpha 1-AT is a glycoprotein with serine protease inhibition activity that interacts with and inhibits elastases, cathepsin G and chymase, and plays a role in reducing tissue inflammation. There are several investigations about the role of α1-AT in the pathogenesis of psoriasis (22, 23). In our study, two variants of α1-AT were identified that were only detected in the serum of the psoriatic patients, and were not detectable in healthy controls. One of these variants existed in four isoforms with Mw/pI about 22000/6-7 which contained 197 amino acids. All previously reportedα1-ATs are different from this new variant in Mw and pI and there are no reports on previous studies about this variant. As shown in Figure 2, in patient serum depleted with Aurum serum protein mini kit, compared to TCA/acetone depletion, four spots were eliminated. The main cause of this condition is the elimination of immunoglobulins from serum by using Aurum serum protein mini kit, and therefore the α1-ATs isoforms that create complexes with them are also removed from the serum. Thus, new complexes of Ig- α1-AT were found in psoriatic serum but the existence of Ig- α1-AT complexes were also reported in relatively longstanding studies in different joint diseases such as ankylosing spondylitis and rheumatoid arthritis (24).
Vitamin A (retinol) and its derivatives play different roles in the body (25, 26), and within the blood, free retinol binds to retinol binding protein (RBP), its serum transport protein. Some investigators have indicated that RBP is present in the intercellular spaces of the epidermis (27). Retinol can store in keratinocytes and convert to all-trans retinoic acid (ATRA) (28). RBP is involved in the regulation of intracellular retinoid concentrations and a decrease in patient serum RBP concentration, as seen in our study, causes psoriatic lesions.
One protein with pI/MW about 4.8/40000 existed in the serum of one patient, which was not observed in healthy human serum and by MS analysis, was identified as keratin 10 (K10). Keratins are intermediate filament-forming proteins that provide mechanical support and fulfill a variety of additional functions in epithelial cells (29). These proteins are grouped into two classes, the neutral-basic type II (K1-K8) and the acidic type I (K9-K20) (30). Proliferative basal cells express the K5-K14 pair. However, when keratinocytes begin terminal differentiation, they move upward, become postmitotic, and switch to the expression of a keratin pair K1-K10 (31). Under hyperproliferative conditions, such as psoriasis, downregulation of K10 occurs in keratinocytes, but there is one report of expression of K10 increase in lesional and symptomless skin of spreading psoriasis (32). In other previous studies, K10 was never found in the serum. However in this study, for the first time, K10 in the serum of patients with psoriasis vulgaris was found. This increase may be due to epidermal apoptosis that causes K10 leaking to the serum of psoriatic patients.
Conclusion
With an analysis of sera 2D maps, it has been possible to describe a pattern of proteins up- and down-regulation upon disease conditions. In conclusion, we here provide serum proteome analysis of patients with psoriasis. Nine proteins were differentially expressed in the psoriasis vulgaris group, as compared to the normal group of these proteins. Using specific ELISA or western blot assay for all of these proteins will provide a more complete evaluation of these valuable changes.
Acknowledgment
The results described in this paper were part of student thesis. This study was conducted as a specialty thesis funded by the Vice-chancellor for Research, University of Sistan & Baluchestan, Zahedan, Iran. We are grateful to all the staff of the Toohid Laboratory of Zahedan for their interest and enthusiasm in doing this research.
References
- 1.Raychaudhuri SP, Farber EM. The prevalence of psoriasis in the world. J Eur Acad Dermatol Venereol. 2001;15:16–17. doi: 10.1046/j.1468-3083.2001.00192.x. [DOI] [PubMed] [Google Scholar]
- 2.Sticherling M. Mechanisms of psoriasis. Drug Discov Today Dis Mech. 2005;2:275–281. [Google Scholar]
- 3.Carlen LM, Sanchez F, Bergman AC, Becker S, Hirschberg D, Franzen B, et al. Proteome analysis of skin distinguishes acute guttate from chronic plaque psoriasis. J Invest Dermatol . 2004;124:63–69. doi: 10.1111/j.0022-202X.2004.23501.x. [DOI] [PubMed] [Google Scholar]
- 4.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol . 2003;112:1195–1202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 5.Oestreicher J, Walters I, Kikuchi T, Gilleaudeau P, Surette J, Schwertschlag U, et al. Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. PharmacogenomicsJ. 2001;1:272–287. doi: 10.1038/sj.tpj.6500067. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, Krueger JG, Kao MCJ, Lee E, Du F, Menter A, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics . 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 7.Broome AM, Ryan D, Eckert RL. S100 protein subcellular localization during epidermal differentiation and psoriasis. J Histochem Cytochem . 2003;51:675–685. doi: 10.1177/002215540305100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Mutairi N, EL Eassa B, Nair V. Measurement of vitamin D and cathelicidin (LL-37) levels in patients of psoriasis with co-morbidities. Indian J Dermatol Venereol Leprol . 2013;79:492–496. doi: 10.4103/0378-6323.113077. [DOI] [PubMed] [Google Scholar]
- 9.Ding Y, Yi X, Yu N. Serum IgE levels are increased in patients with generalized pustular psoriasis. Clin Exp Dermatol . 2013;38:549–552. doi: 10.1111/ced.12086. [DOI] [PubMed] [Google Scholar]
- 10.Pivarcsi A, Meisgen F, Xu N, Stahle M, Sonkoly E. Changes in the level of serum microRNAs in psoriasis patients after anti‐tumor necrosis factor‐α therapy. Br J Dermatol . 2013; 169:563–570. doi: 10.1111/bjd.12381. [DOI] [PubMed] [Google Scholar]
- 11.Nakajima H, Nakajima K, Tarutani M, Sano S. Clear association between serum levels of adipokines and T‐helper 17‐related cytokines in patients with psoriasis. Clin Exp Dermatol. 2013;38:66–70. doi: 10.1111/j.1365-2230.2012.04465.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaur S, Zilmer K, Leping V, Zilmer M. Serum methylglyoxal level and its association with oxidative stress and disease severity in patients with psoriasis. Arch Dermatol Res. 2013; 305:489–494. doi: 10.1007/s00403-013-1362-5. [DOI] [PubMed] [Google Scholar]
- 13.Williamson JC, Scheipers P, Schw mmle V, Zibert JR, Beck HC, Jensen ON. A proteomics approach to the identification of biomarkers for psoriasis utilisingkeratome biopsy. J Proteomics . 2013;94:176–185. doi: 10.1016/j.jprot.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Cowen EW, Liu CW, Steinberg SM, Kang S, Vonderheid EC, Kwak HS, et al. Differentiation of tumor-stage mycosis fungoides, psoriasis vulgaris and normal controls in a pilot study using serum proteomic analysis. Br J Dermatol . 2007;157:946–953. doi: 10.1111/j.1365-2133.2007.08185.x. [DOI] [PubMed] [Google Scholar]
- 15.Kalenka A, Feldmann RE, Otero K, Maurer MH, Waschke KF, Fiedler F. Changes in the serum proteome of patients with sepsis and septic shock. Anesth Analg . 2006;103:1522–1526. doi: 10.1213/01.ane.0000242533.59457.70. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem . 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Jiang L, He L, Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J Chrom A. 2004;1023:317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed N, Barker G, Oliva K, Garfin D, Talmadge K, Georgiou H, et al. An approach to remove albumin for the proteomic analysis of low abundance biomarkers in human serum. Proteomics. 2003;3:1980–1987. doi: 10.1002/pmic.200300465. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature . 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Kazemipour N, Condemine G, Hugouvieux‐Cotte‐Pattat N. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics. 2004;4:3177–3186. doi: 10.1002/pmic.200300814. [DOI] [PubMed] [Google Scholar]
- 21.Harvima IT, Haapanen L, Ackermann L, Naukkarinen A, Harvima RJ, Horsmanheimo M. Decreased chymase activity is associated with increased levels of protease inhibitors in mast cells of psoriatic lesions. Acta Dermatoven-Stockholm . 1999;79:98–104. doi: 10.1080/000155599750011282. [DOI] [PubMed] [Google Scholar]
- 22.Heng M, Moy R, Lieberman J. Alpha 1‐Antitrypsin deficiency in severe psoriasis. Br J Dermatol . 1985;112:129–133. doi: 10.1111/j.1365-2133.1985.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 23.Nini G, Bianchi L, Angelini E, Corleto V, Gatti S, Carrozzo A. Evaluation of serine alpha 1-antitrypsin and polymorphonuclear leukocyte elastase contents and their immunogenetic correlation in psoriasis. Acta Dermatoven Supp. 1994;186:143–145. [PubMed] [Google Scholar]
- 24.Scott L, Evans E, Dawes P, Russell G, Mattey D. Comparison of IgA-alpha1-antitrypsin levels in rheumatoid arthritis and seronegative oligoarthritis: complex formation is not associated with inflammation per se. Br J Rheumatol. 1998;37:398–404. doi: 10.1093/rheumatology/37.4.398. [DOI] [PubMed] [Google Scholar]
- 25.Pavez Lorie E, Cools M, Borgers M, Wouters L, Shroot B, Hagforsen E, et al. Topical treatment with CYP26 inhibitor talarozole (R115866) dose dependently alters the expression of retinoid‐regulated genes in normal human epidermis. Br J Dermatol . 2009;160:26–36. doi: 10.1111/j.1365-2133.2008.08895.x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor GA, Shalita AR. Retinoid therapy of acne and sebocyte-related disorders. Basic Clin Dermatol . 2007;39:103–123. [Google Scholar]
- 27.Rabilloud T, Asselineau D, Bailly C, Miquel C, Tarroux P, Darmon M. Study of the human epidermal differentiation by two dimensional electrophoresis. In: Schaefer-Nielsen C, editor. Electrophoresis. 88th ed. Munich: Verlag Chemie; 1989. [Google Scholar]
- 28.Roos TC, Jugert FK, Merk HF, Bickers DR. Retinoid metabolism in the skin. Pharmacol Rev . 1998;50:315–333. [PubMed] [Google Scholar]
- 29.Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, et al. New consensus nomenclature for mammalian keratins. J Cell Biol . 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller FB, Huber M, Kinaciyan T, Hausser I, Schaffrath C, Krieg T, et al. A human keratin 10 knockout causes recessive epidermolytic hyperkeratosis. Hum Mol Gen. 2006;15:1133–1141. doi: 10.1093/hmg/ddl028. [DOI] [PubMed] [Google Scholar]
- 31.Paramio JM, Segrelles C, Ruiz S, Jorcano JL. Inhibition of protein kinase B (PKB) and PKCζ mediates keratin K10-induced cell cycle arrest. Mol Cell Biol . 2001;21:7449–7459. doi: 10.1128/MCB.21.21.7449-7459.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mommers J, Van Rossum M, Van Erp P, Van De Kerkhof P. Changes in keratin 6 and keratin 10 (co-) expression in lesional and symptomless skin of spreading psoriasis. Dermatol. 2000;201:15–20. doi: 10.1159/000018422. [DOI] [PubMed] [Google Scholar]