Abstract
Background: The CCR5 is a chemokine receptor that serves as a co-receptor for HIV-1 attachment and entry to T lymphocytes. A 32bp deletion (∆32) in this gene is believed to be associated with resistance to infection and delay disease progression. The aim of this study was to determine the∆32 allele frequency in healthy individuals and HIV-infected individuals with AIDS.
Methods: In this experiment, 530 normal individuals from healthy Iranian population and 40 HIV-infected samples from Western Clinic of Tehran were examined for∆32 in CCR5 gene using polymerase chain reaction (PCR) techniques followed by agarose gel electrophoresis.
Results: Allele frequencies of the CCR5∆32 in normal individuals were calculated to be 1.1% for heterozygous genotype and 0.19% for homozygous genotype. None of the co-receptor gene in HIV cases was found to be mutated in this study.
Conclusion: Based on the findings of this study and the literature in Iran, we could conclude that Iranian people similar to neighbor countries such as Arabs are susceptible to HIV virus infection.
Keywords: CCR5∆32, Chemokine receptor, HIV-1, AIDS, Iran
Introduction
Over the past quarter-century, the global spread of HIV virus has become one of the deadliest epidemics in modern times. In 2007 a total of 2.1 million men, women, and children died of AIDS. The death toll will remain high in the future because 33.2 million individuals are currently infected and about 2.5 million new HIV infections occur each year (1).
Among various AIDS restriction genes (ARGs) only CCR5∆32 prevents HIV infection (1-3 , 9-12). The CCR5 chemokine receptor acts as a co-receptor on CD4+ T lymphocytes for HIV-1 virus attachment and penetration (4). People with a 32-bp deletion in T lymphocytes CCR5 (CCR5-∆32) receptor have shown to be nearly resistant to HIV-1 strains (5). HIV heterozygous individuals with CCR5-delta32 deletion also have better therapeutic response to highly active antiretroviral drugs (6). Nevertheless, survival analyses have shown that progression to AIDS is slower in CCR5-∆32 heterozygous individuals (7). Various studies attributed normal function for peripheral blood (PB) T cells, homozygous for the CCR5-∆32 mutation. Besides, no clinical abnormalities have been observed to be associated with this genotype (8). Genetic retardation of HIV progression could introduce new cellular targets for anti-HIV therapy that completes the current available antiretroviral compounds. Different inhibitors designed for blocking the CCR5 chemokine receptor have shown relatively high protective effect in HIV infection (13).
The aim of this study was to determine the ∆32 allele frequency in healthy individuals and HIV-infected individuals with AIDS. The present study explored the effect of the gene variants that can be useful for treatment and predicting response of drugs that target this chemokine receptor in order to block the HIV entrance to the target cells.
Methods
Sample collection
A total of 530 unrelated healthy Iranian individuals were enrolled. The healthy individuals were randomly selected among the university hospital staffs and students from different provinces. Randomized stratification was used for selection of provinces and Gilan, East Azarbaijan, West Azarbaijan, Ghazvin, Tehran, Semnan, Kurdistan, Ghom, Isfahan, Khorasan, Yazd, Lorestan, Hormozgan were selected. Forty blood samples from HIV infected individuals were collected from Tehran University of Medical Sciences clinics, Tehran, Iran. All samples were confirmed to be HIV positive or negative by western blotting and real time PCR.
DNA preparation
DNA was prepared from blood leukocytes by standard methods using QIAamp DNA Mini Kit (Biorain Company. Iran) (14).
PCR
PCR amplification was carried out in 25 μl reactions containing 50–100 ng of template DNA, 12.5 pM of each oligonucleotide primers, 0.2 mM dNTPs, in 1.5 mM MgCl2, 100 mM Tris–HCl (pH 8.8) and 1U Taq polymerase (Qiagen supplied by Biorain company, Iran) using the Corbet CG1-96 thermocycler (Armin teb. Iran). The specific segment of CCR5 gene was amplified by PCR using the following primers:
CCR5-F (5´-ATCACTTGGGTGGCTG TGTTTGCGTCTC-3´) and CCR5-R(5´-AGTAGCAGATGACCATGACAAGCAG CGGCAG-3´).
These primers were used to amplify a 193bp fragment for the normal allele, and 161bp fragment for the mutant allele. The reaction was subjected to 30 cycles: an initial denaturation of 5min at 94◦C, 30s denaturation at 94◦C, 45s at the annealing temperature 58◦C, extension at 72◦C for 30s. Following the amplification cycles a final extension was performed at 72◦C for 10 min. The polymorphism was detected by electrophoresis of 2% MS agarose gel (Applied Roche).
Statistical analysis
The allele frequency was calculated by the allele-counting method. The Hardy-Weinberg equilibrium was tested by chi-square analysis, and Pearson’s correlation coefficient used to find possible correlations with latitude and longitude.
Inclusion Criteria
The healthy subjects included the University hospital staffs and students of different provinces aged between 18 and 35 years, and their sexes notwithstanding. This target age had the highest prevalence of HIV-1 and AIDS infections in the country. The inclusion criteria for patients were HIV infected individuals with AIDS.
Exclusion Criteria
The individual with autoimmune liver disease, thyroid disease or diabetes and serious illness requiring systemic treatment or hospitalization for at least 14 days prior to study were excluded.
Results
We found 523 homozygous for the wild-type CCR5 allele from 530 mononuclear cells of normal Iranian population from various provinces, 6 heterozygous for the CCR5Δ32 allele, giving a CCR5-Δ32 allelic frequency of 1.1% and only one homozygous genotype for the Δ32/Δ32 allele (Fig. 1, Table 1 & 2 ). Almost all of the heterozygous genotypes and the only one homozygous genotype were determined in individuals from the northern parts of Iran (Table 1). The frequency of wt/wt, Δ32/wt, and Δ32/Δ32 genotypes were determined as 523 (98.7%), 6 (1.1%) and 1 (0.19%), respectively (Table 2). The mutant allele was not detected in the HIV-positive samples. The observed genotype frequencies were also in accordance with Hardy–Weinberg equilibrium (p=0.003)
Fig. 1 .
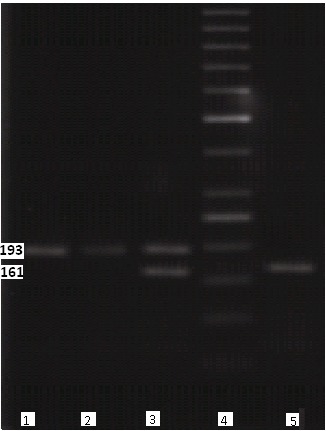
Analysis of the PCR product of CCR5 alleles Lane 1 and2: wild type (w/w), lane 3: heterozygous (CCR5∆32/wt), lane 4: DNA ladder, lane 5: homozygous (CCR5∆32/CCR5∆32)
Table 1 . Samples collected from different provinces from south to the north of Iran .
|
Province from north to south |
Number of sample | Δ32/wt frequency | Δ32/Δ32 frequency |
| Gilan | 20 | 1 | 0 |
| Azarbaijan-e- sharghy | 50 | 1 | 0 |
| Azarbaijan-e-gharbi | 50 | 1 | 0 |
| Ghazvin | 45 | 1 | 0 |
| Tehran | 100 | 2 | 1 |
| Semnan | 30 | 0 | 0 |
| Kurdistan | 35 | 0 | 0 |
| Ghom | 30 | 0 | 0 |
| Isfahan | 40 | 0 | 0 |
| Khorasan | 40 | 0 | 0 |
| Yazd | 30 | 0 | 0 |
| Lorestan | 30 | 0 | 0 |
| Hormozgan | 30 | 0 | 0 |
| Total | 530 | 6 | 1 |
Table2 . Allele and genotype frequency of CCR5Δ32 polymorphism in Iranian healthy individuals .
| Chemokine | Allele | F(%) | Genotype | F(%) | E Genotype |
Pearson’s Chi-Square |
p |
| Homozygous CCR5 | + | 1046(99.25) | wt/wt | 523(98.5%) | 522.03 | 0.03 | 0.99 |
| Heterozygous CCR5 | D32 | 6(0.57) | Δ32/wt | 6(1.1%) | 7.94 | ||
| D32/D32 | 1(0.19%) | 0.03 |
We found a strong positive significant correlation with latitude (r=0.72) and a somewhat weaker negative correlation with longitude (r=–0.34) of the geographic location within Iran.
Discussion
The description of natural CCR5 polymorphisms in human populations is an approach to identify immunologic and genetic factors implicated in innate resistance to HIV-1 infection and in the delay of progression to AIDS (3). In a study by Martinson et al., it was reported that CCR5-Δ32 allele frequency was quite high in North European populations, but it reduced while moving toward the south and east of Europe and almost it becomes rare in Africa, Oceania, the Middle East, or West Asia (28). The highest frequency of heterozygous individuals (%26.6) was reported in Finland from Europe. While increasing HIV prevalence in parts of Asia and Africa may be attributed to social and demographic factors, as well as differences in the phenotype of circulating viruses (26). The racial distribution of HIV risk raises the possibility that differences in the distribution of the CCR5-Δ32 allele or other heritable host factors/mutations may influence the rate of transmission or the speed of the epidemic in different racial groups (22). The present study was the first research conducted with subjects selected from all geographical regions of Iran. A total of 530 individuals with 30–100 from each region were analyzed for the CCR5- Δ32 allele. Results of this study indicate that the CCR5-Δ32 allele exists in Iranian individuals at a very low frequency (1.1%). This result is even lower than previous reports from Iran. The Gharagozloo et al (2005) (21) reported 2.8% for CCR5-Δ32 allele in Fars province located in southern part of Iran. Though we could not find mutant alleles in that part of the country. This probably is because of studies population and limitations. The Omrani et al (2009) (16) reported 2.1% for CCR5-Δ32 allele in Uromia province in North-West of Iran. According to our finding, all of the mutant alleles were found in north and north-west of Iran. This may confirm the correlation between CCR5-Δ32 allele frequency and climate-geographical factors.
Finding very low frequency of CCR5-Δ32 allele in Iran according to this study is in agreement with the result reported from most countries of the Middle East. The rate reported from some of Middle East countries, including Saudi Arabia, and Yemen (CCR5- Δ32 Frequency= 1%) is lower than that of our result (Table 3). It is reported that the frequency of heterozygote forms of the CCR5-Δ32 in some Middle Eastern countries including Jordan, Lebanon, United Arab Emirates and Iraq is about zero (Table 3). The highest frequencies of heterozygous individuals from this area were reported in Turkey (CCR5- Δ32 Frequency= 13%), Kazakstan (CCR5- Δ32 Frequency= 6%) Syria (CCR5- Δ32 Frequency= 3%) and Uzbekistan (CCR5- Δ32 Frequency 3%) ( Table 3).
Table 3 . Comparison of CCR5+D32 allele frequency (%) in Iranian normal population with other locations in Asia and European populations .
| Population | Sample size | CCR5_ 32 Frequency (%) | Reference | p |
|
Iran South of Iran North-West of Iran Jordan Kazakistan Kazakstan Kuwait Lebanon Pakistan Punjab Saudi Arabia Syria Turkey United Arab Emirates Iraq Uzbakistan China Yemen India Bahrain Egypt European populations |
530 395 376 52 50 50 393 51 36 34 341 106 91 26 13 29 447 124 100 304 200 2522 |
1.1 1.4 1.05 0.0 3 6.0 1.2 0.0 2.9 1.5 1 3 13 0.0 0.0 3 0.0 1 1 2.8 0.5 9.16%. |
This study [21] [16] [19] [20] [25] [19] [25] [18] [18] [20] [17] [23] [24] [17] [18] [18] [24] [1] [27] [27] [17] |
0.99 0.0 0.9 --- 0.2014 ---- ---- ----- 0.2755 1.0000 0.9193 0.9 0.79 0.170 ---- 0.2213 0.0046 0.7440 0.4772 0.2053 ------ 0.001 |
In the south part of Middle East and Arabic countries, the frequency of the mutant allele CCR5-∆32 is very low, and very few existed are probably derived from admixture with the populations from European descent rather than as a result of parallel independent mutations (22). No significant differences in CCR5-Δ32 allele frequency between Iranian and Arab populations were detected (Table 3).
Genetically, Iranians are considered to be close to Northern Indian, Greek and some of the European populations such as Italians, German and English. Recently reported human leukocyte antigen (HLA) class II allelic and haplotypic frequency data have confirmed similarity of Iranians, Greek and Italian’s genetic background (29). Historical evidences have suggested divergence of Iranian population and Europeans from a same ancestor known as Indo-Europeans. Around the year 2000 BC, an Indo-European tribe named Aryans invaded central Asia and occupied Iran, Iraq, north of India and Afghanistan. The great difference between the western and eastern migration of the Indo-Europeans was that in the west they genetically mixed with similar populations while in the east, admixture with genetically dissimilar people created largely mixed race population (30). Moreover, extensively genetic admixture with Mongols, Greeks and Arabs gradually diluted the major traces of Indo-Aryans during centuries.
The frequency of homozygote form of the CCR5-Δ32 chemokine as reported in Garaghozoolo et al and Omrani et al (2005 & 2009) and also in present study is about zero in Iran, therefore we should consider Iranians genetically susceptible to HIV virus during any potential exposure in the future. The CCR5-Δ32 allele frequency in Iran was similar to neighbor countries (Table 3).
When the Chi-square and probability values calculated for all individuals are taken into consideration (p = 0.003), the H0 hypothesis becomes acceptable, and the population would be at Hardy–Weinberg equilibrium.
In this study samples from different parts of Iran were examined. All heterozygous genotypes and the only one homozygous genotype were determined in individuals of the north and north-west of Iran (Ghazwin, Azarbaijan, Gilan, and Tehran) (6/270), and no incidence was not found in other parts (0/270) (Table 1). Our data showed that frequency of that allele was consistent with its reduction from north to south worldwide.
Conclusion
Strong positive correlation with latitude of the geographic location within Iran clarified the positive correlation between CCR5-Δ32 allele frequency and climate-geographical factors. The results presented here are consistent with the idea that climatic factors can play a certain selective role, either directly related to the expression of the CCR5-Δ32 allele, or to the action of a pathogen against which that allele confers some degree of protection.
Based on the findings of this study, we conclude that Iranians compared to Europeans are more susceptible to HIV virus infection. The resistance to HIV virus in Iranian population is similar to neighbor countries such as Arabs. However, since the infection with HIV virus is under the influence of the several different factors, therefore, this conclusion should be assumed as a preliminary statement and further investigation should be carried out to prove this hypothesis.
Acknowledgments
The authors thank Dr Ghaderi from Shiraz University of Medical Sciences for providing control samples of CCR5Δ32 genomic DNA. We are also grateful to Ms. A. Vahedi for technical assistance. This work was supported by grant no. k-127 of Iran University of Medical Sciences.
Cite this article as: Rahimi H, Farajollahi M.M, Hosseini A. Distribution of the mutateddelta 32 allele of CCR5 co-receptor gene in Iranian population. Med J Islam Repub Iran 2014 (29 November). Vol. 28:140.
References
- 1. UNAIDS, UNICEF, WHO. Children and AIDS—A Stocktaking Report. Actions and Progress during the First Year of Unite for Children, Unite against AIDS; 2007. «www.unicef.org/ uniteforchildren».
- 2.Stephen JO, Brien GWN. Human genes that limit AIDS. Nature Genetics. 2004;36(6):565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 3.AsmaJlizi JE, Karima FZ, Frigi S, Debré P, Slim A, Theodorou I. et al. Identification of the CCR5-32 HIV resistance allele and new mutations of the CCR5 gene in different Tunisian populations. Human Immunology. 2007;68:993–1000. doi: 10.1016/j.humimm.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 4. Carrington M, et al. Genetics of HIV-1 infection: Chemokine receptor CCR5 polymorphism and its consequences. Human Molecular Genetics 1999. [DOI] [PubMed]
- 5.De Silva E, Stumpf MPH. HIV and the CCR5-Delta32 resistance allele. FEMS microbiology letters. 2004;241(1):1–12. doi: 10.1016/j.femsle.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 6.Laurichesse JJ, Theodorou AP, Rouzioux C, Delfraissy JF, Meyer L. Improved virological response to highly active antiretroviral therapy in HIV-1-infected patients carrying the CCR5 D32 deletion. HIV Medicine. 2007;8:213–219. doi: 10.1111/j.1468-1293.2007.00455.x. [DOI] [PubMed] [Google Scholar]
- 7.Arenzana-Seisdedos F, Parmentier M. Genetics of resistance to HIV infection: Role of co-receptors and co-receptor ligands. Seminars in Immunology. 2006;18(6):387–403. doi: 10.1016/j.smim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Hua-Xin L, David CM, Dhavalkumar DP. et al. Linkage of the CCR5∆32 mutation with a functional polymorphism of CD45RA. The American Association of Immunologists. 2000;165:148–157;. doi: 10.4049/jimmunol.165.1.148. [DOI] [PubMed] [Google Scholar]
- 9.Anzala AO. et al. CCR2-641 allele and genotype association with delayed AIDS progession in African woman. Lancet. 1998;351:1632–1633. doi: 10.1016/s0140-6736(05)77688-1. [DOI] [PubMed] [Google Scholar]
- 10.Mary Carrington, Stephen J O’Brien. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 11.Martin MPea. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nature Genetics. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 12.Shin HD, Winkler C, Stephens JC. et al. Genetic restriction of HIV-1 infection and AIDS progression by promoter alleles of interleukin 10. Proc Natl Acad Sci. 2000;97:14467–14472. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dimitrov DS, et al. Chabot DJ and Broder CC: HIV Coreceptors. The Journal of Membrane Biology 1998;166: p. 75–90. [DOI] [PubMed]
- 14. Sambrook J, Fritsch EF, Maniatus T. Molecular Cloning: A laboratory Manual, Cold Spring Harbor Laboratory Press, ColdSpring Harbor, NY, 1989.
- 15.Dean MCM, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ. et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 16.Omrani MD, Bagheri M. Frequency of CCR5Δ32 Variant in North-West of Iran. Journal of Sciences, Islamic Republic of Iran. 2009;20(2):105–110. [Google Scholar]
- 17.Lucotte G, Mercier G. ¢32 mutation frequencies of the CCR5 coreceptor in different French regions. Life Sci. 1998;321:409–413. doi: 10.1016/s0764-4469(98)80305-3. [DOI] [PubMed] [Google Scholar]
- 18.Su B, Sun G, Lu D. et al. Distribution of three HIV-1 resistance-conferring polymorphisms (SDF1-3_A, CCR2-641, and CCR5-delta32) in global populations. Eur J Hum Genet. 2000;(8):975–979. doi: 10.1038/sj.ejhg.5200568. [DOI] [PubMed] [Google Scholar]
- 19.Voevodin A, Samilchuk E, Dashti S. A survey for 32 nucleotide deletion in the CCR-5 chemokine receptor gene (deltaccr-5) conferring resistance to human immunodeficiency virus type 1 in different ethnic groups and in chimpanzees. J Med Virol. 1998;(55):147–151. [PubMed] [Google Scholar]
- 20.Dean MCM, O’Brien SJ. Balanced polymorphism selected by genetic versus infectious human disease. Annu Rev Genomics Hum Genet. 2002;3:263–92. doi: 10.1146/annurev.genom.3.022502.103149. [DOI] [PubMed] [Google Scholar]
- 21.Gharagozloo M, Farjadian S. The frequency of CCR532 and CCR2-64I in southern Iranian normal population. Immunology Letters. 2005;96:277–281. doi: 10.1016/j.imlet.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 22. Abdel-Halim S and Mark A. Distribution of the HIV resistance CCR5-- 32 allele among Egyptians and Syrians. Mutation Research 2007;616: p. 175-180. [DOI] [PubMed]
- 23. Libert FCP, Beckman G, Samson M, Aksenova M, et al. The delta CCR5 mutation conferring protection against HIV-1 in Caucasian populations has a single and recent origin in Northeastern Europe. Hum Mol Genet 1998;7: p.399-409. [DOI] [PubMed]
- 24.Aseev MV, Shawi A, Dean M, Baranov VS. Population features of gene mutation frequency of the CKR-5 receptor, determining sensitivity to the AIDS virus. Russian J Genet. 1997;33:1724–1726. [PubMed] [Google Scholar]
- 25.Stephens JC, Reich DE, Goldstein DB. et al. Dating the origin of the CCR5-¢32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet. 1998;62:1507–1515. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soto-Ramirez LE, Renjifo B, McLane MF, Marlink R, O’Hara C, Sutthent R. et al. HIV-1 Langerhans’cell tropism associated with heterosexual transmission of HIV. Science. 1996;271:1291–1293. doi: 10.1126/science.271.5253.1291. [DOI] [PubMed] [Google Scholar]
- 27.Abdel Halim S, Eman F, Raouf F, Marwan A, Wassim A, Kyudong H. et al. Distribution of Four HIV Type 1-Resistance Polymorphisms (CCR5-D32, CCR5-m303, CCR2-64I, and SDF1-30A) in the Bahraini Population. Aids Research and Human retroviruses. 2009;5(10):973–977. doi: 10.1089/aid.2009.0066. [DOI] [PubMed] [Google Scholar]
- 28.Martinson JJ, Chapman NH, Rees DC, Liu Y, Clegg JB. Global distribution of the CCR5 gene 32 base-pair deletion. Nat Genet. 1997;16:100–3. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 29.Amirzargar A, Mytilineos J, Farjadian S, Doroudchi M, Scherer S, Opelz G. et al. Human leukocyte antigen class II allele frequencies and haplotype association in Iranian normal population. Hum Immunol. 2001;2:1234–8. doi: 10.1016/s0198-8859(01)00320-2. [DOI] [PubMed] [Google Scholar]
- 30. Kemp A. March of the titans-A history of white race. Burlington, USA: Ostara Publications 1999; Online version 6, http://www.whitehistory.com/index.htm.


