Abstract
Caldesmon is a negative regulator of cell proliferation, migration, and metalloproteinase release. Caldesmon function is regulated by multiple kinases, targeting multiple phosphorylation sites. Recently, overexpression of caldesmon has been shown to inhibit neointimal formation after experimental angioplasty, suggesting that caldesmon may be a potential therapeutic target for proliferative vascular diseases.
Keywords: Actin, atherosclerosis, caldesmon, calmodulin, restenosis, vascular smooth muscle
INTRODUCTION
Proliferative vascular diseases such as atherosclerosis and restenosis are characterized by inflammation-induced vascular smooth muscle cell proliferation, and migration from the media to intima [1–3]. In addition, metalloproteinase release by vascular smooth muscle cells is essential for the degradation of extracellular matrix, a necessary step for the invasion of vascular smooth muscle cells from the media to intima [4–7]. Caldesmon, a calcium-calmodulin and actin-binding protein found most abundantly in smooth muscle cells, is a negative regulator of cell proliferation, cell migration, and metalloproteinase release (Fig. 1). Caldesmon stabilizes the actin cytoskeleton by multiple molecular mechanisms, including protection of filamentous actin against severing by gelsolin, inhibition of actomyosin interaction, and inhibition of Arp2/3-mediated actin polymerization [8]. Furthermore, multiple kinases regulate the inhibitory function of caldesmon by targeting multiple sites (Table 1). This brief review summarizes findings that suggest caldesmon expression and caldesmon phosphorylation may be potential targets for the treatment of proliferative vascular diseases.
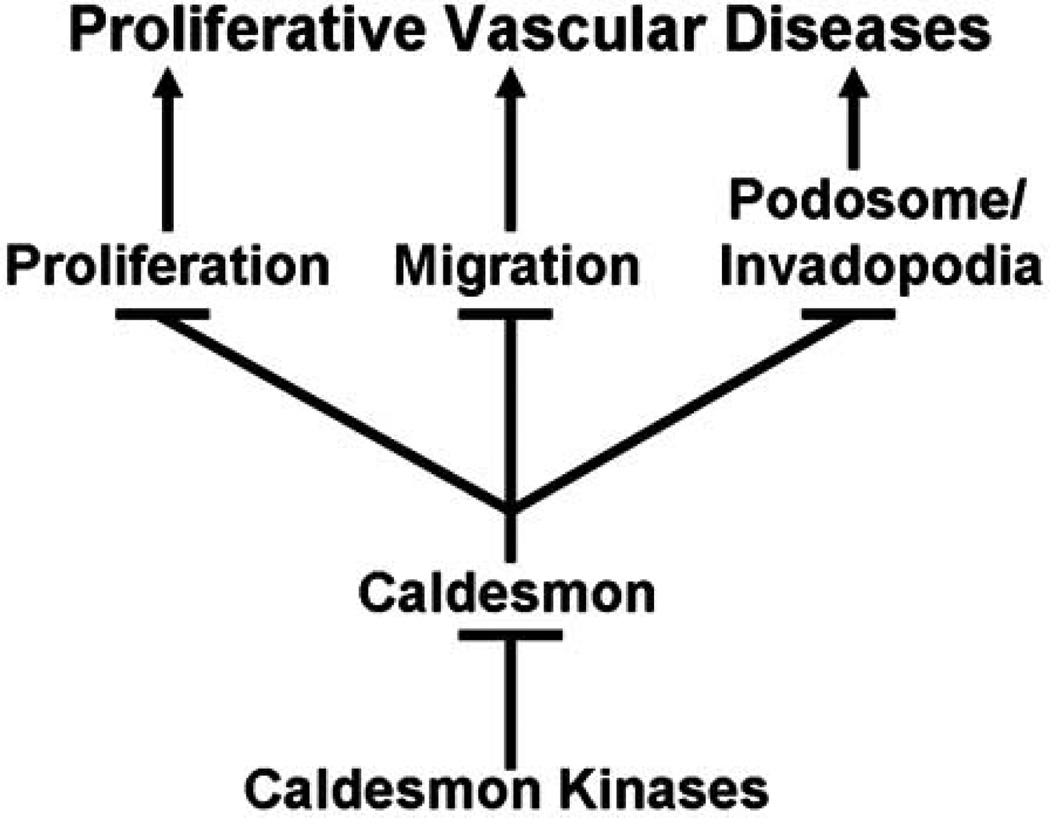
Fig. (1).
Caldesmon is a negative regulator of vascular smooth muscle cell proliferation, migration, and podosome/invadopodia formation, all of which are critical mechanisms underlying proliferative vascular diseases. Multiple caldesmon kinases phosphorylate caldesmon at multiple residues at the amino and carboxyl termini, thereby, attenuating the inhibitory function of caldesmon.
Table 1.
Caldesmon Kinases
| Kinase | # Sites | C or N Terminus | Cell or Tissue | References |
|---|---|---|---|---|
| CaM Kinase II | 2 | C and N | Duck Gizzard | [61] |
| 8 | C and N | Turkey Gizzard | [60] | |
| Casein Kinase II | 1 | N | Chicken Gizzard | [66, 68] |
| 2 | N | Chicken Gizzard | [67] | |
| 1 | N | Duck Gizzard | [65] | |
| Cdc2 Kinase | 5 | C | Chicken Gizzard | [54] |
| 7 | C | Rat Non-Muscle | [38] | |
| Erk1/2 MAPK | 2 | C | Bovine Aorta | [55] |
| 2 | C | Chicken Gizzard | [56] | |
| 2 | C | Pig Carotid Artery | [57] | |
| 2 | C | Rat A7r5 | [19] | |
| PAK | 2 | C | Chicken Gizzard | [58] |
| 2 | C | CHO-K1 | [50] | |
| PKC | 8 | Unknown | Chicken Gizzard | [62] |
| 3 | C | Chicken Gizzard | [64] | |
| 3 | C | Duck Gizzard | [61] | |
| 3 | C | Pig Stomach | [63] |
Abbreviations: CaM Kinase II - Calmodulin-dependent kinase II; MAPK – Mitogen-Activated Protein kinase; PAK – P21-Activated Kinase; PKC – Protein Kinase C.
VASCULAR SMOOTH MUSCLE CELL PROLIFERATION AND MIGRATION IS A CENTRAL PROBLEM IN PROLIFERATIVE VASCULAR DISEASES
Vascular smooth muscle cells normally function as the contractile component of blood vessels in the regulation of vascular resistance and blood flow. However, under pathophysiological conditions such as atherosclerosis and restenosis, vascular smooth muscle cells undergo dedifferentiation to become a proliferative, non-muscle-like phenotype, which proliferate and migrate from the media towards the intima [9, 10]. For example, in atherosclerotic human aorta, proliferative vascular smooth muscle cells in the subendothelial intima express non-muscle protein isoforms of actin, caldesmon, and vinculin [10]. In addition, proliferative vascular smooth muscle cells release metalloproteinases for the degradation of their surrounding extracellular matrix in order to migrate out of the media to invade the intima [11, 12]. Metalloproteinases may also potentially cause plaque destabilization in atherosclerotic arteries. Therefore, cell proliferation, cell migration, and metalloproteinase release are the fundamental processes underlying proliferative vascular diseases. These three processes, despite their differences in regulation, all utilize actin cytoskeletal reorganization as a common mechanism. For example, cell division requires dynamic reorganization of the actin cytoskeleton during cytokinesis; and cell migration requires dynamic actin polymerization and depolymerization [13]. Similarly, cellular release of metalloproteinases is regulated by podosomes and invadopodia, which are actin cytoskeletal, focal adhesionlike structures [14, 15]. Podosomes are filamentous actin cytoskeletal columns arising from the ventral surface of a cell. A large number of signaling and structural proteins are concentrated at the core and ring of podosome columns, associated with the release of metalloproteinases at the bottom. Podosomes are dynamic structures that reorganize constantly over time and space. Monocyte-derived cells such as macrophages and osteoclasts utilize podosomes exclusively for the organization of filamentous actin and metalloproteinase release in extracellular matrix invasion and remodeling. Invadopodia are ventral protrusions of invasive tumor or transformed cells, associated with the release of metalloproteinases for the degradation of extracellular matrix. Recent studies have documented the formation of podosomes and invadopodia in both primary and immortalized vascular smooth muscle cells [16–20].
CALDESMON IS A NEGATIVE REGULATOR OF CELL PROLIFERATION, MIGRATION, CONTRACTILITY, AND METALLOPROTEINASE RELEASE
Caldesmon is a calcium-calmodulin, and actin-binding protein found most abundantly in smooth muscle cells. In mammalian cells, caldesmon is expressed in two isoforms by differential transcription of a single gene [21–23]. The higher molecular-weight isoform (h-caldesmon) is expressed in differentiated, contractile smooth muscle cells, whereas the lower molecular-weight isoform (l-caldesmon), lacking the central region, is expressed in proliferative vascular smooth muscle cells, and non-muscle cells [24–28]. As an exception, mouse T-lymphoma cells express h-caldesmon, and utilize the molecule in the regulation of receptor capping [29]. Actin-binding domains of caldesmon are located predominantly at the carboxyl terminus, whereas myosin-binding domain of caldesmon is located at the amino terminus of the molecule. Since the major functional domains of caldesmon are located at the two ends of the molecule, the two isoforms of caldesmon, with or without the central region, appear to perform similar functions in cells [30, 31].
Caldesmon stabilizes the actin cytoskeleton by multiple molecular mechanisms, which include: a) protection of filamentous actin against severing by gelsolin, b) inhibition of actomyosin interaction, and c) inhibition of Arp2/3-mediated actin polymerization. Ishikawa et al. [32, 33] were the first to report that caldesmon, by potentiating the effect of tropomyosin, protects actin filaments against severing by gelsolin, and also enhances tropomyosin-mediated annealing of gelsolin-severed actin fragments. Based on these findings, they further discovered the dissociation of caldesmon from the actin cytoskeleton during mitosis, and showed that cdc2 kinase-dependent caldesmon phosphorylation is the underlying mechanism [34–38]. The functional significance of caldesmon phosphorylation in mitosis was further confirmed by Yamashiro et al. [39], who showed that expression of mutant caldesmon lacking cdc2 kinase-dependent phosphorylation sites delayed M-phase entry and inhibited cytokinesis in CHO cells. Altogether, these studies established the functional significance of cdc2 kinase-dependent caldesmon phosphorylation in the regulation of cytokinesis during cell division. Recently, Kordowska et al. [40] showed that caldesmon phosphorylation at the Ser497 and Ser527 sites by both cdc2 kinase and Erk1/2 MAPK was critical for the disassembly of actin stress fibers and post-mitotic spreading in non-muscle and vascular smooth muscle cells.
Gelsolin plays a critical role in the assembly of podosomes and motility in monocyte-derived cells such as osteoclasts, which utilize podosomes instead of focal adhesions for attachment to and degradation of the extracellular matrix in bone remodeling. Gelsolin deficiency has been shown to block podosome assembly by osteoclasts, resulting in the increase of bone mass in gelsolin-null mice [41]. Caldesmon protects the actin filament against severing by gelsolin [42], which may be a mechanism by which caldesmon regulates the assembly of podosomes in vascular smooth muscle cells [19, 43].
Caldesmon is found most abundantly in smooth muscle cells [44], and inhibits actomyosin interactions [45]. Although it is generally recognized that phosphorylation of the 20,000 dalton myosin light chain is the central mechanism of smooth muscle contraction, it has been hypothesized that caldesmon may be an additional thin filament-based regulatory mechanism of smooth muscle contraction [46]. The validity of this hypothesis remains controversial. An alternative hypothesis is that caldesmon-dependent inhibition of actomyosin contractility may decrease mechanical stress at the focal adhesions, thereby inducing the disassembly of focal adhesions [9]. This alternative hypothesis appears to be supported by the finding that caldesmon overexpression inhibits the formation of focal adhesions and cell motility in nonmuscle cells [47]. Caldesmon interacts directly with the actin-binding protein cortactin [48], but the physiological function of this interaction has not been elucidated. Caldesmon inhibits the binding of Arp2/3 complex to actin, thereby inhibiting Arp2/3-mediated actin nucleation and polymerization [49]. Arp2/3 complex initiates the branching of actin filaments, an essential process for the formation of membrane protrusions such as lamellopodia, filopodia, podosomes, and invadopodia [13]. Kaverina et al. [18] have demonstrated the essential role of Arp2/3-mediated actin polymerization in the formation of podosomes in vascular smooth muscle cells. Eppinga et al. [50] have demonstrated the essential role of caldesmon phosphorylation by PAK for the formation of lamellopodia in non-muscle cells [50]. The inhibitory effect of caldesmon on the formation of podosomes and lamellopodia may be explained, in part, by the inhibitory effect of caldesmon on Arp2/3-mediated actin polymerization.
CALDESMON PHOSPHORYLATION BY MULTIPLE KINASES
As shown in Table 1, multiple kinases phosphorylate caldesmon, thereby attenuating the inhibitory effect of caldesmon on multiple cellular processes (Fig. 1). Dephosphorylation of caldesmon is catalyzed by protein phosphatase(s), but relatively little has been published on caldesmon phosphatases, except that one caldesmon phosphatase has been identified as a type 2A protein phosphatase [51]. As shown in Table 1, majority of the caldesmon kinases phosphorylate sites at the actin-binding carboxyl terminus of caldesmon, which may lead to the detachment of caldesmon from F-actin, and removing the inhibitory effect of caldesmon [52, 53]. Among the six caldesmon kinases shown in Table 1, cdc2 kinase, Erk1/2 MAPK, and PAK have been well documented as regulators of caldesmon function on the actin cytoskeleton. For example, cdc2 kinase-dependent phosphorylation of caldesmon is essential for the normal progression of cytokinesis during mitosis of non-muscle cells [39]. Since proliferative vascular smooth muscle cells are functionally similar to non-muscle cells, cdc2 kinase may play an important role in proliferative vascular diseases. Cdc2 kinase-dependent phosphorylation of h-caldesmon has been demonstrated in chicken gizzard smooth muscle [54], but the physiological function of cdc2 kinase in differentiated smooth muscle remains unknown.
Erk1/2 MAPK and PAK-dependent caldesmon phosphorylation have been observed in both differentiated and proliferative smooth muscle cells [19, 50–58]. Since Erk1/2 MAPK is a substrate of PKC, and caldesmon is an inhibitor of actomyosin interactions, it has been hypothesized that the PKC/Erk1/2 MAPK/caldesmon phosphorylation cascade is a regulatory mechanism of smooth muscle contraction [46]. The validity of this hypothesis remains controversial. Recent findings suggest that Erk1/2 MAPK and PAK-dependent caldesmon phosphorylation regulate the formation of podosomes and invadopodia in vascular smooth muscle cells, leading to the release of metalloproteinase release and extracellular matrix degradation [19, 20, 59]. Altogether, available data suggest the alternative hypothesis that a physiological function of caldesmon in vascular smooth muscle cells is stabilization of the actin cytoskeleton against cell proliferation and migration.
The two caldesmon kinases, CaM kinase II and PKC, are unique among the multiple caldesmon kinases, as shown in Table 1, in being activated by Ca2+ and diacylglycerol, respectively, which are second messengers for activating smooth muscle contraction. CaM kinase II phosphorylates both the actin-binding carboxyl and myosin-binding amino termini of caldesmon in differentiated smooth muscle cells [60, 61], and therefore may regulate the tethering of actin and myosin filaments for cell contractility and migration. As shown in Table 1, PKC phosphorylates up to eight sites in caldesmon [62], of which only three sites have been identified at the carboxyl terminus of caldesmon in differentiated smooth muscle cells [61, 63, 64]. It is noteworthy that PKC activates Erk1/2 MAPK, and therefore may indirectly phosphorylate the Erk1/2 MAPK-dependent sites on caldesmon [46]. Recently, one study suggests that the PKC/Erk1/2 MAPK/caldesmon phosphorylation cascade regulates podosome size and lifetime in vascular smooth muscle cells [19]. Casein kinase II phosphorylates up to two sites at the myosin-binding amino terminus of caldesmon in differentiated smooth muscle cells [65–68]. Given the recent recognition that caldesmon regulates actin cytoskeletal dynamics, it may be worthwhile investigating the function of CaM kinase II, PKC, and casein kinase II in the regulation of vascular smooth muscle proliferation, migration, and metalloproteinase releases (Fig. 1).
Table 1 reveals the tremendous redundancy in the regulation of caldesmon phosphorylation by multiple kinases, whereas Fig. (1) highlights the function of caldesmon as a common mechanism for the regulation of the three essential cellular processes underlying proliferative vascular diseases. Thus, caldesmon appears to be well positioned as a molecular target for the treatment of proliferative vascular diseases.
CALDESMON AS A THERAPEUTIC TARGET FOR PROLIFERATIVE VASCULAR DISEASES
Gene delivery of anti-proliferative proteins to the site of arterial injury has been proposed as a therapeutic approach to proliferative vascular diseases [69]. The unique function of caldesmon as a negative regulator of the three essential processes underlying proliferative vascular diseases (Fig. 1) suggests that gene delivery of caldesmon to the site of arterial injury is a potential therapeutic approach to the treatment of proliferative diseases such as restenosis. It is noteworthy that ectopic expression of caldesmon has been shown to regulate the formation of podosome/invadopodium in cancer cells, and suppress cancer invasion [70]. Recently, overexpression of l-caldesmon has been shown to suppress cell growth and survival of vascular smooth muscle cells, and inhibit neointimal formation after experimental angioplasty [71]. These encouraging findings suggest that overexpression of caldesmon by gene delivery may be a potential therapeutic approach to proliferative vascular diseases such as atherosclerosis and restenosis.
The incorporation of pharmacologic antagonists of cell cycle proteins into vascular stent is becoming a standard in the prevention of vascular restenosis after angioplasty [72]. Similarly, drug-eluted stents loaded with anti-inflammatory, anti-migratory, anti-proliferative drugs have been used successfully for maintaining the patency of coronary arteries after angioplasty [73–77]. Accordingly, the incorporation of modulators of caldesmon phosphorylation, for example, caldesmon kinase inhibitor(s) or caldesmon phosphatase activator(s), into drug-eluted stents may be a novel approach to suppressing vascular restenosis. It is noteworthy that topical application of antisense cdc2 oligonucleotide has been shown to suppress neointimal smooth muscle accumulation in rat carotid artery [78]; however, it is not clear how much caldesmon contributed to the suppression. Relatively specific inhibitors of Casein Kinase II and Erk1/2 MAPK are available, which include U0126 and PD098059 for Erk1/2 MAPK [79], and TBB for Casein Kinase II [80]. Similarly, relatively specific PKC isoform-specific inhibitors are available, which include Go6976 for targeting conventional PKC-α, and LY333531 for targeting the PKCβ [81]. Small-molecule inhibitors have been developed to target PAK1 at the ATP-binding (CEP-1347) and cdc42-binding (Emodin) sites; however, the potency and specificity of these inhibitors are relatively low [82]. Commonly used CaM Kinase II inhibitors such KN-92 and KN-93 are relatively non-specific. However, discovery of the endogenous CaMK II inhibitor, CaM-KIIN, has led to development of tatCN21, a cell-permeable tat-fusion peptide that appears to be CaMK II-specific [83]. Cdc2 Kinase is a member of the family of cyclin-dependent kinases, which have been targeted for cancer treatment. However, available inhibitors of cyclin-dependent kinases are generally non-selective. For example, AZ703 and NU6102 inhibit both Cdc2 Kinase and cdk2 with similar potencies [84]. The existence of multiple caldesmon kinases, as shown in Table 1, suggests a high degree of redundancy in the phosphorylation of caldesmon. Therefore, simultaneous targeting of multiple caldesmon kinases may be necessary to regulate caldesmon phosphorylation in the treatment of proliferative vascular diseases.
ACKNOWLEDGEMENTS
This study was supported by National Heart, Lung, and Blood Institute Grant HL-52714.
RFEFERFENCES
- 1.Kaperonis EA, Liapis CD, Kakisis JD, Dimitroulis, Papavassiliou VG. Eur. J. Vasc. Endovasc. Surg. 2006;31:386. doi: 10.1016/j.ejvs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Rzucidlo EM, Martin KA, Powell RJ. J. Vasc. Surg. 2007;45:25A. doi: 10.1016/j.jvs.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Weintraub WS. Am. J. Cardiol. 2007;100(suppl):3K. doi: 10.1016/j.amjcard.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Dollery CM, Libby P. Cardiovasc. Res. 2006;69:625. doi: 10.1016/j.cardiores.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Touchard A, Henry TD, Sangiorgi G, Spagnoli LG, Mauriello A, Conover C, Schwartz RS. Arterioscler. Throb. Vasc. Biol. 2005;25:1119. doi: 10.1161/01.ATV.0000164311.48592.da. [DOI] [PubMed] [Google Scholar]
- 6.Newby AC. Phyisol. Rev. 2005;85:1. doi: 10.1152/physrev.00048.2003. [DOI] [PubMed] [Google Scholar]
- 7.Newby AC. Cardiovas. Res. 2006;69:614. doi: 10.1016/j.cardiores.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Hai C-M, Gu Z. Eur. J. Cell Biol. 2006;85:305. doi: 10.1016/j.ejcb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Gerthoffer WT. Circ. Res. 2007;100:607. doi: 10.1161/01.RES.0000258492.96097.47. [DOI] [PubMed] [Google Scholar]
- 10.Glukhova MA, Kabakov AE, Frid MG, Ornatsky OI, Belkin AM, Mukhin DN, Orekhov AN, Koteliansky VE, Smirnov VN. Proc. Natl. Acad. Sci. USA. 1988;85:9542. doi: 10.1073/pnas.85.24.9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzuya M, Iguchi A. J. Atheroscler. Thrombo. 2003;10:275. doi: 10.5551/jat.10.275. [DOI] [PubMed] [Google Scholar]
- 12.Lijnen HR. Pathophysiol. Haemost. Thrombo. 2004;33:275. doi: 10.1159/000083814. [DOI] [PubMed] [Google Scholar]
- 13.Small JV, Stradal T, Vignal E, Rottner K. Trends Cell Biol. 2002;12:112. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- 14.Buccione R, Orth JD, McNiven M. Nat. Rev. Mol. Cell Biol. 2004;5:647. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- 15.Linder S, Kopp P. J. Cell Sci. 2005;118:2079. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- 16.Burgstaller G, Gimona M. Am. J. Physiol. Cell Physiol. 2005;288:H3001. doi: 10.1152/ajpheart.01002.2004. [DOI] [PubMed] [Google Scholar]
- 17.Hai C-M, Hahne P, Harrington EO, Gimona M. Exp. Cell Res. 2002;280:64. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- 18.Kaverina I, Stradal TEB, Gimona M. J. Cell Sci. 2003;116:4915. doi: 10.1242/jcs.00818. [DOI] [PubMed] [Google Scholar]
- 19.Gu Z, Kordowska J, Williams GL, Wang C-LA, Hai C-M. Exp. Cell Res. 2007;313:849. doi: 10.1016/j.yexcr.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furmaniak-Kazmierczak E, Crawley SW, Carter RL, Maurice DH, Cote GP. Circ. Res. 2007;100:1328. doi: 10.1161/CIRCRESAHA.106.147744. [DOI] [PubMed] [Google Scholar]
- 21.Haruna M, Hayashi K, Yano H, Takeuchi O, Sobue K. Biochem. Biophys. Res. Comm. 1993;197:145. doi: 10.1006/bbrc.1993.2453. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi K, Yano H, Hashihda T, Takeuchi R, Takeda O, Asada K, Takahashi E-I, Kato I, Sobue K. Proc. Natl. Acad. Sci. USA. 1992;89:12122. doi: 10.1073/pnas.89.24.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yano H, Hayashi K, Haruna M, Sobue K. Biochem. Biophys. Res. Comm. 1994;201:618. doi: 10.1006/bbrc.1994.1746. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi K, Fujio Y, Kato I, Sobue K. J. Biol. Chem. 1991;266:355. [PubMed] [Google Scholar]
- 25.Kashiwada K, Nishida W, Hayashi K, Ozawa K, Yamanaka Y, Saga H, Yamashita T, Tohyama M, Shimada S, Sato K, Sobue K. J. Biol. Chem. 1997;272:15396. doi: 10.1074/jbc.272.24.15396. [DOI] [PubMed] [Google Scholar]
- 26.Owada MK, Hakura A, Iida K, Yahara I, Sobue K, Kakiuchi S. Proc. Natl. Acad. Sci. USA. 1984;81:3133. doi: 10.1073/pnas.81.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobue K, Tanaka T, Kanda K, Ashino N, Kakiuchi S. Proc. Natl. Acad. Sci. USA. 1985;82:5025. doi: 10.1073/pnas.82.15.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueki N, Sobue K, Kanda K, Hada T, Higashino K. Proc. Natl. Acad. Sci. USA. 1987;84:9049. doi: 10.1073/pnas.84.24.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker G, Kerrick GL, Bourguignon LYW. J. Biol. Chem. 1989;264:496. [PubMed] [Google Scholar]
- 30.Yamakita Y, Yamashiro S, Matsumura F. J. Cell Biol. 1990;111:2487. doi: 10.1083/jcb.111.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marston SB, Redwood CS. Biochem. J. 1991;279:1. doi: 10.1042/bj2790001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa R, Yamashiro S, Matsumura F. J. Biol. Chem. 1989;264:7490. [PubMed] [Google Scholar]
- 33.Ishikawa R, Yamashiro S, Matsumura F. J. Biol. Chem. 1989;264:16764. [PubMed] [Google Scholar]
- 34.Hosoya N, Hosoya H, Yamashiro S, Yamashiro S, Mohri H, Matsumura F. J. Cell Biol. 1993;121:1075. doi: 10.1083/jcb.121.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamakita Y, Yamashiro S, Matsumura F. J. Biol. Chem. 1992;267:12022. [PubMed] [Google Scholar]
- 36.Yamashiro S, Yamakita Y, Ishikawa R, Matsumura F. Nature. 1990;344:675. doi: 10.1038/344675a0. [DOI] [PubMed] [Google Scholar]
- 37.Yamashiro S, Yamakita Y, Hosoya H, Matsumura F. Nature. 1991;349:169. doi: 10.1038/349169a0. [DOI] [PubMed] [Google Scholar]
- 38.Yamashiro S, Yamakita Y, Yoshida K-S, Takiguchi K, Matsumura F. J. Biol. Chem. 1995;270:4023. doi: 10.1074/jbc.270.8.4023. [DOI] [PubMed] [Google Scholar]
- 39.Yamashiro S, Chern H, Yamakita Y, Matsumura F. Mol. Biol. Cell. 2001;12:239. doi: 10.1091/mbc.12.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kordowska J, Hetrick T, Adam LP, Wang C-LA. Exp. Cell Res. 2006:95. doi: 10.1016/j.yexcr.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 41.Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA. J. Cell Biol. 2000;148:665. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dabrowska R, Hinssen H, Galazkiewicz, Nowak E. Biochem. J. 1996;315:753. doi: 10.1042/bj3150753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eves R, Webb BA, Zhou S, Mak AS. J. Cell Sci. 2006;119:1691. doi: 10.1242/jcs.02881. [DOI] [PubMed] [Google Scholar]
- 44.Kakiuchi R, Inui M, Morimoto K, Kanda K, Sobue K, Kakiuchi S. FEBS Lett. 1983;154:351. doi: 10.1016/0014-5793(83)80181-1. [DOI] [PubMed] [Google Scholar]
- 45.Ngai PK, Walsh MP. J. Biol. Chem. 1984;259:13656. [PubMed] [Google Scholar]
- 46.Morgan KG, Gangopadhyay SS. J. Appl. Physiol. 2001;91:953. doi: 10.1152/jappl.2001.91.2.953. [DOI] [PubMed] [Google Scholar]
- 47.Helfman DM, Levy ET, Berthier C, Shtutman M, Riveline D, Grosheva I, Lachish-Zalait A, Elbaum M, Bershadsky AD. Mol. Biol. Cell. 1999;10:3097. doi: 10.1091/mbc.10.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang R, Cao G-J, Guo H, Kordowska J, Wang C-LA. Arch. Biochem. Biophys. 2006;456:175. doi: 10.1016/j.abb.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamakita Y, Oosawa F, Yamashiro S, Matsumura F. J. Biol. Chem. 2003;278:17937. doi: 10.1074/jbc.M208739200. [DOI] [PubMed] [Google Scholar]
- 50.Eppinga RD, Li Y, Lin JL-C, Mak AS, Lin JJ-C. Cell Motil. Cytoskeleton. 2006;63:543. doi: 10.1002/cm.20144. [DOI] [PubMed] [Google Scholar]
- 51.Pato MD, Sutherland C, Winder SJ, Walsh MP. Biochem. J. 1993;293:35. doi: 10.1042/bj2930035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foster DB, Huang R, Hatch V, Craig R, Graceffa P, Lehman W, Wang C-LA. J. Biol. Chem. 2004;279:53387. doi: 10.1074/jbc.M410109200. [DOI] [PubMed] [Google Scholar]
- 53.Greenberg MJ, Wang C-L, Lehman W, Moore JR. Cell Motil. Cytoskeleton. 2008;65:156. doi: 10.1002/cm.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mak AS, Carpenter M, Smillie LB, Wang JH. J. Biol. Chem. 1991;266:19971. [PubMed] [Google Scholar]
- 55.Adam LP, Hathaway DE. FEBS Lett. 1993;322:56. doi: 10.1016/0014-5793(93)81110-l. [DOI] [PubMed] [Google Scholar]
- 56.Childs TJ, Watson MH, Sanghera JS, Campbell DL, Pelech SL, Mak AS. J. Biol. Chem. 1992;267:22853. [PubMed] [Google Scholar]
- 57.D’Angelo G, Graceffa P, Wang C-LA, Wrangle J, Adam LP. J. Biol. Chem. 1999;274:30115. doi: 10.1074/jbc.274.42.30115. [DOI] [PubMed] [Google Scholar]
- 58.Foster DB, Shen L-H, Kelly J, Thibault P, Van Eyk JE, Mak AS. J. Biol. Chem. 2000;275:1959. doi: 10.1074/jbc.275.3.1959. [DOI] [PubMed] [Google Scholar]
- 59.Morita T, Mayanagi T, Yoshio T, Sobue K. J. Biol. Chem. 2007;282:8454. doi: 10.1074/jbc.M609983200. [DOI] [PubMed] [Google Scholar]
- 60.Ikebe M, Reardon S. J. Biol. Chem. 1990;265:17607. [PubMed] [Google Scholar]
- 61.Vorotnikov AV, Shrinsky VP, Gusev NB. FEBS Lett. 1988;236:321. doi: 10.1016/0014-5793(88)80047-4. [DOI] [PubMed] [Google Scholar]
- 62.Umekawa H, Hidaka H. Biochem. Biophys. Res. Commun. 1985;132:56. doi: 10.1016/0006-291x(85)90987-8. [DOI] [PubMed] [Google Scholar]
- 63.Adam LP, Gapinski CJ, Hathaway DR. FEBS Lett. 1992;302:223. doi: 10.1016/0014-5793(92)80446-n. [DOI] [PubMed] [Google Scholar]
- 64.Tanaka T, Ohta H, Kanda K, Tanaka T, Hidaka H, Sobue K. Eur. J. Biochem. 1990;188:495. doi: 10.1111/j.1432-1033.1990.tb15427.x. [DOI] [PubMed] [Google Scholar]
- 65.Bogatcheva NV, Vorotnikov AV, Birukov KG, Shirinsky P, Gusev NB. Biochem. J. 1993;290:437. doi: 10.1042/bj2900437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vorotnikov AV, Gusev NB, Hua S, Collins JH, Redwood CS, Marston SB. FEBS Lett. 1993;334:18. doi: 10.1016/0014-5793(93)81671-l. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Yang Z-Q. Biochemistry. 2000;39:11114. doi: 10.1021/bi0006767. [DOI] [PubMed] [Google Scholar]
- 68.Wawrzynow A, Collins JH, Bogatcheva NV, Vorotnikow AV, Gusev NB. FEBS Lett. 1991;289:213. doi: 10.1016/0014-5793(91)81072-g. [DOI] [PubMed] [Google Scholar]
- 69.Kishore R, Losordo DW. J. Mol. Cell. Cardiol. 2007;42:461. doi: 10.1016/j.yjmcc.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 70.Yoshio T, Morita T, Kimura Y, Tsujii M, Hayashi N, Sobue K. FEBS. Lett. 2007;581:3777. doi: 10.1016/j.febslet.2007.06.073. [DOI] [PubMed] [Google Scholar]
- 71.Yokouchi K, Numaguchi Y, Kubota R, Ishii M, Imai H, Murakami R, Ogawa Y, Kondo T, Okumura K, Ingber DE, Murohara T. Arterioscler. Thromb. Vasc. Biol. 2006;26:2231. doi: 10.1161/01.ATV.0000239441.29687.97. [DOI] [PubMed] [Google Scholar]
- 72.Sriram V, Patterson C. Circulation. 2001;103:2414. doi: 10.1161/01.cir.103.19.2414. [DOI] [PubMed] [Google Scholar]
- 73.Anis RR, Karsch KR, Oberhoff M. Cardiovasc. Haematol. Disord. Drug Targets. 2006;6:245. doi: 10.2174/187152906779010755. [DOI] [PubMed] [Google Scholar]
- 74.Indolfi C, Mongiardo A, Curio A, Torella D. Trends Cardiovasc. Med. 2003;13:142. doi: 10.1016/s1050-1738(03)00038-0. [DOI] [PubMed] [Google Scholar]
- 75.Min S-K, Kenagy RD, Clowes AW. J. Vasc. Surg. 2007;47:662. doi: 10.1016/j.jvs.2007.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moreno R, Macaya C. Curr. Med. Chem. Cardiovasc. Hematol. Agents. 2005;3:221. doi: 10.2174/1568016054368160. [DOI] [PubMed] [Google Scholar]
- 77.Slavin L, Chhabra A, Tobis JM. Cardiol. Rev. 2007;15:1. doi: 10.1097/01.crd.0000200844.16899.fc. [DOI] [PubMed] [Google Scholar]
- 78.Abe J, Zhou W, Taguchi J, Takuwa N, Miki K, Okazaki H, Kurokawa K, Kumada M, Takuwa Y. Biochem. Biophys. Res. Commun. 2007;198:16. doi: 10.1006/bbrc.1994.1003. [DOI] [PubMed] [Google Scholar]
- 79.Duan W, Wong WSF. Curr. Drug Targets. 2006;7:691. doi: 10.2174/138945006777435353. [DOI] [PubMed] [Google Scholar]
- 80.Sarno S, Moro S, Meggio F, Zagotto G, Dal Ben D, Ghisellini P, Battistutta R, Zanotti G, Pinna LA. Pharmacol. Ther. 2002;93:159. doi: 10.1016/s0163-7258(02)00185-7. [DOI] [PubMed] [Google Scholar]
- 81.Mackay HJ, Twelves CJ. Nat. Rev. Cancer. 2007;7:554. doi: 10.1038/nrc2168. [DOI] [PubMed] [Google Scholar]
- 82.Kumar R, Gururaj AE, Barnes CJ. Nat. Rev. Cancer. 2006;6:459. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 83.Vest RS, Davies KD, O’Leary H, Port JD, Bayer KU. Mol. Biol. Cell. 2007;18:5024. doi: 10.1091/mbc.E07-02-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shapiro GI. J. Clin. Oncol. 2006;24:1770. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]