Abstract
The years 2000 and 2007 witnessed milestones in current understanding of G protein-coupled receptor (GPCR) structural biology. In 2000 the first GPCR, bovine rhodopsin, was crystallized and the structure was solved, while in 2007 the structure of β2-adrenergic receptor, the first GPCR with diffusible ligands, was determined owing to advances in microcrystallization and an insertion of the fast-folding lysozyme into the receptor. In parallel with those crystallographic studies, the biological and biochemical characterization of GPCRs has advanced considerably because those receptors are molecular targets for many of currently used drugs. Therefore, the mechanisms of activation and signal transduction to the cell interior deduced from known GPCRs structures are of the highest importance for drug discovery. These proteins are the most diversified membrane receptors encoded by hundreds of genes in our genome. They participate in processes responsible for vision, smell, taste and neuronal transmission in response to photons or binding of ions, hormones, peptides, chemokines and other factors. Although the GPCRs share a common seven-transmembrane α-helical bundle structure their binding sites can accommodate thousands of different ligands. The ligands, including agonists, antagonists or inverse agonists change the structure of the receptor. With bound agonists they can form a complex with a suitable G protein, be phosphorylated by kinases or bind arrestin. The discovered signaling cascades invoked by arrestin independently of G proteins makes the GPCR activating scheme more complex such that a ligand acting as an antagonist for G protein signaling can also act as an agonist in arrestin-dependent signaling. Additionally, the existence of multiple ligand-dependent partial activation states as well as dimerization of GPCRs result in a ‘microprocessor-like’ action of these receptors rather than an ‘on-off’ switch as was commonly believed only a decade ago.
Keywords: G protein-coupled receptors, rhodopsin, β-adrenergic receptors, chemokine receptors, G protein, arrestin
INTRODUCTION
In 2012 the Nobel Prize in chemistry was awarded jointly to two American scientists — Robert J. Lefkowitz of Duke University in Durham, North Carolina and Brian K. Kobilka of Stanford University School of Medicine in Palo Alto, California “for studies of G protein-coupled receptors (GPCRs)”. The scientific achievements of Lefkowitz and Kobilka have allowed the pharmaceutical industry to develop more selective drugs with the hope of fewer side effects through improved understanding of the signaling mechanisms initiated by endogenous substances such as adrenalin, serotonin, histamine, dopamine and many other hormones and neurotransmitters. Lefkowitz started his work on GPCRs by identifying adrenergic receptors (α and β) affected by adrenaline in the 1980s (Shorr et al., 1981; Dixon et al., 1986). With the help of his student, Kobilka, he noticed that the genes for β-adrenergic receptors shows remarkable similarities to the one for rhodopsin, known then as a light-sensing receptor of the eye (Filipek et al., 2003; Palczewski, 2006), and concluded that there likely exists an entire protein family of such receptors with similar structure and function (Lefkowitz, 2000). Another key advance in Lefkowitz’s research was identification of the β-adrenergic receptor kinase (Benovic et al., 1986) followed by studies on β-arrestins (Luttrell & Lefkowitz, 2002; Lefkowitz & Shenoy, 2005). Continuing his work at Stanford, Kobilka together with Stevens from the Scripps Research Institute in California solved the crystal structure of β2-adrenergic receptor stabilized by lysozyme (Cherezov et al., 2007) and together with Schertler from MRC (Cambridge, UK) the structure of the same receptor stabilized by an antibody in the same year (Rasmussen et al., 2007). The structure of the second GPCR (after vertebrate rhodopsin (Palczewski et al., 2000)) confirmed the hypothesis about the common folding of all GPCRs comprising a seven-transmembrane helical bundle (Fig. 1). The structure of β2-adrenergic receptor was followed by several crystal structures of other GPCRs solved by e.g. the Stevens’ group (Jaakola et al., 2008). Those investigators employed lysozyme, apocytochrome BRIL, and nanobody molecules (Cherezov, et al., 2007) to stabilize not only the inactive state of these highly flexible receptors but also their extremely unstable active state in complex with trimeric G protein (Rasmussen et al., 2011). Other investigators successfully employed thermo-stabilizing mutations and truncations (Tate & Schertler, 2009; Lebon et al., 2012).
Figure 1. A scheme of shapes and tilts of transmembrane helices of GPCRs based on the representative crystal structure of β2-adrenergic receptor (PDB id: 2RH1).
Location of a ligand is marked by a red sphere whereas the location of protein G by a blue sphere.
Seven-transmembrane spanning GPCRs are critically involved in transmitting extracellular stimuli including light, hormones and neurotransmitters into specific cellular responses (Muller et al., 2008). GPCRs are also pharmacologically important because they are the targets of about 30% of commercially available drugs (Salon et al., 2011). The structure of vertebrate rhodopsin determined in 2000 marked the first three-dimensional atomic structure of any native GPCR. Significant advances in the field include the structural determination of truncated invertebrate rhodopsin, adrenergic, histamine H1, adenosine A2A, opioid, muscarinic acetylcholine, CXCR4, dopamine D3 and other receptors as well. In many cases GPCRs were modified with fusion proteins such as lysozyme. Whereas non-rhodopsin GPCRs require heterologous expression and extensive protein engineering to enable their stabilization and crystallization, bovine rhodopsin remains the only native, intact GPCR with a determined structure (Palczewski, 2012). GPCRs contain the seven-transmembrane helical bundle that provides a binding site for their ligands. The ligand binding triggers a slight change in GPCR conformation that is propagated through the whole protein, ultimately causing alterations at the receptor’s cytoplasmic surface that permit binding to its cognate G protein. Today, crystal structures of all photoactivated intermediates of rhodopsin and several agonist- and antagonist-bound GPCRs have been determined (Palczewski, 2012). Moreover, the critically important GPCR-G protein complex structure also has become better characterized by low- and high-resolution experimental methods (Jastrzebska et al., 2011b; Rasmussen, et al., 2011; Orban et al., 2012). Research on GPCRs still remains of a high priority. Hopefully, knowing better the connection between structure and function of GPCRs will improve our understanding of these molecular machines as well as will promote their potential pharmacological applications.
GPCRs comprise seven transmembrane α-helices (7TMH) connected by three extracellular loops (ECL1-3) and three intracellular loops (ICL1-3) (Fig. 1). The ex-tracellular (EC) region, which is responsible for ligand binding, also includes the N-terminus that can range from relatively short sequences in rhodopsin-like receptors to large extracellular domains in other classes of GPCRs, e.g. the hormone-binding domain (HBD) in adhesion receptors. The intracellular (IC) region interacts with G proteins, arrestins and other downstream effectors. It includes cytoplasmic helix H8 and a C-terminus that may provide sites for palmitoylation. The 7TM helical bundle contains a number of kinks (Fig. 1), mostly induced by Pro residues, that roughly divide the receptor into the EC and IC regions. The EC module responsible for ligand binding features a high structural diversity but small movement during activation. In contrast, the IC region, which is involved in binding downstream proteins including G proteins and arrestins, is more conserved in the GPCR family but is subjected to much larger conformational changes upon receptor activation than EC (Katritch et al., 2012).
The structures of GPCRs exhibit only limited differences despite the fact that the receptors were crystallized differently: in different crystal packing orientations, with different antagonists and inverse agonists bound and also in two different activation states, either activated (agonist bound) or inactivated (antagonist or inverse-agonist bound) (Rasmussen et al., 2011). Even the conformations of extracellular loops (the most divergent part of GPCR structures) can also be similar as is the case of crystal structures of the following receptors: dopamine receptor D3R (Chien et al., 2010), muscarinic receptors M2R (Haga et al., 2012) and M3R (Kruse et al., 2012), CXCR4 (Wu et al., 2010), μOR (Manglik et al., 2012), δOR (Granier et al., 2012), κOR (Wu et al., 2012), and nociceptin receptor (NOP) (Thompson et al., 2012). In the more distant (by sequence homology) CXCR4 and opioid receptors, ECL2 forms a β-hairpin which is oriented nearly vertically to the membrane surface which is much different than that of rhodopsin, which is oriented horizontally and entirely covers the retinal. However, the ECL2 in rhodopsin is responsible for keeping the ligand in place, whereas in CXCR4 it is crucial for binding of either the small molecule IT1t, or the peptide antagonist CVX15 (they are both ligands in crystal structures of that receptor) that mimics the V3 loop of the HIV envelope glycoprotein gp120. In CXCR4 and opioid receptors, the ligand pocket is much larger than in other solved GPCR structures, and binding of peptide antagonists involves extensive interactions with ECL2. The N-terminus of GPCRs probably also participates in the binding of larger ligands, however, to date its structure has only been resolved in rhodopsin and partially in CXCR4 and also in crystallized peptide neurotensin receptor (White et al., 2012) which is the first receptor structure in the β subfamily of rhodopsin-like GPCRs. Another important region in a GPCR structure, the cytoplasmic helix H8, is anchored to the membrane by palmitoylation and this feature is essential for receptor stabilization and its proper functioning (Maeda et al., 2010). In a group of known GPCR structures, only in the case of CXCR4 and neurotensin receptor the helix H8 shows a disordered behavior. Nevertheless, it does not preclude the possibility of its formation in the cell membrane because this helix is present in all crystallized opioid receptor structures from the same g subfamily of rhodopsin-like GPCRs as CXCR4.
Our current understanding of the GPCR functioning has changed from a simple hypothesis of ‘on-off’ switches to the microprocessor-like action (Kenakin & Miller, 2010). Especially the phenomenon called ‘functional selectivity’, whereby certain ligands initiate only portions of the signaling mechanisms mediated by a given receptor, has opened new horizons for drug discovery. What should be discovered in the nearest future is a new receptor-ligand behavior with quantification of the drug effect in such complex systems. For example, some agonists selectively activate cellular pathways associated with a specific cell type and some antagonists actively induce receptor internalization without its activation. There are also allosteric modulators which can be linked to the co-binding ligands. Agonists are now known to have multiple efficacies that are associated with selected signaling pathways coupled to the receptor. So called ‘functional selectivity’, defined as biased agonism and biased antagonism, is especially interesting in terms of its mechanism and potential therapeutic applications (see an extensive review by Park and colleagues (2008)).
CLASSIFICATION OF GPCRS
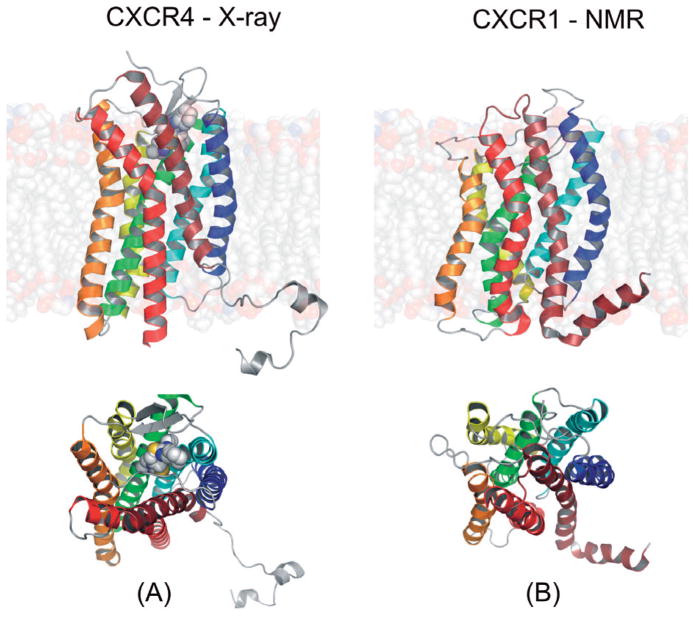
Human GPCRs form a large family of about 800 membrane receptors with sequence lengths between 289 (Mas-related GPCR — uniprot ref. no. Q86SM5) and 3312 (EGF-like protein 1 — uniprot ref. no. Q9NYQ7) residues (Nov. 2012, reviewed Uniprot entries only), with most GPCRs consisting of 300–500 amino acid residues (Mirzadegan et al., 2003). The disparity of sequence lengths is mainly due to the extracellular portion of GP-CRs involved in ligand recognition and/or cell signaling (Fig. 1) such as the N-terminus, extracellular loop 2 (ECL2) and ECL3 (Mirzadegan et al., 2003). The intracellular region of GPCRs, less varied in length than the extracellular part, is involved in signal transduction by interactions with G protein and arrestin. Despite the high degree of sequence variability (Trzaskowski et al., 2012) GPCRs share a common 7TMH core of a size exactly fitted to the cell membrane thickness. Interestingly, the transmembrane helices of GPCRs are frequently tilted with varied tilt and rotation angles that depend not only on the receptor type but also on its activation state. Unfortunately, precise prediction of kinks in the TMHs of GPCRs is still limited as these helical deformations cannot be explained only by the presence of the well-known helix-breakers such as Pro, Ser, Thr or Gly residues. Most likely these deformations are introduced by tertiary interactions which are difficult to capture without a 3D structure of the receptor (Yohannan et al., 2004; Meruelo et al., 2011). In the most commonly used GRAFS classification system (Schioth & Fredriksson, 2005) the GPCR superfamily is divided into five main families: glutamate (former class C), rhodopsin (former class A), adhesion (part of former class B), frizzled/taste2 (former class F), and secretin (part of former class B). Other former classes D (fungal mating pheromone receptors) and E (cyclic AMP receptors) do not contain human receptors and they are not included in the GRAFS classification. So far, only members of the rhodopsin-like receptor family have been crystallized. As of November 2012 there are 15 unique GPCRs deposited through nearly 60 entries in the Protein Data Bank (PDB) involving a variety of ligands, activation states and point mutations. There also have been a few attempts to study GPCR structures by NMR in the ‘membrane-mimicking’ environment of phospholipid bilayers, e.g. CXCR1 by Park et al. (2012), but the resolution of such structures might be still too low to use them for drug discovery.
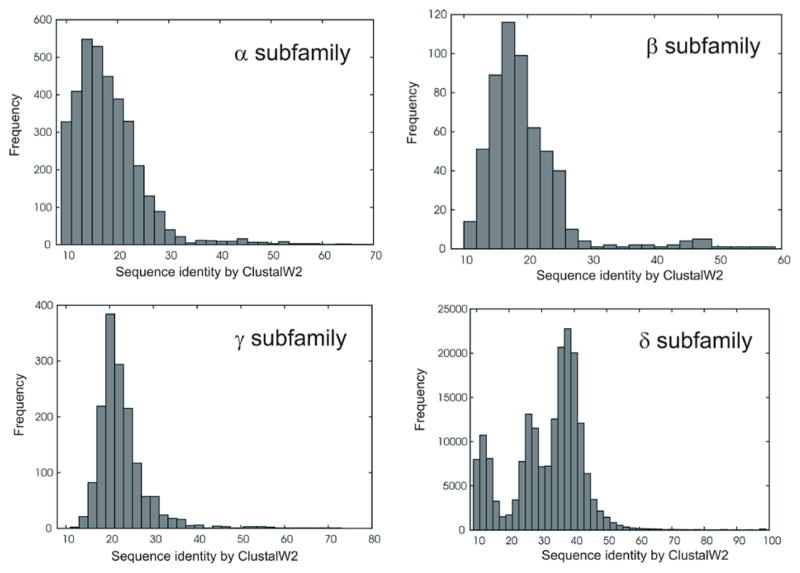
The rhodopsin family can be further divided into four subfamilies: α, β, γ and δ according to the classification of Fredriksson et al. (2003). The α subfamily has five main branches: prostaglandin, amine, opsin, melatonin and adenosine receptors. Currently in the PDB there are crystal structures of amine receptors (histamine H1R, dopamine D3R, muscarinic M2R and M3R, β1- and β2-adrenergic receptors), opsins (rhodopsin), adenosine A2AR and lipid S1P1R receptors). The one-branch β subfamily includes hypocretin receptors, neuro-peptide FF, tachykinin, cholecystokinin, neuropeptide Y, endothelin-related, gastrin-releasing peptide, neuromedin B, uterinbombesin, neurotensin, growth hormone secre-tagogue, neuromedin, thyrotropin releasing hormone, ghrelin, arginine vasopressin, gonadotropin-releasing hormone, oxytocin and orphan receptors. In the β subfamily only neurotensin receptor has been crystallized so far. The γ subfamily consists of three main branches: SOG receptors (including crystallized μOR, δOR, κOR and nociceptin opioid receptors), MCH receptors, and chemokine receptors (including crystallized CXCR4). The last δ subfamily of rhodopsin-like GPCRs has four main branches: Mas-related (oncogene) receptors, glycoprotein receptors, nucleotide receptors and olfactory receptors. However, the δ subfamily has no representative in the PDB so far and only the P2Y12 nucleotide receptor has been selected for crystallization in the near future by the Stevens’ group (see http://gpcr.scripps.edu/tracking_status.htm). The above classification of the rhodopsin family is still under discussion as other methods have provided different shapes for its phylogenetic tree (Surgand et al., 2006; Deville et al., 2009). For example, Pele et al. (2011) split the rhodopsin family into only four subfamilies: G0 — peptide receptors, opsin and melatonin receptors; G1 — somatostatin, opioid, chemokine and nucleotide receptors; G2 — amine and adenosine receptors; and G3 — including melanocortin, S1P and cannabinoid receptors, leucine-rich repeat (LRR)-containing receptors, prostaglandin and Mas-related receptors. The Pele’s classification is not fully consistent with the previous one from Frederiksson et al. as members of G0 are included in both α and β subfamilies, G1 is split between δ and γ, G2 — only α, and finally G3 corresponds to members of both α and δ subfamilies. Even if two GPCRs are classified as members of the same subfamily, they can significantly differ in their amino acid composition (see Fig. 2). A notable exception is the highly populated group of olfactory receptors belonging to the δ subfamily in which most sequences are similar to each other (the two highest peaks in the δ subfamily sequence identity histogram in Fig. 2). In general, sequence diversity is the highest within the extra- and intracellular loop regions, whereas the 7TMH core contains well conserved fragments (motifs) characteristic of GPCRs, for example: ‘D/ERY’ (TMH3), ‘CwxP’ (TMH6) and ‘nPxxy’ (TMH7). The high sequence diversity inside the rhodopsin family corresponds to the high diversity of kinks and bulges in the TM helices and distinct conformations of loops. Even for members of the same subfamily (such as rhodopsin and β2-adrenergic receptor (subfamily α) presented in Fig. 3), their structural diversity still makes homology modeling challenging.
Figure 2.
Histograms of sequence identity between members of four branches of rhodopsin-like family of GPCRs.
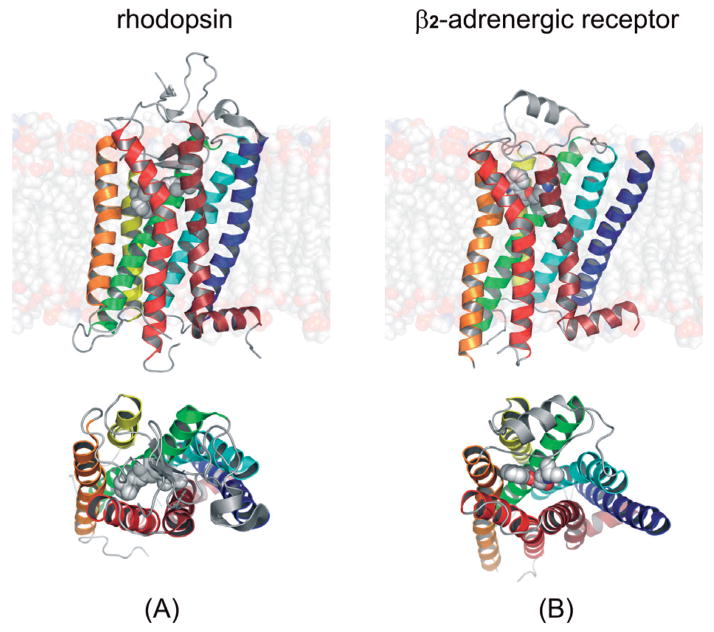
Figure 3. Crystal structures of rhodopsin (PDB id: 1F88) and β2-adrenergic receptor (PDB id: 2RH1).
Top, view along the membrane plane, bottom, from the extracellular side.
CRYSTALLIZATION OF THE FIRST GPCRS — RHODOPSIN AND β2-ADRENERGIC RECEPTOR
The first X-ray crystal structure of any GPCR was that of ground-state rhodopsin (Palczewski, et al., 2000). Refinement of crystallization conditions yielded higher resolution data, extending the model to 2.2 Å, the highest resolution of all rhodopsin structures determined to date (Okada et al., 2004). The first structure of a diffusible ligand-responsive GPCR resulted from work of Kobilka and Stevens, who reported the crystal structure of a human β2-adrenergic receptor-T4 lysozyme fusion protein bound to the partial inverse agonist carazolol at 2.4 Å resolution (Cherezov et al., 2007; Rosenbaum et al., 2007). Several structures of mutant and fusion GPCRs followed (Mustafi & Palczewski, 2009) culminating with a 1.8 Å resolution structure of an engineered human A2A adenosine receptor with its third intracellular loop replaced with apocytochrome b(562)RIL (Liu et al., 2012). This is the highest resolution structure of any GPCR with well-defined Na+ ions and water molecules. Water molecules play critical roles in the GPCR activation process by stabilizing intramolecular interactions (Wikstrom et al., 2003; Osyczka et al., 2005; Garczarek & Gerwert, 2006). Observed in key structurally sensitive areas of many GPCRs, water molecules along with amino acid side chains can form a signal transmission network extending from the ligand-binding site to the cytoplasmic surface (Angel et al., 2009a; 2009b; Orban et al., 2010). Despite their low sequence similarities, the overall folds of structurally determined GPCRs are remarkably similar. For all rhodopsin structures, RMSDs for transmembrane regions are within 1.8 Å, and when adrenergic receptors are compared with rhodopsin, these deviations are 3.3–3.5 A for the β2-adrenergic receptor and 4.3–4.7 Å for the β1-adrenergic receptor (Jaakola, et al., 2008; Lodowski et al., 2009). Remarkably, preservation of only a few essential regions of a GPCR is required for activation of its cognate G protein (Mirzadegan et al., 2003).
Rhodopsin in its inactive, ground state undergoes a series of photointermediate steps upon absorption of a photon and isomerization of its chromophore 11-cis-retinylidene. These photointermediate states exhibit unique absorption maxima and can be isolated by trapping alone or with a G protein-derived peptide analog. Ultimately, the photoisomerized chromophore is hydrolyzed and released from the binding pocket yielding opsin and free all-trans-retinal (Jastrzebska et al., 2011a). Our laboratory expanded upon this work with the structure which exhibits spectral qualities of Meta II, the activated state (Salom et al., 2006; Lodowski et al., 2007). For rhodopsin, only the Meta II intermediate is capable of activating Gt and it differs chemically from other photo-intermediates only by deprotonation of the Schiff base and uptake of a proton from bulk solvent (Salom et al., 2006). By now, the structures of most photo-intermediates have been solved by X-ray and electron crystallographic methods (Breitman et al., 1989; Park et al., 2008; Lodowski et al., 2009). Interestingly, the structures of those rhodopsin photo-intermediates did not exhibit large-scale movements of entire helices. Rather they showed that photoactivation was accomplished with just small-scale, local changes propagated to the cytoplasmic loops, especially the ends of helices V and VI. The 2–8 Å structural shift observed for GPCRs upon activation suggests that such subtle changes directly lead to different receptor activities or indirectly affect the key residues, e.g. the D(E)RY motif on the cytoplasmic surface responsible for the efficacy of G protein coupling along with further conformational changes. This observation is consistent with activation of other GPCRs (summarized by Sprang (2011)).
Structures of opsin revealed marked similarities to photoactivated rhodopsin (Topiol & Sabio, 2009). An additional structure of a photoactivated rhodopsin, obtained by regenerating opsin crystals with all-trans-retinal, superposed well with opsin structures and is spectrally indistinguishable from either our photoactivated rhodopsin structure or Meta II in solution (Choe et al., 2011) (summarized in (Breitman et al., 1989)). Moreover, structures of constitutively active mutants of rhodopsin have also been reported (Standfuss et al., 2007; Standfuss et al., 2011; Xie et al., 2011; Deupi et al., 2012), revealing changes around helices V and VI as compared with rhodopsin that are consistent with proposed models. When one considers the agonist-bound GPCR structures, it becomes readily apparent that the dynamics of the molecule (which make it both a difficult structural target and play a key role in its activation) are recapitulated. The inherent flexibility of GPCRs permits dynamic and conformational changes triggered by only a fraction of the energy derived from ligand binding. Because various agonists can bind to a GPCR leading to varying levels of activity, the small changes induced by agonist binding must account for the differences in the efficacy of such ligands in activating a given G protein (Rosenbaum et al., 2009), suggesting that interaction with signaling proteins may induce further changes on the cytoplasmic surface of these receptors (Sprang 2011). NMR studies of GPCRs, particularly rhodopsin, further advanced and refined those activation models (Smith 2010; 2012; Struts et al., 2011; Eilers et al., 2012).
INTERACTIONS WITH G PROTEIN AND ARRESTIN — PASSING ON THE SIGNAL
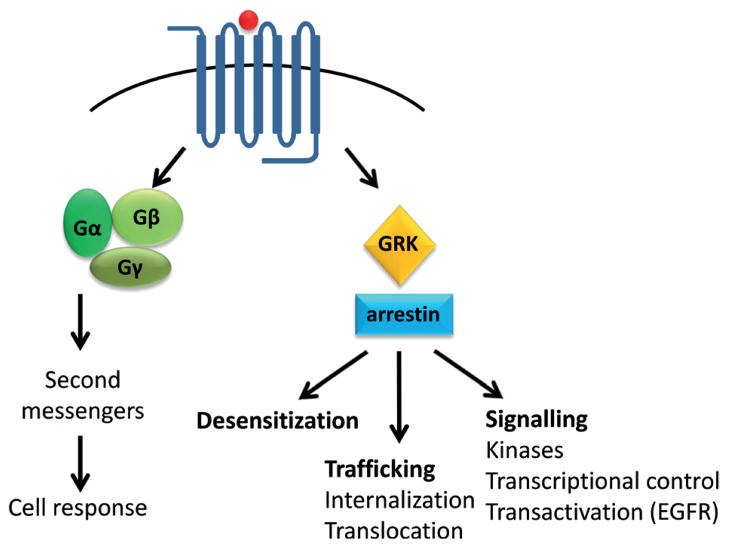
Activation of a GPCR triggers binding of the associated heterotrimeric G protein and nucleotide (GDP) release from its α-subunit. This G protein activation is required for subsequent activation of the cascade of reactions, processes required to advance stepwise signal transduction. The G protein is released from the GPCR by GTP and both its α- and βγ-subunits can activate the effector molecules as adenylyl cyclases and cation channels. Rather than to photon (as in rhodopsin), most GP-CRs respond to molecules called ligands that upon binding to a particular GPCR cause a ligand-specific cellular response. The signal is attenuated by receptor phosphorylation and binding of a capping protein arrestin.
G protein
Heterotrimeric G proteins are composed of a nucleotide-binding α-subunit (Gα) and a dimer consisting of the β- and γ-subunits (Gβγ). In their inactive form, Gα-subunits are bound to GDP and tightly associated with Gβγ. Interactions of β2-adrenergic receptor and Gs (the stimulatory G protein that activates adenylyl cyclase) formed the foundation of the ternary complex model of GPCR activation (Ross et al., 1977; De Lean et al., 1980). In the ternary complex consisting of agonist, receptor and G protein, the affinity of the receptor for the agonist is enhanced and the specificity of the G protein for guanine nucleotides changes in favor of GTP over GDP. Agonist binding to the receptor promotes interactions with the GDP-bound Gsαβγ heterotrimer, leading to the exchange of GDP for GTP and the functional dissociation of G protein into Gα-GTP and Gβγ subunits. These separate subunits can modulate the activity of different cellular effectors (channels, kinases or other enzymes). The intrinsic GTPase activity of Gα leads to hydrolysis of GTP to GDP and the re-association of Gα-GDP and Gβγ subunits with termination of signaling. The active state of a GPCR can be defined as that conformation that couples to and stabilizes a nucleotide-free G protein. In the agonist-β2-adrenergic receptor–Gs ternary complex, Gs has a higher affinity for GTP than for GDP, and the β2-adrenergic receptor has a roughly 100-fold higher affinity for agonists than does β2-adrenergic receptor alone (Rasmussen et al., 2011).
The α-subunit (Gα) of heterotrimeric G proteins mediates signal transduction in a variety of cell signaling pathways. These α-subunits can be divided into four families: Gαs, Gαi/Gαo, Gαq/Gα11, and Gα12/Gα13. Each family comprises various members that often show specific expression patterns. The βγ-complex of mammalian G proteins is assembled from a repertoire of five G protein β-subunits and twelve γ-subunits (Wettschureck & Offermanns, 2005). Most GPCRs are able to activate more than one G protein subtype. Therefore, the activation of a GPCR usually results in the activation of several signal transduction cascades via G protein α-subunits as well as through the freed βγ-complex. G proteins of the Gi/Go family are widely expressed and have been shown to mediate receptor-dependent inhibition of various types of adenylyl cyclases (Sunahara et al., 1996). Because the expression levels of Gi and Go are relatively high, their receptor-dependent activation results in the release of relatively high amounts of βγ-complexes. Activation of Gi/Go is therefore believed to be the major coupling mechanism that results in the activation of βγ-mediated signaling (Clapham & Neer, 1997; Robishaw 2004). The Gq/G11 family of G proteins couples receptors to β-isoforms of phospholipase C (Exton 1996; Rhee 2001). The G proteins G12 and G13 are often activated by receptors coupling to Gq/G11 (Strathmann & Simon, 1990; Dhanasekaran & Dermott, 1996). Analysis of cellular signaling processes regulated through G12 and G13 has been difficult because specific inhibitors of these G proteins are not available. In addition, G12/G13-coupled receptors usually activate other G proteins as well. The ubiquitously expressed G protein Gs couples many receptors to adenylyl cyclase and mediates activation increasing intracellular cAMP concentration (Beavo & Brunton, 2002; Chin et al., 2002). β-adrenergic receptors couple primarily to Gs. The cAMP produced in response to Gs activation directly modulates the gating of hyperpolarization-activated, cyclic nucleotide-gated channels and activates protein kinase A (PKA). PKA in turn phosphorylates many proteins involved in excitation-contraction coupling including L-type Ca2+ channels, phospholamban, and/or troponin I (Bers 2002). Rhodopsin is coupled to rod-transducin (Gt, a homolog of Go), a member of the Gαi/Gαo family. Transducin couples the receptor in a stimulatory manner to cGMP-phosphodiesterase (PDE) by binding and sequestering the inhibitory γ-subunit of the retinal type 6 PDE (PDE6). Activation of PDE lowers cytosolic cGMP levels leading to a decreased probability of cGMP-regulated cation channels in the plasma membrane being open, which eventually causes hyperpolarization of photoreceptor cells (Arshavsky et al., 2002).
The signal captured by photoactivated rhodopsin or an agonist-occupied GPCR propagates along a cell’s plasma membrane to activate a G protein located 40 Å or more away to cause a cellular response. Perhaps our most advanced understanding of this activation process is derived from rhodopsin and the visual system. Absorption of a photon triggers a change in the conformation of rhodopsin’s bound retinal chromophore which is propagated through this receptor, ultimately causing alterations at the cytoplasmic surface that permits binding of G protein transducin, its cognate G protein. This triggers nucleotide release from G protein and its subsequent activation, processes required to advance visual signal transduction. Most GPCRs respond to molecular signals in the form of ligands which upon binding elicit a ligand-specific cellular response. For most GPCRs, the ligand-binding site coincides with the retinylidene-binding pocket in rhodopsin. Thus, ligand binding causes similar conformational changes as those triggered by chromophore photoisomerization in rhodopsin, and the remaining molecular mechanisms for signal transduction are similar for all GPCRs. A model for the photoactivated rhodopsin-G protein complex was described as a 22 Å low-resolution structure from single particle analysis (Jastrzebska et al., 2011b). Its molecular envelope is consistent with dimeric rhodopsin molecules together with one G protein heterotrimer, yielding a 2:1 molar ratio of photoactivated rhodopsin to G protein (Jastrzebska et al., 2011b). The heteropentameric structure for this complex was obtained from native proteins, both rhodopsin and G protein (see also (Jastrzebska et al., 2006)).
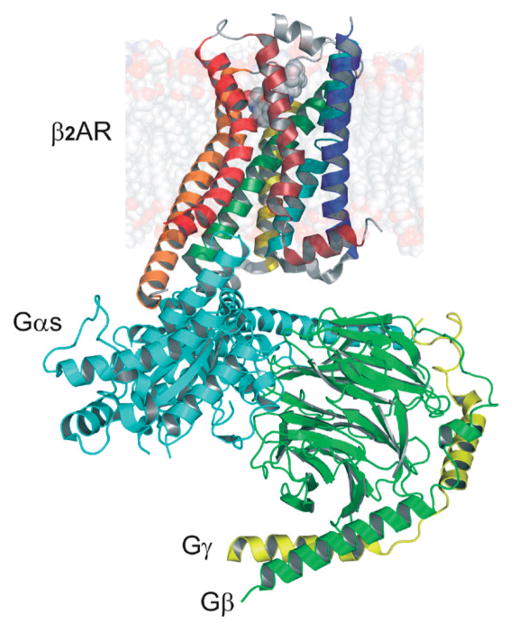
The crystal structure of an active state complex composed of agonist-occupied monomeric β2-adrenergic receptor-T4-lysozyme fusion, a nucleotide-free Gs heterotrimer and a nanobody (Rasmussen et al., 2011) (Fig. 4) may not represent a physiologically relevant complex. First, it was surprising that the receptor was in a monomeric state, as single particle analyses of a similar preparation indicated that the receptor exists to some degree in a dimeric form in solution (Westfield et al., 2011). Second, it was unexpectedly observed that the entire α-helical domain (AH) of Gα was so largely displaced relative to the Ras-like GTPase domain (Rasmussen et al., 2011). This displacement could result from the nano-body stabilization of the β2-adrenergic receptor-Gs complex in the crystal or from the crystallization conditions used. But these discrepancies cannot be explained by differences in the inherent structure of either this GPCR (as mentioned earlier) or its G protein. Both the visual G protein and Gs display a relatively small root-mean-square deviation (~0.5–1.0 A) between structures for the all three subunits. Future research should elucidate this discrepancy.
Figure 4.
Crystal structure of β2-adrenergic receptor in complex with heterotrimeric Gaβ protein (PDB id: 3SN6). View along the membrane plane.
A wide range of different types of studies (Tesmer, 2010) have provided details of the conformational states of Gα, but the mechanism by which the GPCR interaction leads to release of the bound GDP from the Gα subunit and the structure of the resulting empty complex remain a major target for research in the field. The Gα subunit has two structural domains, namely a nucleotide binding or ras-like domain (Ras) and an α-helical domain (AH) that partially occludes the bound nucleotide. Numerous studies indicate that the C-terminus of Gα is bound tightly to the receptor in the nucleotide-free complex (Oldham et al., 2006). In addition, the N-terminal helix of Gα is associated with Gβγ and with the membrane via N-terminal myristoylation (Resh, 1999). Together, these constraints fix the position of the nucleotide domain with respect to the membrane. The helical domain is connected to the ras-like domain through two flexible linkers. The receptor-catalyzed nucleotide exchange in G proteins suggested a large-scale reorientation of domains in the α-subunit (Van Eps et al., 2011; Westfield, et al., 2011). As part of that, binding to a GPCR requires a movement of two helices in Gα, the so called αN and α5 helices at the Gα N- and C-terminus, respectively. A possible sequence of interactions during formation of the nucleotide-free complex has been proposed for the β2-adrenergic receptor-Gs structure (Rasmussen et al., 2011). The first interaction of the β2-adrenergic receptor with the Gs heterotrimer would require a movement of the α5-helix to permit interactions with the β2-adrenergic receptor. The formation of more extensive interactions between the receptor and the amino terminus of Gαs requires a rotation of GαsRas relative to the receptor. This is associated with further conformational changes in both the β2-adrenergic receptor and GαsRas. One cannot say when GDP is released during the formation of the complex. However, it is suggested that the GDP release precedes the uncoupling of the two Gαs domains.
Although much progress has been made in understanding how Gα subunits interact with and regulate the activity of their downstream targets, it is less clear how activated GPCRs initiate this process by catalyzing nucleotide exchange on Gαβγ. The question is of great importance because it represents an essential, pharmacologically relevant interaction that can regulate nearly all aspects of eukaryotic cell physiology. Moreover, an atomic-resolution understanding will explain the GPCR functional selectivity, namely the ability of different agonists to elicit distinct downstream effects from a single GPCR.
Arrestin
Additional knowledge about GPCRs and their interactions with desensitizing proteins has emanated from visual research (Kuhn & Dreyer, 1972; Kuhn, 1974; Weyand & Kuhn, 1990). Photoactivated rhodopsin is specifically phosphorylated by G protein-coupled receptor kinases (GRKs) (Maeda et al., 2003; Singh et al., 2008) and preferential binding of arrestin to activated rhodopsin blocks further G protein activation (Palczewski et al., 1991; Freedman & Lefkowitz, 1996). Lefkowitz, as many other investigators, was interested in the mechanism of signal termination by GPCRs. He demonstrated that other GPCRs are phosphorylated in an agonist-specific manner and that the phosphorylated receptors bind β-arrestin (Lohse et al., 1990), revealing a mechanism of receptor desensitization shared among rhodopsin and most GPCRs.
Mammals have four arrestin subtypes that demonstrate over 50% amino acid conservation and similar structures in their basal state. Arrestin-1 (also known as visual or rod arrestin) and arrestin-4 (cone arrestin) are predominantly expressed in photoreceptors, whereas arrestin-2 and -3 (also known as β-arrestin-1 and -2) are present in virtually every cell in the body with the greatest expression in mature neurons (Palczewski, 1994). Non-visual arrestins bind the great majority of GPCRs found in different mammalian species (Xiao et al., 2007). Visual arrestin shows high selectivity for its cognate receptor rhodopsin, even though several other proteins have been identified to be bound by this arrestin subtype (Gurevich et al., 2011).
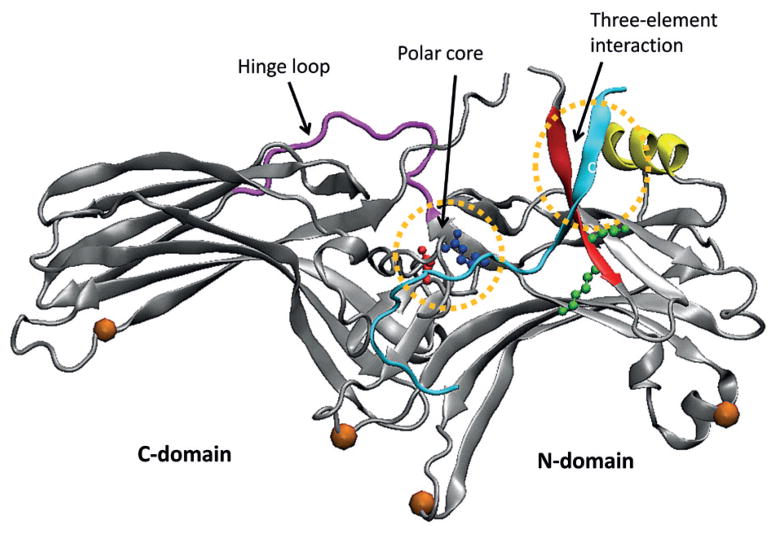
The structures of arrestin (Fig. 5) determined by Granzin et al. (1998) followed by other structures of arrestin and homologs (Hirsch et al., 1999; Han et al., 2001; Sutton et al., 2005; Zhan et al., 2011) consist of two concave lobed regions, termed the C-domain and N-domain. The receptor-binding surface is mainly localized to the side of arrestin containing these two cavities. Several exposed residues in the C- and N-domains of arrestin have been identified as being responsible for GPCR recognition (Hanson, 2006; Vishnivetskiy et al., 2010; 2011). The two domains are linked together by a polar core of charged residues that form a network of salt bridges which stabilize their relative orientation. It seems that non-visual arrestins can form functionally different complexes with the same receptor depending on the number of receptor-attached phosphates and their positions. These receptor-attached phosphates play a major role in arrestin recruitment, whereas their positions apparently determine the functional consequences of arrestin binding to the phosphoreceptor (Tobin et al., 2008; Gimenez et al., 2012). Phosphorylation sites on rhodopsin were well defined a decade ago in vitro and in vivo (Ohguro et al., 1993; 1994; Ohguro & Palczewski, 1995; Ohguro et al., 1995; Kennedy et al., 2001). Lefkowitz’s group has correlated the phosphorylation sites of β2-adrenergic receptor with β-arrestin functions (Nobles et al., 2011).
Figure 5. Crystal structure of arrestin (PDB id: 1CF1) with characteristic elements indicated.
Orange balls indicate regions that change upon GPCR binding but are not directly involved in the interaction with receptor. Colored residues are important for arrestin stability (a salt bridge in blue and red in polar core region) or initial recognition of receptor (two Lys residues in green).
Arrestin-1 preferentially binds to activated and phosphorylated rhodopsin. It also specifically binds inactive phosphorylated and active non-phosphorylated forms, but with much lower affinition. This observation was the first indication that arrestin-1 can recognize activation and phosphorylation of rhodopsin independently of each other. A sequential multi-site binding model (Gurevich & Gurevich, 2004) posits that arrestin-1 first binds rhodopsin either via its structural elements that specifically interact with the light-activated rhodopsin conformation, or via residues that directly bind to the rhodopsin-attached phosphates. If the ‘activated state’ or the phosphates represent the only “attraction factor”, then arrestin-1 binds with low affinity. However, when arrestin-1 encounters phosphorylated photoactivated rhodopsin, the engagement of both primary sites allows arrestin to switch into the high-affinity rhodopsin-binding state, bringing additional arrestin elements into contact with rhodopsin. The new contact surface provides extra energy to encourage the interaction, which accounts for arrestin-1’s much greater affinity for phosphorylated photoactivated rhodopsin.
The “hinge” region of all arrestins (residues 179–191) is flexible, allowing movement of the N- and C-domains relative to each other. However, there is no large relative motion of arrestin domains during complex formation, but a concerted movement of multiple flexible loops most probably helps arrestin mold itself onto the receptor (Kim et al., 2012) (Fig. 5). Lefkowitz’s studies on the conformation of non-visual arrestins in their active state revealed that although the overall activation mechanism is the same for all arrestin types, the final conformations might differ (Nobles et al., 2007). Moreover, β-arrestins might adopt multiple “active” conformations and, depending on their conformation, accomplish different functions (Shukla et al., 2008).
The stoichiometry of rhodopsin complexes with its cognate proteins is a matter of debate. Historically, one-to-one binding was usually assumed (Hanson et al., 2007; Gurevich & Gurevich, 2008). However, it has also been proposed by Palczewski that a single arrestin molecule could accommodate two receptors (Liang et al., 2003; Modzelewska et al., 2006). Monomeric activated and phosphorylated rhodopsin in nanodiscs can bind arrestin (Tsukamoto et al., 2010; Bayburt et al., 2011). At the same time, it has been reported that although arrestin requires at least a single phosphorylated photoactivated rhodopsin to bind to the membrane, a single arrestin can actually interact with a pair of receptors composed of two different photo-intermediate states. The binding stoichiometry depends on the percentage of active receptors (Sommer et al., 2011). From a physiological standpoint, the different binding modes of arrestin correspond well to the functional needs of the cell at different light intensities. In the single-photon range, arrestin binds monomeric phosphorylated photoactivated rhodopsin to quench signaling. But as the lighting level increases and photoactivates more rhodopsin, arrestin also binds dimeric photoactivated rhodopsin with only one phosphorylated protomer. Further studies have revealed that differentiated binding preferences of the two domains of arrestin allow it to accommodate the different functional forms of phosphorylated rhodopsin (Sommer et al., 2012). A general hypothesis is formed that the N-domain of arrestin mediates binding to agonist-activated receptor, whereas the less specific C-domain may serve various functions depending on the requirements of the biological system. The asymmetric ability of arrestin to stimulate ligand binding within receptor dimers is in line with studies on β2-adrenergic receptors (Gurevich et al., 1997) and N-formyl-peptide receptor (Key et al., 2001).
In addition to their role in GPCR desensitization, β-arrestins also participate in receptor internalization (Fig. 6). Lefkowitz’s group identified internalization as a critical initial step in recycling of desensitized receptors (Sibley et al., 1986) that also activates key mitogenic pathways within the cell (Daaka et al., 1998). β-arrestin binds to clathrin via the adaptor protein AP2 (Goodman et al., 1996) that causes arrestin-bound receptors to cluster in clathrin-coated pits. The clathrin-coated pit is pinched off from the plasma membrane by the motor protein dynamin, causing the desensitized receptor to enter an endosomal pool. After internalization, β-arrestin-mediated GPCR trafficking is regulated by both the location and variable binding affinities of different arrestin isoforms to GPCRs (Oakley et al., 2000). Arrestin-2 is found in both the cytoplasm and the nucleus, whereas arrestin-3 is localized only in the cytoplasm. β-arrestins interact with GPCRs with differing affinities. Class A GPCRs, such as β1-adrenergic receptors, μ opioid receptors, and D1 dopamine receptors bind arrestin-3 with a greater affinity than arrestin-2 and their interactions are lost during internalization. Class B receptors, such as angiotensin AT1a receptor, neurotensin receptor 1 and vasopressin V2 receptor, bind arrestin-2 and -3 with equal affinity and their interaction remains intact during internalization. Internalized receptors are then sorted for degradation or recycling; trafficking is regulated by ubiquitination of β-arrestin as revealed by Shenoy et al. (2001) in the case of the β2-adrenergic receptor. Receptors targeted for degradation traffic to lysosomes and are enzymatically degraded, whereas receptors for recycling traffic to acidified vesicles where they are de-phosphorylated and recycled back to the plasma membrane (Tan et al., 2004).
Figure 6. GPCR signaling.
In response to ligand binding a stimulation signaling can occur via G-protein-mediated pathway terminated by subsequent GRK/arrestin binding, or/and via β-arrestin-mediated pathway.
β-arrestins can also initiate a second wave of signaling which is independent of G-protein coupling and activation (Fig. 6). Here they serve as adaptor or scaffold molecules that bring crucial molecular components of specific signaling pathways in close proximity to an activated GPCR. Interestingly, depending on the type of activated GPCR, either both β-arrestin isoforms are required to activate the second wave of signaling (termed co-dependent), or only one isoform is required, whereas the other serves to inhibit the pathway (termed reciprocal regulation) (DeWire et al., 2007). The group of Lefkowitz first found that arrestin-2 is complexed with the tyrosine kinase c-Src. This associates c-Src with the β2-adrenergic receptor, resulting in activation of c-Src which initiates a tyrosine phosphorylation signaling cascade leading to stimulation of the Ras-ERK1/2 pathway (Luttrell et al., 1999). Contributions of the Lefkowitz’s group to understanding β-arrestin-mediated signaling have remained strong since their initiation.
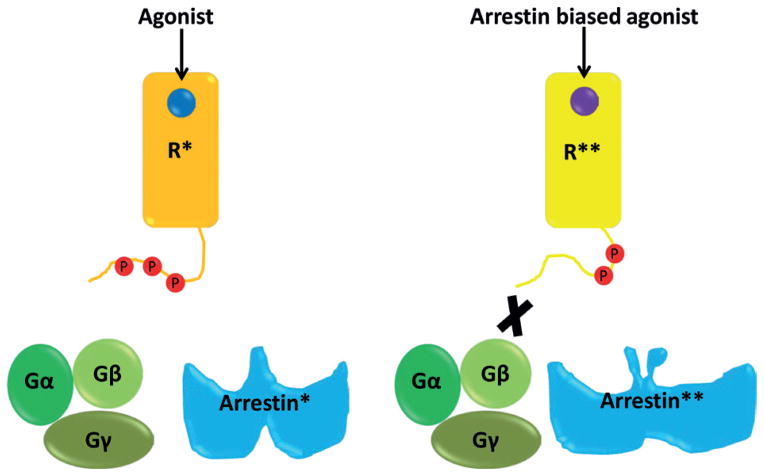
In many cases, signals transmitted by arrestin binding are demonstrably independent of heterotrimeric G protein activation. This observation has triggered the concept of biased agonists, pathway-selective ligands that activate only a subset of the GPCR signaling repertoire (Kenakin, 2005). Biased agonism is best understood by a model in which different GPCR’s active conformations are either competent for the full range of receptor activities or only for a subset of them. Thus, balanced ligands stabilize the conformations that are competent for signaling to all downstream pathways, whereas biased ligands stabilize only those conformations that are capable of promoting a subset of signaling effects (Rajagopal et al., 2010) (Fig. 7). The latter ligands could more selectively target beneficial signaling and even block or negate detrimental or unwanted actions of full receptor activation (e.g. side effects, toxicity or tolerance). Over the last decade a diversity of biased ligands for GPCRs have been identified that selectively activate G proteins or β-arrestins (Whalen et al., 2011), and several of these seem to have distinct functions when compared with traditional ligands with broad range efficacy.
Figure 7. A schematic representation of arrestin-biased signaling.
Binding of standard agonist to receptor induces an active conformation (R*) whereas binding of arrestin-biased agonist induces a different active conformation (R**). Distinct active conformations of receptor are coupled to different active conformation of arrestin which govern different functional outcomes.
Treatment with GPCR agonists can be limited by the development of tachyphylaxis, a decrease in responsiveness to the same dose of a drug, together with tolerance, whereby higher drug doses are required to obtain the same effect. Both processes, which limit the utility of therapeutics, are largely thought to be regulated by β-arrestin-dependent receptor desensitization and down-regulation. For example, arrestin-2 is involved in cardiac β2-adrenergic receptor desensitization (Conner et al., 1997). Clinically the β-agonist, dobutamine is often used to provide inotropic support for patients with severe heart failure, but is associated with the development of tachyphylaxis. Thus a G-protein-biased ligand that does not promote β-arrestin recruitment would cause less tachyphylaxis and would be a more effective therapeutic agent.
Several disease states associated with alterations in receptor trafficking could benefit from therapies that modulate β-arrestin-mediated functions. HIV requires cell-surface co-receptors, either the CCR5 or CXCR4 chemokine receptor, to attach and gain entry into target cells. CCR5-tropic viruses are the predominant species in the early stages of infection and there has been a significant interest in targeting this receptor for the treatment of HIV infection. Indeed, modified CCR5 ligands promote receptor internalization. As β-arrestins regulate CCR5 trafficking, recycling and degradation (Oppermann, 2004), the modified ligands probably regulate CCR5 trafficking via changes in β-arrestin activity. Thus, use of a ligand that modifies β-arrestin-regulated CCR5 trafficking could represent an attractive therapy for the treatment of HIV.
Cardiac β1-adrenergic receptors can stimulate arrestin-2- and -3-dependent signaling in the heart that results in transactivation of the cardioprotective epidermal growth factor receptor (EGFR) (Noma et al., 2007). It is also thought that chronic β2-adrenergic receptor activation is cardiotoxic and that this primarily involves Gs signaling (Xiao, 2001). These combined observations suggest that a β-arrestin-biased ligand acting as a classical antagonist of cardiotoxic G protein signaling while stimulating cardioprotective β-arrestin signaling could be therapeutically beneficial.
Arrestins regulate GPCR signaling by controlling desensitization, endocytosis and recycling/degradation of most GPCRs. They also function as ligand-regulated scaffolds that recruit functionally diverse proteins to GPCRs to confer novel signaling properties. Arrestin-dependent signals are involved in different processes in vivo, such as cell migration, neurotransmission, cardiac muscle contraction, and apoptosis. These signals can be initiated or antagonized independently of G protein activation. Although arrestins have been extensively studied by different groups, the molecular mechanisms of receptor-arrestin complex formation/function remain unclear and much additional work will be required for advances in this field.
IMPORTANCE OF OTHER GPCRS — CASE OF CHEMOKINE RECEPTORS
Chemokine receptors (CRs) are one of the most interesting families of GPCRs due to their key role in a number of diseases that affect millions of people worldwide. Chemokines are small chemotactic cytokines that regulate the trafficking of immune cells by binding to cell surface chemokine receptors (CRs). Chemokines coordinate the homeostatic circulation of leukocytes as well as their movement to sites of inflammation or injury (Murdoch & Finn, 2000). Structurally similar, they are small (8–10 kDa) proteins that share a relatively high sequence identity (20–50%). About 50 human chemokines that interact with 22 different receptors have been identified to date. Disregulated expression of chemokines and their receptors has been implicated in the development of many human diseases (see Table 1). As a result, considerable effort has been made to solve the three-dimensional structure of chemokine receptors and to develop drugs to modulate their activities. By the end of 2012 we have learned the connection between a disease and target protein for at least 15 CRs, developed agonists and antagonists for at least 10 different CRs and solved the 3D structures of two chemokine receptors, CXCR4 and CXCR1 (see Fig. 8). Here we review the status of our knowledge about CRs — their structures, involvement in human diseases and known agonists/antagonists. We also present computational approaches to model CR structures and perform rational drug design.
Table 1.
Major diseases linked to chemokine receptors (adopted from Allen et al., 2007).
| Disease | Chemokine | Chemokine receptor |
|---|---|---|
| Allograft | CCL3, CCL4, CCL5 | CCR5 |
| Asthma | CCL1, CCL17, CCL22 | CCR3, CCR4, CCR8 |
| Atherosclerosis | CCL2, CCL5 | CCR2, CCR5, CXCR1, CXCR2, CX3CR1 |
| Atopic dermatitis | CCL1, CCL13, CCL17-18, CCL27 | CCR4, CCR8, CCR10 |
| Crohn’s | CCL28 | CCR9 |
| Chronic hepatitis | CCL3, CCL4 | CCR5 |
| Gut cancer | CCL25 | CCR9 |
| HIV | CCL3, CCL4, CCL5, CXCL12 | CCR5, CXCR4 |
| Ischemia-reperfusion | CCL2 | CCR2 |
| Lymph node cancer | CCL19, CCL21 | CCR7 |
| Multiple sclerosis | CCL2, CCL4, CCL5, CXCL9 | CCR2, CCR5, CXCR3 |
| Psoriasis | CCL4, CCL20, CCL27 | CCR5, CCR6, CXCR3 |
| Rheumatoid arthritis | CCL2, CCL3, CCL5, CXCL9, CXCL10 | CCR1, CCR2, CCR5, CXCR3 |
Figure 8. Crystal structures of chemokine receptors.
(A) Crystal structure of CXCR4 (PDB id: 3ODU) with small molecule antagonist IT1t. (B) NMR structure of CXCR1 (PDB id: 2LNL). Top, view along the membrane plane, bottom, from the extracellular side.
The largest subfamily of CRs is named after their ability to bind CC chemokines — a subfamily of chemokines with four or six Cys residues forming two or three di-sulfide bonds, with two conserved Cys residues always forming a CC motif. Members of this subfamily share substantial homology with the exception of CCR10. CCR1 was the first CC chemokine receptor identified. It shares a 62.3% sequence identity with CCR3 and binds similar chemokines, but is involved in different diseases than its close relative. Whereas CCR1 has been implicated in multiple sclerosis, rheumatoid arthritis, psoriasis, transplant rejection, cancer and kidney disease, CCR3 has been connected only to asthma and allergic rhinitis. The first antagonists of CCR1 were reported by Hesselgesser et al. (1998) and Brown et al. (1998). Over 100 different small molecule CCR1 antagonists/agonists derived from at least 15 different scaffolds have been described (Pease & Horuk, 2009a; 2009b). Some reported drug candidates are potent antagonists with reported Ki values of around 1 nM, e.g. BX 471 (Liang et al., 2000) and MLN3897 (Carson & Harriman, 2004), and highly selective against CCR1. A few of the most potent and selective CCR1 inhibitors have progressed to clinical development, but despite their high therapeutic potential none have yet passed clinical trials (Gladue et al., 2010).
The story of CCR3 antagonists is similar, although the first chemical compound showing high activity against it (SB-328437) was reported later (White et al., 2000). As of the end of 2012 more than thirty different compounds interacting with CCR3 derived from at least ten different scaffolds have been identified, some with high CCR3 affinity (EC 50 values of 1–5 nM (Naya et al., 2003; Ting et al., 2005; Morokata et al., 2006)). Interestingly, both SB-328437 and another early CCR3 inhibitor, UCB 35625, are bi-specific, with nanomolar inhibitory activity towards both CCR3 and CCR1 (Naya et al., 2001). The high homology and similarity in the transmembrane region allowed the design in this case of ‘dual’ antagonists affecting both receptors. More dual inhibitors have been reported (Dhanak et al., 2001a), some with subnanomolar activities against both CCR1 and CCR3 (Dhanak et al., 2001b). Though some of those inhibitors were tested in patients with various diseases, none have yet succeeded.
CCR2 and CCR5 constitute another interesting pair of similar chemokine receptors, sharing 63.4% sequence identity. They also interact with some of the same chemokines and both have been linked to the same immunologic and cardiovascular diseases (Zhao, 2010). Of this pair, however, only CCR5 has achieved fame for serving as an entry factor for macrophage-tropic strains of HIV-1 (Moore et al., 1997), a role that has instigated a search for small molecular antagonists of this receptor that could block viral entry.
The first CCR2 antagonists were described in the literature in 2000, six years after the successful cloning of this receptor (Forbes et al., 2000; Mirzadegan et al., 2000). By the end of 2012 several pharmaceutical companies have disclosed more than fifty with different molecular scaffolds including piperidine, spiropiperidine, aminopyrrolidine, compounds with bisubstituted cyclohexane groups, and others. Some of the optimized ligands evidenced CCR2 inhibition in the nanomolar range, with the lowest IC50 (subnanomolar) value reported by Teijin company for one of their optimized homopiperazine derivatives (Moree et al., 2008). But as in the CCR1/CCR3 case, no drug has yet been approved for use against this receptor despite considerable effort.
Most CCR5 inhibitors were developed to prevent the cellular invasion of HIV. The first CCR5 inhibitor reported was TAK-779 synthesized by Takeda Chemical Industries in 1999 (Baba et al., 1999). This compound bound to the receptor at nanomolar concentrations and inhibited HIV cellular entry in vitro. Unfortunately, it also exhibited a poor oral bioavailability, but still was used as a CCR5 inhibitor model by other pharmaceutical companies. In the last ten years at least forty different CCR5 antagonists have been identified and, due to the importance of AIDS, many were optimized to display inhibitory efficacy at nanomolar or subnanomolar concentrations. Based on favorable HIV inhibition data, a relatively large number of these compounds entered clinical trials for treatment of AIDS. Unfortunately, most CCR5 inhibitors experienced multiple problems in stage II or III clinical trials (Wilkin & Gulick, 2012) and only one (maraviroc) was cleared and approved as an anti-HIV drug targeting CCR5 (Hitchcock, 2005).
The second largest subfamily of CRs was named for their ability to bind CXC chemokines that possess the conserved CXC motif. CXCR4 is one of the two chemokine receptors (together with CCR5) used by HIV to enter human cells, a finding that has greatly accelerated structural research aimed at this protein (Oberlin et al., 1996). The first small molecular antagonist of CXCR4 was described in 1992 (De Clercq et al., 1992) and since then a number of potent antagonists (with binding in the nanomolar or even subnanomolar range) have been described (Ichiyama et al., 2003; Zhan et al., 2007). Moreover, the relatively large amount of data for this receptor provided insights into the binding modes of a number of antagonists (Vabeno et al., 2006).
A breakthrough in CXCR4-based anti-HIV research occurred with the experimental solving of crystal structures of CXCR4 bound to either a small-molecular ligand or a cyclic peptide antagonist (Wu et al., 2010) (Fig. 8A). Earlier it was suggested that both those ligands block the interaction between CXCR4 and its natural ligand CXC4, and also inhibit CXCL12 interactions with the HIV-1 glycoprotein gp120. Indeed, both ligands found in those crystal structures interacted with the receptor’s extracellular loops and N-terminus and most likely altered their conformation, making the interaction with CXCL12 and gp120 energetically unfavorable.
The structure of CXCR4 was rather surprising, especially because it revealed a number of relatively large differences from the crystal structures of the β2-adrenergic receptor and other GPCRs obtained earlier. The most striking differences were the positions and rotations of helices 1, 2 and 6 that resulted in a much looser packing of all helices, and also the lengths of helices 5 and 7. Surprisingly, the binding cavity of CXCR4 also was larger, more open and located much closer to the extracellular surface than that of other known GPCRs. This investigation not only provides invaluable information about the structure of CXCR4 and possibly other CR-family members, but it also serves as a platform for rational drug design, contributing to understanding of the HIV-1antagonists entry process along with the elucidated structures of native HIV-1 gp120 trimers (Liu et al., 2008). This progress should lead to a series of new scaffolds and compounds targeting CXCR4 as well as dual inhibitors able to bind to both CXCR4 and CCR5 (Murray et al., 2010). Interestingly, despite the known structure of CXCR4, no anti-HIV drug targeting this receptor has yet been approved for use. However, one of the first CXCR4 inhibitors found has been approved to be used to mobilize hematopoietic stem cells in cancer patients (AnorMED, 2007).
CXCR1 and CXCR2 form a pair of chemokine receptors which are most closely related to each other, with 76.6% sequence identity. The number of publications on the role of both receptors has increased exponentially within the last 10 years, in part due to the discovery of potent and selective CXCR1/CXCR2 dual antagonists. The first dual antagonist for this pair of proteins was shown to inhibit acute and chronic models of arthritis in the rabbit (Podolin et al., 2002). There is also an interest in developing new CXCR1/CXCR2 antagonists as therapeutic agents for chronic obstructive pulmonary disease (COPD), asthma and various forms of cancer. Two such dual antagonists have passed all clinical trials and are now marketed — reparixin which attenuates inflammatory responses and promotes recovery of function after spinal cord injury (Gorio et al., 2007) and navarixin, an anti-COPD drug.
CXCR1 is also the second chemokine receptor with a known three-dimensional structure. In 2012 its structure was obtained by using rotationally aligned solid-state NMR (Park et al., 2012) (Fig. 8B). That novel method involved the use of NMR for the first time in GPCR structural studies and provided important information about this receptor in its natural phospholipid bilayer environment. A comparison of its structure with that of CXCR4 (32.9% sequence identity) shows a high homology and overall similarity. Four charged residues in the helical region of CXCR1 form a polar cluster in the ligand-binding site of this receptor. They most likely are important for ligand binding, similar to the three polar residues found in CXCR4. There also are some potentially important differences in positions and rotations of helices 1, 2 and 6, which alter the overall structure of the receptor’s core and could contribute to the different biological activities exhibited by CXCR1 and CXCR4. As with CXCR4, solution of the CXCR1 structure hopefully will lead to the rational design of new CXCR1 and CXCR2 antagonists with improved properties.
Chemokine receptors are a family of GPCRs that have always attracted much attention from researchers and health professionals. Immediately after their recognition as potential targets in various diseases, these proteins became the focus of numerous programs run by pharmaceutical companies. Development of novel experimental and theoretical techniques has been crucial in finding and/or designing new CR antagonists. Of these recent innovations, the possibility of obtaining three-dimensional structures by using crystallography or NMR was one of the most important. This has led to the crystal structure of CXCR4 and the NMR structure of CXCR1 described above (Wu et al., 2010; Park et al., 2012). There are still many questions about the structure of other CRs, their probable oligomerization and dynamics in human cells, but our knowledge of this family of proteins is now increasing exponentially.
FUTURE DIRECTIONS
The GPCR research will further shape the field of pharmacology and medicine ne a functional channel mediating activation in the decades ahead. Thus it will be challenging but necessary to determine high resolution structures of native GPCRs alone and in complexes with their G protein, receptor kinase and arrestin (Jastrzebska et al., 2010). Moreover, GPCRs and G proteins are critically altered by several post-translational modifications which should be taken into account in structural studies. In addition to these modifications, water molecules have been shown to play a key role in all proteins, including transmembrane ones. It will be essential to determine the exact location of water molecules in these receptors and their re-arrangements during GPCR activation. Understanding the energy landscape of GPCRs corresponding to their folding pathway and activation is highly anticipated. Identification of key residues responsible for folding and membrane insertion is needed to explain the etiology of many human diseases associated with receptor mutations. Moreover, GPCRs are not monomeric — they have a propensity to interact not only with each other but also with other transmembrane proteins. In recent years, more than 50% of published papers in the field have explored the oligomerization of GPCRs. But we are still far from understanding “the logic” of oligomerization at both structural and functional levels. Understanding the structural complementarities of GPCR homo- and hetero-oligomerization offers several novel pharmacological opportunities to explore the high specificity of these interactions. A large number of GPCRs communicate with only a limited number of G proteins and even a smaller number of effectors such as enzymes and channels. Although the subcellular localization of specific sets of receptors and interacting proteins is clear, diffusible ligands such as cAMP or Ca2+ allow a cross-talk between many specialized pathways. It will be important to determine the intersections of different relevant GPCR signaling pathways in native tissues of interest. Modern 3D structural electron microscopy (cryo-electron microscopy and tomography) and hybrid microscopic techniques will facilitate obtaining high resolution structures of signaling complexes between GPCRs, their cognate G proteins and effector molecules in native tissues. Such studies can determine how the GPCR signaling complexes are compartmentalized within cells to evoke their local effects. We are confident that the discovery of mutations responsible for genetic diseases will be dramatically accelerated due to the DNA and RNA sequencing of whole personal genomes as well as tissue transcriptomes. This new methodology will enable evaluation of the impact of mutations and polymorphisms affecting GPCR expression levels on human conditions. Hence, GPCRs will certainly play a central role in drug research in the foreseeable future.
Acknowledgments
This work was supported by funding from the National Institutes of Health, USA (EY008061) and the National Center of Science, Poland (DEC2011/03/B/NZ1/03204). K.P. is John H. Hord Professor of Pharmacology.
Abbreviations
- A2AR
adenosine A2A receptor
- BRIL
apocytochrome b(562)RIL
- CRs
chemokine receptors
- CXCR4
chemokine receptor CXCR4
- D3R
dopamine D3 receptor
- EC
extracellular
- ECL
extracellular loop
- GPCRs
G protein-coupled receptors
- GRK
G protein-coupled receptor kinase
- H1R
histamine H1 receptor
- HBD
hormone-binding domain
- IC
intracellular
- ICL
intracellular loop
- M2R
M3R, muscarinic M2 and M3 receptors
- MCH
melanin-concentrating hormone
- NOP
nociceptin opioid receptor
- OR
opioid receptor
- PDB
protein data bank
- PDE
phosphodiesterase
- PKA
protein kinase A
- SOG
somatostatin, opioid and galanin receptors
- TM
transmembrane
- 7TMH
seven transmembrane helices
References
- Allen SJ, Crown SE, Handel TM. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci USA. 2009a;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. of Structuralwaters the GPCR, rhodopsin. Proc Natl Acad Sci USA. 2009b;106:14367–14372. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anor MED. Plerixafor: AMD 3100, AMD3100, JM 3100, SDZ SID 791. Drugs R D. 2007;8:113–119. doi: 10.2165/00126839-200708020-00006. [DOI] [PubMed] [Google Scholar]
- Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavo JA, Brunton LL. Cyclic nucleotide research — still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- Benovic JL, Strasser RH, Caron MG, Lefkowitz RJ. Beta-adrenergic receptor kinase: identification of a novel protein kinase that phosphorylates the agonist-occupied form of the receptor. Proc Natl Acad Sci USA. 1986;83:2797–2801. doi: 10.1073/pnas.83.9.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- Breitman ML, Bryce DM, Giddens E, Clapoff S, Goring D, Tsui LC, Klintworth GK, Bernstein A. Analysis of lens cell fate and eye morphogenesis in transgenic mice ablated for cells of the lens lineage. Development. 1989;106:457–463. doi: 10.1242/dev.106.3.457. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K-V, Yang W-L, Ravatn R, Kita T, Reitman E, Vettori D, Cvijic ME, Shin M, Iacono L. Reinventing the wheel of cyclic AMP: novel mechanisms of cAMP signaling. Ann N Y Acad Sci. 2002;968:49–64. doi: 10.1111/j.1749-6632.2002.tb04326.x. [DOI] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Neer EJ. G protein beta gamma subunits. Annu Rev Pharmacol Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- Conner DA, Mathier MA, Mortensen RM, Christe M, Vatner SF, Seidman CE, Seidman JG. beta-Arrestin1 knockout mice appear normal but demonstrate altered cardiac responses to beta-adrenergic stimulation. Circ Res. 1997;81:1021–1026. doi: 10.1161/01.res.81.6.1021. [DOI] [PubMed] [Google Scholar]
- Daaka Y, Luttrell LM, Ahn S, Della Rocca GJ, Ferguson SS, Caron MG, Lefkovitz RJ. Essential role for G protein-coupled receptor endocytosis in the activation of mitogen-activated protein kinase. J Biol Chem. 1998;273:685–688. doi: 10.1074/jbc.273.2.685. [DOI] [PubMed] [Google Scholar]
- De Clercq E, Yamamoto N, Pauwels R, Baba M, Schols D, Nakashima H, Balzarini J, Debyser Z, Murrer BA, Schwartz D, et al. Potent and selective inhibition of human immunodeficiency virus (HIV)-1 and HIV-2 replication by a class of bicyclams interacting with a viral uncoating event. Proc Natl Acad Sci USA. 1992;89:5286–5290. doi: 10.1073/pnas.89.12.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J Biol Chem. 1980;255:7108–7117. [PubMed] [Google Scholar]
- Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville J, Rey J, Chabbert M. An indel in transmembrane helix 2 helps to trace the molecular evolution of class A G-protein-coupled receptors. J Mol Evol. 2009;68:475–489. doi: 10.1007/s00239-009-9214-9. [DOI] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- Dhanak D, Christmann LT, Darcy MG, Jurewicz AJ, Keenan RM, Lee J, Sarau HM, Widdowson KL, White JR. Discovery of potent and selective phenylalanine derived CCR3 antagonists. Part 1. Bioorg Med Chem Lett. 2001a;11:1441–1444. doi: 10.1016/s0960-894x(01)00248-7. [DOI] [PubMed] [Google Scholar]
- Dhanak D, Christmann LT, Darcy MG, Keenan RM, Knight SD, Lee J, Ridgers LH, Sarau HM, Shah DH, White JR, Zhang L. Discovery of potent and selective phenylalanine derived CCR3 receptor antagonists. Part 2. Bioorg Med Chem Lett. 2001b;11:1445–1450. doi: 10.1016/s0960-894x(01)00249-9. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran N, Dermott JM. Signaling by the G12 class of G proteins. Cell Signal. 1996;8:235–245. doi: 10.1016/0898-6568(96)00048-4. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Kobilka BK, Strader DJ, Benovic JL, Dohlman HG, Frielle T, Bolanowski MA, Bennett CD, Rands E, Diehl RE, Mumford RA, Slater EE, Sigal IS, Caron MG, Lefkowitz RJ, Strader CD. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- Eilers M, Goncalves JA, Ahuja S, Kirkup C, Hirshfeld A, Simmerling C, Reeves PJ, Sheves M, Smith SO. Structural transitions of transmembrane helix 6 in the formation of metarhodopsin I. J Phys Chem B? 2012 doi: 10.1021/jp3019183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exton JH. Regulation of phosphoinositide phospholipases by hormones, neurotransmitters, and other agonists linked to G proteins. Annu Rev Pharmacol Toxicol. 1996;36:481–509. doi: 10.1146/annurev.pa.36.040196.002405. [DOI] [PubMed] [Google Scholar]
- Filipek S, Stenkamp RE, Teller DC, Palczewski K. G protein-coupled receptor rhodopsin: A prospectus. Annu Rev Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes IT, Cooper DG, Dodds EK, Hickey DM, Ife RJ, Meeson M, Stockley M, Berkhout TA, Gohil J, Groot PH, Moores K. CCR2B receptor antagonists: conversion of a weak HTS hit to a potent lead compound. Bioorg Med Chem Lett. 2000;10:1803–1806. doi: 10.1016/s0960-894x(00)00347-4. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Freedman NJ, Lefkowitz RJ. Desensitization of G protein-coupled receptors. Recent Prog Horm Res. 1996;51:319–351. [PubMed] [Google Scholar]
- Garczarek F, Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. Role of receptor-attached phosphates in binding of visual and non-visual arrestins to G protein-coupled receptors. J Biol Chem. 2012;287:9028–9040. doi: 10.1074/jbc.M111.311803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladue PR, Brown FM, Zwillich HS. CCR1 Antagonists: what have we learned from clinical trials. Curr Top Med Chem. 2010;10:1268–1277. doi: 10.2174/156802610791561237. [DOI] [PubMed] [Google Scholar]
- Goodman OBJ, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:477–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Gorio A, Madaschi L, Zadra G, Marfia G, Cavalieri B, Bertini R, Di Giulio AM. Reparixin, an inhibitor of CXCR2 function, attenuates inflammatory responses and promotes recovery of function after traumatic lesion to the spinal cord. J Pharmacol Exp Ther. 2007;322:973–981. doi: 10.1124/jpet.107.123679. [DOI] [PubMed] [Google Scholar]
- Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. The functional cycle of visual arrestins in photoreceptor cells. Prog Retin Eye Res. 2011;30:405–430. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci USA. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SMFD, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci USA. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselgesser J, Ng HP, Liang M, Zheng W, May K, Bauman JG, Monahan S, Islam I, Wei GP, Ghannam A, Taub DD, Rosser M, Snider RM, Morrissey MM, Perez HD, Horuk R. Identification and characterization of small molecule functional antagonists of the CCR1 chemokine receptor. J Biol Chem. 1998;273:15687–15692. doi: 10.1074/jbc.273.25.15687. [DOI] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- Hitchcock CA. The Discovery and Exploratory Development of Maraviroc (UK-427,857): A Novel CCR5 antagonist for the treatment of HIV. Retrovirology. 2005;2:S11. [Google Scholar]
- Ichiyama K, Yokoyama-Kumakura S, Tanaka Y, Tanaka R, Hirose K, Bannai K, Edamatsu T, Yanaka M, Niitani Y, Miyano-Kurosaki N, Takaku H, Koyanagi Y, Yamamoto N. A duodenally absorbable CXC chemokine receptor 4 antagonist, KRH-1636, exhibits a potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 2003;100:4185–4190. doi: 10.1073/pnas.0630420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and structural characterization of rhodopsin oligomers. J Biol Chem. 2006;281:11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Tsybovsky Y, Palczewski K. Complexes between photoactivated rhodopsin and transducin: progress and questions. Biochem J. 2010;428:1–10. doi: 10.1042/BJ20100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Palczewski K, Golczak M. Role of bulk water in hydrolysis of the rhodopsin chromophore. J Biol Chem. 2011a;286:18930–18937. doi: 10.1074/jbc.M111.234583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Ringler P, Lodowski DT, Moiseenkova-Bell V, Golczak M, Muller SA, Palczewski K, Engel A. Rhodopsin-transducin heteropentamer: three-dimensional structure and biochemical characterization. J Struct Biol. 2011b;176:387–394. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T. New concepts in drug discovery: collateral efficacy and permissive antagonism. Nat Rev Drug Discov. 2005;4:919–927. doi: 10.1038/nrd1875. [DOI] [PubMed] [Google Scholar]
- Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Lee KA, Niemi GA, Craven KB, Garwin GG, Saari JC, Hurley JB. Multiple phosphorylation of rhodopsin and the in vivo chemistry underlying rod photoreceptor dark adaptation. Neuron. 2001;31:87–101. doi: 10.1016/s0896-6273(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Key TA, Bennett TA, Foutz TD, Gurevich VV, Sklar LA, Prossnitz ER. Regulation of formyl peptide receptor agonist affinity by reconstitution with arrestins and heterotrimeric G proteins. J Biol Chem. 2001;276:49204–49212. doi: 10.1074/jbc.M109475200. [DOI] [PubMed] [Google Scholar]
- Kim M, Vishnivetskiy SA, Van Eps N, Alexander NS, Cleghorn WM, Zhan X, Hanson SM, Morizumi T, Ernst OP, Meiler J, Gurevich VV, Hubbell WL. Conformation of receptor-bound visual arrestin. Proc Natl Acad Sci USA. 2012;109:18407–18412. doi: 10.1073/pnas.1216304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H, Dreyer WJ. Light dependent phosphorylation of rhodopsin by ATP. FEBS Lett. 1972;20:1–6. doi: 10.1016/0014-5793(72)80002-4. [DOI] [PubMed] [Google Scholar]
- Kuhn H. Light-dependent phosphorylation of rhodopsin in living frogs. Nature. 1974;250:588–590. doi: 10.1038/250588a0. [DOI] [PubMed] [Google Scholar]
- Lebon G, Warne T, Tate CG. Agonist-bound structures of G protein-coupled receptors. Curr Opin Struct Biol. 2012;22:482–490. doi: 10.1016/j.sbi.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ. The superfamily of heptahelical receptors. Nat Cell Biol. 2000;2:E133–136. doi: 10.1038/35017152. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Liang M, Mallari C, Rosser M, Ng HP, May K, Monahan S, Bauman JG, Islam I, Ghannam A, Buckman B, Shaw K, Wei GP, Xu W, Zhao Z, Ho E, Shen J, Oanh H, Subramanyam B, Vergona R, Taub D, Dunning L, Harvey S, Snider RM, Hesselgesser J, Morrissey MM, Perez HD. Identification and characterization of a potent, selective, and orally active antagonist of the CC chemokine receptor-1. J Biol Chem. 2000;275:19000–19008. doi: 10.1074/jbc.M001222200. [DOI] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, Ijzerman AP, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodowski DT, Salom D, Le Trong I, Teller DC, Ballesteros JA, Palczewski K, Stenkamp RE. Crystal packing analysis of Rhodopsin crystals. J Struct Biol. 2007;158:455–462. doi: 10.1016/j.jsb.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodowski DT, Angel TE, Palczewski K. Comparative analysis of GPCR crystal structures. Photochem Photobiol. 2009;85:425–430. doi: 10.1111/j.1751-1097.2008.00516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, Caron MG, Lefkowitz RJ. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Lefkowitz RJ. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci. 2002;115:455–465. doi: 10.1242/jcs.115.3.455. [DOI] [PubMed] [Google Scholar]
- Maeda A, Okano K, Park PS, Lem J, Crouch RK, Maeda T, Palczewski K. Palmitoylation stabilizes unliganded rod opsin. Proc Natl Acad Sci USA. 2010;107:8428–8433. doi: 10.1073/pnas.1000640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Imanishi Y, Palczewski K. Rhodopsin phosphorylation: 30 years later. Prog Retin Eye Res. 2003;22:417–434. doi: 10.1016/s1350-9462(03)00017-x. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meruelo AD, Samish I, Bowie JU. TMKink: a method to predict transmembrane helix kinks. Protein science: a publication of the Protein Society. 2011;20:1256–1264. doi: 10.1002/pro.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadegan T, Diehl F, Ebi B, Bhakta S, Polsky I, McCarley D, Mulkins M, Weatherhead GS, Lapierre JM, Dankwardt J, Morgans D, Jr, Wilhelm R, Jarnagin K. Identification of the binding site for a novel class of CCR2b chemokine receptor antagonists: binding to a common chemokine receptor motif within the helical bundle. J Biol Chem. 2000;275:25562–25571. doi: 10.1074/jbc.M000692200. [DOI] [PubMed] [Google Scholar]
- Mirzadegan T, Benko G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 2003;42:2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewska A, Fillipek S, Palczewski K, Park PS. Arrestin interaction with rhodopsin: conceptual models. Cell Biochem Biophys. 2006;46:1–15. doi: 10.1385/CBB:46:1:1. [DOI] [PubMed] [Google Scholar]
- Moore JP, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- Moree WJ, Kataoka K-i, Ramirez-Weinhouse MM, Shiota T, Imai M, Tsutsumi T, Sudo M, Endo N, Muroga Y, Hada T, Fanning D, Saunders J, Kato Y, Myers PL, Tarby CM. Potent antagonists of the CCR2b receptor. Part 3: SAR of the (R)-3-aminopyrrolidine series. Bioorg Med Chem Lett. 2008;18:1869–1873. doi: 10.1016/j.bmcl.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Morokata T, Suzuki K, Masunaga Y, Taguchi K, Morihira K, Sato I, Fujii M, Takizawa S, Torii Y, Yamamoto N, Kaneko M, Yamada T, Takahashi K, Shimizu Y. A novel, selective, and orally available antagonist for CC chemokine receptor 3. J Pharmacol Exp Ther. 2006;317:244–250. doi: 10.1124/jpet.105.097048. [DOI] [PubMed] [Google Scholar]
- Muller DJ, Wu N, Palczewski K. Vertebrate membrane proteins: structure, function, and insights from biophysical approaches. Pharmacol Rev. 2008;60:43–78. doi: 10.1124/pr.107.07111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch C, Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood. 2000;95:3032–3043. [PubMed] [Google Scholar]
- Murray EJ, Leaman DP, Pawa N, Perkins H, Pickford C, Perros M, Zwick MB, Butler SL. A low-molecular-weight entry inhibitor of both CCR5- and CXCR4-tropic strains of human immunodeficiency virus type 1 targets a novel site on gp41. J Virol. 2010;84:7288–7299. doi: 10.1128/JVI.00535-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi D, Palczewski K. Topology of class A G protein-coupled receptors: insights gained from crystal structures of rhodopsins, adrenergic and adenosine receptors. Mol Pharmacol. 2009;75:1–12. doi: 10.1124/mol.108.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya A, Kobayashi K, Ishikawa M, Ohwaki K, Saeki T, Noguchi K, Ohtake N. Discovery of a novel CCR3 selective antagonist. Bioorg Med Chem Lett. 2001;11:1219–1223. doi: 10.1016/s0960-894x(01)00176-7. [DOI] [PubMed] [Google Scholar]
- Naya A, Kobayashi K, Ishikawa M, Ohwaki K, Saeki T, Noguchi K, Ohtake N. Structure-activity relationships of 2-(benzothiazolylthio)acetamide class of CCR3 selective antagonist. Chem Pharm Bull. 2003;51:697–701. doi: 10.1248/cpb.51.697. [DOI] [PubMed] [Google Scholar]
- Nobles KN, Guan Z, Xiao K, Oas TG, Lefkowitz RJ. The active conformation of beta-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of beta-arrestins 1 and -2. J Biol Chem. 2007;282:21370–21381. doi: 10.1074/jbc.M611483200. [DOI] [PubMed] [Google Scholar]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, Shenoy SK, Gygi SP, Lefkowitz RJ. Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma T, Lemaire A, Naga Prasad SV, Barki-Harrington L, Tilley DG, Chen J, Le Corvoisier P, Violin JD, Wei H, Lefkowitz RJ, Rockman HA. Beta-arrestin-mediated beta1-adrenergic receptor transactivation of the EGFR confers cardioprotection. J Clin Invest. 2007;117:2445–2458. doi: 10.1172/JCI31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley RH, Laporte SA, Holt JA, Caron MG, Barak LS. Differential affinities of visual arrestin, beta arrestin1, and beta arrestin2 for G protein-coupled receptors delineate two major classes of receptors. J Biol Chem. 2000;275:17201–17210. doi: 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler DF, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- Ohguro H, Palczewski K, Ericsson LH, Walsh KA, Johnson RS. Sequential phosphorylation of rhodopsin at multiple sites. Biochemistry. 1993;32:5718–5724. doi: 10.1021/bi00072a030. [DOI] [PubMed] [Google Scholar]
- Ohguro H, Johnson RS, Ericsson LH, Walsh KA, Palczewski K. Control of rhodopsin multiple phosphorylation. Biochemistry. 1994;33:1023–1028. doi: 10.1021/bi00170a022. [DOI] [PubMed] [Google Scholar]
- Ohguro H, Palczewski K. Separation of phospho- and non-phosphopeptides using reverse phase column chromatography. FEBS Lett. 1995;368:452–454. doi: 10.1016/0014-5793(95)00710-q. [DOI] [PubMed] [Google Scholar]
- Ohguro H, Van Hooser JP, Milam AH, Palczewski K. Rhodopsin phosphorylation and dephosphorylation in vivo. J Biol Chem. 1995;270:14259–14262. doi: 10.1074/jbc.270.24.14259. [DOI] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal. 2004;16:1201–1210. doi: 10.1016/j.cellsig.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Orban T, Gupta S, Palczewski K, Chance MR. Visualizing water molecules in transmembrane proteins using radiolytic labeling methods. Biochemistry. 2010;49:827–834. doi: 10.1021/bi901889t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, Chance MR, Palczewski K. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20:826–840. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osyczka A, Moser CC, Dutton PL. Fixing the Q cycle. Trends Biochem Sci. 2005;30:176–182. doi: 10.1016/j.tibs.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Palczewski K, McDowell JH, Jakes S, Ingebritsen TS, Hargrave PA. Regulation of rhodopsin dephosphorylation by arrestin. J Biol Chem. 1991;264:15770–15773. [PubMed] [Google Scholar]
- Palczewski K. Structure and functions of arrestins. Protein Sci. 1994;3:1355–1361. doi: 10.1002/pro.5560030901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PS, Lodowski DT, Palczewski K. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu Rev Pharmacol Toxicol. 2008;48:107–141. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, Opella SJ. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012 doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JE, Horuk R. Chemokine receptor antagonists: Part 1. Expert Opin Ther Pat. 2009a;19:39–58. doi: 10.1517/13543770802641346. [DOI] [PubMed] [Google Scholar]
- Pease JE, Horuk R. Chemokine receptor antagonists: Part 2. Expert Opin Ther Pat. 2009b;19:199–221. doi: 10.1517/13543770802641353. [DOI] [PubMed] [Google Scholar]
- Pele J, Abdi H, Moreau M, Thybert D, Chabbert M. Multidimensional scaling reveals the main evolutionary pathways of class A G-protein-coupled receptors. PLoS One. 2011;6:e19094. doi: 10.1371/journal.pone.0019094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolin PL, Bolognese BJ, Foley JJ, Schmidt DB, Buckley PT, Widdowson KL, Jin Q, White JR, Lee JM, Goodman RB, Hagen TR, Kajikawa O, Marshall LA, Hay DW, Sarau HM. A potent and selective nonpeptide antagonist of CXCR2 inhibits acute and chronic models of arthritis in the rabbit. J Immunol. 2002;169:6435–6444. doi: 10.4049/jimmunol.169.11.6435. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Crystal structure of the human beta(2) adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;145:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robishaw JD. Specificity of G protein beta-gamma dimer signaling. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. Academic; Boston, MA: 2004. pp. 623–629. [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Maguire ME, Sturgill TW, Biltonen RL, Gilman AG. Relationship between the beta-adrenergic receptor and adenylate cyclase. J Biol Chem. 1977;252:5761–5775. [PubMed] [Google Scholar]
- Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salon JA, Lodowski DT, Palczewski K. The significance of G protein-coupled receptor crystallography for drug discovery. Pharmacol Rev. 2011;63:901–937. doi: 10.1124/pr.110.003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schioth HB, Fredriksson R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen Comp Endocrinol. 2005;142:94–101. doi: 10.1016/j.ygcen.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- Shorr RG, Lefkowitz RJ, Caron MG. Purification of the beta-adrenergic receptor. Identification of the hormone binding subunit. J Biol Chem. 1981;256:5820–5826. [PubMed] [Google Scholar]
- Shukla AK, Violin JD, Whalen EJ, Gesty-Palmer D, Shenoy SK, Lefkowitz RJ. Distinct conformational changes in beta-arrestin report biased agonism at seven-transmembrane receptors. Proc Natl Acad Sci USA. 2008;105:9988–9993. doi: 10.1073/pnas.0804246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley DR, Strasser RH, Benovic JL, Daniel K, Lefkowitz RJ. Phosphorylation/dephosphorylation of the beta-adrenergic receptor regulates its functional coupling to adenylate cyclase and subcellular distribution. Proc Natl Acad Sci USA. 1986;83:9408–9412. doi: 10.1073/pnas.83.24.9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJG. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SO. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 2010;39:309–328. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- Smith SO. Insights into the activation mechanism of the visual receptor rhodopsin. Biochem Soc Trans. 2012;40:389–393. doi: 10.1042/BST20110751. [DOI] [PubMed] [Google Scholar]
- Sommer ES, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286:7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR. Cell signalling: Binding the receptor at both ends. Nature. 2011;469:172–173. doi: 10.1038/469172a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann M, Simon MI. G protein diversity: a distinct class of alpha subunits is present in vertebrates and invertebrates. Proc Natl Acad Sci USA. 1990;87:9113–9117. doi: 10.1073/pnas.87.23.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struts AV, Salgado GF, Martinez-Mayorga K, Brown MF. Retinal dynamics underlie its switch from inverse agonist to agonist during rhodopsin activation. Nat Struct Mol Biol. 2011;18:392–394. doi: 10.1038/nsmb.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunahara RK, Dessauer CW, Gilman AG. Complexity and diversity of mammalian adenylyl cyclases. Annu Rev Pharmacol Toxicol. 1996;36:461–480. doi: 10.1146/annurev.pa.36.040196.002333. [DOI] [PubMed] [Google Scholar]
- Surgand JS, Rodrigo J, Kellenberger E, Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins. 2006;62:509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 23A: evolution of receptor specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Tan CM, Brady AE, Nickols HH, Wang Q, Limbird LE. Membrane trafficking of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:559–609. doi: 10.1146/annurev.pharmtox.44.101802.121558. [DOI] [PubMed] [Google Scholar]
- Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Tesmer JJ. The quest to understand heterotrimeric G protein signaling. Nat Struct Mol Biol. 2010;17:650–652. doi: 10.1038/nsmb0610-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, Liu W, Chun E, Katritch V, Wu H, Vardy E, Huang XP, Trapella C, Guerrini R, Calo G, Roth BL, Cherezov V, Stevens RC. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting PC, Umland SP, Aslanian R, Cao J, Garlisi CG, Huang Y, Jakway J, Liu Z, Shah H, Tian F, Wan Y, Shih N-Y. The synthesis of substituted bipiperidine amide compounds as CCR3 ligands: Antagonists versus agonists. Bioorg Med Chem Lett. 2005;15:3020–3023. doi: 10.1016/j.bmcl.2005.04.054. [DOI] [PubMed] [Google Scholar]
- Tobin AB, Butcher AJ, Kong KC. Location, location, location... site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol Sci. 2008;29:413–420. doi: 10.1016/j.tips.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topiol S, Sabio M. X-ray structure breakthroughs in the GPCR transmembrane region. Biochem Pharmacol. 2009;78:11–20. doi: 10.1016/j.bcp.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs — theoretical and experimental studies. Curr Med Chem. 2012;19:1090–1109. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabeno J, Nikiforovich GV, Marshall GR. Insight into the binding mode for cyclopentapeptide antagonists of the CXCR4 receptor. Chem Biol Drug Des. 2006;67:346–354. doi: 10.1111/j.1747-0285.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- Van Eps N, Preininger AM, Alexander N, Kaya AI, Meier S, Meiler J, Hamm HE, Hubbell WL. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci USA. 2011;108:9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Francis D, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, Gurevich VV. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci USA. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1154–1209. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- Weyand I, Kuhn H. Subspecies of arrestin from bovine retina. Equal functional binding to photoexcited rhodopsin but various isoelectric focusing phenotypes in individuals. Eur J Biochem. 1990;193:459–467. doi: 10.1111/j.1432-1033.1990.tb19360.x. [DOI] [PubMed] [Google Scholar]
- Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Lee JM, Dede K, Imburgia CS, Jurewicz AJ, Chan G, Fornwald JA, Dhanak D, Christmann LT, Darcy MG, Widdowson KL, Foley JJ, Schmidt DB, Sarau HM. Identification of potent, selective non-peptide CC chemokine receptor-3 antagonist that inhibits eotaxin-, eotaxin-2-, and monocyte chemotactic protein-4-induced eosinophil migration. J Biol Chem. 2000;275:36626–36631. doi: 10.1074/jbc.M006613200. [DOI] [PubMed] [Google Scholar]
- Wikstrom M, Verkhovsky MI, Hummer G. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim Biophys Acta. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- Wilkin TJ, Gulick RM. CCR5 Antagonism in HIV infection: current concepts and future opportunities. Annu Rev Med. 2012;63:81–93. doi: 10.1146/annurev-med-052010-145454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates R, Jr, Lefkowitz RJ. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao RP. Beta-adrenergic signaling in the heart: dual coupling of the beta2-adrenergic receptor to G(s) and G(i) proteins. Sci STKE. 2001;104:re15. doi: 10.1126/stke.2001.104.re15. [DOI] [PubMed] [Google Scholar]
- Xie G, D’Antona AM, Edwards PC, Fransen M, Standfuss J, Schertler GF, Oprian DD. Preparation of an activated rhodopsin/transducin complex using a constitutively active mutant of rhodopsin. Biochemistry. 2011;50:10399–10407. doi: 10.1021/bi201126r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors. Proc Natl Acad Sci USA. 2004;101:959–963. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan W, Liang Z, Zhu A, Kurtkaya S, Shim H, Snyder JP, Liotta DC. Discovery of small molecule CXCR4 antagonists. J Med Chem. 2007;50:5655–5664. doi: 10.1021/jm070679i. [DOI] [PubMed] [Google Scholar]
- Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q. Dual targeting of CCR2 and CCR5: therapeutic potential for immunologic and cardiovascular diseases. J Leukoc Biol. 2010;88:41–55. doi: 10.1189/jlb.1009671. [DOI] [PubMed] [Google Scholar]