Abstract
Exposure to inorganic arsenic increases the risk of basal cell carcinoma (BCC). Arsenic metabolism is a susceptibility factor for arsenic toxicity, and specific haplotypes in arsenic (+3 oxidation state) methyltransferase (AS3MT) have been associated with increased urinary fractions of the most toxic arsenic metabolite, methylarsonic acid (MMA). The aim of this study is to elucidate the association of AS3MT haplotypes with arsenic metabolism and the risk of BCC. Four AS3MT polymorphisms were genotyped in BCC cases (N = 529) and controls (N = 533) from Eastern Europe with low to moderate arsenic exposure (lifetime average drinking water concentration: 1.3 µg/L, range 0.01–167 µg/L). Urinary metabolites [inorganic arsenic (iAs), MMA, dimethylarsinic acid (DMA)] were analyzed by HPLC-ICPMS. Five AS3MT haplotypes (based on rs3740400 A/C, rs3740393 G/C, rs11191439 T/C and rs1046778 T/C) had frequencies >5%. Individuals with the CCTC haplotype had lower %iAs (P = 0.032) and %MMA (P = 0.020) in urine, and higher %DMA (P = 0.033); individuals with the CGCT haplotype had higher %MMA (P < 0.001) and lower %DMA (P < 0.001). All haplotypes showed increased risk of BCC with increasing arsenic exposure through drinking water (ORs 1.1–1.4, P values from <0.001 to 0.082), except for the CCTC haplotype (OR 1.0, CI 0.9–1.2, P value 0.85). The results suggest that carriage of AS3MT haplotypes associated with less-efficient arsenic methylation, or lack of AS3MT haplotypes associated with a more-efficient arsenic methylation, results in higher risk of arsenic-related BCC. The fact that AS3MT haplotype status modified arsenic metabolism, and in turn the arsenic-related BCC risk, supports a causal relationship between low-level arsenic exposure and BCC. Environ. Mol. Mutagen. 56:60–69, 2015. © 2014 The Authors. Environmental and Molecular Mutagenesis published by Wiley Periodicals, Inc. on behalf of Environmental Mutagen Society
Keywords: polymorphism, methylarsonic acid, dimethylarsinic acid, metabolism
Introduction
Inorganic arsenic occurs globally in drinking water, particularly ground water, and elevated exposure has been associated with increased risk of nonmelanoma skin cancers, including basal cell carcinoma (BCC) [Hughes et al., 2011; Karagas et al., 2001; IARC, 2012; Leonardi et al., 2012]. Humans, however, show marked variation in susceptibility to arsenic toxicity. The main known susceptibility factors are gender, nutrition, and the efficiency of metabolizing arsenic [Vahter, 2002]. Inorganic arsenic (iAs) is methylated in the body to methylarsonic acid (MMA) and dimethylarsinic acid (DMA). In humans, like most mammals, efficient methylation from iAs to DMA is associated with decreased reactivity and increased rate of urinary arsenic excretion [Vahter, 2002; Gardner et al., 2011]. Individuals and populations, however, show major differences in arsenic metabolism. Incomplete and less-efficient arsenic metabolism, with higher urinary fractions of iAs and MMA and lower fractions of DMA, seems to be a marker of sensitivity to certain arsenic-related diseases, including cancer [Lindberg et al., 2008b; Chung et al., 2009].
Genetic variants in the main arsenic-methylating enzyme, arsenic (+3 oxidation state) methyltransferase (AS3MT), have been shown to modify arsenic metabolism in several populations [Sumi and Himeno, 2012]. For eight AS3MT single nucleotide polymorphisms (SNPs) combined into haplotypes, four AS3MT haplotypes had similar effects on the arsenic metabolite pattern in two different populations, one from Argentina and one from Bangladesh [Engstrom et al., 2011]. Associations between AS3MT polymorphisms and toxic effects, such as skin lesions, have also been reported [Valenzuela et al., 2009; Pierce et al., 2012]. Our hypothesis is that AS3MT haplotypes that are associated with the arsenic metabolite pattern also are associated with the arsenic-related risk of BCC.
Materials and Methods
Study Population
Within a European case–control study that previously identified arsenic exposure in drinking water to be associated with risk of BCC [Leonardi et al., 2012] we evaluated the impact of AS3MT haplotypes on: (1) the urinary pattern of arsenic metabolites (controls only) and (2) the association between lifetime arsenic concentration and BCC risk (cases and controls). BCC cases (N = 529) and controls (N = 533) were recruited as part of a large case–control study designed to evaluate the risk of various cancers in relation to low environmental arsenic exposure in Hungary, Romania, and Slovakia between 2002 and 2004 [Leonardi et al., 2012]. Study areas were defined as certain counties (total population 2.8 million) in Hungary (Bacs, Békés, Csongràd, and Jàzs-Nagykun-Szolnok), Romania (Bihor and Arad) and Slovakia (Banska Bystrica and Nitra) [Lindberg et al., 2006]. These areas were selected because of past, and in some places ongoing, arsenic exposure through drinking water, including public water supplies. Exposure assessment was based on measurements of water supplies in the study, and data from routine monitoring of water in the past [Hough et al., 2010]. Overall, 25% of the population had a lifetime average water concentration above 10 µg/L and 8% above 50 µg/L [Hough et al., 2010]. The large majority of the study participants were of European ancestry.
Table1 shows descriptive data of the study subjects. Incident cases of BCC (ICD-10 code C44) were identified and invited on the basis of histopathological examinations by pathologists. These were compared with hospital-based controls (general surgery, orthopedic and trauma patients with, for example, appendicitis, abdominal hernias, duodenal ulcers, cholelithiasis, and fractures), which were selected and included in the study based on a published set of criteria [Leonardi et al., 2012]. They were frequency-matched with the cases for age, gender, and county of residence. Patients with malignant tumors, diabetes or cardiovascular diseases were excluded as study controls. Cases and controls were included if they had resided in the study area for at least 1 year during their lifetime. Clinicians and pathologists were blinded to the exposure status of cases and controls.
Table 1.
Characteristics of the Study Population
| Controls |
Cases |
|||||||
|---|---|---|---|---|---|---|---|---|
| Variable | N | Median | Min | Max | N | Median | Min | Max |
| Age (years) | 533 | 61 | 28 | 83 | 529 | 66 | 30 | 85 |
| BMI (kg/m2) | 510 | 27 | 13 | 48 | 501 | 27 | 16 | 48 |
| Lifetime water arsenic conc. (per 10 µg/L) | 529 | 1.8 | 0.01 | 167 | 523 | 1.06 | 0.09 | 140 |
| Gender (% women) | 533 | 49 | 529 | 55 | ||||
| Smoking (% yes) | 502 | 53 | 496 | 65 | ||||
| Country (N) | 533 | 529 | ||||||
| Hungary | 242 | 160 | ||||||
| Romania | 156 | 158 | ||||||
| Slovakia | 135 | 211 | ′ | |||||
BMI (body mass index), N = number of individuals.
Cases and controls were interviewed by trained personnel and they completed a general questionnaire, which included information on detailed residential history focused on identification of drinking water sources at home, individual cumulative sun exposure in summer, skin complexion, effects of sun exposure on skin and age(s) at diagnosis of BCC. Skin sensitivity to burns (effects of sun exposure) is an index based on the reported intensity of the cutaneous reaction to 1 hr midday sun exposure of the upper trunk, and skin complexion as the self-reported complexion of the skin as light, medium or dark. Clinicians collected venous blood and urine samples from cases and controls after the study participants had signed consent forms. The blood samples were kept frozen at −80°C until analysis. Further details about the study populations and sampling have been described previously [Lindberg et al., 2006; Thirumaran et al., 2006; Leonardi et al., 2012; Surdu et al., 2013].
Ethical approval was obtained from the Ethical Committee of the National Health Research Council and the Regional Ethical Committee of the Szent-Gyorgyi Albert University of Szeged (Hungary), from local hospitals and Public Health Departments (Romania), and from Ethical Committees established in hospitals and State Health Institutes (Slovakia).
Arsenic Exposure Assessment
All water samples were analyzed by one laboratory, using hydride generation-atomic absorption spectrometry (HG-AAS), with a limit of detection (LOD) of 0.2 µg/L [Hough et al., 2010]. The exposure assessment details are described in [Hough et al., 2010]. In short, for each individual, the concentrations of arsenic in drinking water at addresses over their lifetime were derived from measurements at the time of the study and historical data provided by national water authorities. If historical data were not available, concentrations of arsenic in village, and private wells were assumed to have been the same as the currently measured concentrations. The lifetime average arsenic concentration (µg/L) constitutes the time-weighted average concentration of arsenic in residential drinking water over all the years with nonmissing arsenic concentrations over a participant's lifetime. This variable was used as a measure of lifetime arsenic exposure in this study. Ten individuals (four controls and six cases) had missing data for lifetime average arsenic concentration and were thus excluded from the BCC analyses.
Arsenic Metabolism
Urine samples were collected for determination of individual arsenic metabolite patterns. Only urine samples from the controls were included in this study, as disease status may affect arsenic metabolism. Each participant was asked to provide an early morning urine sample. The samples were frozen at −20°C within 15 min of collection. Urinary metabolites iAs, MMA, and DMA were measured with HPLC online with inductively coupled plasma-mass spectrometry (ICPMS) [Lindberg et al., 2006]. Total urinary arsenic metabolites (U-As), computed as the sum of iAs, MMA, and DMA, were used as a measure of the current exposure to iAs. The fractions (%) of the different metabolites in urine were used to assess the efficiency of arsenic metabolism. Concentrations of arsenic in urine were adjusted to the average specific gravity of the urine (1.018 g/mL) in the study population. There was a cluster of values close to 100% DMA at the lowest water arsenic concentrations, which likely was due to contribution of DMA from certain foods [Lindberg et al., 2006]. Additionally, there were uncertainties in the speciation data of urine samples with very low concentrations, as DMA is often the only species well above the limit of detection. Because such uncertainties would invalidate the urinary arsenic metabolite pattern as marker of arsenic methylation efficiency, we decided to include only individuals with total urinary arsenic metabolite concentrations >5 μg/L (N = 285) in the genetic association analyses. Another eight individuals were excluded because they had high urinary arsenic concentrations (above 20 µg/L) and low water arsenic concentrations (below 1 µg/L) in combination with a high percentage of DMA in the urine (above 80%), suggesting food as a major source of DMA. The sample size after these exclusions was 277 individuals. For %iAs and %MMA, the numbers were somewhat lower (N = 269 and 245, respectively) since individuals with fractions of 0% were excluded when natural log (ln) transforming the data.
Genotyping
Several SNPs in AS3MT have been associated with arsenic metabolite pattern [review in Sumi and Himeno, 2012]. To evaluate if genetic variation in AS3MT influences the risk of arsenic-related BCC risk, we genotyped four SNPs, all of which were present in haplotypes that we previously have shown are associated with arsenic metabolism in populations from Argentina and Bangladesh [Engström et al., 2011]. The selection of SNPs to be genotyped was based on the following observations. Rs11191439 is a nonsynonymous SNP and carriers of the C-allele (coding for Thr) have higher %iAs, higher %MMA and lower %DMA compared to TT carriers [Drobna et al., 2004; Lindberg et al., 2007; Hernandez et al., 2008; Engstrom et al., 2011], i.e., less efficient arsenic metabolism. Carriers of the intronic rs3740400 A- and rs3740393 G-alleles, as well as rs1046778 (located in the 3′-untranslated region) T-alleles have higher %iAs, higher %MMA and lower %DMA compared with reference genotypes [Meza et al., 2005; Agusa et al., 2009; Chung et al., 2009; Engstrom et al., 2011], indicating less-efficient metabolism. Rs3740400 and rs1046778 have been associated with gene expression of AS3MT as well [Engström et al., 2011, 2013].
DNA was isolated from peripheral blood using Qiagen mini-preparation kits (Qiagen GmbH, Hilden, Germany). The four AS3MT SNPs (rs11191439, rs3740400, rs3740393, and rs1046778) were genotyped by allelic discrimination with Taqman assays (Applied Biosystems Foster City, CA) on an ABI 7900 instrument (Applied Biosystems). Haplotypes were constructed on the combination of these four SNPs using PHASE [Stephens and Donnelly, 2003]. Haplotype frequencies are presented in Table2 together with the associations between haplotype and arsenic metabolism reported in other populations [Engstrom et al., 2011]. Although the haplotypes in Engstrom et al. [2011] were based on eight SNPs, the haplotype frequencies were similar when based on the four SNPs included in the present study. The genomic region where AS3MT is situated (10q24) has been shown to have a long linkage disequilibrium (LD) block spanning over the AS3MT gene [Engstrom et al., 2013].
Table 2.
AS3MT Haplotype Frequencies by Country
| Haplotypea | Hungary | Romania | Slovakia | All | More efficient arsenic metabolismb |
|---|---|---|---|---|---|
| Haplotype 1—AGTT | 51%c | 57% | 52% | 53% | 0 copies |
| 0/1/2 copies | 25/47/28 | 23/42/36 | 21/54/25 | 23/48/29 | |
| Haplotype 2—CCTC | 10% | 12% | 14% | 12% | 1–2 copies |
| 0/1/2 copies | 82/17/1 | 78/20/2 | 75/24/2 | 78/20/2 | |
| Haplotype 3—CGTC | 18% | 14% | 16% | 16% | 1–2 copies |
| 0/1/2 copies | 66/31/3 | 75/22/3 | 71/26/3 | 70/27/3 | |
| Haplotype 4—CGCT | 11% | 11/% | 10% | 11% | 0 copies |
| 0/1/2 copies | 78/21/1 | 79/20/1 | 82/17/1 | 80/19/1 | |
| Haplotype 5—ACTC | 7% | 5% | 5% | 6% | NAd |
| 0/1/2 copies | 87/12/0.5 | 89/11 | 87/13/0.5 | 88/12/0.3 | |
| Haplotype 6—CGTT | 2% | 1% | 2% | 2% | NA |
| 0/1/2 copies | 96/4/0 | 98/2/0 | 96/4/0 | 96/4/0 | |
| Haplotype 7—AGTC | <1% | <1% | <1% | <1% | NA |
| 0/1/2 copies | 100/<1%/0 | 100/<1%/0 | 100/<1%/0 | 100/<1%/0 |
The SNPs are, in the 5' to 3' direction: rs3740400, rs3740393, rs11191439 and rs1046778.
Denotes the haplotype previously associated with a more efficient arsenic methylation (lower fractions of iAs or MMA and/or higher DMA in urine) [Engstrom et al. 2011].
Frequency of the haplotype.
NA = not available.
Statistical Analyses
Deviation from Hardy–Weinberg equilibrium was tested using chi-square analysis. The LD pattern was evaluated in each country by Haploview [Barrett et al., 2005]. Individuals from all three study areas were pooled in the statistical analyses. Groups with one or two haplotype copies were pooled, since there were few individuals with two haplotype copies in all groups, with exception for haplotype 1.
Associations between haplotypes (independent variable) and urinary arsenic metabolites were evaluated by multivariable-adjusted regression analyses with %iAs, %MMA, and %DMA as dependent variables. These variables were used as continuous variables, and in order to achieve normally distributed residuals, %iAs and %MMA were natural log (ln) transformed in all analyses. For covariate inclusion, univariate models were performed, evaluating the association of the biologically relevant variables with each metabolite (dependent variables). Variables with P < 0.10 were included in the final multivariable-adjusted regression model together with haplotype (as categorical variable). The following variables were tested: U-As (µg/L, continuous variable), age, gender, cigarette smoking (yes/no), county, and BMI. The final models for AS3MT and metabolites are presented in Table3. Arsenic exposure (measured as U-As) has been shown to influence the arsenic metabolite pattern in high-exposed individuals in Bangladesh and Argentina [Engström et al., 2011]. We therefore evaluated the influence of U-As on the arsenic metabolite pattern in this population. However, including U-As into the models did not impact on the associations between haplotype and metabolite pattern (data not shown) and we did not adjust for arsenic exposure in the final models.
Table 3.
Geometric Mean Metabolite Fractions (Arithmetic Mean for %DMA) for each AS3MT Haplotype and P values from Multivariate Regression Analyses (Controls Only)a
| Controls with total urinary arsenic >5 µg/L |
|||
|---|---|---|---|
| Haplotype/haplotype combinations | %iAs (N) | %MMA (N) | %DMA (N) |
| Haplotype 1 | |||
| 0 copies | 8.2 (75) | 15.3 (68) | 72.2 (78) |
| 1/2 copies | 8.6 (194) | 16.1 (177) | 72.4 (199) |
| P valueb | 0.21 | 0.49 | 0.88 |
| Haplotype 2 | |||
| 0 copies | 8.9 (210) | 16.6 (189) | 71.3 (215) |
| 1/2 copies | 6.9 (59) | 13.5 (56) | 76.0 (62) |
| P value | 0.032 | 0.020 | 0.033 |
| Haplotype 3 | |||
| 0 copies | 8.1 (173) | 15.9 (160) | 73.1 (180) |
| 1/2 copies | 9.2 (96) | 15.7 (85) | 70.9 (97) |
| P value | 0.24 | 0.92 | 0.25 |
| Haplotype 4 | |||
| 0 copies | 8.2 (210) | 14.8 (192) | 73.9 (216) |
| 1/2 copies | 9.5 (59) | 20.5 (53) | 66.9 (61) |
| P value | 0.14 | <0.001 | <0.001 |
| Haplotype 5 | |||
| 0 copies | 8.6(234) | 16.0(215) | 72(240) |
| 1/2 copies | 7.8(35) | 14.9(30) | 74.5(37) |
| P value | 0.26 | 0.30 | 0.11 |
| Haplotypes 2 and 4c | |||
| 1/2 and 0 copies | 6.6 (48) | 12.5 (45) | 77.3 (50) |
| 0 and 0 copies | 8.7 (162) | 15.5 (147) | 72.8 (166) |
| 1/2 and 1/2 copies | 8.7 (11) | 18.4 (11) | 70.7 (12) |
| 0 and 1/2 copies | 9.7 (48) | 21.1 (42) | 66.0 (49) |
| P value | 0.032 | <0.001 | <0.001 |
The multivariate models was as follows: %iAs (natural log (ln) transformed) = β1 × haplotype + β2 × gender + β3 × smoking + β4 × county + β5 × BMI;
%MMA (ln) = β1 × haplotype + β2 × gender + β3 × smoking + β4 × BMI;
%DMA = β1 × haplotype + β2 × gender + β3 × smoking + β4 × county + β5 × BMI;
P values presented are for haplotype term as a whole in the multivariate regression (β1 × haplotype).
Number of copies of haplotype 2 are denoted first. The groups are in order of expected effect (the groups with the lowest expected %MMA are first), based on the analysis of the haplotypes separately.
We stratified the study population according to haplotype and used unconditional logistic regression to evaluate the association between average lifetime water arsenic concentration (per 10 µg/L increase, continuous variable) and BCC risk. A separate model, not stratified for haplotype, provided the P value for the interaction term (haplotype × lifetime arsenic concentration). Case/control status was used as the dependent variable in all analyses.
The following covariates were included in the logistic regression models: age, gender, years of education (three groups, defined as “below 8 years”, between 8 and 12 years” and “above 12 years”), county (since the number of controls per case varied by county [Leonardi et al., 2012], as well as the two indices of sun exposure that most strongly predicted BCC risk: skin complexion (three groups, defined as “light”, “medium”, and “dark”) and skin response to 1 hr of midday sun (five groups, defined as “blistered”, “sunburnt”, “mild burn”, “tan”, and “no change”). As neither smoking, nor body mass index (BMI), are likely confounders for BCC, and these covariates did not alter the magnitude of associations (data not shown), we did not include them in the final models.
We also evaluated combinations of two haplotypes, giving four combined groups (for example, the combinations of haplotypes 1 and 2 gave the following groups: (a) zero copies of haplotype 1 and zero copies of haplotype 2; (b) one or two copies of haplotype 1 and zero copies of haplotype 2; (c) zero copies of haplotype 1 and one or two copies of haplotype 2; and (d) one or two copies of haplotype 1 and one or two copies of haplotype 2). We combined haplotypes based on the results from the analyses encompassing a single haplotype. In the analysis of metabolite pattern, we combined haplotypes that were significantly associated with arsenic metabolites. For the analysis of BCC risk, we combined haplotypes that were either significantly associated with metabolites or haplotypes that showed a significant effect modification with BCC risk (based on the hypothesis that bcc risk could be influenced by both metabolite pattern or directly by the haplotype). Haplotypes were combined regardless of direction (e.g., efficient or nonefficient methylator).
Results
Genetic Background
All SNPs were in Hardy–Weinberg equilibrium. The LD values (R2) were similar in all countries (Fig. 1) and to those of the Hapmap European reference population (CEPH, derived from US residents with northern and western European ancestry (CEU) [Thorisson et al., 2005]). Rs1046778 was in moderate LD with both rs3740400 and rs3740393 (R2 = between 0.36 and 0.46, the C-alleles of both SNPs were associated to each other), whereas rs11191439 was not in LD with any SNP.
Figure 1.
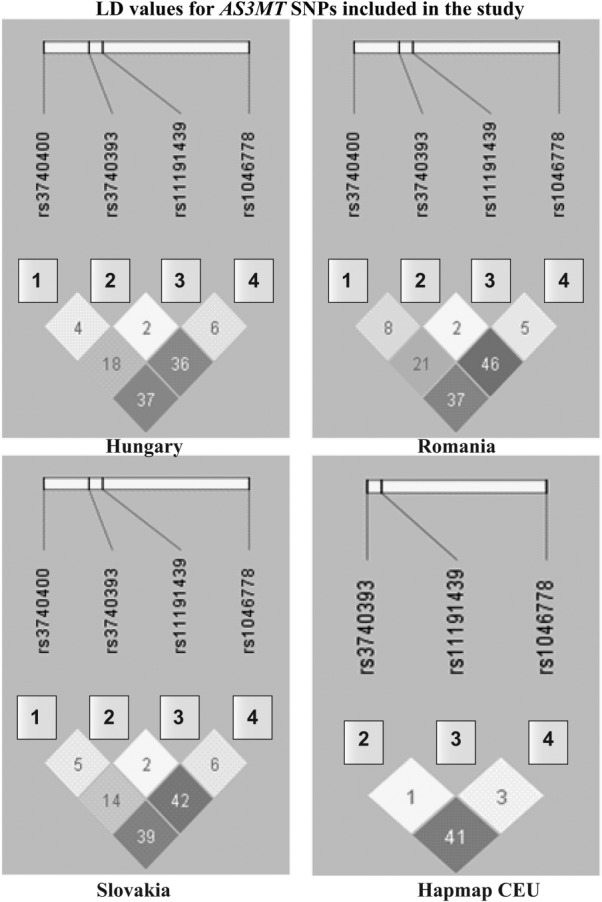
LD values (R2) for the AS3MT SNPs in the different study populations as well as the reference CEU population from Hapmapa. aSNPs are shown in the 5′ to 3′ direction. CEU denotes the reference Hapmap population (CEPH, derived from US residents with northern and western European ancestry, http://www.hapmap.org, no data available for rs3740400) [Thorisson et al., 2005].
Seven haplotypes were found in this population, five of which had frequencies of >5% and were included in the subsequent statistical analyses (Table2). The AGTT (denoted haplotype 1) was the most common (SNPs are in 5′–3′ order based on their position on the gene: rs3740400, rs3740393, rs11191439, and rs1046778).
AS3MT Haplotypes and the Metabolism of Arsenic
The control individuals included in the assessment of associations between AS3MT haplotype and arsenic metabolizing phenotype had a geometric mean U-As concentration of 15.0 µg/L (N = 277, range 5.0–140 µg/L), and geometric mean fraction of iAs of 8.5% (N = 269, range 1.1–88%), MMA of 15% (N = 245, range 1.5–63%) and DMA of 72% (arithmetic mean) (range 9.8–100%).
Two haplotypes, 2 and 4, were significantly associated with the arsenic metabolite pattern (Table3). Individuals with one or two copies of haplotype 2 had lower %iAs (P = 0.032), lower %MMA (P = 0.020) and higher %DMA (P = 0.033). Individuals with one or two copies of haplotype 4 had particularly high %MMA (P < 0.001) and low %DMA (<0.001). No statistically significant effects were seen for the most frequently occurring haplotype 1.
When combining the haplotypes that influenced arsenic metabolite pattern (haplotypes 2 and 4) (Table3) we found that individuals with one or two copies of haplotype 2 and zero copies of haplotype 4 had the lowest %iAs (P = 0.032) and %MMA (P < 0.001), and the highest %DMA (P < 0.001). Conversely, individuals with zero copies of haplotype 2 and one or two copies of haplotype 4 had the highest %iAs and %MMA, and the lowest %DMA. Individuals with zero copies of both haplotypes 2 and 4 had somewhat lower %MMA and higher %DMA than individuals with one or two copies of both haplotypes 2 and 4. However, the group with one or two copies of both haplotypes 2 and 4 comprised only 11 or 12 individuals so the results for this group have low power.
AS3MT Haplotypes and Arsenic-Related BCC Risk
Based on cases and controls, the risk of BCC increased with increasing lifetime concentration of arsenic in drinking water (OR 1.2 per 10 µg/L increase, 95% CI 1.1–1.3), as previously presented [Leonardi et al., 2012]. Results of the logistic regression analysis of haplotype-specific associations between lifetime water arsenic concentrations and BCC are shown in Table4. All haplotypes showed increased risk of BCC with increasing arsenic exposure. The OR:s were above 1.0 for all analyses, but some analyses were not statistically significant. ORs ranged from 1.1 to 1.4, P values ranged from <0.001 to 0.082, except for among individuals with one or two copies of haplotype 2 (OR 1.0, CI 0.9–1.2, P value 0.85).
Table 4.
Association Between Lifetime Arsenic Concentration and Risk of Basal Cell Carcinoma, Stratified for Haplotype (Logistic Regression)
| Effect estimatesa |
||||||
|---|---|---|---|---|---|---|
| Haplotype | N | P | OR | 95% CI | Effect modificationb | |
| Haplotype 1 | 0.036 | |||||
| 0 copies | 239 | 0.001 | 1.4 | 1.1 | 1.8 | |
| 1/2 copies | 782 | 0.045 | 1.1 | 1.0 | 1.2 | |
| Haplotype 2 | 0.098 | |||||
| 0 copies | 805 | <0.001 | 1.2 | 1.1 | 1.4 | |
| 1/2 copies | 216 | 0.85 | 1.0 | 0.9 | 1.2 | |
| Haplotype 3 | 0.43 | |||||
| 0 copies | 719 | 0.072 | 1.1 | 1.0 | 1.2 | |
| 1/2 copies | 302 | <0.001 | 1.4 | 1.2 | 1.8 | |
| Haplotype 4 | 0.58 | |||||
| 0 copies | 809 | 0.004 | 1.2 | 1.1 | 1.3 | |
| 1/2 copies | 212 | 0.025 | 1.3 | 1.0 | 1.6 | |
| Haplotype 5 | 0.054 | |||||
| 0 copies | 894 | 0.006 | 1.1 | 1.0 | 1.2 | |
| 1/2 copies | 127 | 0.082 | 1.3 | 1.0 | 1.8 | |
| Haplotypes 1 and 2 | 0.010 | |||||
| 0 and 0 copies | 140 | 0.001 | 2.7 | 1.5 | 4.8 | |
| 0 and 1/2 copies | 99 | 0.99 | 1.0 | 0.7 | 1.4 | |
| 1/2 and 0 copies | 665 | 0.040 | 1.1 | 1.0 | 1.3 | |
| 1/2 and 1/2 copies | 117 | 0.95 | 0.99 | 0.8 | 1.2 | |
| Haplotypes 1 and 4 | 0.020 | |||||
| 0 and 0 copies | 139 | 0.012 | 1.6 | 1.1 | 2.2 | |
| 0 and 1/2 copies | 100 | 0.014 | 1.5 | 1.1 | 2.0 | |
| 1/2 and 0 copies | 670 | 0.041 | 1.1 | 1.0 | 1.2 | |
| 1/2 and 1/2 copies | 112 | 0.79 | 1.1 | 0.7 | 1.6 | |
| Haplotypes 2 and 4 | 0.26 | |||||
| 0 and 0 copies | 618 | 0.003 | 1.2 | 1.1 | 1.3 | |
| 0 and 1/2 copies | 187 | 0.006 | 1.5 | 1.1 | 1.9 | |
| 1/2 and 0 copies | 191 | 0.52 | 1.1 | 0.9 | 1.3 | |
| 1/2 and 1/2 copiesc | 25 | |||||
CI = confidence interval, OR = odds ratio, N = number of individuals.
Model included life time arsenic concentration (per 10 µg/L increase), skin complexion, skin response to 1-hr midday sun, age, gender, education, and county.
A separate model that included all individuals provided the P value for the effect modification (the interaction term haplotype × life time arsenic concentration).
Too few individuals to conduct any analyses.
Individuals with zero copies of haplotype 1 had a higher BCC risk with increasing lifetime concentration of arsenic, compared to those with those with one or two copies [zero copies: OR 1.4, CI 1.1–1.8, P value 0.001; one copy: OR 1.2, CI 0.9–1.2, P value 0.36 (N = 492); and two copies OR 1.2, CI 1.0–1.4 (N = 290), P value 0.054]. There was a significant effect modification (P = 0.036). Similarly, individuals with zero copies of haplotype 2 had a significantly increased BCC risk (OR 1.2, CI 1.1–1.4, P value <0.001; P value for interaction 0.098). Haplotypes 3, 4, and 5 all showed higher risk with one or two copies than with zero copies.
AS3MT Haplotype Combinations and Arsenic-Related BCC Risk
We found a significant effect modification on BCC risk for the combination of haplotypes 1 and 2 (P = 0.010) and haplotypes 1 and 4 (P = 0.020; Table4). The highest risk among all the haplotype combinations was seen for individuals with zero copies of both haplotype 1 and 2, who had 2.7 times higher risk in relation to arsenic exposure (CI 1.5–4.8). Individuals with one or two copies of haplotype 2 did not show any increased risk of BCC with increasing arsenic exposure, irrespective of the number of copies of haplotype 1. Also, for the combination of haplotype 1 and 4, individuals with zero copies of both haplotypes had the highest BCC risk (OR 1.6, CI 1.1–2.2). Individuals with zero copies of haplotype 1 in combination with one or two copies of haplotype 4, which was associated with very high urinary fraction of MMA (Table3), showed almost as high risk (OR 1.5). Individuals with one or two copies of both haplotypes had a lower risk of BCC (OR of 1.1, CI 0.7–1.6, P value 0.79), mainly due to the protective effects of haplotype 1. When combining haplotypes 2 and 4, the interaction term was not statistically significant and the analysis was hampered by having too few individuals with one or two copies of both haplotypes.
Discussion
This study, carried out in a European population with low to moderate arsenic exposure, confirms earlier findings of an association between genetic variation in AS3MT and arsenic metabolism in Asian or American populations exposed to high levels of arsenic. The study also showed that this gene, which encodes a key arsenic-metabolizing enzyme, modifies the previously reported association between arsenic exposure and BCC [Leonardi et al., 2012]. Importantly, the observed gene–environment interaction supports the hypothesis that the association between low-level arsenic exposure and BCC risk reflects a causal relationship.
The directions of the results suggest that polymorphisms in AS3MT that associated with less-efficient arsenic methylation (especially lack of copies of haplotypes 1 and 2, particularly in combination, and one or two copies of haplotype 4) are associated with a higher risk of arsenic-related BCC. However, only haplotype 1 demonstrated a statistically significant effect modification, where individuals with one or two copies of haplotype 1 had lower BCC risk with increasing exposure compared to individuals with no copies of haplotype 1, but this haplotype did not seem to affect arsenic metabolism. Thus, AS3MT may influence the susceptibility to BCC via other mechanisms not related to arsenic metabolism. Other functions of AS3MT in the human body apart from arsenic metabolism have not yet been elucidated. An interesting finding was that individuals lacking both haplotype 1 and 2 had an almost three times increase in BCC risk per 10 µg/L increase in lifetime drinking water arsenic. Similarly, those lacking both haplotypes 1 and 4 had a 60% increase in BCC risk for the same degree of arsenic exposure. It should be noted that the speciation of arsenic metabolites in urine was only performed in the controls and we cannot be sure if the effects found are the same in the individuals that developed cancer. This fact stresses that prospective studies are needed to follow individual arsenic metabolism and relate that to the development of cancer.
The AS3MT haplotypes found in this population were the same as those previously found in Argentina and Bangladesh [Engstrom et al., 2011]. We also found three haplotypes that were not found in the studies in Argentina or Bangladesh: one haplotype with a frequency above 5% (haplotype 5), and two minor haplotypes. The haplotype frequencies were similar in all European countries studied here and included all four major haplotypes that have been shown to affect arsenic metabolism [Engstrom et al., 2011]. However, in the current study, the associations between AS3MT haplotypes and metabolism differed somewhat from the studied Asian and South American populations in that haplotypes 1 and 3 did not modify arsenic metabolism. This suggests that the SNPs analyzed are not functional themselves, but are in LD with functional variants that have a different LD pattern in these European populations compared with the Argentinean and Bangladeshi populations.
This study has several strengths. The study population is fairly large and several of the variant alleles had relatively high frequency. The LD analysis indicated a similar genetic structure for the AS3MT gene in the three different subpopulations and those could thus be pooled to increase the power of the analyses. Differences in unmeasured confounders between countries were partly addressed by including county, accounting for regional differences in arsenic exposure and BCC risk, as a variable in the models. All arsenic measurements and genotyping analyses were made by the same method and in the same laboratory, using adequate quality control. However, the low levels of arsenic exposure and the uncertainties in lifetime arsenic exposure assessments have likely hampered the evaluation of associations between AS3MT haplotypes and arsenic metabolism. The urinary arsenic concentrations were substantially lower in this study (median 6 µg/L for all individuals) compared with previous studies in Argentina and Bangladesh (medians 200 and 100 µg/L, respectively [Engstrom et al., 2011]. At such low levels of exposure, food may also be an important source of arsenic exposure, and food may contain both iAs, DMA, and arsenosugars, which are metabolized to DMA in the body [Lindberg et al., 2006]. Also, other urinary metabolites than DMA were often below the limit of detection, distorting the true metabolite distribution. Although we excluded individuals with arsenic in urine below 5 µg/L in these analyses, the arsenic metabolites in urine may, for some individuals, not represent exposure to exclusively inorganic arsenic. However, a limitation of this study is that for some of the analyses (e.g., the combination of haplotypes 2 and 4) there are a small number of individuals with certain haplotype/haplotype combinations. We attempted to find out if gender-specific effects of AS3MT could explain gender differences in arsenic metabolism [Lindberg et al., 2008a]. However, this appears not to be the case, as we observed similar directions of the associations for both men and women, and there were in general no big differences in genetic effect modification between the sexes (data not shown).
The association between genetic variation in AS3MT and cancer is less studied than the association between genetic variation in AS3MT and arsenic metabolism. A recent genome-wide association study [Pierce et al., 2012] confirmed our previous results of strong associations between AS3MT genotype and arsenic metabolism [Schlawicke Engstrom et al., 2007; Engstrom et al., 2011] and two SNPs in the chromosome 10 region near AS3MT (rs9527 and rs11191659) were also associated with increased risk for arsenic-related skin lesions [Pierce et al., 2012]. We have previously found that AS3MT haplotypes associated with levels of premalignant DNA damage in women exposed to arsenic [Hossain et al., 2012, Li et al., 2012]. To our knowledge, no studies have evaluated rs3740400 and arsenic-related cancer risk. Rs11191439, rs1046778 and rs3740393 have recently been evaluated in relation to arsenic-related bladder cancer. Individuals with one or more copies of the C allele (coding for Thr) in rs11191439 (here in haplotype 4) had an elevated risk of bladder cancer [Beebe-Dimmer et al., 2012]. No significant effects were found for rs1046778 and rs3740393 [Lesseur et al., 2012], but many of the analyses lacked statistical power. One study found that the Thr-allele of the rs11191439 polymorphism associated with a marginally increased risk of arsenic-related skin lesions [Valenzuela et al., 2009], whereas no effect was found in another study [De Chaudhuri et al., 2008] or in the present study. There was no association between rs3740393 and cancer risk (all types of cancer combined) in a population from Taiwan [Chung et al., 2009]. Also, two polymorphisms not evaluated in this study, rs11191453 and rs7085104, did not affect the risk of skin lesions in a Mexican population [Valenzuela et al., 2009]. Polymorphisms in genes other than AS3MT may also influence BCC risk, such as SNPs in DNA repair genes NBS1 and XRCC3, which showed effect modulation for BCC [Thirumaran et al., 2006]. Other candidates include SNPs in fibrous sheath interacting protein 1 (FSIP1) and solute carrier family 39, member 2 (SLC39A2) that influence the risk of arsenic-related bladder cancer, as shown in a study scanning 10,000 SNPs [Karagas et al., 2012] or SNPs in p21 or glutathione S-transferase omega 1 (GSTO1) and GSTO2 that influence the risk of arsenic-related urothelial carcinoma [Chung et al., 2008, 2011].
In conclusion, AS3MT haplotypes associated with the arsenic metabolite profile, and possibly also other AS3MT haplotypes, modified the risk of BCC among individuals with low exposure to arsenic. These results may explain part of the observed variation in susceptibility to arsenic-related cancer.
Acknowledgments
We thank Karin Paulsson for help with the genetic analyses. The study was supported by the Swedish Council for the Working Life and Social Research, Medical faculty of Lund University, the Karolinska Institutet, European Commission project No. QLK4-CT-2001-00264 (ASHRAM) and Erik Philip-Sörensens stiftelse. The authors have no competing financial interests.
Author Contributions
Dr. Broberg and Prof. Vahter designed the study. Dr. Engström analyzed the data and drafted the manuscript. Drs Fletcher, Leonardi, Goessler, Gurzau, Koppova, Rudnai, and Kumar have been involved in sample collection, clinical data acquisition and exposure assessment. All authors have revised the manuscript and approved the final manuscript.
References
- Agusa T, Iwata H, Fujihara J, Kunito T, Takeshita H, Minh TB, Trang PT, Viet PH, Tanabe S. Genetic polymorphisms in AS3MT and arsenic metabolism in residents of the Red River Delta, Vietnam. Toxicol Appl Pharmacol. 2009;236:131–141. doi: 10.1016/j.taap.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beebe-Dimmer JL, Iyer PT, Nriagu JO, Keele GR, Mehta S, Meliker JR, Lange EM, Schwartz AG, Zuhlke KA, Schottenfeld D, Cooney KA. Genetic variation in glutathione S-transferase omega-1, arsenic methyltransferase and methylene-tetrahydrofolate reductase, arsenic exposure and bladder cancer: A case–control study. Environ Health. 2012;11:43. doi: 10.1186/1476-069X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CJ, Hsueh YM, Bai CH, Huang YK, Huang YL, Yang MH, Chen CJ. Polymorphisms in arsenic metabolism genes, urinary arsenic methylation profile and cancer. Cancer Causes Control. 2009;20:1653–1661. doi: 10.1007/s10552-009-9413-0. [DOI] [PubMed] [Google Scholar]
- Chung CJ, Huang CJ, Pu YS, Su CT, Huang YK, Chen YT, Hsueh YM. Polymorphisms in cell cycle regulatory genes, urinary arsenic profile and urothelial carcinoma. Toxicol Appl Pharmacol. 2008;232:203–209. doi: 10.1016/j.taap.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Chung CJ, Pu YS, Su CT, Huang CY, Hsueh YM. Gene polymorphisms of glutathione S-transferase omega 1 and 2, urinary arsenic methylation profile and urothelial carcinoma. Sci Total Environ. 2011;409:465–470. doi: 10.1016/j.scitotenv.2010.10.053. [DOI] [PubMed] [Google Scholar]
- De Chaudhuri S, Ghosh P, Sarma N, Majumdar P, Sau TJ, Basu S, Roychoudhury S, Ray K, Giri AK. Genetic variants associated with arsenic susceptibility: Study of purine nucleoside phosphorylase, arsenic (+3) methyltransferase, and glutathione S-transferase omega genes. Environ Health Perspect. 2008;116:501–505. doi: 10.1289/ehp.10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Waters SB, Walton FS, LeCluyse EL, Thomas DJ, Styblo M. Interindividual variation in the metabolism of arsenic in cultured primary human hepatocytes. Toxicol Appl Pharmacol. 2004;201:166–177. doi: 10.1016/j.taap.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Engstrom K, Vahter M, Mlakar SJ, Concha G, Nermell B, Raqib R, Cardozo A, Broberg K. Polymorphisms in arsenic (+III oxidation state) methyltransferase (AS3MT) predict gene expression of AS3MT as well as arsenic metabolism. Environ Health Perspect. 2011;119:182–188. doi: 10.1289/ehp.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom KS, Hossain MB, Lauss M, Ahmed S, Raqib R, Vahter M, Broberg K. Efficient arsenic metabolism—The AS3MT haplotype is associated with DNA methylation and expression of multiple genes around AS3MT. PLoS One. 2013;8:e53732. doi: 10.1371/journal.pone.0053732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RM, Nermell B, Kippler M, Grander M, Li L, Ekstrom EC, Rahman A, Lonnerdal B, Hoque AM, Vahter M. Arsenic methylation efficiency increases during the first trimester of pregnancy independent of folate status. Reprod Toxicol. 2011;31:210–218. doi: 10.1016/j.reprotox.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Xamena N, Surralles J, Sekaran C, Tokunaga H, Quinteros D, Creus A, Marcos R. Role of the Met(287)Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutat Res. 2008;637:80–92. doi: 10.1016/j.mrfmmm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Hossain MB, Vahter M, Concha G, Broberg K. Environmental arsenic exposure and DNA methylation of the tumor suppressor gene p16 and the DNA repair gene MLH1: Effect of arsenic metabolism and genotype. Metallomics. 2012;4:1167–1175. doi: 10.1039/c2mt20120h. [DOI] [PubMed] [Google Scholar]
- Hough RL, Fletcher T, Leonardi GS, Goessler W, Gnagnarella P, Clemens F, Gurzau E, Koppova K, Rudnai P, Kumar R. Lifetime exposure to arsenic in residential drinking water in Central Europe. Int Arch Occup Environ Health. 2010;83:471–481. doi: 10.1007/s00420-010-0519-1. , et al. [DOI] [PubMed] [Google Scholar]
- Hughes MF, Beck BD, Chen Y, Lewis AS, Thomas DJ. Arsenic exposure and toxicology: A historical perspective. Toxicol Sci. 2011;123:305–332. doi: 10.1093/toxsci/kfr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. A review of human carcinogens: Arsenic, metals, fibres, and dusts. Lyon, France: IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, International Agency for Research on Cancer, World Health Organization; 2012. [PMC free article] [PubMed] [Google Scholar]
- Karagas MR, Stukel TA, Morris JS, Tosteson TD, Weiss JE, Spencer SK, Greenberg ER. Skin cancer risk in relation to toenail arsenic concentrations in a US population-based case–control study. Am J Epidemiol. 2001;153:559–565. doi: 10.1093/aje/153.6.559. [DOI] [PubMed] [Google Scholar]
- Karagas MR, Andrew AS, Nelson HH, Li Z, Punshon T, Schned A, Marsit CJ, Morris JS, Moore JH, Tyler AL. SLC39A2 and FSIP1 polymorphisms as potential modifiers of arsenic-related bladder cancer. Hum Genet. 2012;131:453–461. doi: 10.1007/s00439-011-1090-x. , et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi G, Vahter M, Clemens F, Goessler W, Gurzau E, Hemminki K, Hough R, Koppova K, Kumar R, Rudnai P, Surdu S, Fletcher T. Inorganic arsenic and basal cell carcinoma in areas of Hungary, Romania, and Slovakia: a case–control study. Environ Health Perspect. 2012;120:721–726. doi: 10.1289/ehp.1103534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesseur C, Gilbert-Diamond D, Andrew ASRME, Li Z, Kelsey KT, Marsit CJ, Karagas MR. A case–control study of polymorphisms in xenobiotic and arsenic metabolism genes and arsenic-related bladder cancer in New Hampshire. Toxicol Lett. 2012;210:100–106. doi: 10.1016/j.toxlet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Engström K, Vahter M, Broberg K. Arsenic exposure through drinking water is associated with longer telomeres in peripheral blood. Chem Res Toxicol. 2012;25:2333–2339. doi: 10.1021/tx300222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg AL, Goessler W, Gurzau E, Koppova K, Rudnai P, Kumar R, Fletcher T, Leonardi G, Slotova K, Gheorghiu E, Vahter M. Arsenic exposure in Hungary, Romania and Slovakia. J Environ Monit. 2006;8:203–208. doi: 10.1039/b513206a. [DOI] [PubMed] [Google Scholar]
- Lindberg AL, Kumar R, Goessler W, Thirumaran R, Gurzau E, Koppova K, Rudnai P, Leonardi G, Fletcher T, Vahter M. Metabolism of low-dose inorganic arsenic in a central European population: influence of sex and genetic polymorphisms. Environ Health Perspect. 2007;115:1081–1086. doi: 10.1289/ehp.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg AL, Rahman M, Persson LA, Vahter M. The risk of arsenic induced skin lesions in Bangladeshi men and women is affected by arsenic metabolism and the age at first exposure. Toxicol Appl Pharmacol. 2008a;230:9–16. doi: 10.1016/j.taap.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Lindberg AL, Rahman M, Persson LA, Vahter M. The risk of arsenic induced skin lesions in Bangladeshi men and women is affected by arsenic metabolism and the age at first exposure. Toxicol Appl Pharmacol. 2008b;230:9–16. doi: 10.1016/j.taap.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Meza MM, Yu L, Rodriguez YY, Guild M, Thompson D, Gandolfi AJ, Klimecki WT. Developmentally restricted genetic determinants of human arsenic metabolism: Association between urinary methylated arsenic and CYT19 polymorphisms in children. Environ Health Perspect. 2005;113:775–781. doi: 10.1289/ehp.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, Paul-Brutus R, Rahaman R, Rakibuz-Zaman M, Parvez F, Ahmed A, Quasem I, Hore SK, Alam S, Islam T, Slavkovich V, Gamble MV, Yunus M, Rahman M, Baron JA, Graziano JH, Ahsan H. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in bangladesh. PLoS Genet. 2012;8:e1002522. doi: 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlawicke Engstrom K, Broberg K, Concha G, Nermell B, Warholm M, Vahter M. Genetic polymorphisms influencing arsenic metabolism: Evidence from Argentina. Environ Health Perspect. 2007;115:599–605. doi: 10.1289/ehp.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi D, Himeno S. Role of arsenic (+3 oxidation state) methyltransferase in arsenic metabolism and toxicity. Biol Pharm Bull. 2012;35:1870–1875. doi: 10.1248/bpb.b212015. [DOI] [PubMed] [Google Scholar]
- Surdu S, Fitzgerald EF, Bloom MS, Boscoe FP, Carpenter DO, Haase RF, Gurzau E, Rudnai P, Koppova K, Fevotte J, Vahter M, Leonardi G, Goessler W, Kumar R, Fletcher T. Occupational exposure to arsenic and risk of nonmelanoma skin cancer in a multinational European study. Int J Cancer. 2013;133:2182–2191. doi: 10.1002/ijc.28216. [DOI] [PubMed] [Google Scholar]
- Thirumaran RK, Bermejo JL, Rudnai P, Gurzau E, Koppova K, Goessler W, Vahter M, Leonardi GS, Clemens F, Fletcher T, Hemminki K, Kumar R. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006;27:1676–1681. doi: 10.1093/carcin/bgi381. [DOI] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahter M. Mechanisms of arsenic biotransformation. Toxicology. 2002;181–182:211–217. doi: 10.1016/s0300-483x(02)00285-8. [DOI] [PubMed] [Google Scholar]
- Valenzuela OL, Drobna Z, Hernandez-Castellanos E, Sanchez-Pena LC, Garcia-Vargas GG, Borja-Aburto VH, Styblo M, Del Razo LM. Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol Appl Pharmacol. 2009;239:200–207. doi: 10.1016/j.taap.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


