Abstract
Cancer stem cells (CSCs) are rare, tumour-initiating cells that exhibit stem cell properties: capacity of self-renewal, pluripotency, highly tumorigenic potential, and resistance to therapy. Cancer stem cells have been characterised and isolated from many cancers, including breast cancer. Developmental pathways, such as the Wnt/β-catenin, Notch/γ-secretase/Jagged, Shh (sonic hedgehog), and BMP signalling pathways, which direct proliferation and differentiation of normal stem cells, have emerged as major signalling pathways that contribute to the self-renewal of stem and/or progenitor cells in a variety of organs and cancers. Deregulation of these signalling pathways is frequently linked to an epithelial-mesenchymal transition (EMT), and breast CSCs often possess properties of cells that have undergone the EMT process. Signalling networks mediated by microRNAs and EMT-inducing transcription factors tie the EMT process to regulatory networks that maintain “stemness”. Recent studies have elucidated epigenetic mechanisms that control pluripotency and stemness, which allows an assessment on how embryonic and normal tissue stem cells are deregulated during cancerogenesis to give rise to CSCs. Epigenetic-based mechanisms are reversible, and the possibility of “resetting” the abnormal cancer epigenome by applying pharmacological compounds targeting epigenetic enzymes is a promising new therapeutic strategy. Chemoresistance of CSCs is frequently driven by various mechanisms, including aberrant expression/activity of ABC transporters, aldehyde dehydrogenase and anti-oncogenic proteins (i.e. BCL2, B-cell lymphoma-2), enhanced DNA damage response, activation of pro-survival signalling pathways, and epigenetic deregulations. Despite controversy surrounding the CSC hypothesis, there is substantial evidence for their role in cancer, and a number of drugs intended to specifically target CSCs have entered clinical trials.
Keywords: cancer stem cells, breast cancer, signalling pathways
Introduction
The “cancer stem cell theory” has attracted a great deal of attention following the identification of a rare population of leukaemia-initiating cells exhibiting stem-like features [1]. It has been further strengthened by the isolation and characterisation of cancer stem cells (CSCs, named also tumour-initiating cells or stem-like cells) from solid tumours such as breast [2], glioblastoma [3], colon [4], lung [5], ovary [6], and thyroid [7]. Besides the properties shared with adult, tissue-specific stem cells, such as self-renewal and the ability to differentiate into other cells, a subpopulation of candidate CSCs must meet certain criteria: 1) the strong ability to engraft; 2) the ability to recapitulate the tumour of origin both morphologically and immunophenotypically in xenografts; and 3) the ability to be serially transplanted.
To distinguish CSCs from other tumour cells and normal stem cells, a search for specific surface and intracellular biomarker phenotypes has been ongoing in recent years. The most common method used to identify CSCs is fluorescence-activated cell sorting (FACS). Breast cancer stem cells (BCSC) have been identified as CD44+, CD24-/low, and ESA+ (epithelial specific antigen) and lacking expression of specific lineage markers (ESA+CD44+CD24−/low Lin−) [2]. Amongst primary breast tumours, there is an association between the metastatic status and a high prevalence of markers such as CD44+/CD24-/low, ESA+, CD133+, CXCR4+, and PROCR+ in tumour cells.
Wnt (wingless), Shh (sonic hedgehog), Notch, and BMP/TGF-β (bone morphogenetic proteins/transforming growth factor β) signalling pathways contribute to the self-renewal of stem and/or progenitor cells in a variety of organs. When deregulated, these pathways can contribute to oncogenesis.
The Wnt/β-catenin signalling pathway modulates a balance between stemness and differentiation in several adult stem cell niches, such as the hair follicles in the skin, the mammary gland, and the intestinal crypt. Constitutive Wnt signalling activation resulting from mutations in genes encoding its downstream components underlies tumorigenesis in these tissues. Loss of adenomatous polyposis coli (APC) tumour suppressor function or oncogenic β-catenin mutations occur in the majority of sporadic colorectal cancers and melanoma [8]. Monoclonal antibodies against the Wnt cascade and several inhibitors of Wnt signalling compounds are under investigation in several cancers [9].
The Hedgehog (Hh) family of secreted proteins includes Sonic (Shh), Indian (Ihh), and Desert (Dhh). The Hh proteins exert their activity by binding to a transmembrane protein, Patched (PTCH), which constitutively represses Hh pathway activity through its interaction with a transmembrane protein Smoothened (SMO). Several groups have exploited cyclopamine (SMO signaling inhibitor), to inhibit the Hh cascade, and thereby inhibiting the growth, invasion, and metastasis of breast, prostatic, pancreatic, and brain malignancies both in vitro and in vivo [10, 11].
Notch signalling is initiated through the interaction of a receptor on the signal-receiving cell and a ligand on the neighbouring cell. Upon binding to Delta-Serrate LAG2 (DSL) ligand, the Notch receptor is activated by an ordered proteolytic cleavage. Release of the Notch intracellular domain from the cell membrane mediated by γ-secretase results in its translocation to the nucleus where it interacts with DNA-binding proteins of the CSL family (CBF1 or RBPJ in humans) and induces target gene transcription. The best-characterised Notch target genes are the basic helix-loop-helix (bHLH) transcriptional repressors of the Hairy enhancer of split (Hes) and Hairy-related (Hrt) protein families [12]. Inhibition of Notch1 with specific antibodies significantly reduced the CD44+CD24-/low subpopulation (BCSC) and diminished the incidence of brain metastases from breast cancer cells [13].
Bone morphogenetic proteins (BMPs), TGF-β and GDFs (growth and differentiation factors), belong to the TGF-β superfamily and are pluripotent factors involved in the regulation of embryonic development and postnatal homeostasis of various organs and tissues by controlling cellular differentiation, proliferation, and apoptosis [14]. TGF-β and BMP/GDF form homo- and hetero-dimers that interact with heterodimers of type I and type II receptor to produce signalling complexes, leading to the activation of SMAD transcription factors [15]. Stimulation of an epithelial-to-mesenchymal transition (EMT) by TGF-β is accompanied by the generation of breast CSCs [16]. Many of the genes actively transcribed by CD44+/CD24-/low BCSCs are classical TGF-β targets, associated with a mesenchymal, migratory phenotype. In a breast cancer model of MDA-MB-231 cells injected to athymic mice, BMP7 or BMP2/7 heterodimer antagonised the pro-tumorigenic and pro-metastatic actions of TGF-β, and reduced TGF-β-driven Smad signalling and cancer cell invasiveness. The maintenance of a subpopulation of ALDHhi/CD44hi/CD24–/low BCSCs and formation of bone metastases by MDA-MB-231 cells growing in nude mice was strongly reduced by heterodimeric BMP2/7 [17].
In addition, pro-survival and anti-apoptotic pathways are frequently overactivated in cancer stem cells. STAT (signal transducers and activators of transcription) proteins are activated in response to extracellular ligands that bind to appropriate receptors and activate receptor-associated tyrosine kinases (i.e. as Janus kinase – JAK) and non-receptor tyrosine kinases (i.e. as Src kinase). Phosphorylated STAT proteins form dimers and translocate to the nucleus where they activate target genes [18]. Increased levels of STAT3 were found in CSCs comparing to bulk cells in brain, breast, colon, and liver cancers. Blocking STAT3 function in BCSC correlated with lower proliferation and viability of stem-like cells, suggesting the involvement of this factor in the maintenance of CSCs [19].
Nuclear factor-κB (NF-κB) transcription factors are constitutively active in many solid tumours, including breast, colon, and liver cancers [20]. Nuclear factor-κB activation is regulated by the IκB kinase (IKK) complex composed of IKKα and IKKβ catalytic subunits. IKKα activity is required for self-renewal of ErbB2/Her2-transformed mammary tumour-initiating cells [21]. IKKα phosphorylates p27/Kip1, the cyclin-dependent kinase inhibitor, and stimulates its nuclear export or exclusion. Reduced p27 expression restored mammary tumorigenesis in IKKα knockout mice and self-renewal of mammary tumour-initiating cells.
Mechanisms that regulate self-renewal of breast cancer stem cells
The best characterised signalling pathways controlling self-renewal and differentiation in normal stem cells, such as Wnt/β-catenin, Notch, Hedgehog, and TGF-β/BMP pathways, are frequently deregulated in breast cancer cells, which leads to acquisition of the stem-cell phenotype [22, 23] (Fig. 1). Furthermore, networks of co-ordinately working proto-oncogenes and tumour suppressors have evolved to control self-renewal of stem cells throughout their life. For example, the Polycomb group (PcG) protein, Bmi-1, a proto-oncogene consistently required for the self-renewal of diverse adult stem cells, is also essential for the proliferation of cancer stem cells in the same tissues [24–26]. Moreover, EMT also may impart a self-renewal capability to cancer cells [16]. This process enables reprogramming of polarised epithelial cells towards a mesenchymal motile phenotype, and a growing body of evidence links EMT to the acquisition of stem cell properties by breast cancer cells [27, 28]. While overexpression of OCT3/4, SOX2, KLF4, and c-MYC genes in somatic cells leads to dedifferentiation into induced pluripotent stem cells (iPSCs) [29, 30], the activation of the molecular targets of these pluripotency-associated genes is frequently observed in poorly differentiated breast tumours and other cancers [31–33]. Of interest, accumulating evidence indicates that the expression of Oct3/4, Nanog, and Sox2 transcription factors have a strong correlation with CSCs; knockdown of these genes decreased tumour sphere formation and inhibited tumour formation in xenograft tumour models [34–38].
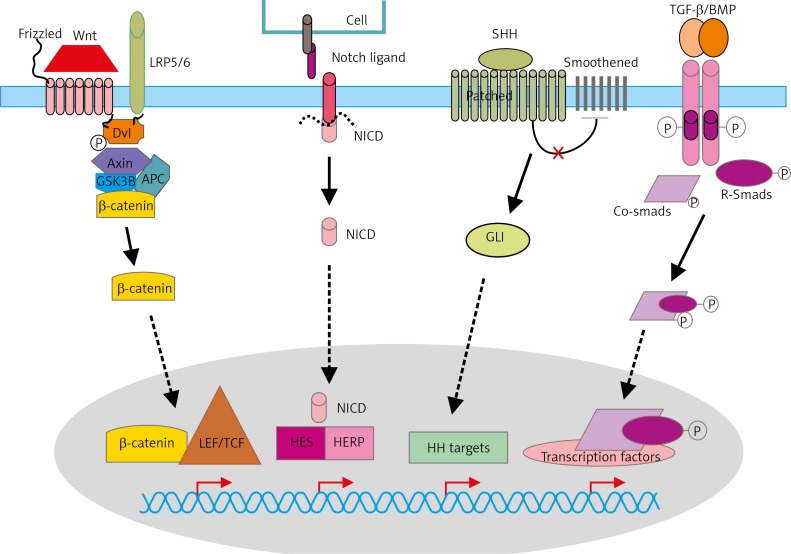
Fig. 1.
Schematic representation of signalling pathways that control maintenance of BCSCs
Wnt binds its co-receptors: Frizzled (Fz) and LRP5/LRP6. Various molecules that interact with the receptors and GSK-3 and CK1 (casein kinase 1). In the presence of Wnt, β-catenin is stabilised and induces gene expression by complexing with various transcription factors such as TCF/LET. The Delta-like and the Jagged proteins produced by signal-sending cells serve as ligands for Notch receptors. Upon ligand binding, the receptor fragment is cleaved by TACE (TNF-α ADAM metalloprotease converting enzyme) then γ-secretase to create NICD (Notch intracellular domain), which translocates to the nucleus, forms a complex with transcription factors RBPJ and CSL (CBF1/Suppressor of Hairless/LAG-1), and activates the expression of target genes (f.e. Hes1 and Herp). Sonic Hedgehog (SHH) binds to a transmembrane protein Patched (PTCH), which constitutively represses Hh pathway activity through its interaction with a transmembrane protein Smoothened (SMO). Shh-bound PTCH activates SMO, and activated SMO releases GLI1 (Glioma-Associated Oncogene Homolog 1) from cytoplasmic sequestration, and, in turn, GLI1 translocates into the cell nucleus to regulate gene expression. Activation of TGF-β type I and type II receptors leads to activation of receptor kinases and phosphorylation of the R-Smads, forming a complex with co-Smad 4, which translocates to the nucleus
Epigenetic regulation of the expression of pluripotency markers in breast cancer stem cells
Recent studies have elucidated epigenetic mechanisms that control pluripotency and stemness, thus allowing an assessment of how embryonic and normal tissue stem cells are deregulated in cancer to give rise to CSCs. Levels of transcription factors acting in embryonic stem cells (ESCs), such as Oct3/4, Nanog, or Sox2, strongly correlate with acquisition and maintenance of CSC phenotype. The expression of these factors in cancer stem cells is regulated by epigenetic mechanisms [37, 38]. Wang et al. demonstrated that DNA methylation acts synergistically with histone modifications in regulation of NANOG, OCT3/4, and c-MYC gene expression and contribute to the metastatic potential of CSCs. Results from this work suggest that the reactivation of pluripotency circuits by aberrant epigenetic alterations is one of the key events of CSC initiation [39]. Furthermore, Rivenbark et al. have shown that active expression of the SOX2 gene in breast cancer cells is critically controlled by its promoter demethylation. Direct methylation of this region by zinc-finger (ZF)-based artificial transcription factors (ATFs) having a methyltransferase activity led to repression of the promoter activity and down-regulation of SOX2 expression [40]. Further examples of epigenetic alterations affecting the expression of pluripotency markers are presented in Table 1.
Table 1.
Epigenetic regulation of selected pathways and mechanisms in cancer stem cells
| Mechanism/Pathway | Target | Epigenetic modification | Epigenetic modulator | Reference |
|---|---|---|---|---|
| Wnt/β-catenin | APC | Promoter DNA hypermethylation | DNMTs | 41 |
| miRNA targeting | miR-135a, miR-135b | 42 | ||
| β-catenin | miRNA targeting | miR-200a | 43 | |
| miR-203 | 44 | |||
| miR-214 | 45 | |||
| miR-1826 | 46, 47 | |||
| miR-320 | 48, 49 | |||
| WNT3A | miRNA targeting | miR-15a cluster | 50 | |
| miR-16-1 | 50 | |||
| AXIN2 | H3K27me3 repressive mark | EZH2 | 51 | |
| WIF1 | Promoter DNA hypermethylation | DNMT1, DNMT3b | 52–54 | |
| SFRP1-5 | Promoter DNA hypermethylation | DNMTs | 52 | |
| DKK1 | Promoter DNA hypermethylation | DNMTs | 52 | |
| Decreased H4K16Ac and increased H3K27me3 | Polycomb group proteins | 55 | ||
| miRNA targeting | miR-371-373 cluster | 56 | ||
| DKK3 | Promoter DNA hypermethylation | DNMTs | 54, 57 | |
| DACT1 | Promoter DNA hypermethylation | DNMTs | 58 | |
| DACT3 | Bivalent H3K27me3 and H3K4me3 histone modifications | Polycomb group proteins | 59 | |
| NOTCH | NOTCH1 | miRNA targeting | miR-34a | 60 |
| NOTCH2 | miRNA targeting | miR-34a | 60 | |
| NOTCH4 | miRNA targeting | miR-34c | 61 | |
| JAGGED 1 | miRNA targeting | miR-200c | 62 | |
| miR-141 | 62 | |||
| miR-34a | 63, 64 | |||
| JAGGED 2 | Acetylated histone | Down-regulated HDACs activity | 65 | |
| miRNA targeting | miR-34a | 63, 64 | ||
| NUMB | miRNA targeting | miR-146a | 66 | |
| Hedgehog | Gli1 | miRNA targeting | miR-324-5p | 67 |
| TGF-β | TGF-β | miRNA targeting | miR-106b-25 cluster; miR-179-92 cluster | 68 |
| Pluripotency factors | Sox2 | miRNA targeting | miR-200c | 69, 70 |
| Promoter DNA hypomethylation | 39 | |||
| Oct3/4 | Promoter DNA hypomethylation | 39 | ||
| H3K4me3 active mark | Trithorax group proteins | 71 | ||
| Klf4 | miRNA targeting | miR-200c | 69, 70 | |
| miR-7 | 72 | |||
| Promoter DNA hypomethylation | 39 | |||
| Nanog | H3K27me3 repressive mark | EZH2 | 73 | |
| Promoter DNA hypomethylation | 39 | |||
| H3K4me3 active mark | Trithorax group proteins | 71 | ||
| EMT | CDH1 (E-cadherin) | Promoter DNA hypermethylation | DNMTs | 74 |
| H3K27me3 repressive mark | EZH2/PRC2 | 75, 76 | ||
| Histone H3 and H4 deacetylation | HDAC1/Snai1; HDAC2/Snai1 | 77 | ||
| miRNA targeting | miR-495 | 78 | ||
| Decreased H3K4me2 | LSD1/Snai1 | 79, 80 | ||
| CDH2 (N-cadherin) | H4K20me1 active mark | SET8/Twist | 81 | |
| ZEB1 | miRNA targeting | miR-200 family | 82–84 | |
| ZEB2 | miRNA targeting | miR-200 family | 82–84 | |
| miRNA | miR-34c | DNA methylation | DNMTs | 61 |
| miR-200c | DNA methylation | DNMTs | 85 | |
| miR-203 | DNA methylation | DNMTs | 44 |
Deregulation of Wnt/β-catenin pathway by altered methylation and aberrant histone modifications
Wnt/β-catenin is an important pathway involved in the regulation of stem cells as well as cancer stem cells. In breast cancer, many genes coding for components of Wnt/β-catenin signalling, including the Wnt inhibitor – WIF1 (Wnt inhibitory factor1), SFRP1-5 (Secreted Frizzled-Related Protein 1), and DKK1 (Dickkopf-related protein 1), are methylated and silenced, which leads to aberrant activation of the Wnt-pathway and acquisition of stem-cell phenotype [52–54, 57]. Among various antagonists of the Wnt pathway, DACT1 (Dapper/Frodo) has been identified as a protein interacting with Dishevelled (Dvl), a central mediator of Wnt signalling. As demonstrated by Yin et al., DACT1 expression was silenced in breast tumour through promoter methylation leading to overactivation of Wnt signalling and enhanced migratory behaviour of breast cancer cells [58]. Upregulated Wnt signalling was also observed when the APC gene promoter was found methylated. Loss of APC favours β-catenin accumulation and stimulates TCF/LEF-induced transcription [52, 86]. Further studies demonstrated aberrant histone modifications such as enrichment of EZH2-mediated H3K27me3 histone mark on Wnt genes in parathyroid tumour cells, which leads to deregulated Wnt signalling [87]. Furthermore, miRNAs have also been implicated in the regulation of different players of Wnt/β-catenin in different types of cancer modulating the acquisition of stem cell properties [43, 45, 48], as recently reviewed elsewhere [22, 23, 88].
Posttranscriptional modifications impair Notch signalling in BCSCs
The Notch pathway is associated with the regulation of cell fate at several distinct developmental stages of the mammary gland and has been implicated in cancer initiation and progression. Recent reports have shown that Notch signalling is essential for maintaining the CSC population in breast cancer cell lines [89, 90]. In a large proportion of breast carcinomas, epigenetic mechanisms that activate Notch signalling were related to the role of miR-146a, which targets NUMB, a negative regulator of Notch. Numb is known to down-regulate Notch signalling through direct interaction and the subsequent ubiquitin-mediated protein degradation. Recent reports demonstrate a role of miR-146a in mediating the induction and maintenance of breast CSCs during EMT and provide new insights into the mechanisms for breast cancer progression [65, 91, 92]. Furthermore, an epigenetic enzyme EZH2, which belongs to the Polycomb group, was demonstrated to regulate NOTCH signalling in breast carcinoma. Gonzalez et al. have identified EZH2 as a direct regulator of NOTCH1, inducing activation of Notch1 expression and signalling, leading to stem cell expansion in triple-negative breast cancer [93].
Epigenetic regulation of epithelial-to-mesenchymal transition
Epigenetic changes may influence acquisition of stem cell-like phenotype by cancer cells via methylation of genes implicated in EMT. Taube et al. identified microRNAs, which are regulated by DNA methylation and which regulate EMT-derived stemness properties [44]. The promoter of microRNA-203 (miR-203) – a known regulator of skin cell differentiation – was methylated significantly in cells that had undergone EMT due to Twist overexpression, and its down-regulation facilitated the gain of mesenchymal/stemness properties. Thus, activating miR-203 – either epigenetically or by other means – may inhibit invasion and metastasis [44]. Furthermore, miR-200 family members are also down-regulated due to epigenetic alteration in breast CSCs in comparison to non-tumorigenic cancer cells [69, 94]. Demethylation of the miR-200 promoter was strongly inhibited by another group of miRNAs, miR-22, expression of which correlates with tumour invasiveness and metastatic properties. Down-regulation of miR-200 expression expanded the stem cell compartment and promoted breast cancer progression. Therefore, miR-22 is a crucial epigenetic modifier and promoter of EMT and cancer stemness toward metastasis [69, 94].
A rapidly growing body of research demonstrates that EMT is also epigenetically regulated by chromatin remodelling, DNA methylation, and changes to histone modification levels. Yang et al. demonstrated that TWIST, a master modulator of EMT process, is physically associated with SET8, a methyltransferase specifically targeting H4K20 for monomethylation in breast cancer cells. SET8 was recruited by TWIST to the CDH2 (N-cadherin) gene promoter and its H4K20 monomethylation activity contributed to activation of the CDH2 expression. On the other hand, TWIST protein functions as a transcriptional repressor and cooperates with SET8 to repress CDH1 (E-cadherin) expression, during which SET8 acts as a co-repressor by establishing H4K20me1 mark on the CDH1 gene promoter [81]. The expression of CDH1 could be also regulated by Snai1 transcription factor. It was previously shown that Snai1 induces repressive histone modifications at the CDH1 gene promoter through recruitment of histone deacetylases (HDACs) and H3K27 methyltransferase EZH2 [75, 76, 95]. Furthermore, Lin et al. demonstrated that Snai1 directly interact with LSD1, a histone demethylase, recruiting LSD1 complex to the CDH1 and other epithelial gene promoters, resulting in down-regulation of the active H3K4me2 mark and promoter activity. Down-regulation of epithelial gene promoters correlates with acquisition of cancer stem-cell properties, so targeting the enzymatic components of the LSD1 complex with therapeutic agents may offer a new way to halt tumour progression and dissemination [79]. The protein KAP1 (KRAB-associated protein 1), an interaction partner of members of the family of KRAB (Krüppel-associated box) domain-containing zinc finger transcription factors, may play a role in regulation of EMT. Venkov et al. demonstrated formation of a ternary complex composed of the CArG box-binding factor-A (CBF-A) and the KAP-1 protein at the fibroblast transcription site-1 (FTS-1) within the promoter of FSP1 gene (also known as S100A4), which can activate other known transcriptional regulators of EMT, including Snai1 and Twist. However, the exact role of KAP1 protein in acquisition mesenchymal-properties by EMT is unknown [96–98].
Mechanisms of CSC resistance to anti-cancer therapy
The numerous mechanisms of chemoresistance have been identified in CSCs of different origin, and these mechanisms include the following: aberrant ABC transporter expression/activity, augmented aldehyde dehydrogenase (ALDH) activity, enhanced DNA damage response, activation of self-renewal signalling pathways, and epigenetic deregulations (for a review see [99]). Cancer stem cells also have a slow rate of cell turnover and therefore can escape from chemotherapeutic agents that target rapidly proliferating cells. Adenosine triphosphate-binding cassette (ABC) transporters belong to a family of transmembrane transporters, amongst which at least 15 of these genes are implicated in drug resistance. Some of them have specific and narrow substrate recognition (ABCA3 for anthracyclines) while others exhibit resistance to a wide group of chemotherapeutic drugs (ABCC1 for anthracyclines, mitoxantrone, vinca alkaloids, imatinib, epipodophyllotoxins, camptothecins, colchicine, saquinivir, and methotrexate) [100]. Many studies demonstrate that ABCB1 and ABCG2 proteins cooperate at the blood-brain and blood-tumour barriers to restrict penetration of various anti-cancer drugs. Tissue-specific stem cells and cancer stem cells express a higher level of specific ABC pumps in comparison to their differentiated progeny that protect stem cells against toxins, but none of these efflux pumps operate as a regulator of “stemness”. The feature of higher activity/expression of ABC transporters is used to isolate a side population (SP), which could be sorted using fluorescent rhodamine 123 (ABCB1 specific) or Hoechst 33342 (ABCG2 specific) dyes and contains cells with high capability for efflux anti-mitotic drugs.
The expression of ABC proteins in cancer stem cells is regulated mostly at the transcriptional level. Transcription factors that directly regulate the expression of the genes coding for ABC pumps include TP53 [101], liver X receptor/retinoid X receptor – LXR/RXR [102], and thyroid hormone receptor [103]. The expression of ABCG2 gene coding for breast cancer resistance protein is regulated by progesterone and oestrogen receptors, nuclear factor-κB (NF-κB), hypoxia-inducible factors (HIFs), nuclear factor erythroid 2-related factor 2 (Nrf2), aryl hydrocarbon receptor (AhR), peroxisome proliferator-activated receptors (PPARs), and Krüppel-like factor 5 (KLF5) [104]. Oncogenic miR451 and miR-27a up-regulate ABCB1/MDR1/p-glycoprotein expression in multidrug-resistant cancer cell lines [105, 106].
Aldehyde dehydrogenases (ALDH) are a family ofNAD(P) + -dependent enzymes which catalyse irreversible oxidation of endogenous and exogenous aldehydes generated during cellular metabolism processes. The human ALDH superfamily consists of 19 known genes grouped in 11 families and 4 subfamilies (www.aldh.org). Aldefluor assay is based on staining living cells with aldefluor substrate – BODIPY aminoacetate (BAAA), which is converted to a negatively charged compound (BAA–) and becomes highly fluorescent, which allows sorting of ALDH bright cells (ALDHbri). Aldehyde dehydrogenases 1 activity is higher in human progenitor cells (there is a lower level of ALDH activity in primitive stem cells) and CSCs [107]. Aldehyde dehydrogenases bright cells of breast cancer are highly tumorigenic in NOD-SCID mice [108]. Although ALDH has been detected in CSCs from various tumour types, it is not a universal CSCs marker and could be used for CSCs derived from tumours that do not express ALDH1 at a high level, such as breast, lung, or colon, but not of CSCs from tumours that normally express a high level of ALDH1 (liver and pancreatic cancers).
Overexpression of ALDH1 in many drug-resistant cancer cell lines, tissues derived from chemotherapy-resistant patients, and in various CSCs makes it a promising target for anticancer treatment. The best-known inhibitors of ALDH enzymes are ATRA (all trans retinoic acid) and DEAB (diethylaminobenzaldehyde). All trans retinoic acid is a differentiation agent with the ability to indirectly down-regulate ALDH expression, while DEAB is a small molecule which directly inhibits enzymatic activity of ALDH1 [109]. Treatment of breast cancer patients with ATRA in combination with tamoxifen gave a more promising outcome than ATRA alone [110]. Disulfiram (DSF commercially known as Antabuse), which is a potent inhibitor of ALDH1A1 and has cytotoxic effects on glioblastoma stem-like cells inhibiting the growth of temozolamide-resistant tumour cells and blocking self-renewal [111], has entered a phase II clinical trial for newly diagnosed glioblastoma (ClinicalTrials.gov Identifier: NCT01777919).
Tissue-specific stem cells are equipped with multiple protective mechanisms to ensure the lifetime function of tissues. For example, epidermal stem cells show greater resistance to DNA-damaging agents than other cells of the epidermis, due to higher expression of anti-apoptotic molecules, shorter p53 activation, and enhanced non-homologous end-joining (NHEJ) activity (this repair system has lower fidelity of repair than other systems). These mechanisms are shared by other tissue-resident stem cells, suggesting that stem cells have evolved highly efficient repair mechanisms [112]. Current evidence suggests that CSCs have evolved even more efficient repair mechanisms. Comparison of breast tumour biopsies showed an increase in CSCs with mammosphere-forming capacity following chemotherapy with the EGFR/HER2 inhibitors lapatinib [113] and cisplatin [114]. In addition, mammary gland CSCs harboured lower levels of reactive oxygen species (ROS) compared to the rest of the tumour cells. This was due to increased levels of genes regulating free radical scavenging systems, such as the glutathione metabolism, which may contribute to radioresistance [115]. It has been shown that subsets of CSCs in human, and murine breast tumours, contain lower ROS levels than corresponding non-tumorigenic cells. As ROS are critical mediators of ionising-radiation-induced cell killing; CSCs in these tumours exhibit less DNA damage and are preferentially spared after irradiation compared to normal counterparts. Pharmacological depletion of ROS scavengers in CSCs decreases their clonogenicity and results in radiosensitisation [115]. In general, mechanisms related to DNA repair and repair other than DNA (in particular low proliferation, low ROS levels, and activation of the DNA damage checkpoint response) could be responsible for CSC chemoresistance.
Conclusions
Despite the controversy surrounding the CSC hypothesis, arising due to inconsistencies in phenotypic and functional markers, there is growing evidence for their role in carcinogenesis and cancer progression. Cancer stem cells share many mechanisms described for tissue-specific stem cells, but due to oncogenic deregulations these cells have evolved even more efficient repair mechanisms, antioxidant and drug-intoxicating systems that may contribute to tumour recurrence and enhanced resistance to chemo- and/or radiotherapy. Wnt, BMP/TGF-β, Shh, and Notch signalling pathways contribute to the self-renewal of stem and/or progenitor cells in a variety of organs, but deregulation of these pathways can contribute to oncogenesis and maintenance of CSCs. A number of drugs affecting properties specific to CSCs and intended to specifically target these cells have entered clinical trials. Understanding intrinsic properties of CSCs resulting in their resistance to chemotherapy will help to develop more personalised approaches to treating cancer and improve clinical outcomes for cancer patients.
The authors declare no conflict of interest.
References
- 1.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. PNAS U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 5.Eramo A, Lotti F, Sette G. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Balch C, Chan MW, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todaro M, Iovino F, Eterno V, et al. Tumorigenic and metastatic activity of human thyroid cancer stem cells. Cancer Res. 2010;70:8874–85. doi: 10.1158/0008-5472.CAN-10-1994. [DOI] [PubMed] [Google Scholar]
- 8.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–8. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen L, De Sousa E, Melo F, van der Heijden M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–76. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 10.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–33. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X, Shapiro CL, Majumder S. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res. 2012;72:5048–59. doi: 10.1158/0008-5472.CAN-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 13.McGowan PM, Simedrea C, Ribot EJ, Foster PJ, Palmieri D, Steeg PS, Allan AL, Chambers AF. Notch1 inhibition alters the CD44hi/CD24lo population and reduces the formation of brain metastases from breast cancer. Mol Cancer Res. 2011;9:834–44. doi: 10.1158/1541-7786.MCR-10-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye L, Bokobza SM, Jiang WG. Bone morphogenetic proteins in development and progression of breast cancer and therapeutic potential (review) Int J Mol Med. 2009;24:591–7. doi: 10.3892/ijmm_00000269. [DOI] [PubMed] [Google Scholar]
- 15.Kaminska B, Kocyk M, Kijewska M. TGF beta signaling and its role in glioma pathogenesis. Adv Exp Med Biol. 2013;986:171–87. doi: 10.1007/978-94-007-4719-7_9. [DOI] [PubMed] [Google Scholar]
- 16.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buijs JT, van der Horst G, van den Hoogen C, et al. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene. 2012;31:2164–74. doi: 10.1038/onc.2011.400. [DOI] [PubMed] [Google Scholar]
- 18.Kaminska B, Swiatek-Machado K. Targeting signaling pathways with small molecules to treat autoimmune disorders. Expert Rev Clin Immunol. 2008;4:93–112. doi: 10.1586/1744666X.4.1.93. [DOI] [PubMed] [Google Scholar]
- 19.Marotta LL, Almendro V, Marusyk A, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24– stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 21.Cao Y, Luo JL, Karin M. IkappaB kinase alpha kinase activity is required for self-renewal of ErbB2/Her2-transformed mammary tumor-initiating cells. PNAS U S A. 2007;104:15852–7. doi: 10.1073/pnas.0706728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuna M, Amos CI. The Role of MicroRNAs in Regulating Cancer Stem Cells. In: Gholamrezanezhad A, editor. Stem Cells in Clinic and Research. 2011. pp. 185–210. InTech. [Google Scholar]
- 23.Muñoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol Oncol. 2012;6:620–36. doi: 10.1016/j.molonc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 25.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–85. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- 26.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–8. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 27.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:1–12. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leis O, Eguiara A, Lopez-Arribillaga E, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–65. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 33.Basu-Roy U, Seo E, Ramanathapuram L, Rapp TB, Perry JA, Orkin SH, Mansukhani A, Basilico C. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene. 2012;31:2270–82. doi: 10.1038/onc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang ML, Chiou SH, Wu CW. Targeting cancer stem cells: emerging role of Nanog transcription factor. Onco Targets Ther. 2013;4:1207–20. doi: 10.2147/OTT.S38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levings PP, McGarry SV, Currie TP, Nickerson DM, McClellan S, Ghivizzani SC, Steindler DA, Gibbs CP. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res. 2009;69:5648–55. doi: 10.1158/0008-5472.CAN-08-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stolzenburg S, Rots MG, Beltran AS, Rivenbark AG, Yuan X, Qian H, Strahl BD, Blancafort P. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012;40:6725–40. doi: 10.1093/nar/gks360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33:2655–2664. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo W, Li S, Peng B, Ye Y, Deng X, Yao K. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS One. 2013;8:e56324. doi: 10.1371/journal.pone.0056324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang XQ, Ng RK, Ming X, et al. Epigenetic regulation of pluripotent genes mediates stem cell features in human hepatocellular carcinoma and cancer cell lines. PLoS One. 2013;8:e72435. doi: 10.1371/journal.pone.0072435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350–60. doi: 10.4161/epi.19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SY, Kwon HJ, Choi Y, et al. Distinct patterns of promoter CpG island methylation of breast cancer subtypes are associated with stem cell phenotypes. Mod Pathol. 2012;25:185–96. doi: 10.1038/modpathol.2011.160. [DOI] [PubMed] [Google Scholar]
- 42.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 43.Xia H, Cheung WK, Sze J, et al. miR-200a regulates epithelial-mesenchymal to stem-like transition via ZEB2 and beta-catenin signaling. J Biol Chem. 2010;285:36995–7004. doi: 10.1074/jbc.M110.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taube JH, Malouf GG, Lu E, et al. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Sci Rep. 2013;3:2687–96. doi: 10.1038/srep02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia H, Ooi LL, Hui KM. MiR-214 targets β-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7:e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirata H, Hinoda Y, Ueno K, Shahryari V, Tabatabai ZL, Dahiya R. MicroRNA-1826 targets VEGFC, beta-catenin (CTNNB1) and MEK1 (MAP2K1) in human bladder cancer. Carcinogenesis. 2012;33:41–8. doi: 10.1093/carcin/bgr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirata H, Hinoda Y, Ueno K, Nakajima K, Ishii N, Dahiya R. MicroRNA-1826 directly targets beta-catenin (CTNNB1) and MEK1 (MAP2K1) in VHL-inactivated renal cancer. Carcinogenesis. 2012;33:501–8. doi: 10.1093/carcin/bgr302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh IS, Chang KC, Tsai YT, et al. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–8. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 49.Sun JY, Huang Y, Li JP, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun. 2012;420:787–92. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 50.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 51.Cheng AS, Lau SS, Chen Y, et al. EZH2-mediated concordant repression of Wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–39. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- 52.Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2008;3:59–63. doi: 10.4161/epi.3.2.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ai L, Tao Q, Zhong S, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–8. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 54.Veeck J, Wild PJ, Fuchs T, Schüffler PJ, Hartmann A, Knüchel R, Dahl E. Prognostic relevance of Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter methylation in human breast cancer. BMC Cancer. 2009;9:217–30. doi: 10.1186/1471-2407-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–8. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou AD, Diao LT, Xu H, Xiao ZD, Li JH, Zhou H, Qu LH. beta-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/beta-catenin-signaling pathway. Oncogene. 2012;31:2968–78. doi: 10.1038/onc.2011.461. [DOI] [PubMed] [Google Scholar]
- 57.Xiang T, Li L, Yin X, Zhong L, Peng W, Qiu Z, Ren G, Tao Q. Epigenetic silencing of the WNT antagonist Dickkopf 3 disrupts normal Wnt/(β-catenin signalling and apoptosis regulation in breast cancer cells. J Cell Mol Med. 2013;17:1236–46. doi: 10.1111/jcmm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin X, Xiang T, Li L, et al. DACT1, an antagonist to Wnt/β-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2013;15:R23. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang X, Tan J, Li J, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–41. doi: 10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266–78. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu F, Jiao Y, Zhu Y, et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem. 2012;287:465–73. doi: 10.1074/jbc.M111.280768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30:756–69. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–76. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, Lee KF, Yeung WS. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–44. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- 65.Ghoshal P, Nganga AJ, Moran-Giuati J, et al. Loss of the SMRT/NCoR2 corepressor correlates with JAG2 overexpression in multiple myeloma. Cancer Res. 2009;69:4380–7. doi: 10.1158/0008-5472.CAN-08-3467. [DOI] [PubMed] [Google Scholar]
- 66.Kuang W, Tan J, Duan Y. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem Biophys Res Commun. 2009;378:259–63. doi: 10.1016/j.bbrc.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 67.Ferretti E, De Smaele E, Miele E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–27. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–4. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 69.Shimono Y, Zabala M, Cho RW, et al. Down-regulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang CJ, Yang JY, Xia W, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Okuda H, Xing F, Pandey PR, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–44. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villasante A, Piazzolla D, Li H, Gomez-Lopez G, Djabali M, Serrano M. Epigenetic regulation of Nanog expression by Ezh2 in pluripotent stem cells. Cell Cycle. 2011;10:1488–98. doi: 10.4161/cc.10.9.15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koizume S, Tachibana K, Sekiya T, Hirohashi S, Shiraishi M. Heterogeneity in the modification and involvement of chromatin components of the CpG island of the silenced human CDH1 gene in cancer cells. Nucleic Acids Res. 2002;30:4770–80. doi: 10.1093/nar/gkf593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herranz N, Pasini D, Díaz VM, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–81. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao Q, Yu J, Dhanasekaran SM, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–84. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–19. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang-Verslues WW, Chang PH, Wei PC, et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–74. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 79.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, Zhang Y, Yan ZS. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109:994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31:110–23. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 84.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 85.Neves R, Scheel C, Weinhold S, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prasad CP, Mirza S, Sharma G, Prashad R, DattaGupta S, Rath G, Ralhan R. Epigenetic alterations of CDH1 and APC genes: relationship with activation of Wnt/beta-catenin pathway in invasive ductal carcinoma of breast. Life Sci. 2008;83:318–25. doi: 10.1016/j.lfs.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 87.Svedlund J, Koskinen Edblom S, Marquez VE, Åkerström G, Björklund P, Westin G. Hypermethylated in cancer 1 (HIC1), a tumour suppressor gene epigenetically deregulated in hyperparathyroid tumours by histone H3 lysine modification. J Clin Endocrinol Metab. 2012;97:E1307–15. doi: 10.1210/jc.2011-3136. [DOI] [PubMed] [Google Scholar]
- 88.Sethi A, Sholl LM. Emerging Evidence for MicroRNAs as Regulators of Cancer Stem Cells. Cancers (Basel) 2011;3:3957–71. doi: 10.3390/cancers3043957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harrison H, Farnie G, Brennan KR, Clarke RB. Breast cancer stem cells: something out of notching? Cancer Res. 2010;70:8973–6. doi: 10.1158/0008-5472.CAN-10-1559. [DOI] [PubMed] [Google Scholar]
- 90.Hoey T, Yen WC, Axelrod F, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–77. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, Li Y, Kong D, Ahmad A, Banerjee S, Sarkar FH. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010;292:141–8. doi: 10.1016/j.canlet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Lu H, Li T, Yu L, Liu G, Peng X, Zhao J. Krüppel-like factor 8 promotes tumorigenic mammary stem cell induction by targeting miR-146a. Am J Cancer Res. 2013;3:356–73. [PMC free article] [PubMed] [Google Scholar]
- 93.Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, Kidwell KM, Kleer CG. EZH2 expands breast stem cells through activation of NOTCH1 signaling. PNAS U S A. 2014;111:3098–103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song SJ, Poliseno L, Song MS, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via tet-family-dependent chromatin remodeling. Cell. 2013;154:311–24. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peinado H, Ballestar E, Esteller M, Cano A, et al. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–19. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Venkov CD, Link AJ, Jennings JL, et al. A proximal activator of transcription in epithelial-mesenchymal transition. J Clin Invest. 2007;117:482–91. doi: 10.1172/JCI29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venkov CD, Plieth D, Ni T, Karmaker A, Bian A, George AL, Jr, Neilson EG. Transcriptional networks in epithelial-mesenchymal transition. PLoS One. 2011;6:e25354. doi: 10.1371/journal.pone.0025354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Teng Y, Zeisberg M, Kalluri R. Transcriptional regulation of epithelial-mesenchymal transition. J Clin Invest. 2007;117:304–6. doi: 10.1172/JCI31200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaminska B, Kulesza DW, Ramji K. Overview of Mechanisms of Cancer Stem Cell Drug Resistance. Current Signal Transduction Therapy. 2013;8:180–92. [Google Scholar]
- 100.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 101.Chin KV, Ueda K, Pastan I, Gottesman MM. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science. 1992;255:459–62. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 102.Thymiakou E, Zannis VI, Kardassis D. Physical and functional interactions between liver X receptor/retinoid X receptor and Sp1 modulate the transcriptional induction of the human ATP binding cassette transporter A1 gene by oxysterols and retinoids. Biochemistry. 2007;46:11473–83. doi: 10.1021/bi700994m. [DOI] [PubMed] [Google Scholar]
- 103.Huuskonen J, Vishnu M, Pullinger CR, Fielding PE, Fielding CJ. Regulation of ATP-binding cassette transporter A1 transcription by thyroid hormone receptor. Biochemistry. 2004;43:1626–32. doi: 10.1021/bi0301643. [DOI] [PubMed] [Google Scholar]
- 104.Nakanishi T, Ross DD. Breast cancer resistance protein (BCRP/ABCG2): its role in multidrug resistance and regulation of its gene expression. Chin J Cancer. 2012;31:73–99. doi: 10.5732/cjc.011.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–8. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Z, Ma T, Huang C, Zhang L, Lv X, Xu T, Hu T, Li J. MiR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/β-catenin pathway in hepatocellular carcinoma cells. Cell Signal. 2013;25:2693–701. doi: 10.1016/j.cellsig.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 107.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 108.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koppaka V, Thompson DC, Chen Y, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64:520–39. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Budd GT, Adamson PC, Gupta M, et al. Phase I/II trial of all-trans retinoic acid and tamoxifen in patients with advanced breast cancer. Clin Cancer Res. 1998;4:635–42. [PubMed] [Google Scholar]
- 111.Triscott J, Lee C, Hu K, et al. Disulfiram, a drug widely used to control alcoholism, suppresses the self-renewal of glioblastoma and over-rides resistance to temozolomide. Oncotarget. 2012;3:1112–23. doi: 10.18632/oncotarget.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blanpain C, Mohrin M, Sotiropoulou PA, Passegué E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8:16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 113.Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 114.Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, Stanbridge EJ, Lee EY. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–50. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–3. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]