Abstract
The Cancer Genome Atlas (TCGA) is a public funded project that aims to catalogue and discover major cancer-causing genomic alterations to create a comprehensive “atlas” of cancer genomic profiles. So far, TCGA researchers have analysed large cohorts of over 30 human tumours through large-scale genome sequencing and integrated multi-dimensional analyses. Studies of individual cancer types, as well as comprehensive pan-cancer analyses have extended current knowledge of tumorigenesis. A major goal of the project was to provide publicly available datasets to help improve diagnostic methods, treatment standards, and finally to prevent cancer. This review discusses the current status of TCGA Research Network structure, purpose, and achievements.
Keywords: The Cancer Genome Atlas (TCGA), cancer genomics, big data analysis
New roads to conquer cancer
Cancer is considered the most complex disease that mankind has to face. More than 200 forms of cancer have been described and each type can be characterised by different molecular profiles requiring unique therapeutic strategies. Cancer involves dynamic changes in the genome [1]. The architecture of occurring genetic aberrations such as somatic mutations, copy number variations, changed gene expression profiles, and different epigenetic alterations, is unique for each type of cancer. The demand for better diagnosis, treatment, and prevention of cancer has appeared, and strongly correlates with a better understanding of genetic changes in the tumour. The latest progress in the technological development of genome-wide sequencing and bioinformatics has shed new light on the cancer genome [2–4]. In 2005, The Cancer Genome Atlas (TCGA) and in 2008 the International Cancer Genome Consortium (ICGC) were launched as the two main projects accelerating the comprehensive understanding of the genetics of cancer using innovative genome analysis technologies, helping to generate new cancer therapies, diagnostic methods, and preventive strategies [5, 6].
The National Institute of Health (NIH) launched TCGA Pilot Project to create a comprehensive “atlas” of cancer genomic profiles. The TCGA is a public funded project that aims to catalogue and discover major cancer-causing genome alterations in large cohorts of over 30 human tumours through large-scale genome sequencing and integrated multi-dimensional analyses. Providing publicly available cancer genomic datasets will allow the improvement of diagnostic methods, treatment standards, and finally cancer prevention. Phase I of the project (a 3-year pilot study) aimed to develop and test the research infrastructure based on the characterisation of chosen tumours having poor prognosis: brain, lung, and ovarian cancers. Since 2009 (phase II) analyses have expanded to additional types reaching 30 different tumour types analysed by 2014. The TCGA project engaged scientists and managers from NIH's National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI) funded by the US government, as well as cooperating with institutions across the USA and Europe. To run the project, the NCI as well as the NHGRI each invested $50 million for the 3-year pilot study. Additional funding was also provided from different sources, such as the American Recovery and Reinvestment Act (ARRA), to help stimulate the US economy in the context of biomedicine [5–7].
In this review, we provide a short description of TCGA structure and the major goals of the project. Furthermore, we intend to expound on current knowledge of platforms, analytical tools, and visualisation methods that were applied for TCGA data generation. As it would be overwhelming to discuss all the updates of the new discoveries in cancer profiling, we have focused on the updates of the main tumour types with poor overall prognosis in patients. We hope that an understanding of some of the fundamentals, recent updates of cancer genomic profiles, and new discoveries utilising open access TCGA data will afford each researcher to extend their current knowledge in this area and therefore help to find new roads for cancer treatment and prevention.
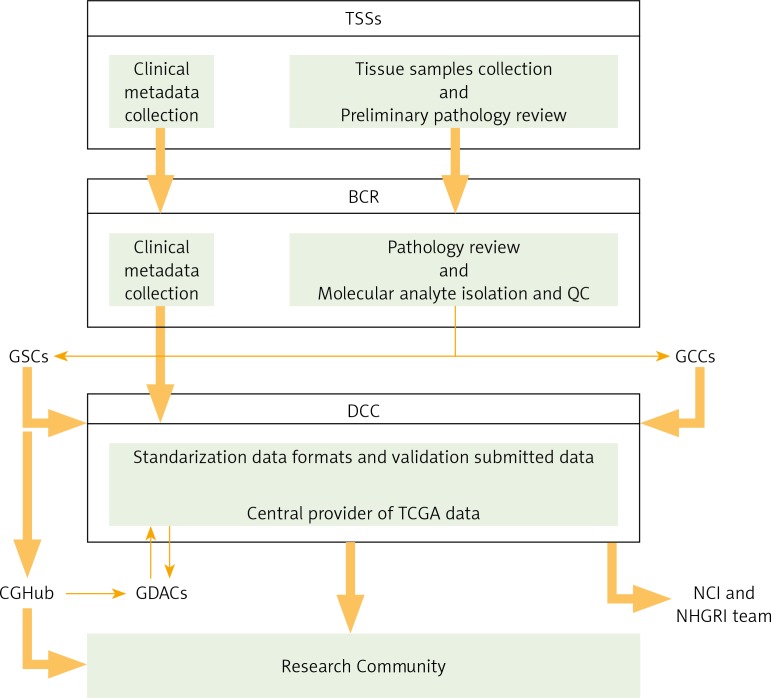
The Cancer Genome Atlas Research Network
The structure of TCGA is well organised and involves several cooperating centres responsible for collection and sample processing, followed by high-throughput sequencing and sophisticated bioinformatics data analyses (Table 1). First, different Tissue Source Sites (TSSs) collect the required biospecimens (blood, tissue) from eligible cancer patients and deliver them to the Biospecimen Core Resource (BCR). Next, the BCR catalogue, process, and verify the quality and quantity of samples, and then submit clinical data and metadata to the Data Coordinating Center (DCC) and provide molecular analytes for the Genome Characterization Centers (GCCs) and Genome Sequencing Centers (GSCs) for further genomic characterisation and high-throughput sequencing. Then, sequence-related data are deposited in the DCC. The Genome Characterisation Centers also submit trace files, sequences, and alignment mappings to NCI's Cancer Genomics Hub (CGHub) secure repository. The generated genomic data is made available to the research community and Genome Data Analysis Centers (GDACs). The GDACs provide new information-processing, analysis, and visualisation tools to the entire research community to facilitate broader use of TCGA data. Furthermore, the information generated by the TCGA Research Network is centrally managed at the DCC and entered into public free-access databases (TCGA Portal, NCBI's Trace Archive, CGHub), allowing scientists to continually access the cancer datasets and to speed advancements in cancer biology and linked technologies (Fig. 1) [8].
Table 1.
The Cancer Genome Atlas (TCGA) organisation centres. Based on [7]
| Centre Name | Centre Description | Localisation |
|---|---|---|
| Tissue Source Sites (TSSs) | Collection of the samples (blood and tissue from tumour and normal controls) and clinical metadata from patients (donors) Shipment of the annotated biospecimens to Biospecimen Core Resources (BCR) https://wiki.nci.nih.gov/display/TCGA/Tissue+Source+Site |
https://tcga-data.nci.nih.gov/datareports/codeTablesReport.htm?codeTable=tissue%20source%20site |
| Biospecimen Core Resource (BCR) | Coordination of sample delivery and data collection, cataloguing, processing, and verifying the quality and quantity Isolation and distribution of RNA and DNA from biospecimens to other institutions for genomic characterisation and high-throughput sequencing http://cancergenome.nih.gov/abouttcga/overview/howitworks/bcr http://www.nationwidechildrens.org/biospecimen-core-resource-about-us |
Research Institute at Nationwide Children's Hospital in Columbus, Ohio |
| Genome Sequencing Centers (GSCS) | High-throughput sequencing (data are available in TCGA Data Portal or at NIH's database of Genotype and Phenotype) Identification of the DNA alterations http://cancergenome.nih.gov/abouttcga/overview/howitworks/sequencingcenters |
Broad Institute Sequencing Platform in Cambridge Human Genome Sequencing Center, Baylor College of Medicine in Houston The Genome Institute at Washington University |
| Cancer Genome Characterisation Centers (GCCs) | Utilisation of novel technologies and multiple platforms Comprehensive description of the genomic changes: alterations in miRNA and gene expression, SNP, CNV, and others http://cancergenome.nih.gov/abouttcga/overview/howitworks/characterizationcenters |
Copy Number Alteration (Brigham and Women's Hospital and Harvard Medical School in Boston, The Broad Institute in Cambridge) Epigenomics (University of Southern California in Los Angeles, Johns Hopkins University in Baltimore) Gene (mRNA) Expression (University of North California at Chapel Hill) miRNA Analysis (British Columbia Cancer Agency in Vancouver) Targeted Sequencing Center (Baylor College of Medicine in Houston) Functional Proteomics (MD Anderson Cancer Center) |
| Proteome Characterization Centres (PCCs) | Identification of cancer-specific proteins http://cancergenome.nih.gov/abouttcga/overview/howitworks/proteomecharacterization |
Cancer Proteomic Center Center for Application of Advanced Clinical Proteomic Technologies for Cancer Proteo-Genomic Discovery Prioritization and Verification of Cancer Biomarkers Proteome Characterisation Centre and Vanderbilt Proteome Characterization Center |
| Data Coordinating Center (DCC) | Management of all generated data and transfer them to public databases (TCGA Data Portal and Cancer Genomics Hub) http://cancergenome.nih.gov/abouttcga/overview/howitworks/datasharingmanagement |
|
| Cancer Genomics Hub (CGHub) | Storage, catalogue, and access to lower levels of cancer genome sequences and alignments http://cancergenome.nih.gov/abouttcga/overview/howitworks/SharingAndManagingLowerLevelSeqData |
University of California Santa Cruz |
| Genome Data Analysis Centers (GDACs) | Development of novel informatics tools to facilitate with processing and integrating data analyses across the entire genome http://cancergenome.nih.gov/abouttcga/overview/howitworks/dataanalysiscenters |
Broad Institute, Cambridge, Massachusetts Institute for Systems Biology, Seattle, Washington, University of Texas MD Anderson Cancer Center, Houston, Texas Memorial Sloan-Kettering Cancer Center, New York, New York Oregon Health and Science University, Portland, Oregon University of California, Santa Cruz, California Buck Institute for Research on Aging, Novato, California University of North Carolina at Chapel Hill, Chapel Hill, North Carolina University of Texas MD Anderson Cancer Center, Houston, Texas |
Fig. 1.
The Cancer Genome Atlas (TCGA) Research Network Centres flowchart. Based on [6]
The TCGA structure involves several cooperating centres for processing the samples and managing all the obtained datasets. First, different Tissue Source Sites (TSSs) collect clinical metadata and biospecimens from eligible cancer patients. After preliminary pathology review, TSSs deliver biospecimens and metadata to the Biospecimen Core Resource (BCR), where they are approved. Next, the BCR catalogues and submits metadata to the Data Coordinating Centre (DCC), as well as processing and verifying the quality and quantity of isolated molecular analytes, which are further provided to Genome Characterisation Centres (GCCs) and Genome Sequencing Centres (GSCs) for further genomic characterisation and high-throughput sequencing. Then, sequence-related data are deposited in DCC. The GSCs also submit trace files, sequences, and alignment mappings to NCI's Cancer Genomics Hub (CGHub) secure repository. Generated genomic data submitted to the DCC and CGHub are made available to the research community and Genome Data Analysis Centres (GDACs). The GDACs provide new information-processing, analysis, and visualisation tools to the entire research community. Furthermore, the information generated by the TCGA Research Network is centrally managed at the DCC and entered into public free-access databases (TCGA Portal, NCBI's Trace Archive, CGHub), allowing scientists to continually access the cancer datasets
Platforms and data types
To provide comprehensive analysis of cancer genome profiles, TCGA applied high-throughput technologies based on microarrays (to test nucleic acids and proteins) and next-generation sequencing methods (for global analysis of nucleic acids). The research network structure includes many centres utilising different platforms to provide global information of cancer genomics. Some of the applied methods are briefly described below.
RNA sequencing (RNAseq) is a high-throughput technology for transcriptome (total RNA) profiling, deriving strand information with very high precision. RNAseq is able to rapidly identify and quantify rare and common transcripts, isoforms, novel transcripts, gene fusions, and non-coding RNAs, among a wide range of samples, including low-quality samples [9]. For transcriptome analysis TCGA uses a platform based on the Illumina system. The TCGA deposited data contains information about both nucleotide sequence and gene expression. RNA sequence alignment provides different levels of information such as RNA sequence coverage, sequence variants (e.g. fusion genes), expression of genes, exon, or junction. The NCBI dbGaP database is the official repository for the actual sequence data [10].
MicroRNA sequencing (miRNAseq) is a type of RNA-Seq, utilising material enriched in small RNAs, allowing the detection of specific sets of short, noncoding RNAs (miRNAs) that have the capacity to regulate hundreds of genes within and across diverse signalling pathways. Moreover, miRNA-sequencing defines tissue-specific miRNA expression profiles, their isoforms, connection with diseases, and the discovery of unreported miRNAs [11–15].
DNA sequencing (DNAseq) is a high-throughput method for determining the nucleotides within a DNA molecule, providing information about DNA alterations, such as insertions, deletions, polymorphism as well as copy number variation, mutation frequencies, or viral infection events. To catalogue the genomic diversity across cancer types, TCGA Genome Sequencing Centers utilise DNA sequencing systems based on Sanger Sequencing [16–18].
SNP-based platforms are used to analyse genome-wide structural variation across multiple cancer genomes. The TCGA researchers have chosen the most powerful genotyping tools. Array-based detection of single nucleotide polymorphisms (SNPs) included platforms able to define SNP, CNV, and loss of LOH across multiple samples [19, 20].
Array-based DNA methylation sequencing is a high-throughput, genome-wide analysis of DNA methylation profile providing information of epigenetic changes in the genome. Abnormal profile of DNA methylation of CpG sites is among the earliest and most frequent alterations in cancer [21, 22]. The TCGA utilises DNA methylation assay mainly based on the Illumina platform, assuring single-base-pair resolution, high accuracy, easy workflows, and low input DNA requirements. Methylation profiling technologies are based on highly multiplexed genotyping of bisulphite-converted genomic DNA. The TCGA DNA methylation data files contain information of signal intensities (raw and normalised), detection confidence, and calculated beta values for methylated (M) and unmethylated (U) probes [23].
Reverse-phase protein array (RPPA) is a highly sensitive (detecting nanograms of proteins), reproducible, high-throughput, functional and quantitative proteomic method for large-scale protein expression profiling, biomarker discovery, and cancer diagnostics. Reverse-phase protein array is an antibody-based technique allowing for the analysis of > 1000 samples with up to 500 different antibodies at a time. Protein arrays contain data of protein expression and concentration. The data archives are deposited to the TCGA DCC and include original images of protein arrays, calculated raw signals, relative concentrations of proteins, and normalised protein signals [24–28].
Each platform can potentially produce many kinds of data (data types), such as the following: gene expression, exon expression, miRNA expression, copy number variation (CNV), single nucleotide polymorphism (SNP), loss of heterozygosity (LOH), mutations, DNA methylation, and protein expression. Generated data are categorised not only by data type but also by data level. Raw, non-normalised data (Level I), processed data (Level II), and segmented/interpreted data (Level III) apply to individual samples, while summarised data (Level IV) refer to analyses across sample sets. Importantly, data of level III and IV are freely available from the publicly accessible databases, but to access lower level (Level I and II) data, specific permissions must be acquired and granted [29].
Visualisation and analysis of the genomic data
Nowadays, next-generation sequencing (NGS) and array-based profiling methods generate large amounts of diverse types of genomic data enabling researchers to study the cancer genome at an advanced level. Integrated multi-dimensional data visualisation is an essential component of cancer genomic data analysis. Therefore, demand for advanced comprehensive visualisation tools has appeared allowing the emergence of numerous useful imaging tools and databases, examples of which with a short description are provided below [30, 31].
The Cancer Imaging Archive, TCIA (http://www.cancerimagingarchive.net), is a service created by the NCI to collect and share with the public a large number of medical images of cancer (radiological imaging data), from TCGA cases, thus e.g. supporting imaging phenotype-genotype research [32].
Berkeley Morphometric Visualisation and Quantification from H&E sections (http://tcga.lbl.gov/biosig/tcgadownload.do) is a data repository of computed histology-based images of different tumour samples from TCGA cases, and is sponsored by the Lawrence Berkeley National Laboratory [33].
The Cancer Digital Slide Archive, CDSA (http://cancer.digitalslidearchive.net/), is an on-line interactive tool for viewing and annotating diagnostic and tissue slide images of different tumour types from TCGA project. The CDSA was created by Dr. David Gutman and Dr. Lee Cooper of Emory University in an effort to facilitate the broader access to TCGA data [34].
The Broad GDAC Firehose (https://confluence.broadinstitute.org/display/GDAC/Home) is an analytical infrastructure created at the Broad Institute based on the needs of TCGA project to coordinate the flow of terabyte-scale cancer datasets, providing a large amount of different quantitative algorithms such as GISTIC, MutSig, Clustering, and Correlation [35].
The MD Anderson GDAC's MBatch (http://bioinformatics.mdanderson.org/tcgabatcheffects) is a website that enables scientists to identify and quantify the batch effects accompanying TCGA data set, currently according to hierarchical clustering and enhanced PCA plots [36].
Cancer Genome Workbench, CGWB (https://cgwb.nci.nih.gov/), is an application developed by the NCI to integrate and display sample-level genomic and transcription alterations in various cancers, from data from several cancer projects, including TCGA. The major viewers in CGWB are Integrated tracks view, Heatmap view, and an alignment viewer called Bambino [37].
UCSC Cancer Genomics Browser (https://genome-cancer.soe.ucsc.edu/) is a suite of an open-access web-based tools developed and maintained by the UCSC Cancer Genomics Group to host, visualise, and analyse cancer genomics together with clinical data by utilising genomic coordinate heatmaps. The browser also provides interactive views of genomic regions with annotated biological pathways, as well as allowing for quantitative analysis within all available datasets through access to integrated statistical tools [38].
Integrative Genomics Viewer, IGV (http://www.broadinstitute.org/igv) is a freely-to-download, high-performance visualisation tool created by the Broad Institute for interactive exploration of large, heterogeneous, integrated data sets. Integrative Genomics Viewer allows easy analysis of user-prepared data or data from the IGV server, including some TCGA data. To facilitate viewing genomes, the IGV has coordinate-type data providing some genome annotations with specific labels [39, 40].
The cBioPortal for Cancer Genomics (http://cbioportal.org) is an open-access resource developed at the Memorial Sloan-Kettering Cancer Centre (MSKCC) for visualisation, analysis, and download of large-scale cancer genomics data sets. Additionally, the portal also allows for interactive exploration of custom datasets by access to OncoPrinter or MuttationMapper web tools. Currently, the portal stores data from 69 cancer genomics studies (datasets from literature and TCGA portal) including DNA copy-number data, mRNA and miRNA expression data, mutations, RPPA data, DNA methylation data, and limited clinical data related to survival. Visualisation type involves networks, matrices, and heatmaps. The cBio portal complements existing tools, such as the TCGA and ICGC data portals, the IGV, the UCSC Cancer Genomics Browser, and IntOGen [41, 42].
Regulome Explorer (http://explorer.cancerregulome.org/) is a web tool for the integrative exploration of associations between clinical and molecular features of TCGA data. Regulome enables users to search and visualise analytical data filtered according to user-specified parameters. Visualisation data types include circular and linear genomic coordinates and networks. Regulome Explorer is an effort by the Center for Systems Analysis of the Cancer Regulome (CSACR), linked to TCGA project, as well as a collaboration between the Institute for Systems Biology and The University of Texas MD Anderson Cancer Center [43].
New discoveries with The Cancer Genome Atlas data
The Cancer Genome Atlas is an unprecedented and comprehensive publicly available collection of cancer genomic data providing researchers with a great possibility to expand current knowledge of carcinogenesis. As of 2014 more than 30 tumours have been analysed and the results published in prestigious articles such as Cell or Nature. Moreover, multidimensional analyses performed on distinct platforms provide scientists with better understanding of cancer biology, leading to improved cancer classification, development of new diagnostic methods and therapeutic approaches. A brief description of novel discoveries is provided below.
Glioblastoma
Glioblastoma (World Health Organization grade IV) was the first cancer studied by TCGA in a pilot study. This program led to the development of important principles in biospecimen banking and collection, and the establishment of the highly organised infrastructure that served similar efforts in further studies. Integrative analysis of genomic DNA copy number arrays, gene expression, and DNA methylation patterns in 206 cancer samples as well as nucleotide sequence aberrations in almost half of the samples pinpointed deregulation of RB, p53, and RTK/RAS/PI3K pathways as obligatory events in virtually all glioblastoma tumours. Furthermore, the analysis of multidimensional genomic data suggests benefits from several therapeutic strategies: treatment with CDK inhibitors, PI3K, or PDK1 inhibitor or anti-RTK therapeutic cocktails, according to the presence of specific genomic alterations. Another observation with potential clinical implications is the link between the methylation status of MGMT promoter and MMR-defective hypermutator phenotype in glioblastomas treated with alkylating agents [44].
Moreover, in 2010 Verhaak et al. reported the molecular classification of glioblastoma tumours based on gene expression profiles and defined four subtypes of GBM: Proneural, Neural, Classical, and Mesenchymal. The importance of this classification lies in the specific therapeutic strategies that different subtypes require. Each class was associated with distinct DNA copy-number aberrations and somatic mutations. Alterations in EGFR, NF1, and PDGFRA/IDH1 each define the Classical, Mesenchymal, and Proneural subgroups, respectively. Survival analysis of aggressively treated patients demonstrates a clear treatment effect in the Classical and Mesenchymal subtypes and no survival advantage in the Proneural subgroup. Therefore, improved molecular understanding of GBM could ultimately result in beneficial personalised therapies [45].
Furthermore, profiling of promoter DNA methylation alterations in 272 glioblastoma tumours from TCGA database lead to identification of a glioma-CpG island methylator phenotype (G-CIMP). Noushmehr et al. identified a subgroup of GBM tumours with specific promoter DNA methylation status, which are more prevalent among lower-grade gliomas [46]. In addition, patients with G-CIMP are younger at time of diagnosis and display significantly improved survival. G-CIMP gliomas belong to the Proneural subgroup and are characterised by distinct copy-number alterations and a high frequency of IDH1 mutations. The identification of individual subsets of gliomas with specific clinical features has implications for differential therapeutic strategies for glioma patients.
In 2013, Brennan CW et al. confirmed that the survival advantage of the Proneural subgroup is associated with the G-CIMP phenotype, and the methylation status of MGMT promoter may serve as a predictive biomarker for treatment outcome only in the Classical subtype of GBM [47]. Although this work points out the limitations of TCGA data, e.g. the inability to map genetic and protein changes to the single cells or distinct cell populations within the tumour, the authors robustly highlight the importance of TCGA resource that would expand our understanding of this lethal disease.
Furthermore, cancer genomics researchers all around the world are intensively using TCGA data to develop and test hypotheses about how GBM evolves, leading to great discoveries suggesting potential drug targets in GBM and creating sophisticated approaches to select GBM patients that are most likely to respond to developed drug trials [48–52].
Together, those results emphasise the value and power of TCGA project, demonstrating how unbiased and systematic cancer genome analyses of large sample cohorts can rapidly expand our knowledge of the molecular basis of cancer.
Breast cancer
Integrated information from genomic DNA copy number arrays, DNA methylation, exome sequencing, mRNA arrays, microRNA sequencing, and RPPA was utilised to characterise molecular portraits of human breast tumours [53]. As expected, results from different platforms confirmed the existence of four main breast cancer classes. Besides identifying nearly all genes previously implicated in breast cancer, several novel, significantly mutated genes were identified, including TBX3, RUNX1, CBFB, AFF2, PIK3R1, PTPN22, PTPRD, NF1, SF3B1, and CCND3. The overall mutation rate was the lowest in the luminal A subtype and highest in the basal-like and HER2-positive subtypes. Applied genomic characterisations also indicated potential druggable targets. In luminal/ER-positive cancers, inhibitors of PI3K pathway may be beneficial due to the high frequency of PIK3CA mutations. Correspondingly, in HER2-positive tumours somatic mutations, including a high frequency of PIK3CA mutations, a lower frequency of PTEN and PIK3R1 mutations, and genomic losses of PTEN and INPP4B, represent potential therapeutic targets. Other possible targets include druggable mutations within the HER receptor family. On the other hand, the somatic mutation analysis for basal-like breast cancers has not provided a common drug targeted mutation apart from BRCA1 and BRCA2. However, comparison of basal-like breast cancers with high-grade serous ovarian tumours showed many molecular similarities, indicating a related aetiology and common therapeutic approaches, which is supported by the activity of platinum analogues and taxanes in breast basal-like and serous ovarian tumours.
Taken together, the integrated molecular analyses of breast carcinomas by TCGA Network significantly extends our knowledge base, which may result in enhanced therapeutic strategies.
Ovarian cancer
Ovarian serous cystadenocarcinoma is a major type of ovarian cancer. The high mortality of ovarian cancer patients (only 31% of patients are expected to live for five years or more) is attributed to a lack of methods for early detection and treatment [54]. Recently TCGA researchers performed a wide-range analysis of the genomic and epigenetic changes that occur in high-grade serous ovarian carcinoma (HGS-OvCa) and demonstrated several potential therapeutic targets. In their work published in 2011 in Nature, TCGA scientists analysed 489 tumour samples and determined the presence of TP53 mutation in almost all specimens (96%) and a low but significant frequency of somatic mutations in nine further genes, including BRCA1 and BRCA2 (mutated in 22% of tumours). Integrated multidimensional analyses led to the identification of four ovarian cancer transcriptional subtypes, three miRNA subtypes, four promoter methylation subtypes, and a transcriptional signature that is associated with survival outcome. However, the main goal of TCGA research is to identify new therapeutic approaches. Therefore, TCGA scientists imply opportunities for therapeutic attack in commonly dysregulated pathways: RB, RAS/PI3K, FOXM1, and NOTCH. Moreover, the research group from Johns Hopkins Medical Institution identified an amplified region in chromosome 19, containing a NACC1 gene known to contribute to chemoresistance. Analysing TCGA data, they demonstrated the correlation of amplified NACC1 with early tumour reoccurrence in ovarian cancer patients [55]. Furthermore, TCGA data have helped to shed light on the effect of BRCA1/2 mutations on ovarian cancer patients’ survival [56, 57]. Recent findings from analyses of the ovarian cancer dataset have the potential to enhance the therapeutic management of this deadly disease.
Lung cancer
Until 2012, genomic and epigenomic alterations in squamous cell lung cancers (SQCC) have not been comprehensively characterised. Therefore, TCGA network has undertaken the challenge to identify molecularly targeted agents for lung SQCC treatment based on genomic and epigenomic profiles of about 180 lung SQCCs [58]. Except for confirmation of complex genomic alterations characteristic for this cancer type and statistically recurrent mutations in previously reported signalling pathways, the effort of TCGA network has revealed thus far undiscovered loss-of-function mutations in the HLA-A class MHC I gene, which suggests a possible role for genotypic selection of patients for immunotherapy. Lung adenocarcinoma is treated with targeted kinase inhibitors; however, they do not succeed in lung SQCC therapy. The observations presented in TCGA work suggest the demand for detailed analysis of clinical tumour specimens for a panel of specific mutations, which can help to select patients for appropriately targeted therapeutic strategies.
Colon and rectal cancer
Initially, colon and rectal cancers were considered as distinct groups and examined separately. However, excluding hypermutated tumours (16% of the studied samples), colon and rectal cancers were found to have remarkably similar patterns of genomic and epigenetic alterations: DNA copy number mutations, mRNA expression profile, promoter methylation status, and changes in miRNA expression [59]. Analysis of 276 colorectal carcinoma (CRC) samples led to the identification of frequent mutations in ARID1A, SOX9, and FAM123B. Interestingly, APC and TP53 mutations were more frequent in the non-hypermutated tumours than the hypermutated ones, suggesting different development of these tumours on a genetic level. The TCGA researchers found significant differences between tumours from the right/ascending colon and all other sites. Right/ascending colon tumours were more hypermethylated, and nearly 75% of hypermutated samples came from this site. Although these discrepancies are not clear, the origins of the colon from embryonic midgut and hindgut may provide an explanation.
Moreover, frequent amplification of ERBB2 gene, a potential therapeutic target, was identified. Furthermore, integrated molecular analyses provided more insights into the pathways that are dysregulated in CRC. In 94% of analysed samples, a mutation in one or more members of the WNT signalling pathway occurred, mainly the APC gene. Therefore, WNT-signalling inhibitors as well as small-molecule β-catenin inhibitors may serve as therapeutic approaches to treating CRC [60–62]. Moreover, several proteins in the RTK-RAS and PI3K pathways, including IGF2, IGFR, ERBB3, MEK, AKT, and MTOR could be targets for inhibition.
Clear cell renal cell carcinoma
Complex molecular characterisation of clear cell renal cell carcinoma (ccRCC) revealed correlation between metabolic shift and tumour aggressiveness. Cellular metabolism in ccRCC is remodelled by downregulation genes involved in the TCA (tricarboxylic acid) cycle, decreasing AMPK, and PTEN protein, and by upregulation of the pentose phosphate pathway and glutamine transporter genes, increasing acetyl-CoA carboxylase protein, and changing promoter methylation of MIR21 and GRB10. Thus, all those changes support tumour growth and result in worse survival outcome. Renal carcinomas are known for chemotherapy-resistance that can be defined by histopathological features and gene mutations [63]. Now, researchers highlight potential therapeutic targets, including significantly mutated genes in PI3K/AKT pathway and genes coding for the components of the SWI/SNF chromatin remodelling complex (PBRM1, ARID1A, SMARCA4), which could have a great impact on other cellular pathways, to treat advanced kidney cancer [64].
Acute myeloid leukaemia
The TCGA researchers have identified new genomic alterations that underlie the development of acute myeloid leukaemia (AML). Acute myeloid leukaemia is a relatively rare disease, still not fully understood, and difficult to treat. Surprisingly, the landscape of mutated genes across all studied cases revealed that AML cancers present the lowest mutation level among other adult types of cancer. The average of mutated genes account only for 13 mutations per case, of which 5 were recurrently mutated, indicating potential targeted therapy. Furthermore, each of the analysed samples showed at least one non-synonymous substitution of nine functionally correlated genes with pathogenesis, including the following: transcription-factor fusions (18% of cases), the gene encoding nucleophosmin (NPM1) (27%), tumour-suppressor genes (16%), DNA-methylation–related genes (44%), signalling genes (59%), chromatin modifying genes (30%), myeloid transcription-factor genes (22%), cohesin-complex genes (13%), and spliceosome-complex genes (14%). These data highlight the importance of looking into individual mutations for disease classification and prognostication [65].
Endometrial carcinoma
Integrated genomic and proteomic analysis of endometrial carcinoma has contributed to the identification of four types of the endometrioid tumours. Previous classification delineated only two major groups being insufficient overall for successful treatment, and contributing to placing the endometrial carcinoma as the sixth most common malignancy among women worldwide [66]. New genomic classification dividing endometrial cancer into four groups: (1) POLE ultramutated (exhibiting high mutation rates and hotspot mutations in the POLE gene involved in DNA replication and repair), (2) microsatellite instability hypermutated (showing a high mutation rate, few copy number alterations, not exhibiting mutations in the POLE gene), (3) copy-number low (presenting mutation in CTNNB1 gene critical for maintaining endometrium), and (4) copy-number high tumours (showing molecular landscape characteristic for serous tumours), will complement existing pathology methods with new potential treatment strategies. Moreover, endometrial cancer sharing similarities with breast, ovarian, and colorectal cancers may benefit from a similar course of treatment [67].
Urothelial bladder carcinoma
Comprehensive molecular characterisation of a major form of bladder cancer has provided new insights into the molecular basis of the disease and revealed new potential therapeutic targets for relevant altered genes and pathways. Bladder cancer is the leading major cause of morbidity and mortality worldwide [68]. Current treatments for muscle-invasive bladder carcinoma are still limited to cisplatin-based combination chemotherapy, radiotherapy, or surgery, without any second-line treatment, or any defined molecularly targeted factors [69]. Recently, the whole molecular landscape of bladder carcinoma has confirmed and extended current knowledge, highlighting 32 significantly mutated genes, along with nine new genes not previously reported. Most of the mutation events were observed in genes engaged in cell cycle regulation, cell growth, and development, indicating potential drug targets in the PI3K/AKT/mTOR pathway, targets (including ERBB2) in the RTK/MAPK pathway, as well as chromatin regulatory genes, which showed the highest mutation rate comparing to other cancers. Recurring fusion of FGFR3-TACC3 associated with papillary morphology is also a promising therapeutic target. Moreover, four expression subtypes of bladder cancer were identified, with some subtypes similar to subtypes of breast, head and neck, and lung cancers, assuming the same ways of development, and similar drugs to apply [70].
Gastric adenocarcinoma
Complex statistical analyses of molecular data from 295 gastric tumours revealed new genetic subtypes of gastric adenocarcinoma. So far, classification of gastric cancers assumed the existence of two major types: intestinal or diffuse, according to Lauren classification [71]. Unfortunately, such classification is not sufficient for clinical utility and results in overall ineffective treatment. Surprisingly, utilisation of sophisticated bioplatforms in genetic, epigenetic, and protein alterations led to classification of gastric cancers into four subtypes. The first subtype, EBV-positive tumours (EBV), has been correlated with PIK3CA mutations, immense level of DNA hypermethylation, and amplification of JAK2, PD-L1, and PDCD1LG2. The second subtype, microsatellite unstable tumours (MSI), displays characteristic hypermutation phenotype, and down-regulation of MLH1 gene. The third subtype, genomically stable tumours (GS), has been associated with diffuse tumours, mutations of RHOA and CDH1, or fusions involving RHO-family GTPase-activating proteins. The last subtype of gastric adenocarcinoma, chromosomally unstable tumours (CIN), has been related with marked aneuploidy and focal amplification of receptor tyrosine kinases, as well with mutation of TP53. This novel classification of gastric cancer has opened a new road for drug discoveries, as well as better diagnosis and personalised treatment [72].
Pan-cancer project
The TCGA researchers have so far collected a broad range of genomic data of individual cancer types, yielding a better understanding of the biology and pathology of each tumour, and resulting in the development of specific treatment strategies. Furthermore, TCGA Pan-Cancer project, which aims to run new comprehensive integrated analysis of genomic data across multiple cancers, has been set up [73]. Increasing the number of tumour sample data sets in the project enhanced the statistical power and thus also the ability to detect and analyse molecular defects in cancers. Data of this project provide scientists with a lot of information concerning similarities and differences among the genomic and cellular changes in tumours, and help to cluster and develop cancer group-related therapy. Data and results of the Pan-cancer project are shared through the Synapse platform (http://sagebase.org/synapse/) [74].
In October 2013, researchers published the first set of papers related to multiple cancer-integrated analysis. One of the first cross-tumour analyses investigating the mechanisms underlying cancer initiation and progression was performed by Kandoth et al., showing the mutational landscape across 12 major cancer types already analysed by TCGA. The integrated data sets revealed 127 significantly mutated genes (SMGs) from various cellular processes involved in tumorigenesis. Moreover, common tumour-driving mutations and related mutations in BAP1, FBXW7, and TP53 were correlated with bad phenotypes across several cancer types. Furthermore, breast, head and neck, and ovarian clusters of TP53-driven cancers have been linked with a lack of other mutations in SMGs, suggesting application of basic therapy to treat this group of tumours [75]. New avenues to better understand the mechanisms of tumorigenesis also allowed Tamborero et al. to combine different complementary methods to define a reliable list of 291 high-confidence cancer driver genes among 12 cancer types [76]. Lawrence et al. complemented previous studies with the list of “true” genes responsible for the initiation and progression of cancer, by developing a novel analytical methodology (MutSigCV) eliminating the problem with false positive findings [77]. Another cross-tumour study utilising TCGA data published by Ciriello et al. indicated the landscape of oncogenic signature [78]. By inventing a new method combining specific algorithms and biological knowledge, they derived a tissue-independent hierarchical classification of thousands of tumours from 12 cancer types, identifying major classes based on large number of mutations (M class) or copy number alteration (C class). Although there are still limitations to the current data, this research provides deeper insight into the mechanisms of oncogenesis and potential class-specific combination therapy. Furthermore, Zack et al. expanded cancer studies to somatic copy number alteration (SCNA) patterns, delivering insights into mechanisms of generation and functional consequences of cancer-related SCNAs [79]. Moreover, a broad analysis of microRNA combining TCGA data and microRNA target atlas composed of publicly available Argonaute Crosslinking Immunoprecipitation (AGO-CLIP) data performed by Hamilton et al. revealed a pan-cancer co-regulated oncogenic microRNA “superfamily”[80]. Reimand et al. demonstrated identification of SNVs (single nucleotide variants) in known phosphorylated sites of specific proteins utilising the newly developed ActiveDriver method [81]. Another work by Tang et al. demonstrated a reference viral-tumour map emphasising the importance of coadaptation between host and viral gene expression and extending current knowledge of viral aetiology in several cancers [82]. Besides looking into RNA and DNA changes across cancers, Li et al. focused on proteomics as a powerful new way to understand the pathophysiology and therapy of cancer. Utilising and developing RPPA technology created The Cancer Proteome Atlas (TCPA) database [83]. A recent multiplatform analysis of thousands of tumours from different cancer types performed by Hoadley et al. revealed molecular classification into 11 major subtypes within and across tissues of origin [84]. Although they found that five subtypes were very close to their tissue-of-origin counterparts, several unconnected cancer types grouped into common subtypes. Clusters of cancers including lung, head and neck, and a subset of bladder cancers each showed common TP53 alteration, TP63 amplification, and increased expression of immune and proliferation encoding genes. Importantly, three pan-cancer subtypes were discovered among bladder cancers. This new molecular taxonomy gives independent information for predicting clinical outcomes and might also provide new insights for personalised medicine.
Future perspectives
Systematic advances in cancer genomics provided by TCGA have revealed a new comprehensive picture of the molecular biology of cancer. The application of sophisticated high-throughput technology together with well-developed bioinformatics tools has contributed to highlighting the similarities and differences in the genomic architecture of each cancer and across multiple types. The culmination of this effort has been a series of manuscripts published recently. The TCGA has provided a huge amount of publicly available data giving researchers around the world an immeasurable source of knowledge about cancer genetic and epigenetic profiles, highlighting candidate cancer biomarkers and drug targets. Moreover, translation of cancer genomics into therapeutics and diagnostics will provide a great potential to develop personalised cancer medicine. Furthermore, the next goal for scientists is to develop even better bioinformatics tools to eliminate potential noise and improve the resolution of the analysis, then look carefully into the data sets for new discoveries. In the near future, all novel findings will facilitate diagnosis, treatment, and cancer prevention. Progress in technology comes with progress in analysis, contributing to the expansion of knowledge of diseases, and which finally results in improvements in medicine. Recently researchers have gone further and are attempting to “teach” a machine – an artificially intelligent computer, called Watson – to support doctors in diagnosing patients [85, 86]. However, only time will show how fast advances will be incorporated into clinics.
The authors declare no conflict of interest.
TCGA project in the Wiznerowicz laboratory was supported by the United States National Institutes of Health contract No: HHSN261201000026I and HHSN261200800001E through SAIC-Frederick, Inc and the Greater Poland Cancer Center intramural grant No: 1/2012(43), KT was supported by the Foundation for Polish Science Welcome grant No: 2010-3/3 to MW. PC is supported by the National Science Centre grants No: 2012/06/A/NZ1/00089 and 3342/B/P01/2010/39 (MW).
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Samur MK, Yan Z, Wang X, Cao Q, Munshi NC, Li C, Shah PK. canEvolve: a web portal for integrative oncogenomics. PLoS One. 2013;8:e56228. doi: 10.1371/journal.pone.0056228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas homepage. http://cancergenome.nih.gov/abouttcga.
- 6.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 7. https://wiki.nci.nih.gov/display/TCGA/The+Cancer+Genome+Atlas.
- 8. http://cancergenome.nih.gov/abouttcga/overview.
- 9.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. https://wiki.nci.nih.gov/display/TCGA/RNASeq.
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. The Journal of Pathology. 2011;223:102–5. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandhu S, Garzon R. Potential applications of microRNAs in cancer diagnosis, prognosis, and treatment. Semin Oncol. 2011;38:781–7. doi: 10.1053/j.seminoncol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Gunaratne PH, Coarfa C, Soibam B, Tandon A. miRNA data analysis: next-gen sequencing. Methods Mol Biol. 2012;822:273–88. doi: 10.1007/978-1-61779-427-8_19. [DOI] [PubMed] [Google Scholar]
- 15. https://wiki.nci.nih.gov/display/TCGA/miRNASeq#miRNASeq-Definition.
- 16.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–8. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 17.Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628–37. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 18.Shendure J, Ji H. Next generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 19.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 20. http://www.broadinstitute.org/collaboration/gcc/methods/technology.
- 21.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 22. http://res.illumina.com/documents/products/datasheets/datasheet_dna_methylation_analysis.pdf.
- 23. https://wiki.nci.nih.gov/display/TCGA/DNA+methylation.
- 24.Stanislaus R, Carey M, Deus HF, Coombes K, Hennessy BT, Mills GB, Almeida JS. RPPAML/RIMS: a metadata format and an information management system for reverse phase protein arrays. BMC Bioinformatics. 2008;9:555. doi: 10.1186/1471-2105-9-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akbani R, Becker KF, Carragher N, et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (Reverse Phase Protein Array) society. Mol Cell Proteomics. 2014;13:1625–43. doi: 10.1074/mcp.O113.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. http://www.mdanderson.org/education-and-research/resources-for-professionals/scientific-resources/core-facilities-and-services/functional-proteomics-rppa-core/index.html.
- 27.Spurrier B, Ramalingam S, Nishizuka S. Reverse-phase protein microarrays for cell signaling analysis. Nat Protoc. 2008;3:1796–808. doi: 10.1038/nprot.2008.179. [DOI] [PubMed] [Google Scholar]
- 28.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 29. https://tcga-data.nci.nih.gov/tcga/tcgaDataType.jsp.
- 30. https://tcga-data.nci.nih.gov/tcga/tcgaAnalyticalTools.jsp.
- 31.Schroeder MP, Gonzalez-Perez A, Lopez-Bigas N. Visualizing multidimensional cancer genomics data. Genome Med. 2013;5:9. doi: 10.1186/gm413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–57. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang H, Han J, Borowsky A, Loss L, Gray JW, Spellman PT, Parvin B. Invariant delineation of nuclear architecture in glioblastoma multiforme for clinical and molecular association. IEEE Trans Med Imaging. 2013;32:670–82. doi: 10.1109/TMI.2012.2231420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutman DA, Cobb J, Somanna D, Park Y, Wang F, Kurc T, Saltz JH, Brat DJ, Cooper LA. Cancer Digital Slide Archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J Am Med Inform Assoc. 2013;20:1091–8. doi: 10.1136/amiajnl-2012-001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. http://www.broadinstitute.org/cancer/cga/Firehose.
- 36. https://wiki.nci.nih.gov/display/TCGA/MD+Anderson+GDAC+MBatch.
- 37.Zhang J, Finney R, Edmonson M, et al. The Cancer Genome Workbench: identifying and visualizing complex genetic alterations in tumors. NCI Nature Pathway Interaction Database. 2010 [Google Scholar]
- 38.Sanborn JZ, Benz SC, Craft B, et al. The UCSC Cancer Genomics Browser: update 2011. Nucleic Acids Res. 2011;39:D951–9. doi: 10.1093/nar/gkq1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pI1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madhavan S, Gusev Y, Natarajan TG, et al. Genome-wide multi-omics profiling of colorectal cancer identifies immune determinants strongly associated with relapse. Front Genet. 2013;4:236. doi: 10.3389/fgene.2013.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh D, Chan JM, Zoppoli P, et al. Transforming Fusions of FGFR and TACC Genes in Human Glioblastoma. Science. 2012;337:1231–5. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masica D, Karchin K. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 2011;71:4550–61. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci U S A. 2010;107:2183–8. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stegh AH, Brennan C, Mahoney JA, et al. Gliomaoncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24:2194–204. doi: 10.1101/gad.1924710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LaFramboise T, Dewal N, Wilkins K, Pe'er I, Freedman ML. Allelic selection of amplicons in glioblastoma revealed by combining somatic and germline analysis. PLoS Genet. 2010;6:e1001086. doi: 10.1371/journal.pgen.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ying H, Zheng H, Scott K, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci U S A. 2010;107:6912–7. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shih IM, Nakayama K, Wu G, Nakayama N, Wang TL. Amplification of the ch19p13.2 NACC1 locus in ovarian high-grade serous carcinoma. Mod Path. 2011;24:638–45. doi: 10.1038/modpathol.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolton KL, Chenevix-Trench G, Goh C, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–90. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, Zhang W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–65. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen B, Dodge ME, Tang W, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–7. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ewan K, Pajak B, Stubbs M, et al. A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription. Cancer Res. 2010;70:5963–73. doi: 10.1158/0008-5472.CAN-10-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sack U, Walther W, Scudiero D, et al. S100A4-induced cell motility and metastasis is restricted by the Wnt/β-catenin pathway inhibitor calcimycin in colon cancer cells. Mol Biol Cell. 2011;22:3344–54. doi: 10.1091/mbc.E10-09-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol. 2003;170:2163–72. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- 64.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The Cancer Genome Atlas Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferlay J, Soerjomataram I, Ervik M, et al. Lyon, France: International Agency for Research on Cancer; 2013. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] Available from: http://globocan.iarc.fr, accessed on 13/12/2013. http://www.wcrf.org/cancer_statistics/data_specific_cancers/endometrial_cancer_statistics.php. [Google Scholar]
- 67.The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 69.von der Maase H, Sengelov L, Roberts JT, et al. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–8. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 70.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 72.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Omberg L, Ellrott K, Yuan Y, et al. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nat Genet. 2013;45:1121–6. doi: 10.1038/ng.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–33. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamilton MP, Rajapakshe K, Hartig SM, et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. doi: 10.1038/ncomms3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer. Sci Rep. 2013;3:2651. doi: 10.1038/srep02651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li J, Lu Y, Akbani R, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–7. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. http://www3.mdanderson.org/streams/FullVideoPlayer.cfm?xml=cfg%2FMoon-Shots-IBM-Watson-2013.
- 86. http://www.ibm.com/smarterplanet/us/en/ibmwatson/index.html.