Abstract
Growing knowledge concerning transcriptional control of cellular pluripotency has led to the discovery that the fate of differentiated cells can be reversed, which has resulted in the generation, by means of genetic manipulation, of induced pluripotent stem cells. Overexpression of just four pluripotency-related transcription factors, namely Oct3/4, Sox2, Klf4, and c-Myc (Yamanaka factors, OKSM), in fibroblasts appears sufficient to produce this new cell type. Currently, we know that these factors induce several changes in genetic program of differentiated cells that can be divided in two general phases: the initial one is stochastic, and the subsequent one is highly hierarchical and organised. This review briefly discusses the molecular events leading to induction of pluripotency in response to forced presence of OKSM factors in somatic cells. We also discuss other reprogramming strategies used thus far as well as the advantages and disadvantages of laboratory approaches towards pluripotency induction in different cell types.
Keywords: induced pluripotent stem cells, cellular reprogramming, OKSM
Introduction
Pluripotency is the ability of a given cell to become any cell type of our body. The majority of cells lose this “talent” during development of an organism due to cellular differentiation. Thus, in our bodies very few cells remain pluripotent (e.g. mesenchymal stem cells). This observation puzzled researchers for over a century and prompted them to ask how cells become and stay pluripotent and whether or not cellular fate can be reversed. Answering these questions led to a technology called cellular reprogramming, allowing for the generation of pluripotent cells, so-called induced pluripotent stem cells (iPS cells), from terminally differentiated cells (e.g. fibroblasts). With their characteristics, iPS cells resemble embryonic stem cells found in the early stages of mammalian development. The discovery that cellular fate can be reversed by genetic manipulations was awarded in 2012 with the Nobel Prize in Physiology or Medicine to Prof. Yamanaka. It also raised great hopes for the generation of new, personalised cellular models of several diseases, as well as for progress of regenerative medicine. Here, we briefly review the laboratory approaches and molecular mechanisms leading to induced pluripotency.
Methods for induced pluripotent stem cell generation
Reprogramming factors used for pluripotency induction
In 2006, Takahashi and Yamanaka analysed 24 transcription factors (TFs), which could efficiently transform terminally differentiated cells, like fibroblasts, into induced pluripotent stem cells, which share characteristics with embryonic stem cells (ESCs) [1]. Surprisingly, they discovered that a combination of just four TFs, namely octamer-binding transcription factor 3/4 (Oct3/4, O), sex determining region Y-box 2 (Sox2, S), Krueppel-like factor 4 (Klf4, K), and c-Myc (M) (Yamanaka factors, OKSM), is sufficient to reverse the fate of somatic cells and push them towards pluripotent, embryonic-like stage. However, high expression of Oct3/4, compared to other factors, was required for efficient generation of iPS cells [2]. Yamanaka's success triggered the use of this technology for reprogramming of other somatic cells and the search for other effective combinations of reprogramming factors (RFs), including TFs and proteins with different cellular functions (e.g. RNA-binding protein Lin28).
Shortly after Yamanaka's discovery, Thomson's team reported successful reprogramming of human fibroblasts with a different cocktail of four RFs consisting of Oct3/4 (O), Sox2 (S), Nanog (N), and Lin28 (L) (OSNL) [3]. Further studies included increasing and decreasing number of RFs. Cocktails including five (OSKMN or OSKNL) or six (OSKMNL) transgenes have been tested in order to improve the efficiency of iPS cell generation from keratinocytes and fibroblasts [4, 5], or reprogramming of more problematic cells like vascular smooth muscle cells, neonatal fibroblasts, cord blood, or peripheral blood cells [6–8]. On the other hand, it turned out that endogenous expression of at least one of the reprogramming factors in certain cell types allowed a reduction in the number of overexpressed RFs. The amniotic fluid-derived cells, characterised by high levels of c-Myc, could be transformed with Oct3/4, Sox2, and Nanog only [9]. CD133-positive stem cells from umbilical cord blood required overexpression of only Oct3/4 and Sox2 [10]. Human melanocytes were found to express endogenous Sox2, and were successfully reprogrammed with the three-factor cocktail (Oct3/4, Klf4, and c-Myc) [11]. When foetal neural stem cells, which express high levels of endogenous Sox2, were subjected to reprogramming, the success of the process required just Oct3/4 overexpression [12].
One of the major obstacles of use of Yamanaka's factors is low efficiency of reprogramming. It transpired that the p53 signalling pathway activation by c-Myc and Klf4 accounts for that [13]. Consequently, knockdown of p53 in keratinocytes allowed for iPS cell generation in the absence of either c-MYC (OSK cocktail) or c-MYC and KLF4 (OS cocktail) [14]. A combination of shRNA against p53 with OKSM appeared to be indispensable for successful reprogramming of postmitotic neurons [15].
In addition to OKSM/OSNL cocktails, a variety of other pluripotency-related factors have also been tested, e.g. undifferentiated embryonic cell transcription factor 1 (Utf1), Sal-like protein 4 (Sall4), or telomerase reverse transcriptase (hTERT). Utf1, when overexpressed together with OSKM, increased colony formation during primary fibroblast reprogramming [16]. Sall4 allowed for efficient generation of iPS cells from dermal fibroblasts, without c-Myc overexpression [17]. Finally, telomerase reverse transcriptase (hTERT) when expressed alongside OSKM and SV40 large T antigen, not only increased the number of obtained iPS cell colonies, but also allowed for reprogramming of uncommon “target cells” such as bone marrow mesenchymal stem cells and peripheral blood mononuclear cells [18, 19].
Delivery methods for induced pluripotent stem cells generation
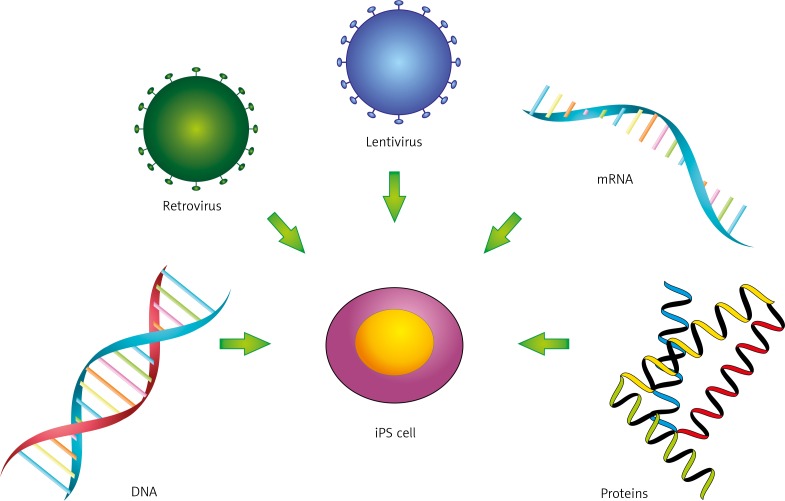
As mentioned above, production of iPS cells requires induction of a pluripotency state that usually is achieved by overexpression of RF cocktails. However, regardless of the cell type and combination of used RFs, the success rate is very low and the reprogramming efficiency on average does not exceed 1% [20]. Thus, a major aim of new strategies is to increase reprogramming efficiency. Delivery methods are typically classified as viral and non-viral [20] (Fig. 1). Both can be further divided to integrative and non-integrative methods as well as to DNA- or RNA-based approaches [20]. Protein-based methods are also used [20].
Fig. 1.
Delivery methods for generation of iPS cells
Up-to-date integrating viral vectors such as recombinant gamma retroviruses (e.g. Moloney murine leukaemia virus) and lentiviruses have been the most intensively used. Originally, each gene for RF was placed under the control of a viral promoter and introduced to somatic cells as a separate retroviral particle [1]. The advantage of this approach is that expression of reprogramming RFs is epigenetically silenced once pluripotency is achieved, lowering the risk of oncogenic transformation of reprogrammed cells [18, 21]. However, reactivation of RF expression was reported upon further differentiation [20]. Moreover, use of separate retroviral vectors leads to multiple insertional mutagenesis, increasing malignancy risk [20]. These safety concerns led to important technological improvements, concerning insert copy number and control of transgene expression. Firstly, viral vectors with single polycistronic expression cassette, encoding RFs, have been developed to decrease the risk of insertional mutagenesis. Both polycistronic retroviruses and lentiviruses have been successfully used for reprogramming [22, 23]. Another important step was replacement of viral promoters, controlling expression of RFs, to genetically engineered, inducible ones, e.g. tetracycline-inducible promoter [20]. Such promoters can be turned on and off pharmacologically (e.g. by doxycycline administration to the culture media); therefore, expression of reprogramming factors is tightly controlled. This approach was used for iPS cell generation in combination with various delivery methods [20].
To further increase the safety of viral technology, excisable transgenes and non-integrating vectors were developed [20]. For the first approach, polycistronic OSKM or OSK cassette, flanked by loxP or frt sequences recognised by Cre or Flp recombinase, respectively, were introduced to fibroblasts [24, 25]. Once reprogramming was finished, appropriate recombinase was transiently introduced to IPS cells and the expression cassette was removed. However, both recombinases leave “genomic scars”. Therefore, as an alternative to widely used Cre-LoxP system, non-integrating vectors such as adenoviruses, integration-deficient lentiviruses, and Sendai viruses were introduced [20]. However, such vectors might be difficult to eliminate from reprogrammed cells, and currently major efforts are focused on resolving this issue.
Non-viral delivery approaches for production of iPS cells can be divided to DNA-, mRNA-, or protein-based, due to the nature of the used “vehicle” [20]. In case of DNA-based delivery, both integration and integration-free methods were developed but often are quite inefficient and require several rounds of transfection [20]. Among the integrative methods, use of transposon-derived excisable vectors encoding a polycistronic cassette (e.g. OKSM) is an attractive option. There are two transposon-based methods currently used, namely piggyback and sleeping beauty, and both were effectively used to obtain mouse and human iPS cells [26, 27]. Importantly, in the case of piggyback, transposase-driven excision of OKSM cassette left the genome scar-free after reprogramming. As an alternative, several integration-free methods for IPS cell generation have been utilised based on polycistronic plasmids, episomal vectors, and DNA minicircles [20].
An important alternative to the above methods is protein- or RNA-based reprogramming. The protein-based approach was the first reported strategy that bypasses the need for exogenous DNA introduction to targeted cells, leading to generation of footprint-free iPS cells. Proteins have limited ability to cross the cellular membrane; therefore, for intracellular delivery reprogramming proteins need to be fused to peptides facilitating protein transmembrane penetration. Polyarginine domains, called cell penetrating peptide (CPP), and human immunodeficiency virus transactivator protein (HIV-TAT), called protein transduction domain (PTD), are commonly used to tag OKSM or OKSMN factors for protein-based reprogramming of somatic cells [28–30]. The protein fusions were produced either in Escherichia coli or in HEK293 cells, and next target cells (usually mouse or human fibroblasts) were exposed to purified proteins or HEK293 cell extract [28–30]. Protein reprogramming, however, required several rounds of exposition to RFs as well as the presence of valproic acid (VPA), and its efficiency varied between 0.001% and 0.01%.
The ability to transfect cells with mRNA encoding RFs offers another method to make footprint-free iPS cells. Using a cocktail of in vitro-generated, modified synthetic mRNA that encodes the reprogramming factors (OKSM), Warren et al., upon several rounds of transfection, reprogrammed human fibroblasts and keratinocytes to iPS cells within 20 days with an efficiency of 1.4%. By adding Lin28 to the Yamanaka reprogramming factor protocol, culturing cells in low oxygen, and including valproic acid in the cell culture medium the efficiency of the reprogramming process could be increased to 4.4% [31]. The RNA-based method has several advantages over the others, yet several hurdles still need to be overcome, e.g. short mRNA lifetime and foreign mRNA cytotoxicity triggered by the interferon-mediated innate immune response [31–33].
Molecular mechanisms underlying successful reprogramming
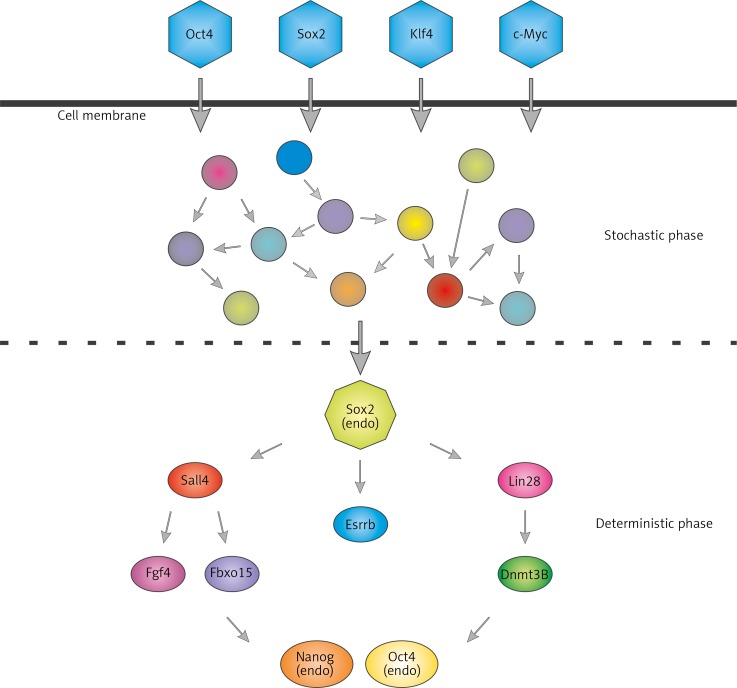
Takahashi and Yamanaka in 2006 showed that exogenous Oct3/4 and Sox2 are essential for generating mature iPS cells, while exogenous Klf-4 and c-Myc increase efficiency and accelerate the process [34]. However, despite a number of published reports on generation of iPS cells, knowledge of molecular actions of key pluripotency-inducing factors and the timeline of reprogramming is surprisingly limited. Global analysis of gene expression during reprogramming suggests that this is a stepwise process [35]. During the initial stage expression of genes that control DNA replication and cell division is enhanced, while the expression of genes responsible for cell adhesion and cell-cell contact is repressed [36]. Originally, it was hypothesised that the reprogramming process is either stochastic or hierarchical, with a strictly defined sequence of molecular changes leading to the formation of iPS cells [37, 38]. However, work by Buganim et al. [39] united both theories and showed that reprogramming consists of both, a stochastic and hierarchical phase [39] (Fig. 2). Single-cell expression analysis revealed that in fibroblasts, exogenous RFs randomly trigger a cascade of events leading to the formation of a fraction of pre-iPS cells, of which only a small proportion become fully functional stem cells. Interestingly, the authors showed that in sister cells from initial colonies the order of induced genes can significantly differ. Nevertheless, their expression needs in turn to activate predictive markers of reprogramming – oestrogen-related receptor beta (Esrrb1) and Utf1. This is a key step for transition from the random phase to the hierarchical one, which starts with activation of endogenous Sox2 [36].
Fig. 2.
Early stochastic and late hierarchical phases of reprogramming. After transfection with reprogramming factors, cells proceed to an early probabilistic, chaotic phase of gene activation, which leads to endogenous Sox2 activation. This is a key step for transition from the random phase to the hierarchical phase, characterised by the highly ordered sequence of ongoing processes (activation of major pluripotency markers)
One of OKSM factors is c-Myc, which is known to activate genes responsible for increased proliferation and cell transformation, many of which are involved in the generation of iPS cells. c-Myc, individually or in combination with other Yamanaka's factors, acts during the initial phase of reprogramming and activates pluripotency markers just a few days after derivation of the OKSM into target cells [36]. It combines with histone acetyltransferase complexes and induces global histone acetylation, allowing for exogenous Oct3/4 and Sox2 binding to DNA [40].
Additionally, Klf4 from Yamanaka's cocktail acts during the initial, stochastic stage of reprogramming. However, the level of genes regulated by Klf4 is changing not exclusively in the initial phase but also during the late phase. Therefore, it seems that Klf4 plays a dual role. It first inhibits the gene expression characteristic for a cell to be reprogrammed during the initiation of reprogramming. On the other hand, during the late phase Klf4 activates pluripotency genes [36], e.g. Klf4 induces expression of NANOG and other genes characteristic for stem cells, by repression of p53 protein [41, 42].
While the first wave of transcriptional activity, driven by c-Myc and Klf-4, occurs within the first days of reprogramming, Oct3/4 and Sox2 are connected with the later stage of the reprogramming. The TFs of Sox family are well-established regulators of cell fate decision during development. Sox2 is one of the TFs involved during all of the stages of the reprogramming process [43]. Initially, exogenous Sox2 is associated with the stochastic phase of reprogramming process, while the activation of endogenous Sox2 starts the hierarchical phase. Once endogenous Sox2 is activated, intracellular cofactors ensure that the proper set of target genes are being expressed. Sox2-dependent activation of SALL4 and LIN28 activates expression of genes associated with pluripotency such as fibroblast growth factor 4 (FGF4), F-box protein 15 (FBXO15), and DNA cytosine-5-methyltransferase 3β (DNMT3B). In turn, activation of endogenous OCT3/4 and NANOG is observed [39]. Moreover, Sox2-dependent induction of the pluripotency marker expression is connected with an active chromatin state. It was previously shown that endogenous Sox2 interacts with a chromatin modifier – Wdr5, an effector of H3K4 trimethylation [44]. As discussed earlier, Sox2 and other pluripotency factors (Oct3/4, Nanog, FGF4, Fbxo15, Lefty) work together, often binding to the same DNA sequence and thus intensifying the regulation of target genes [36, 45, 46].
Oct3/4 was identified as being TF specific to early development [47]. It is considered to be an essential component in all reprograming cocktails [48]. It was shown, however, that exogenous Oct3/4 can be omitted by either use of mesendodermal specifiers such as GATA binding protein 4 (GATA4), GATA binding protein 6 (GATA6), sex determining region Y-box 1 (SOX1), and sex determining region Y-box 1 (SOX3) [49, 50] or selecting appropriate late hierarchical phase factors such as Lin28, Sall4, and Esrrb1 [39]. Activation of gene expression by transgenic Oct3/4 occurs with a reorganisation of chromatin. It recruits not only chromatin remodelling complexes to the regulatory regions [51], but also binds to closed chromatin, thus acting as a pioneer transcription factor [40]. Moreover, activation of endogenous Oct3/4 is a crucial step to obtain fully reprogrammed, mature iPS cells [36, 52].
A cocktail of Yamanka's TFs activates the network of endogenous regulators of pluripotency, from which Nanog, a critical factor for mammalian development [53–55], is vital for achieving a pluripotency [56]. Nanog interactome connects with multiple epigenetic regulators, e.g. (SWItch/Sucrose NonFermentable (Swi/SNF), Nucleosome Remodelling Deacetylase (Nurd), and Polycomb, which regulate the expression of genes important for ESC maintenance and early development (e.g. forkhead box D3 [Foxd3], SET domain bifurcated 1 [Setdb1], or Esrrb [45, 56, 57]). Its expression is regulated at the epigenetic level. For example, Wdr5 is recruited to Nanog promoter in an Oct3/4-dependent manner to stimulate H3K4 trimethylation and activation of Nanog expression [44]. Interestingly, the Nanog promoter undergoes faster demethylation compared with OCT3/4 promoter during reprogramming [56], but the Nanog function reveals itself only at the final stage of reprogramming when other factors are already available. Only then, can Nanog complete its function [56].
While the main research focused on protein effectors downstream of pluripotency factors, it is worth stressing the important role of microRNAs for reprogramming and pluripotency [58, 59]. MicroRNAs are small non-coding RNAs, which have several cellular functions, including translational silencing. Based on sequence similarity, microRNAs are grouped into families and often are found in transcriptionally co-regulated clusters throughout the genome. Several studies have revealed that certain microRNA families are either upregulated (e.g. miR-290/295, miR-302/367, and miR-106a/363) or downregulated (let-7) in human pluripotent stem cells when compared to differentiated cells, suggesting that non-coding RNAs play a role in acquisition of pluripotency [58, 59]. Indeed, mouse embryonic stem cells derived from Dicer knockout mice that are deficient in microRNA biogenesis are not pluripotent [60]. Moreover, sole overexpression of miR-302/367 cluster in fibroblasts induced their full reprogramming to iPS cells without a need for overexpression of OKSM or OSNL [61]. Further studies have revealed that in mouse cells transcription of miR-290/295, miR-302/367, and miR-106a/363 clusters is controlled by Oct3/4, Sox 2, and Nanog, while Lin28 prevents maturation of let-7 [58]. Similar observations were made in human pluripotent stem cells. Experimental and bioinformatic searches for targets of microRNAs expressed during reprogramming as well as phenotypic analysis of pluripotent cells lacking functional microRNA biogenesis pathway revealed that these microRNAs are probably involved in control of cell cycle progression and mesenchymal-epithelial transition, a process vital for reprogramming of fibroblasts [58, 59]. Some evidence suggests also that during acquisition of pluripotency microRNAs might be needed for senesce prevention [58, 59]. Consequently, in addition to hierarchical activation of proteins upon OKSM, the microRNA expression profile is changed as part of the response to the reprogramming trigger.
Growth factors involved in reprogramming
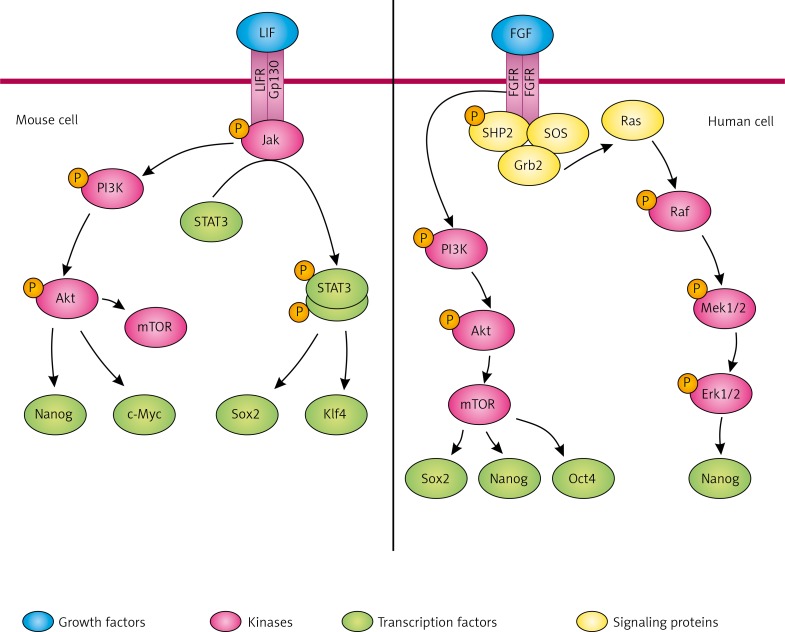
The leukaemia inhibitory factor (LIF)/Janus kinase (Jak)/signal transducer and activator of transcription 3 (Stat3) signalling pathway is the first known pluripotency-related signal transduction in naive mESC [62] (Fig. 3). The leukaemia inhibitory factor is a member of the interleukin-6 (IL-6) cytokine family and binds to the LIF receptor. The leukaemia inhibitory factor together with the membrane protein gp130 forms a heterodimer receptor which phosphorylates Jak leading to activation of the Stat3 pathway and the phosphoinositide 3-kinase (PI3K)/protein kinase Akt pathway [63]. Thereby, LIF is accountable for maintaining pluripotency in two ways: activation of Jak/Stat3 pathway increases the expression of Klf4 and Sox2, while PI3K/Akt cascade activates the expression of c-Myc and Nanog [64]. Phosphorylated Jak also activates mitogen-activated protein kinase (MAPK)/extracellular-signal-regulated kinase (Erk) pathway, which is known to be involved in mESC differentiation [65]. Jak/Stat3 signalling plays a key role in self-renewal of mouse ESC; however, this cascade does not reinforce self-renewal of human ESC [66].
Fig. 3.
Signalling networks in mouse and human iPS cells. Activation of LIF receptor triggers phosphorylation of Jak/STAT and PI3K/Akt. Jak kinase phosphorylates STAT3 leading to its homodimerisation and regulation of the target genes expression. PI3K/Akt pathway upregulates the expression of c-Myc and Nanog in mouse and human iPS cells. An important target of PI3K/Akt cascade is mTOR, which supports self-renewal of iPS by regulating Sox2, Oct4, and Nanog. FGF receptor activation leads to PI3K/Akt and MAPK/Erk phosphorylation and consequently maintains pluripotency
Fibroblast growth factors (FGFs) bind to a family of tyrosine kinase receptors, the FGF receptors (FGFRs) [67] (Fig. 3). Interaction between ligand and receptor triggers dimerisation of the receptor, autophosphorylation of the kinase domains located on the two receptor chains, and subsequently activation of the PI3K/Akt and MAPK/Erk signalling cascades [68]. In MAPK/Erk pathway active (tyrosine-phosphorylated) receptors phosphorylate the Src homology 2 domain tyrosine phosphatase 2 (SHP2). The adapter protein-growth factor receptor-bound protein 2 (Grb2) containing SH2 domain binds to the receptor phosphorylated at tyrosine and to the Son of Sevenless (SOS) protein, which belongs to the guanine nucleotide exchange factors (GEF). Created complex activates Ras protein, which has GTPase activity, and afterwards it initiates phosphorylation cascade of Raf, Mek1/2, and Erk1/2 proteins [69]. FGF/Erk signaling is thought to be responsible for promoting conversion from a naive to a primed pluripotency, and maintenance of the primed pluripotency state in cells by activating Nanog expression [64, 70].
Fibroblast growth factor also activates PI3K/Akt signalling, which is the only cascade playing an important role in promoting self-renewal of both human and mouse pluripotent stem cells [71]. This pathway is activated by bFGF, insulin, and insulin-like growth factor (IGF) in hESC and by LIF in mESC [72]. Chemical inhibition of PI3K pathway in mESC results in downregulation of Nanog, c-Myc, Klf2, Klf4, Esrrb, or T-box 3 (Tbx3) [73]. In hESC, inhibition of PI3K signalling leads to initiation of differentiation and upregulation of Mix paired-like homeobox (MixL1), Eomes, Brachyury, and Goosecoid, mesoderm markers [72]. PI3K/Akt signalling regulates the mammalian target of rapamycin (mTOR), which has been found to suppress differentiation of hESC into endoderm and mesoderm, furthermore mTOR maintains pluripotency by regulating the expression of pluripotency-related transcription factors Sox2, Oct3/4, and Nanog. PI3K pathway seems to modulate connections between different pathways in pluripotent stem cells [74].
Conclusions
The ability to induce cellular pluripotency raised a lot of hope for new disease models and for regenerative medicine. Therefore, over the last eight years enormous progress has been made in the development of reprogramming strategies. Additionally, deeper understanding of molecular aspects of pluripotency has been reached due to studies on Yamanaka's factors activity during reprogramming. Consequently, it is very likely that the discovery made just eight years ago will be used for clinical purposes less than one year from now. This in turn will accelerate further the methodological progress in the field of induced pluripotency with the most attention focused on safety issues.
The authors declare no conflict of interest.
This work was supported by the Foundation for Polish Science: Welcome program grant No: 2010-3/3 to Maciej Wiznerowicz and Homing Plus program grant No: HOMING PLUS/2012-5/6, UT MD Anderson Cancer Center intramural grants, co-financed by the European Union under the European Regional Development Fund (EL), and ERA-NET NEURON/06/2011 “AMRePACELL” (co-financed by NCBiR, JJ).
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Papapetrou EP, Tomishima MJ, Chambers SM, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A. 2009;106:12759–64. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Buecker C, Geijsen N. Different flavors of pluripotency, molecular mechanisms, and practical implications. Cell Stem Cell. 2010;7:559–64. doi: 10.1016/j.stem.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–5. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee TH, Song SH, Kim KL, et al. Functional recapitulation of smooth muscle cells via induced pluripotent stem cells from human aortic smooth muscle cells. Circ Res. 2010;106:120–8. doi: 10.1161/CIRCRESAHA.109.207902. [DOI] [PubMed] [Google Scholar]
- 7.Liao J, Wu Z, Wang Y, et al. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res. 2008;18:600–3. doi: 10.1038/cr.2008.51. [DOI] [PubMed] [Google Scholar]
- 8.Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, Goshima N, Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–66. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 9.Zhao HX, Li Y, Jin HF, et al. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation. 2010;80:123–9. doi: 10.1016/j.diff.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Giorgetti A, Montserrat N, Aasen T, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5:353–7. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122:3502–10. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–3. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 13.Marión RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisúa Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–4. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Lengner CJ, Kirak O, et al. Reprogramming of postnatal neurons into induced pluripotent stem cells by defined factors. Stem Cells. 2011;29:992–1000. doi: 10.1002/stem.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Yin X, Qin H, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–9. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells. 2009;14:683–94. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- 18.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–6. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 19.Kunisato A, Wakatsuki M, Kodama Y, Shinba H, Ishida I, Nagao K. Generation of induced pluripotent stem cells by efficient reprogramming of adult bone marrow cells. Stem Cells Dev. 2010;19:229–38. doi: 10.1089/scd.2009.0149. [DOI] [PubMed] [Google Scholar]
- 20.Bayart E, Cohen-Haguenauer O. Technological overview of iPS induction from human adult somatic cells. Curr Gene Ther. 2013;13:73–92. doi: 10.2174/1566523211313020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Pizà I, Richaud-Patin Y, Vassena R, González F, Barrero MJ, Veiga A, Raya A, Izpisúa Belmonte JC. Reprogramming of human fibroblasts to induced pluripotent stem cells under xeno-free conditions. Stem Cells. 2010;28:36–44. doi: 10.1002/stem.248. [DOI] [PubMed] [Google Scholar]
- 23.Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–9. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–9. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- 25.Voelkel C, Galla M, Maetzig T, et al. Protein transduction from retroviral Gag precursors. Proc Natl Acad Sci U S A. 2010;107:7805–10. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–5. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabundzija I, Wang J, Sebe A, et al. Sleeping Beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Res. 2013;41:1829–47. doi: 10.1093/nar/gks1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Ma Y, Gu J, Liao B, Li J, Wong J, Jin Y. Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials. 2012;33:5047–55. doi: 10.1016/j.biomaterials.2012.03.061. [DOI] [PubMed] [Google Scholar]
- 29.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–6. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Wu S, Joo JY, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–4. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angel M, Yanik MF. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS One. 2010;5:e11756. doi: 10.1371/journal.pone.0011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drews K, Tavernier G, Demeester J, Lehrach H, De Smedt SC, Rejman J, Adjaye J. The cytotoxic and immunogenic hurdles associated with non-viral mRNA-mediated reprogramming of human fibroblasts. Biomaterials. 2012;33:4059–68. doi: 10.1016/j.biomaterials.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Mah N, Wang Y, Liao MC, et al. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS One. 2011;6:e24351. doi: 10.1371/journal.pone.0024351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–32. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–6. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buganim Y, Faddah DA, Cheng AW, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–82. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 42.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–71. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Vega VB, Ng HH. Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:203–9. doi: 10.1101/sqb.2008.73.026. [DOI] [PubMed] [Google Scholar]
- 44.Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 46.Fidalgo M, Shekar PC, Ang YS, Fujiwara Y, Orkin SH, Wang J. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011;29:1705–16. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–72. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- 48.Sterneckert J, Hoing S, Scholer HR. Concise review: Oct4 and more: the reprogramming expressway. Stem Cells. 2012;30:15–21. doi: 10.1002/stem.765. [DOI] [PubMed] [Google Scholar]
- 49.Montserrat N, Nivet E, Sancho-Martinez I, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–50. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 50.Shu J, Wu C, Wu Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–75. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singhal N, Graumann J, Wu G, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–55. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 52.Samavarchi-Tehrani P, Golipour A, David L, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Levasseur DN, Orkin SH. Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2008;105:6326–31. doi: 10.1073/pnas.0802288105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullin NP, Yates A, Rowe AJ, Nijmeijer B, Colby D, Barlow PN, Walkinshaw MD, Chambers I. The pluripotency rheostat Nanog functions as a dimer. Biochem J. 2008;411:227–31. doi: 10.1042/BJ20080134. [DOI] [PubMed] [Google Scholar]
- 55.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–13. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 56.Silva J, Nichols J, Theunissen TW, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–37. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 58.Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol. 2012;14:1114–21. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anokye-Danso F, Snitow M, Morrisey EE. How microRNAs facilitate reprogramming to pluripotency. J Cell Sci. 2012;125:4179–87. doi: 10.1242/jcs.095968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–9. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–22. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 64.Hassani SN, Totonchi M, Gourabi H, Schöler HR, Baharvand H. Signaling roadmap modulating naive and primed pluripotency. Stem Cells Dev. 2014;23:193–208. doi: 10.1089/scd.2013.0368. [DOI] [PubMed] [Google Scholar]
- 65.Burdon T, Chambers I, Stracey C, Niwa H, Smith A. Signaling mechanisms regulating self-renewal and differentiation of pluripotent embryonic stem cells. Cells Tissues Organs. 1999;165:131–43. doi: 10.1159/000016693. [DOI] [PubMed] [Google Scholar]
- 66.Daheron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, Itskovitz-Eldor J, Daley GQ. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–8. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- 67.Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731–42. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Overmeyer JH, Maltese WA. Death pathways triggered by activated Ras in cancer cells. Front Biosci (Landmark Ed) 2011;16:1693–713. doi: 10.2741/3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–7. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh AM, Bechard M, Smith K, Dalton S. Reconciling the different roles of Gsk3beta in “naive” and “primed” pluripotent stem cells. Cell Cycle. 2012;11:2991–6. doi: 10.4161/cc.21110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh AM, Reynolds D, Cliff T, et al. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–26. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Storm MP, Kumpfmueller B, Thompson B, Kolde R, Vilo J, Hummel O, Schulz H, Welham MJ. Characterization of the phosphoinositide 3-kinase-dependent transcriptome in murine embryonic stem cells: identification of novel regulators of pluripotency. Stem Cells. 2009;27:764–75. doi: 10.1002/stem.3. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J, Su P, Wang L, et al. mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106:7840–5. doi: 10.1073/pnas.0901854106. [DOI] [PMC free article] [PubMed] [Google Scholar]