Abstract
Background
Rhipicephalus (Boophilus) microplus evades the host’s haemostatic system through a complex protein array secreted into tick saliva. Serine protease inhibitors (serpins) conform an important component of saliva which are represented by a large protease inhibitor family in Ixodidae. These secreted and non-secreted inhibitors modulate diverse and essential proteases involved in different physiological processes.
Methods
The identification of R. microplus serpin sequences was performed through a web-based bioinformatics environment called Yabi. The database search was conducted on BmiGi V1, BmiGi V2.1, five SSH libraries, Australian tick transcriptome libraries and RmiTR V1 using bioinformatics methods. Semi quantitative PCR was carried out using different adult tissues and tick development stages. The cDNA of four identified R. microplus serpins were cloned and expressed in Pichia pastoris in order to determine biological targets of these serpins utilising protease inhibition assays.
Results
A total of four out of twenty-two serpins identified in our analysis are new R. microplus serpins which were named as RmS-19 to RmS-22. The analyses of DNA and predicted amino acid sequences showed high conservation of the R. microplus serpin sequences. The expression data suggested ubiquitous expression of RmS except for RmS-6 and RmS-14 that were expressed only in nymphs and adult female ovaries, respectively. RmS-19, and -20 were expressed in all tissues samples analysed showing their important role in both parasitic and non-parasitic stages of R. microplus development. RmS-21 was not detected in ovaries and RmS-22 was not identified in ovary and nymph samples but were expressed in the rest of the samples analysed. A total of four expressed recombinant serpins showed protease specific inhibition for Chymotrypsin (RmS-1 and RmS-6), Chymotrypsin / Elastase (RmS-3) and Thrombin (RmS-15).
Conclusion
This study constitutes an important contribution and improvement to the knowledge about the physiologic role of R. microplus serpins during the host-tick interaction.
Keywords: Genome, Protease inhibitor, Rhipicephalus microplus, serpin, Cattle tick
Background
Ticks are worldwide-distributed ectoparasites that have evolved as obligate haematophagous arthropods of animals and humans. These parasites have been categorised after mosquitoes as the second most important group of vectors transmitting disease-causing agents to mammals [1,2]. In particular, the cattle tick (Rhipicephalus microplus) is considered the most economically important ectoparasite of cattle distributed in tropical and subtropical regions of the world. The principal reason for this affirmation is that R. microplus affects beef and dairy cattle producers causing direct economic losses due to host parasitism and tick borne diseases such as anaplasmosis and babesiosis [3,4].
The success of the parasitic cycle of R. microplus begins with the larval capability to overcome haemostatic and immunological responses of the host. Following larval attachment, a great amount of blood is ingested and digested by ticks in order to complete their parasitic cycle. The full-engorged adult females drop off from host to initiate the non-parasitic phase with the laying and hatching of eggs. R. microplus has an intensive production and physiological secretion of proteins during the entire parasitic cycle in order to disrupt host responses such as protease inhibitors which play an important role in tick survival, feeding and development [5-8]. Serpins (Serine Protease Inhibitors) are important regulatory molecules with roles during host- parasite interactions such as fibrinolysis [9], host response mediated by complement proteases [10], and inflammation [11-13] among other tick physiological functions [14,15]. These protease inhibitors conformed a large superfamily that is extensively distributed within bacteria, insects, parasite, animals and plants [16,17]. Serpins differ from Kunitz protease inhibitors by distinctive conformational change during the inhibition of their target proteases. The presence of a small domain designated as the reactive center loop (RCL) constitutes their most notable characteristic. This domain extends outside of the protein and leads to the formation of the firm bond of the serpin with its specific proteinase [18-20]. Members of the tick Serpin family have been studied and recommended as useful targets for tick vaccine development [21]. Consequently, serpin sequences from diverse tick species have been reported such as, Amblyomma americanum [22], Amblyomma variegatum [23], Amblyomma maculatum [24], Dermacentor variabilis [25]; Rhipicephalus appendiculatus [26], R. microplus [6,27], Haemaphysalis longicornis [28], Ixodes scapularis [21,29], and Ixodes ricinus [9,11]. Additionally, an in silico identification of R. microplus serpin was conducted using different databases [30]. However, a great number of tick serpins continue to be functionally uncharacterised which limits the studies related with their function during host – parasite interaction [11,31,32].
In this study serpins from different R. microplus genomic databases were identified and four new serpins molecules were reported. In silico characterization of these serpins was undertaken using bioinformatics methods. Additionally, R. microplus serpins (RmS) were cloned, sequenced, and expressed in order to determine their protease inhibition specificity. The spatial expression of these serpins was carried out by PCR using cDNA from different tick life stages and female adult organs. Finally, this study is an important step forward in uncovering the role of RmS in the physiology of this ectoparasite and their potential use for the future improvement of ticks control methods.
Methods
Bioinformatics and Serpin identification
The identification of R. microplus serpin sequences was performed through a web-based bioinformatics environment called Yabi [33]. The current available tick serpin sequences of Amblyomma americanum [22], A. maculatum [24], A. variegatum [23], A. monolakensis [34] H. longicornis [28,35], I. ricinus [9,36], Ixodes scapularis [21], R. microplus [37], R. appendiculatus [26], and A. monolakensis [34] were retrieved from the National Centre for Biotechnology Information non-redundant protein (NCBI) (http://www.ncbi.nlm.nih.gov). These tick serpin sequences and the human α1-antitrypsin (GenBank, AAB59495) were used as queries against BmiGi V1 [38], BmiGi V2.1 [37], five SSH libraries [39], Australian tick transcriptome libraries [40] and RmiTR V1 [40] using the Basic Local Alignment Search Tool (BLAST) with the tblastX algorithm [41]. The qualified serpin sequences (E-value < 100) were six-frame translated for deduced protein sequences. The presence of the serpin conserved domain (cd00172) was analysed using the batch CD-Search Tool with an expected value threshold cut-off at 1 against NCBI’s Conserved Domain Database (CDD) [42]. SignalP 4.1 [43] was used to predict signal peptide cleavage sites. Also, the amino acid sequences of the R. microplus serpins were scanned for the presence of the C-terminal sequence Lys-Asp-Glu-Leu (KDEL) the endoplasmic reticulum lumen retention signal (KDEL motif, Prosite ID: PS00014) using ScanProSite (http://prosite.expasy.org/scanprosite/) in order to reduce the incidence of false positive results from the SignalP prediction. Putative N-glycosylation sites were predicted using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/).
Tick sources
Hereford cattle at the tick colony maintained by Biosecurity Queensland from the Queensland Department of Agriculture, Fisheries and Forestry (DAFF) [44] were used to collect the acaricide susceptible strain R. microplus NRFS (Non-Resistant Field Strain). All of the eggs (E), larvae (L), nymphs (N), adult males (M) and feeding females (F) were collected from infested animals maintained within a moat pen (DAFF Animal Ethics approval SA2006/03/96). Tick organs were dissected from 17 day-old adult females for cDNA preparation including salivary glands (FSG), guts (FG) and ovaries (Ovr).
Total RNA extraction
RNA was isolated from eggs, nymphs, and the organs (guts, ovaries and salivary glands) dissected from semi-engorged females. Ticks/organs were ground in liquid nitrogen using diethylpyrocarbonate water-treated mortar and pestle prior to processing utilising the TRIzol® reagent (Gibco-BRL, USA). The tissue samples were stored in the ice-cold TRIzol® Reagent immediately after dissection, and then homogenised through a sterile 25-gauge needle. The total RNA was isolated following the manufacture’s protocol (Gibco-BRL, USA) and the mRNA was purified using Poly (A) Purist™ MAG Kit (Ambion, USA) as recommended by the manufacturer.
Isolation, cloning and sequencing of rms genes
cDNA from nymphs, ovaries and salivary glands was synthesised from purified mRNA using the BioScript™ Kit (Clontech, USA) following the manufacturer’s recommended protocol. PCRs were conducted for isolation of the rms genes using gene specific 5′ and 3′ primers, and designed for the amplification of the coding sequences (CDS) of serpin. Following the amplification and confirmation of the PCR products by agarose gel electrophoresis, the PCR products were sub-cloned into the pCR 2.1-TOPO® vector following the manufacturer’s instructions (Invitrogen, USA). The recombinant plasmids obtained were named pCR-rms1, rms2 and pCR-rms (n + 1). Ten individual colonies for each clone were selected and grown in 5 mL of LB broths supplemented with ampicillin (50 μg.mL−1) 18 hours prior to the purification of the plasmid using the QIAprep Spin miniprep kit (Qiagen, USA). The direct sequencing of the plasmid inserts was performed using the BigDye v3.1 technology (Applied Biosystems, USA) and analysed on the Applied Biosystems 3130xl Genetic Analyser at the Griffith University DNA Sequencing Facility (School of Biomolecular and Biomedical Science, Griffith University, Qld, Australia). The sequencing reactions were prepared using M13 primers in a 96-well plate format according to the manufacturer’s instructions (Applied Biosystems, USA). The sequences were visualised, edited and aligned using Sequencher v4.5 (Gene Codes Corporation, USA) to remove vector sequence and to thus confirm the CDS of the rms genes.
Cloning and expression of RmS in the yeast P. pastoris
The coding sequence of rms1, -rms3, -rms6, and rms15 were inserted into the pPICZα A and pPIC-B expression vector (Invitrogen, USA) for intracellular and extracellular expression. The resultant recombinant plasmids were transformed into the yeast P. pastoris GS115 and SMD1168H by electroporation as described in the Pichia Expression Kit manual (Invitrogen). The recombinant protein were purified from yeast pellet and supernatant using a Histrap FF 5 mL column (GE Healthcare, USA) as recommended by the manufacturer following by a gel filtration purification step using a HiLoad™ 16/600 Superdex™ 200 pg column (GE Healthcare, USA).
Expression analysis by semi-quantitative Reverse Transcription (RT)-PCR
Gene specific primers were used to determine the gene expression pattern in eggs, nymphs, female guts, ovaries and salivary gland samples. A total of fifty ticks were dissected to isolate the different organs samples, and 25 nymphs were used on the preparation of the nymph sample. Approximately, five grams of eggs from ten different ticks were processed to conform this experimental sample. Briefly, the densitograms of amplified PCR products were analysed by ImageJ and normalised using the following formula, Y = V + V(H-X)X where Y = normalised mRNA density, V = observed rms PCR band density in individual samples, H = highest tick housekeeping gene PCR band density among tested samples, X = tick housekeeping gene density in individual samples [22]. All experimental samples were processed in triplicated.
Protease inhibition assays
RmS-1, RmS-3, RmS-6 and RmS-15 expressed in P. pastoris yeast using the methodology reported previously [6,45] were used in this assay. The inhibition test was conducted as reported formerly [46] to screen the activity of RmS-1, -3, -6 and -15 against different proteases including bovine Chymotrypsin and Trypsin, porcine Elastase and Kallikrein, human Plasmin, and Thrombin (Sigma-Aldrich, USA). Briefly, 96-well plates were blocked with Blocking buffer (20 mM Tris-HCl, 150 mM NaCl and 5% skim milk, pH 7.6), and washed three times with Wash buffer (20 mM Tris-HCl, 150 mM NaCl, 0.01% Tween 20, pH 7.6) every 5 min. A total of 50 μL containing each protease were incubated with 50 fold molar of RmS-1, RmS-3, RmS-6 and RmS-15 at 37°C for 60 minutes in duplicate. The specific substrate (0.13 mM) was added and substrate hydrolysis was monitored every 30 second using Epoch Microplate Spectrophotometer (BioTek, USA) (see Table 1). The inhibition rate was calculated by comparing the enzymatic activity in the presence and absence of recombinant RmS. The experiments were conducted in triplicate.
Table 1.
The conditions of serpin inhibition reactions against commercially available bovine, porcine and human serine proteases
| Enzymes* | [nM] | Binding buffer | Substrates* | [mM] |
|---|---|---|---|---|
| Chymotrypsin | 10 | 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl2, 0.01 % Triton X-100, pH 8.0 | N-Succinyl-Ala-Ala-Pro-Phe-p-nitroanilide | 0.13 |
| Elastase | 50 | 50 mM Hepes, 100 mM Nacl, 0.01 % Triton X-100, pH 7.4 | N-Succinyl-Ala-Ala-Ala-p-nitroanilide | 0.13 |
| Kallikrein | 50 | 20 mM Tris-HCl, 150 mM NaCl, 0.02 % Triton X-100, pH 8.5 | N-Benzoyl-Pro-Phe-Arg-p-nitroanilide hydrochloride | 0.13 |
| Plasmin | 50 | 20 mM Tris-HCl, 150 mM NaCl, 0.02 % Triton X-100, pH 8.5 | Gly-Arg-p-nitroanilide dihydrochloride | 0.13 |
| Thrombin | 2 | 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl2, 0.01 % Triton X-100, pH 8.0 | Sar-Pro-Arg-p-nitroanilide dihydrochloride | 0.13 |
| Trypsin | 2 | 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl2, 0.01 % Triton X-100, pH 8.0, | N-Benzoyl-Phe-Val-Arg-p-nitroanilide hydrochloride | 0.13 |
*All enzymes and substrates were purchased from Sigma-Aldrich, USA.
Statistical analysis
The semi-quantitative PCR and protease inhibition assay data were evaluated by one-way ANOVA with Bonferroni testing (p ≤ 0.05). All analyses were conducted by the GraphPad Prism version 6.02 (GraphPad Software). Data were represented as the mean ± standard deviation (SD).
Results
Identification of RmS
The analysis of the R. microplus sequence databases revealed twenty-two different putative RmS ultimately identified after the elimination of redundant sequences. The full CDS for RmS-1 to RmS-18 reported by Tirloni and co-workers [30] were found in this study. The percentage identities after the alignment of the RmS was variable for example, RmS-3 and RmS-20 showed a 94% and 31% identities with hypothetical bacterial serpin (Paraphysa parvula) and R. appendiculatus Serpin-3, respectively. The reactive center loop characteristic of serine protease inhibitor family was found in the CDS of RmS- 19 to – 22. These new sequences were deposited in the Genbank with the following Accession Numbers: RmS-19: KP121409, RmS-20: KP121408, RmS-21: KP121411, and RmS-22: KP121414.
There was observed a high variability of the identity among the RmS family that ranged from 29% between RmS-14 and RmS-15 to 62% between RmS-3 and RmS-5. The characteristic reactive center loop (RCL) domain associated with the serpin family members was found in all RmS (Figure 1). The type of amino acid at the P1 site of the RCL showed a high variation, for example in RmS-1, -4, 7, 10, 11, -14, 20 to 22 have a polar uncharged amino acid, but RmS-2, -3, -12 and -19 have hydrophobic amino acids. Basic amino acids such as arginine or lysine at the P1 site were found in RmS-5, -6, -9, -13. -15 to -18 (Figure 1). The consensus amino acid motif VNEEGT [47] and the canonical sequence representing the RCL hinge from P17 to P8 (EEGTIATAVT) [18] which are characteristic of serpins were highly conserved in the RmS aligned (Figure 1). Finally, the data confirmed the conservation of the reactive center loop and the characteristic motif of this proteins family in RmS -19 to RmS-22.
Figure 1.

Amino acid sequence alignment of the characteristic reactive center loops of R. microplus serpins (RmS-1 to –22). Highly conserved residues and motifs were highlighted in gray shade. The P1 regions were highlighted with a dash dot line rectangle over the specific amino acid sequence [47].
RT-PCR analysis
Reverse transcriptase PCR analysis was used to validate the spatial expression of rms-1, rms-3 to -6, rms -14, rms -15, -19 to rms-22 transcripts. Data showed expression of the rms transcripts in different organs and developmental stages of R. microplus (Figure 2A -D). High expression of rms-1, -3, -5 and -15 in adult female compared with nymph stage was observed (Figure 2A and B), and rms-3 transcript was not detected in eggs. Expression of rms -14 and rms -6 was only detected in nymphs and ovaries respectively. The rms -4 was detected only in ovaries and salivary glands (Figure 2A and B). The rms-1, -3, -5, -15, -19 to -22 transcripts were highly expressed in almost all tissues and tick stages analysed. The rms-21 transcript was expressed in all tick samples except in ovaries, no expression of rms-22 transcript was detected in nymph and ovaries (Figure 2C and D).
Figure 2.
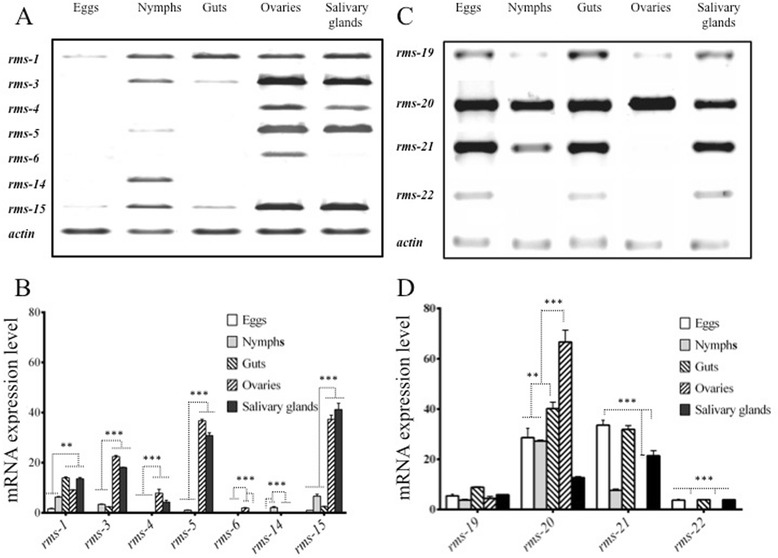
Semi-quantitative analysis of the expression of rms transcripts in R. microplus tick samples . A and C: PCR products obtained from cDNA samples from different tick’ tissues. The PCR products were run in 1% Tris Borate agarose gels. B and D: Normalised mRNA density was obtained after the densitogram analyses of the amplified PCR products. All experiments were conducted in triplicate. Data were represented as the mean ± standard deviation (SD). The symbols ** and *** indicate statistical significance with p < 0.05 and p < 0.001, respectively.
Protease Inhibition of the recombinant R. microplus serpins (rRmS)
The coding sequences for rms-1, -3, -6 and -15 were cloned and expressed in P. pastoris yeast in order to test their inhibitory activity. These serpins were tested against different serine proteases including Chymotrypsin, Elastase, Kallikrein, Thrombin and Trypsin. The protease activity analysis showed that RmS-1 is a strong inhibitor of Chymotrypsin but weak inhibitor of Trypsin and Thrombin. RmS-3 has Chymotrypsin and Elastase as its principal target molecules and some faint inhibition of Trypsin and Thrombin was observed (Figure 3). RmS-15 exhibited strong inhibitory action of Thrombin while RmS-6 only inhibits Chymotrypsin (Figure 3).
Figure 3.
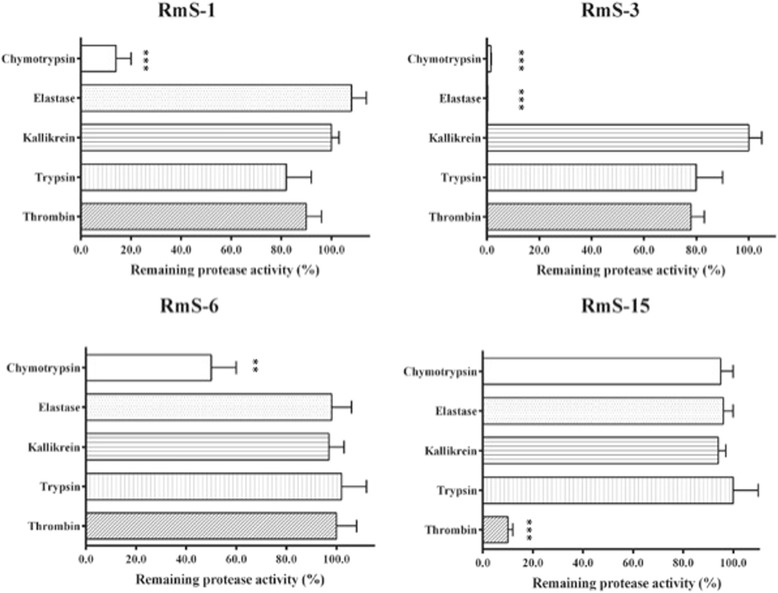
Protease inhibition profiles for the recombinant RmS-1, -3, -6 and RmS-15 obtained from the yeast P. pastoris . The RmS-1 and RmS-16 were expressed intracellular in the yeast P. pastoris. The symbols ** and *** indicate statistical significance with p < 0.05 and p < 0.001, respectively. The experiments were conducted in triplicate. Data were represented as the mean ± standard deviation (SD).
Discussion
The serpin family is conformed by a high and variable number of genes which are found in many different organisms, for example the human genome has approximately 36 serpins genes [48], 29 genes were found in Drosophila melanogaster [49], 45 in I. scapularis, and 17 serpins genes in A. americanum [21,22]. This corroborates a high conservation of the regulatory role of serpins among different species, and their functional versatility suggesting an evolutionary adaptation to confront different and novel proteases [50]. Processes such as host innate immune response regulation [51,52]; tick defences [49,53]; hemolymph coagulation cascade [54] and tick development [55,56] are regulated by serine protease inhibitors. In ixodidae, serpins are an extensive protein family with an important role at the physiological level, particularly during the parasitic periods of attachment and blood feeding [14,21-23,26,28,32,35,52,57,58]. Especially in R. microplus, a single host tick that has a highly efficient and complex combination of proteins in saliva that are useful in order to achieve the successful blood feeding, and serpins have an important role within tick saliva. In this study data obtained from transcriptome studies conducted on different stages of development of R. microplus and stored at CattleTick Base [40] were an important resource for this study to determine the members of the R. microplus serpin family [40]. However, full coverage of R. microplus genome would be necessary to give a precise number of R. microplus serpins [59]. Previous studies have provided important evidence of tick serpin sequences and transcript expression, but research discerning the specific targets or biological functions of these serpins is not forthcoming [6,13,32,52,60,61]. Following the elimination of redundant sequences the data obtained in this research suggested the presence of 22 putative R. microplus serpins from all databases studied. The amino acid sequences of these serpins revealed similar numbers of secreted and non-secreted serpins as described by Tirloni and co-workers [30]. A total of 18 R. microplus serpins showed high amino acid identity (range from 97 to 100%) with serpins reported in the BmGI and RmiTR V1 databases (including USA and Australian R. microplus), and those reported by Tirloni and co-worker (RmINCT-EM database, including Brazilian R. microplus) [30,40,62,63]. This observation confirms the conservation of these serpins in geographically distant populations of R. microplus.
The extracellular secretion of serine protease inhibitors during host – parasite interaction is important for ticks in order to overcome the haemostatic response of the host, blood digestion, and defence [64-70]. Anti-haemostasis serpins have been reported from A. americanum [32], H. longicornis [28,35], I. ricinus [9,11,57], and R. haemaphysaloides [71]. This study identified RmS-15 as an anti-haemostatic serpin that specifically inhibited Thrombin, an important serine protease of the coagulation pathway [72]. The result suggests an important role of RmS-15 to impair host blood coagulation during tick feeding. Similar results specifically related with blood coagulation was previously obtained for a mutant M340R of the I. ricinus serpin (Iris) that gained inhibitory activity against Thrombin and Factor Xa after losing its Elastase affinity through directed mutation [9].
This study improved the P. pastoris culture, expression and purification of previously described RmS-3 [6] demonstrated by significant inhibition of Chymotrypsin and Elastase observed in this study. The neutrophils’ elastase is discharged at the tick bite site which has reported to have an accumulation of this particular group of cells [73]. Additionally, previous studies have reported that neutrophils contribute to local inflammation during tick infestation which is an evasion mechanism employed by the host to resist tick infestation [67,70,74]. RmS-3 showed high levels of recognition by sera obtained from tick resistant cattle corroborating its secretion within tick saliva and an important role of this serpin during the host – parasite interaction [6]. RmS-3 might play an important role in the inhibition of host immune response. Similar results were obtained with the recombinant serpin from I. ricinus –Iris- with Elastase as its principal natural target [9,13]. However, the high expression of the rms-3 gene observed in this study in tick ovaries is related with its possible role to protect tick reproductive cells from digestive proteases released into tick hemocoel. This defensive pathway was attributed to insect serpins that inhibit Chymotrypsin [75].
Serpins without a secretion signal have been reported to have a regulatory role in intracellular pathways such as tick development, intracellular digestion or vitellogenesis [67,68,76]. The predicted intracellular serpin, RmS-14 was only detected in nymphs showing specific expression of this serpin at this particular stage of tick development. RmS-14 was not detected by RT-PCR conducted previously in tissue samples from the Porto Alegro R. microplus strain (Rio Grande do Sur, Brazil) [30], however, nymph samples were not screened in this related study.
Four new serpins are reported in this investigation, two of them, RmS-19 and -20 were expressed in all tissues samples analysed showing their important role in both parasitic and non-parasitic stages of R. microplus development. RmS-21 and -22 were not detected in ovaries suggesting a regulatory role of these serpins in the proteolysis activity during digestion and embryos development in the eggs stage. Additionally, RmS-1 is a serpin that lacks a detectable signal peptide but was found to specifically inhibit Chymotrypsin with comparatively less inhibition of Trypsin and Thrombin. RmS-1 contains two methionines at P4, P5, and cysteines at P1 and P’1 sites of the RCL. The presence of these amino acids sensitive to oxidation (methionine and cysteine) at the RCL is characteristic of human intracellular serpins [77]. Also, RmS-1 clusters together with RAS1 and Lospin7, which are intracellular serpins from R. appendiculatus and A. americanum respectively [22,26]. The secreted and glycosylated RmS-1 expressed in P. pastoris had no significant inhibition against serine proteases tested in this study. However, protease inhibition data were obtained only using an intracellular and nonglycosylated RmS1 expressed in P. pastoris. Data showed a significant inhibition of Chymotrypsin by the nonglycosylated RmS-1. The rms-1 gene was expressed in all tissue samples analysed suggesting a broad regulatory role. Similar behaviour was observed with RmS-6, where only the intracellular and nonglycosylated RmS-6 showed activity against Chymotrypsin (Figure 3). The rms16 was expressed only in the ovary sample suggesting a role for this serpin during tick embryogenesis or vitellogenesis. Further studies should be conducted in order to understand and characterise the activity and role during tick development and host parasite interaction of all R. microplus serpins identified.
Conclusion
The present study provides an insight into the R. microplus serpin family allowing the study of differential expression within specific organs and different developmental stage with four new R. microplus serpins reported. The successful expression of recombinant serpins allowed the determination of their specific host target(s). Finally, the results obtained offer an important source of information to understand R. microplus serpin function and will deepen the knowledge about the role of serpins during tick-host interactions and tick development.
Acknowledgments
The authors acknowledge support from the Cooperative Research Centre for Beef Genetic Technologies, and Meat & Livestock Australia for the scholarship stipend for Tao Xu, as well as research funding support. Thanks to Dr. Louise Jackson (Qld Department of Agriculture, Fisheries and Forestry, Australia) for the provision ticks used in this study. Thanks also to Prof. Matthew Bellgard and Paula Moolhuijzen for their support and access to CattleTickBase (Centre for Comparative Genomics, Murdoch University, Perth, Western Australia, Australia).
Footnotes
Competing interests
The authors have declared that they have no competing interests.
Authors’ contributions
MR has made substantial contributions to conception, design, analysis, and interpretation of data. XT has implemented, developed the experimental design and protocols. AL contributed with the data analysis; helped to draft the manuscript and revising it critically. SK contributed with the bioinformatics analysis. All authors contributed to the final draft, read and approved the manuscript.
Contributor Information
Manuel Rodriguez-Valle, Email: m.rodriguezvalle@uq.edu.au.
Tao Xu, Email: tao.xu@uqconnect.edu.au.
Sebastian Kurscheid, Email: sebastian.kurscheid@gmail.com.
Ala E Lew-Tabor, Email: a.lewtabor@uq.edu.au.
References
- 1.de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. 2008;13:6938–6946. doi: 10.2741/3200. [DOI] [PubMed] [Google Scholar]
- 2.Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis. 2001;32(6):897–928. doi: 10.1086/319347. [DOI] [PubMed] [Google Scholar]
- 3.McCosker PJ. Global aspects of the management and control of ticks of veterinary importance. Recent Adv Acarol. 1979;2:45–53. doi: 10.1016/B978-0-12-592202-9.50012-4. [DOI] [Google Scholar]
- 4.Grisi L, Leite RC, Martins JR, Barros AT, Andreotti R, Cancado PH, et al. Reassessment of the potential economic impact of cattle parasites in Brazil. Brazilian J Vet Parasitol. 2014;23(2):150–156. doi: 10.1590/S1984-29612014042. [DOI] [PubMed] [Google Scholar]
- 5.Pohl PC, Sorgine MH, Leal AT, Logullo C, Oliveira PL, Vaz Ida S, Jr, et al. An extraovarian aspartic protease accumulated in tick oocytes with vitellin-degradation activity. Comp Biochem Physiol B Biochem Mol Biol. 2008;151(4):392–399. doi: 10.1016/j.cbpb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Valle M, Vance M, Moolhuijzen PM, Tao X, Lew-Tabor AE. Differential recognition by tick-resistant cattle of the recombinantly expressed Rhipicephalus microplus serine protease inhibitor-3 (RMS-3) Ticks Tick Borne Dis. 2012;3(3):159–169. doi: 10.1016/j.ttbdis.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Valle M, Moolhuijzen P, Piper EK, Weiss O, Vance M, Bellgard M, et al. Rhipicephalus microplus lipocalins (LRMs): genomic identification and analysis of the bovine immune response using in silico predicted B and T cell epitopes. Int J Parasitol. 2013;43(9):739–752. doi: 10.1016/j.ijpara.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Logullo C, Vaz Ida S, Sorgine MH, Paiva-Silva GO, Faria FS, Zingali RB, et al. Isolation of an aspartic proteinase precursor from the egg of a hard tick, Boophilus microplus, Boophilus microplus. Parasitology. 1998;116(Pt 6):525–532. doi: 10.1017/S0031182098002698. [DOI] [PubMed] [Google Scholar]
- 9.Prevot PP, Adam B, Zouaoui Boudjeltia K, Brossard M, Lins L, Cauchie P, et al. Anti-hemostatic effects of a serpin from the saliva of the tick Ixodes ricinus. J Biol Chem. 2006;281(36):26361–26369. doi: 10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- 10.Mika A, Reynolds SL, Mohlin FC, Willis C, Swe PM, Pickering DA, et al. Novel scabies mite serpins inhibit the three pathways of the human complement system. PLoS One E. 2012;7(7):e40489. doi: 10.1371/journal.pone.0040489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chmelar J, Oliveira CJ, Rezacova P, Francischetti IMB, Kovarova Z, Pejler G, et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaufays J, Adam B, Menten-Dedoyart C, Fievez L, Grosjean A, Decrem Y, et al. Ir-LBP, an Ixodes ricinus tick salivary LTB4-binding lipocalin, interferes with host neutrophil function. PLoS One. 2008;3(12):e3987. doi: 10.1371/journal.pone.0003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prevot PP, Beschin A, Lins L, Beaufays J, Grosjean A, Bruys L, et al. Exosites mediate the anti-inflammatory effects of a multifunctional serpin from the saliva of the tick Ixodes ricinus. FEBS J. 2009;276:3235–3246. doi: 10.1111/j.1742-4658.2009.07038.x. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiro JMC, Labruna MB, Mans BJ, Maruyama SR, Francischetti IMB, Barizon GC, et al. The sialotranscriptome of Antricola delacruzi female ticks is compatible with non-hematophagous behavior and an alternative source of food. Insect Biochem Molec. 2012;42(5):332–342. doi: 10.1016/j.ibmb.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francischetti MI, Valenzuela GJ, Andersen FJ, Mather NT, Ribeiro MJ. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;99:3602–3612. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- 16.Wright TH. The structural puzzle of how serpin serine proteinase inhibitors work. Bioessays. 1996;18(6):453–464. doi: 10.1002/bies.950180607. [DOI] [PubMed] [Google Scholar]
- 17.Gettins WGP. Serpin Structure, Mechanism, and Function. Chem Rev. 2002;102:4751–4803. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 18.Potempa J, Korzus E, Travis J. The Serpins Superfamily of Proteinase Inhibitors: Structure, Funtion, and Regulation. J Biol Chem. 1994;269(23):15957–15960. [PubMed] [Google Scholar]
- 19.Zhou A, Carrell RW, Huntington JA. The serpin inhibitory mechanism is critically dependent on the length of the reactive center loop. J Biol Chem. 2001;276(29):27541–27547. doi: 10.1074/jbc.M102594200. [DOI] [PubMed] [Google Scholar]
- 20.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407(6806):923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 21.Mulenga A, Khumthong R, Chalaire KC. Ixodes scapularis tick serine proteinase inhibitor (serpin) gene family; annotation and transcriptional analysis. BMC Genomics. 2009;10:217. doi: 10.1186/1471-2164-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulenga A, Khumthong R, Blandon MA. Molecular and expression analysis of a family of the Amblyomma americanum tick Lospins. J Exp Biol. 2007;210(18):3188–3198. doi: 10.1242/jeb.006494. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro JM, Anderson JM, Manoukis NC, Meng Z, Francischetti IM. A further insight into the sialome of the tropical bont tick, Amblyomma variegatum. BMC Genomics. 2011;12:136. doi: 10.1186/1471-2164-12-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One. 2011;6(12):e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonenshine DE, Bissinger BW, Egekwu N, Donohue KV, Khalil SM, Roe RM. First transcriptome of the testis-vas deferens-male accessory gland and proteome of the spermatophore from Dermacentor variabilis (Acari: Ixodidae) PLoS One. 2011;6(9):e24711. doi: 10.1371/journal.pone.0024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulenga A, Tsuda A, Onuma M, Sugimoto C. Four serine proteinase inhibitors (serpin) from the brown ear tick, Rhiphicephalus appendiculatus; cDNA cloning and preliminary characterization. Insect Biochem Molec. 2003;33:267–276. doi: 10.1016/S0965-1748(02)00240-0. [DOI] [PubMed] [Google Scholar]
- 27.Kaewhom P, Sirinarumitr T, Chantakru S, Jittapalapong S. Cloning and characterization of cDNA encoding a serine protease inhibitor from salivary glands of Thai cattle tick (Boophilus microplus) Kasetsart J (Nat Sci) 2007;41:74–80. [Google Scholar]
- 28.Imamura S, Da Silva Vaz Junior I, Sugino M, Ohashi K, Onuma M. A serine protease inhibitor (serpin) from Haemaphysalis longicornis as an anti-tick vaccine. Vaccine. 2005;23(10):1301–1311. doi: 10.1016/j.vaccine.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 29.Ribeiro JMC, Francisco AC, Franchischetti IMB, Mans BJ, Mather TN, Valenzuela JG, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Molec. 2006;36:111–119. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Tirloni L, Seixas A, Mulenga A, Da Silva Vaz I, Jr, Termignoni C. A family of serine protease inhibitors (serpins) in the cattle tick Rhipicephalus (Boophilus) microplus. Expl Parasitol. 2014;137:25–34. doi: 10.1016/j.exppara.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Kovarova Z, Chmelar J, Sanda M, Brynda J, Mares M, Rezacova P. Crystallization and diffraction analysis of the serpin IRS-2 from the hard tick Ixodes ricinus. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2010;66(Pt 11):1453–1457. doi: 10.1107/S1744309110032343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulenga A, Kim T, Ibelli AM. Amblyomma americanum tick saliva serine protease inhibitor 6 is a cross-class inhibitor of serine proteases and papain-like cysteine proteases that delays plasma clotting and inhibits platelet aggregation. Insect Mol Biol. 2013;22(3):306–319. doi: 10.1111/imb.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter AA, Macgregor AB, Szabo TO, Wellington CA, Bellgard MI. Yabi: An online research environment for grid, high performance and cloud computing. Source Code Biol Med. 2012;7(1):1. doi: 10.1186/1751-0473-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mans BJ, Andersen JF, Francischetti IMB, Valenzuela JG, Schwan TG, Pham VM, et al. Comparative sialomics between hard and soft ticks: Implications for the evolution of blood-feeding behavior. Insect Biochem Molec. 2008;38(1):42–58. doi: 10.1016/j.ibmb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugino M, Imamura S, Albert M, Nakajima M, Tsuda A, Ohashi K, et al. A serine proteinase inhibitor (serpin) from the ixodid tick, Haemaphysalis longicornis; cloning and preliminary assessment of its suitability as a candidate for a tick vaccine. Vaccine. 2003;20:2844–2851. doi: 10.1016/S0264-410X(03)00167-1. [DOI] [PubMed] [Google Scholar]
- 36.Leboulle G, Rochez C, Louahed J, Ruti B, Brossard M, Bollen A, et al. Isolation of Ixodes ricinus salivary gland mRNA encoding factors induced during blood feeding. Am J Trop Med Hyg. 2002;66(3):225–233. doi: 10.4269/ajtmh.2002.66.225. [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Guerrero FD, Pertea G, Nene VM. Global comparative analysis of ESTs from the southern cattle tick, Rhipicephalus (Boophilus) microplus. BMC Genomics. 2007;8:368. doi: 10.1186/1471-2164-8-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerrero FD, Miller RJ, Rousseau ME, Sunkara S, Quackenbush J, Lee Y, et al. BmiGI: a database of cDNAs expressed in Boophilus microplus, the tropical/southern cattle tick. Insect Biochem Molec. 2005;35(6):585–595. doi: 10.1016/j.ibmb.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 39.Lew-Tabor AE, Moolhuijzen PM, Vance ME, Kurscheid S, Rodriguez-Valle M, Jarrett S, et al. Suppressive subtractive hybridization analysis of Rhipicephalus (Boophilus) microplus larval and adult transcript expression during attachment and feeding. Vet Parasitol. 2010;167(2–4):304–320. doi: 10.1016/j.vetpar.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 40.Bellgard MI, Moolhuijzen PM, Guerrero FD, Schibeci D, Rodriguez-Valle M, Peterson DG, et al. CattleTickBase: an integrated Internet-based bioinformatics resource for Rhipicephalus (Boophilus) microplus. Int J Parasitol. 2012;42(2):161–169. doi: 10.1016/j.ijpara.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40(Database issue):D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen TN, Brunak S, Von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.Stewart NP, Callow LL, Duncalfe F. Biological comparisons between a laboratory-maintained and a recently isolated field strain of Boophilus microplus. J Parasitol. 1982;68:691–694. doi: 10.2307/3280930. [DOI] [PubMed] [Google Scholar]
- 45.Canales M, de la Lastra J, Naranjo V, Nijhof A, Hope M, Jongejan F, et al. Expression of recombinant Rhipicephalus (Boophilus) microplus, R. annulatus and R. decoloratus Bm86 orthologs as secreted proteins in Pichia pastoris. BMC Biotechnol. 2008;8(1):14. doi: 10.1186/1472-6750-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvath AJ, Lu BG, Pike RN, Bottomley SP. Methods to measure the kinetics of protease inhibition by serpins. Methods Enzymol. 2011;501:223–235. doi: 10.1016/B978-0-12-385950-1.00011-0. [DOI] [PubMed] [Google Scholar]
- 47.Han J, Zhang H, Min G, Kemler D, Hashimoto C. A novel Drosophila serpin that inhibits serine proteases. FEBS Lett. 2000;468(2–3):194–198. doi: 10.1016/S0014-5793(00)01224-2. [DOI] [PubMed] [Google Scholar]
- 48.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, et al. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichhart JM. Tip of another iceberg: Drosophila serpins. Trends Cell Biol. 2005;15(12):659–665. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Kruger O, Ladewig J, Koster K, Ragg H. Widespread occurrence of serpin genes with multiple reactive centre-containing exon cassettes in insects and nematodes. Gene. 2002;293(1–2):97–105. doi: 10.1016/S0378-1119(02)00697-2. [DOI] [PubMed] [Google Scholar]
- 51.Gulley MM, Zhang X, Michel K. The roles of serpins in mosquito immunology and physiology. J Insect Physiol. 2013;59(2):138–147. doi: 10.1016/j.jinsphys.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Valle M, Lew-Tabor AE, Gondro C, Moolhuijzen P, Vance M, Guerrero FD, et al. Comparative microarray analysis of Rhipicephalus (Boophilus) microplus expression profiles of larvae pre-attachment and feeding adult female stages on Bos indicus and Bos taurus cattle. BMC Genomics. 2010;11:47. doi: 10.1186/1471-2164-11-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15(18):2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 54.Iwanaga S, Kawabata S. Evolution and phylogeny of defense molecules associated with innate immunity in horseshoe crab. Front Biosci. 1998;3:D973–D984. doi: 10.2741/a337. [DOI] [PubMed] [Google Scholar]
- 55.Coleman S, Drahn B, Petersen G, Stolorov J, Kraus K. A Drosophila male accessory gland protein that is a member of the serpin superfamily of proteinase inhibitors is transferred to females during mating. Insect Biochem Molec. 1995;25(2):203–207. doi: 10.1016/0965-1748(94)00055-M. [DOI] [PubMed] [Google Scholar]
- 56.Charron Y, Madani R, Combepine C, Gajdosik V, Hwu Y, Margaritondo G, et al. The serpin Spn5 is essential for wing expansion in Drosophila melanogaster. Int J Dev Biol. 2008;52(7):933–942. doi: 10.1387/ijdb.072419yc. [DOI] [PubMed] [Google Scholar]
- 57.Leboulle G, Crippa M, Decrem Y, Mejri N, Brossard M, Bollen A, et al. Characterization of a novel salivary immunosuppressive protein from Ixodes ricinus ticks. J Biol Chem. 2002;15:15. doi: 10.1074/jbc.M111391200. [DOI] [PubMed] [Google Scholar]
- 58.Mulenga A, Sugino M, Nakajim M, Sugimoto C, Onuma M. Tick-Encoded serine proteinase inhibitors (serpins); potential target antigens for tick vaccine development. J Vet Med Sci. 2001;63(10):1063–1069. doi: 10.1292/jvms.63.1063. [DOI] [PubMed] [Google Scholar]
- 59.Guerrero FD, Miller RJ, Perez de Leon AA. Cattle tick vaccines: many candidate antigens, but will a commercially viable product emerge? Int J Parasitol. 2012;42(5):421–427. doi: 10.1016/j.ijpara.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 60.Valdes JJ, Schwarz A, Cabeza De Vaca I, Calvo E, Pedra JH, Guallar V, et al. Tryptogalinin is a tick Kunitz serine protease inhibitor with a unique intrinsic disorder. PLoS One. 2013;8(5):e62562. doi: 10.1371/journal.pone.0062562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lai R, Takeuchi H, Jonczy J, Rees HH, Turner PC. A thrombin inhibitor from the ixodid tick, Amblyomma hebraeum. Gene. 2004;342(2):243–249. doi: 10.1016/j.gene.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 62.Wang YH, Reverter A, Kemp D, McWilliam SM, Ingham A, Davis CK, et al. Gene expression profiling of Hereford Shorthorn cattle following challenge with Boophilus microplus tick larvae. Aust J Exp Agricul. 2007;47:1397–1407. doi: 10.1071/EA07012. [DOI] [Google Scholar]
- 63.Saldivar L, Guerrero FD, Miller RJ, Bendele KG, Gondro C, Brayton KA. Microarray analysis of acaricide-inducible gene expression in the southern cattle tick, Rhipicephalus (Boophilus) microplus. Insect Mol Biol. 2008;17(6):597–606. doi: 10.1111/j.1365-2583.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- 64.Heekin AM, Guerrero FD, Bendele KG, Saldivar L, Scoles GA, Dowd SE, et al. The ovarian transcriptome of the cattle tick, Rhipicephalus (Boophilus) microplus, feeding upon a bovine host infected with Babesia bovis. Parasit Vectors. 2013;6:276. doi: 10.1186/1756-3305-6-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heekin AM, Guerrero FD, Bendele KG, Saldivar L, Scoles GA, Dowd SE, et al. Gut transcriptome of replete adult female cattle ticks, Rhipicephalus (Boophilus) microplus, feeding upon a Babesia bovis-infected bovine host. Parasitol Res. 2013;112(9):3075–3090. doi: 10.1007/s00436-013-3482-4. [DOI] [PubMed] [Google Scholar]
- 66.Stutzer C, Van Zyl WA, Olivier NA, Richards S, Maritz-Olivier C. Gene expression profiling of adult female tissues in feeding Rhipicephalus microplus cattle ticks. Int J Parasitol. 2013;43(7):541–554. doi: 10.1016/j.ijpara.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 67.Anderson JM, Sonenshine DE, Valenzuela JG. Exploring the mialome of ticks: an annotated catalogue of midgut transcripts from the hard tick, Dermacentor variabilis (Acari: Ixodidae) BMC Genomics. 2008;9:552. doi: 10.1186/1471-2164-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson JM, Valenzuela JG. Tick saliva: from pharmacology and biochemistry to transcriptome analysis and funtion genomics. In: Nuttall ASBaP, editor. Ticks: Biology, Disease and Control. New York: Cambridge University Press; 2008. pp. 92–107. [Google Scholar]
- 69.Francischetti IMB, Sá-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Front Biosci. 2010;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chalaire KC, Kim TK, Garcia-Rodriguez H, Mulenga A. Amblyomma americanum (L.) (Acari: Ixodidae) tick salivary gland serine protease inhibitor (serpin) 6 is secreted into tick saliva during tick feeding. J Exp Biol. 2011;214(4):665–673. doi: 10.1242/jeb.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y, Cao J, Zhou Y, Zhang H, Zhou J. Isolation and characterization of two novel serpins from the tick Rhipicephalus haemaphysaloides. Ticks Tick Borne Dis. 2013;4(4):297–303. doi: 10.1016/j.ttbdis.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Bode W. The structure of thrombin: a janus-headed proteinase. Semin Thromb Hemost. 2006;32(Suppl 1):16–31. doi: 10.1055/s-2006-939551. [DOI] [PubMed] [Google Scholar]
- 73.Constantinoiu CC, Jackson LA, Jorgensen WK, Lew-Tabor AE, Piper EK, Mayer DG, et al. Local immune response against larvae of Rhipicephalus (Boophilus) microplus in Bos taurus indicus and Bos taurus taurus cattle. Int J Parasitol. 2010;40:865–875. doi: 10.1016/j.ijpara.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 74.Rubin H. Serine protease inhibitors (SERPINS): where mechanism meets medicine. Nat Med. 1996;2(6):632–633. doi: 10.1038/nm0696-632. [DOI] [PubMed] [Google Scholar]
- 75.Ragan EJ, An C, Yang CT, Kanost MR. Analysis of mutually exclusive alternatively spliced serpin-1 isoforms and identification of serpin-1 proteinase complexes in Manduca sexta hemolymph. J Biol Chem. 2010;285(38):29642–29650. doi: 10.1074/jbc.M110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Franta Z, Frantova H, Konvickova J, Horn M, Sojka D, Mares M, et al. Dynamics of digestive proteolytic system during blood feeding of the hard tick Ixodes ricinus. Parasit Vectors. 2010;3:119. doi: 10.1186/1756-3305-3-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, et al. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci. 2004;61(3):301–325. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


