Abstract
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine tumor of the skin currently with no cure. In this study, we have first demonstrated that c-Myc overexpression is common in MCC. By targeting c-Myc, bromodomain inhibitors have demonstrated antitumor efficacy in several preclinical human cancer models. Thus we interrogated the role of c-Myc inhibition in MCC with c-Myc amplification by employing the BET inhibitor JQ1. We have uncovered that c-Myc can be regulated by JQ1 in MCC cells with pathological c-Myc activation. Moreover, JQ1 potently abrogates c-Myc expression in MCC cells and causes marked G1 cell cycle arrest. Mechanistically, JQ1 induced cell cycle arrest coincides with downrgulation of cyclin D1 and upregulation of p21, p27 and p57, whereas JQ1 exerts no effect on apoptosis in MCC cells. Further knockdown of p21, p27 or p57 by shRNA partially protects cells from JQ1 induced cell cycle arrest. Additionally, c-Myc knockdown by shRNA generates significant cell cycle arrest, suggesting that c-Myc overexpression plays a role in MCC pathogenesis. Most importantly, JQ1 significantly attenuates tumor growth in xenograft MCC mouse models. Our results provide initial evidence indicating the potential clinical utility of BET protein inhibitors in the treatment of MCC with pathologic activation of c-Myc.
Keywords: Merkel cell carcinoma, BET protein inhibitors, cell cycle arrest, JQ1
Introduction
Merkel cell carcinoma (MCC) is an aggressive skin tumor of neuroendocrine origin with a rising incidence. Its 5-year mortality rate is 46% and there is no cure for metastatic disease (1, 2). Although a causative link between Merkel cell polyomavirus (MCV) and MCC has been proposed, the cellular mechanisms involved in MCC pathogenesis remains largely unknown (3, 4).
Interrogation of MCC tumors for mutations of both tumor suppressor genes and oncogenes, such as p53, PTEN, Ras, B-RAF, c-kit, β-catenin, which are frequently mutated and dysregulated in many cancers, have failed to reveal a consistent significant role for any of these genes in MCC (5, 6). Intriguingly, one study has shown that the MAP kinase pathway is silent, as demonstrated by lack of pathway activation and no ERK phosphorylation (7). Recently, PI3K/AKT and the mTOR pathway, the most commonly dysregulated pathway in human cancer, is found to be upregulated in MCCs, though low mutation rates of PI3K/Akt have been detected (8, 9). Interestingly, Paulson et al reported amplification of L-Myc in MCC by array-comparative genome hybridization (CGH) in 2008 (10). Moreover, a recent study by Moore’s group suggests that MCV small T antigen stabilizes c-Myc expression by inhibiting the cellular ubiquitin ligase protein complex (11), suggesting that c-Myc plays a role in MCC pathogenesis.
c-Myc is a master regulator of cell proliferation and metabolism and is central to the pathogenesis of many human cancers, by the coordinated upregulation of a transcriptional program influencing metabolic adaptation, cell division and survival (12–14). c-Myc also promotes transformation and maintenance of stem cells in genetically engineered mouse models of glioblastoma (15–17). Furthermore, conditional transgenic models featuring tunable transcriptional suppression have shown that even transient inactivation of Myc results in sustained regression of tumors (17). However, therapies directly targeting Myc hyperactivation are not currently available in the clinic.
Members of the bromodomains and extra-terminal (BET) domain family of proteins (BRD2, BRD3, BRD4 and Brdt) are associated with acetylated chromatin and facilitate transcriptional activation through increasing the effective molarity of recruited transcriptional activators (18). BET proteins primarily bind to the transcriptional start sites of genes expressed during mitosis and affect the transcription of growth- and survival-promoting genes (19, 20). Recently, an RNA interference screen has discovered that knockdown of BRD4 leads to downregulation of c-Myc in acute myeloid leukemia (21). Subsequently, small- molecule compounds with high potency against BET proteins, such as JQ1, I-BET151, iBET762 and MS417, have been developed (22, 23). Through epigenetic mechanism, they repress down-stream gene expression by competitively binding to BET proteins and displacing BET proteins from acetylated lysines on chromatin. Notably, c-Myc transcription is associated locally and globally with increases in histone lysine side-chain acetylation (18, 19, 22). Consistent with this model, inhibition of BET protein with JQ1 results in significant downregulation of c-Myc and antitumor activity in several hematopoietic malignancies as well as in NUT midline carcinoma (24–35). Thus, BET protein inhibitors are currently in Phase I and Phase II clinical trials for advanced malignancies.
Despite two previously published studies, the role of c-Myc in MCC pathogenesis remains poorly defined. In this study, we have revealed that c-Myc overexpression is common in MCC fresh tumors examined and primary human MCC cell lines. c-Myc inhibition by the BET protein inhibitor JQ1 induces cell cycle arrest and decreased MCC cell proliferation. Most importantly, JQ1 significantly attenuated xenograft tumor growth in vivo. Thus, our results establish the therapeutic rationale for BET protein inhibitors in the management of MCC with pathological activation of c-Myc.
Materials and Methods
Cell lines
In accordance with institutional approvals for human study protocol, we have established three primary human Merkel cell carcinoma cell lines (MCC-2, MCC-3 and MCC-5) derived from lymph node metastases of three MCC patients as previously described (36, 37). Primary MCC cells were cultured with RPMI-1640 medium containing 10% fetal bovine serum (FBS) and 10% penicillin-streptomycin-L-glutamine and incubated at 37 °C in a humidified incubator with 5% CO2. Fresh medium was added every other day.
Reagents
Human embryonic kidney (293T/17, ATCC) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and 5 mg/ml of sodium pyruvate. The following antibodies were used for immunoblotting analyses or immunohistochemistry: c-Myc, β-actin, and cleaved caspase-3 (Cell Signaling). BRD4, p21, p27, p57 and cyclinD1 (Santa Cruz). Additional reagents used in the study include: TransIT-LT1 transfection reagent (Mirus), puromycin, polybrene, fibronectin and 1xRIPA buffer (Sigma-Aldrich), enhanced chemiluminescence (ECL) detection reagent (Millipore) and staurosporine (Selleck Chemicals).
Immunohistochemistry
Briefly, 5 µm paraffin sections were deparaffinised with xylene and graded ethanol, and antigen retrieval was performed by microwaving in 0.01 M sodium citrate for 20 min. Endogenous tissue peroxidase activity was blocked with 1% hydrogen peroxide at room temperature (RT) for 1 hour. The sections were further blocked with normal goat or horse serum at RT for 1 hour following incubation with primary antibody dilution at 4°C overnight. Secondary antibody was applied to the slides for 1 hour at RT before developing in HRP detection system and freshly prepared diaminobenzidine as the chromogen (brown). Sections were counterstained with hematoxylin. Immunostained slides were viewed on an Olympus BX51 Research System Microscope by 20× and 40× UPlanApo air objective lenses (Olympus America). Images were photographed using a high-resolution interline CCD camera (CoolSNAP cf, Photometrics), and acquired with automated microscopy acquisition software (MetaMorph version 7.7, Molecular Devices).
Gene expression analysis
Total RNA was isolated from primary MCC cell lines and MCC fresh tissues with RNeasy kit (Qiagen). cDNA was generated from mRNA using Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR (qRT-PCR) was performed with a StepOne Plus Real-Time PCR System (Applied Biosystems). The following TaqMan Gene Expression Assays primers were used: Hs00905030_m1 (c-Myc), Hs00355782_m1 (p21), Hs01597588_m1 (p27), Hs00175938_m1 (p57), Hs00211334_m1 (MRPS2) and Hs00765553_m1 (cyclin D1), Hs01062014_m1(Notch1), Hs00765730_m1 (NFB). Triplicate runs of each sample were normalized to MRPS2 mRNA to determine relative expression.
Immunoblotting
Cultured cells were washed with ice-cold PBS and lysed in 1xRIPA buffer containing 1 mM DTT and Complete Mini EDTA-free protease inhibitor cocktail. After incubation on ice for 30 min, the cell lysates were clarified by centrifugation at 14,000 rpm for 15 min at 4°C. 10–30 µg of total protein was subjected to 8% or 12% SDS-PAGE gels and transferred electrophoretically onto PVDF membrane by a semidry blotting system (Bio-Rad). The membrane was blocked in 5% fat-free milk/Tris-buffered saline-0.1% Tween 20 for 1 hour at RT and incubated with primary antibodies at 4 °C overnight, followed by secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology). Then the membrane was subjected to Western blot analysis with ECL detection reagent. The xenograft tumor tissue was homogenized in 2% SDS lysis buffer and then processed for Western blotting as described above.
MCV detection
DNA was prepared using DNeasy kit (Qiagen). DNA quality was confirmed by GAPDH. PCR was performed with 120ng of genomic DNA using the Taq DNA polymerase (Invitrogen) in a final volume of 50 l for 30–35 cycles. Primer sets for LT3 and MCPVS1 were used as published previously (36).
Cell proliferation and viability assays
Cell proliferation analysis was performed using Cell Counting Kit-8 (CCK-8 kit, Sigma) and manually cell counting. MCC cell lines were plated at 1 × 104 cells per well in 96-well plates, allowed to recover for 3 hours and then exposed to serial concentrations of JQ1 (Selleck Chemicals) for 24, 48, 72 and 120 hours, respectively. CCK-8 (10µl) was added to each well and incubated at 37 °C for another 4 hours before measuring using a spectrophotometer at 450 nm. MCC cell lines were plated at 10 × 104 cells per well in 6-well plates, allowed to recover for 3 hours and then exposed to serial concentration of JQ1 (Selleck Chemicals) for 24, 48, 72 and 120 hours, respectively. Cells were collected at different time points and counted manually with trypan blue exclusion staining (ViCell, Beckman Coulter).
Methylcellulose colony assay
MCC cells clonogenic formation was assayed by culturing MCC cells in complete methylcellulose (Methocult GF+ H4435; Stem Cell Technologies) according to the manufacturer’s protocol. Briefly, MCC cells (3000 cells) were resuspended in 1 ml complete methylcellulose with JQ1 (800nM) or vehicle, and incubated in a humidified incubator. Colony formation was assayed after 21 days in culture by microscopy. Colonies consisting of at least 40 cells were counted.
Apoptosis and cell cycle analysis
Apoptosis was detected by flow cytometry using Annexin V-FITC according to the manufacturer’s protocol (BD, Biosciences). Briefly, MCC cells (1 × 106 cells) were plated in 6 well plates for 3 hours followed by treatment with JQ1 for 72 hours before Annexin-V and propidium iodide (PI) staining (BD Biosciences FACS Aria). Cells were resuspended in the binding buffer with only Annexin-V or PI served as controls. For each dye, appropriate electronic compensation of the instrument was performed to avoid overlapping of the two emission spectra. For cell cycle analysis, MCC cells (1 × 106 cells) were seeded in 6-well plates for 3 hours followed by treatment with JQ1 for 72 hours and then were labeled with 10µM BrdU for 2 hours. BrdU incorporation was detected using Alexa Fluor 488-conjugated mouse anti-BrdU antibody (BD Biosences-Pharmingen) followed by 7AAD staining (BD Biosences-Pharmingen) for cell cycle analysis per the manufacturer’s protocol.
Lentiviral transduction
Lentivector directing expression of shRNA specific to p21 (TRCN0000040123), p27 (TRCN0000009856), p57 (TRCN0000010484) and c-Myc (TRCN0000039642) were purchased from Sigma-Aldrich and nontargeting PLKO.1 scramble shRNA (plasmid 1864) was purchased from Addgene. To generate lenti-virus media, 293T/17 cells were co-transfected with gene transfer vectors and virus packaging vectors ΔH8.2 and VSVG by TransIT-LT1 transfection reagent (Mirus). Two days following transfection, viral supernatants were collected and MCC cells were transduced with viral supernatant for 48 hours in fibronectin-coated 6-well plates in the presence of 8 ug/ml polybrene after spinocculation at 800 g, 32°C for 30 min. Cells were then selected in culture media containing 2 µg/mL puromycin for at least 48 hours.
Xenograft implantation
Five-week old female immunodeficient NOD/SCID/IL2r-γnull (NSG) mice (Jackson Laboratories, Strain #005557) were used for generating xenograft mouse models. Tumor cells were prepared by suspending 2 × 107 MCC cells in 80 µl of media + 120 µl of Matrigel (BD Bioscences, Catalog #354248), and inoculated on right rear flanks. Palpable tumor growth appeared within 3–5 days of inoculation, and treatment per protocol began when tumors reached approximately 100 mm3 in volume.
In vivo drug study
Tumor-bearing mice were randomized into treatment and control groups (n ≥7 for each condition) and began receiving intraperitoneal injection (i.p.) administration of vehicle (10% 2-Hydroxypropyl-β-cyclodextrin in water) or 50 mg/kg/day JQ1 for 3 weeks duration. Mice were monitored daily, tumor xenografts were measured with digital calipers, and tumor volume was calculated as L2×W/2, where L is length and W is width. All animal experiments were done under a protocol approved by the University Institutional Animal Care and Use Committee. In accordance with institutional guidelines on animal care, experimental endpoints were determined by one of the following: (1) completion of twenty-one day treatment course, or (2) attainment of tumor burden exceeding 2 cm in any dimension, or (3) further complications affecting animal welfare. Upon reaching experimental endpoints, mice were humanely euthanized, and tumors were excised and dissected for characterization and mechanistic studies.
Statistical analysis
All the measurements were made in triplicate, and all values are represented as mean ± S.D. Statistical analysis was performed with the Student’s t test or one-way analysis of variance (ANOVA). *P value < 0.05 was considered statistically significant.
Results
c-Myc overexpression is common in MCC tumors and primary MCC cell lines
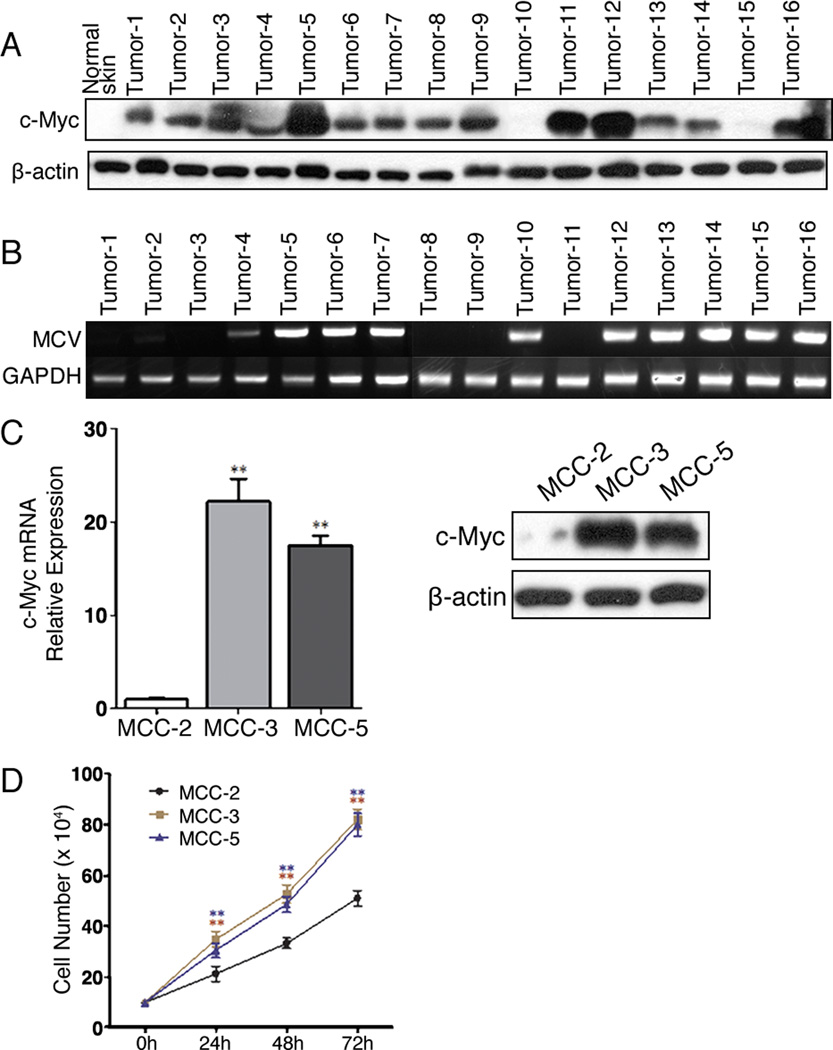
c-Myc is a transcription factor that not only regulates the expression of many genes crucial for cell proliferation and differentiation, it is also one of the most prevalent oncogenes deregulated in human cancers (13, 38). Interestingly, an array-CGH study revealed amplification of a region harboring L-Myc in MCC (10). Thus, we were compelled to examine c-Myc expression in MCC fresh tumors. We found that 87.5% (14/16) of MCCs overexpressed c-Myc as compared to normal skin by immunoblotting (Figure 1A). Our findings confirm a recent publication suggesting that MCV small T antigen contributes to c-Myc overexpression (39). To see if c-Myc overexpression correlates with MCV status, DNAs were extracted from MCC fresh tumors and MCV was detected as described previously (36). As shown in Figure 1B, 6 MCC samples that were negative for MCV also possessed c-Myc overexpression. Intriguingly, two MCV positive MCC (Tumor-10 and Tumor-15) failed to demonstrate c-Myc amplification. Thus, c-Myc overexpression in MCC tumors was independent of MCV status in our study. Next, we assessed c-Myc expression in 3 primary MCC cell lines established in our laboratory. MCC-2, MCC-3 and MCC-5 cell lines have been described previously (37, 40). Primary MCC cells grow in cluster in cultures and display large, round to oval, vesicular nuclei with scant cytoplasm that are characteristic of MCC (Figure S1). As shown in Figure 1C, both MCC-3 and MCC-5 overexpressed c-Myc at the mRNA and protein levels, but discernable expression of c-Myc was detected in MCC-2 cells. We next wanted to determine the cell growth properties among these three primary MCC cell lines. Cell proliferation was assessed by cell counting manually. A shown in Figure 1D, MCC-3 and MCC-5 cells with c-Myc overexpression possessed higher cell proliferation as compared to MCC-2 cells. Therefore, we have demonstrated that c-Myc overexpression is common in MCC tumors and it is independent of MCV. Moreover, primary MCC cells with c-Myc overexpression carry a higher proliferation rate.
Figure 1. c-Myc protein expression in Merkel cell carcinoma fresh tumors and primary cell lines.
A. c-Myc overexpression in fresh MCC tumor tissue by immunoblotting. Tissue lysate from normal skin was employed as a negative control; β-actin was used as a loading control. B. MCV detection by PCR. DNA was extracted from 16 MCC fresh tumor tissues. C. c-Myc expression in MCC cell lines (MCC-2, MCC-3 and MCC-5) by qPCR and immunoblotting. The mRNA expression of target genes was normalized to that of MRPS2 and a value of 1.0 was assigned to the mRNA expression of target genes in the control group (means ± SEM), (*P<0.05, **P<0.01 vs control); β-actin was used as a loading control for immunoblotting. (D) Proliferation rates of primary human MCC cell lines by cell counting (means ± SEM), (**P<0.01 vs MCC2).
BET Inhibitor JQ1 abolishes c-Myc expression and represses primary MCC cell proliferation
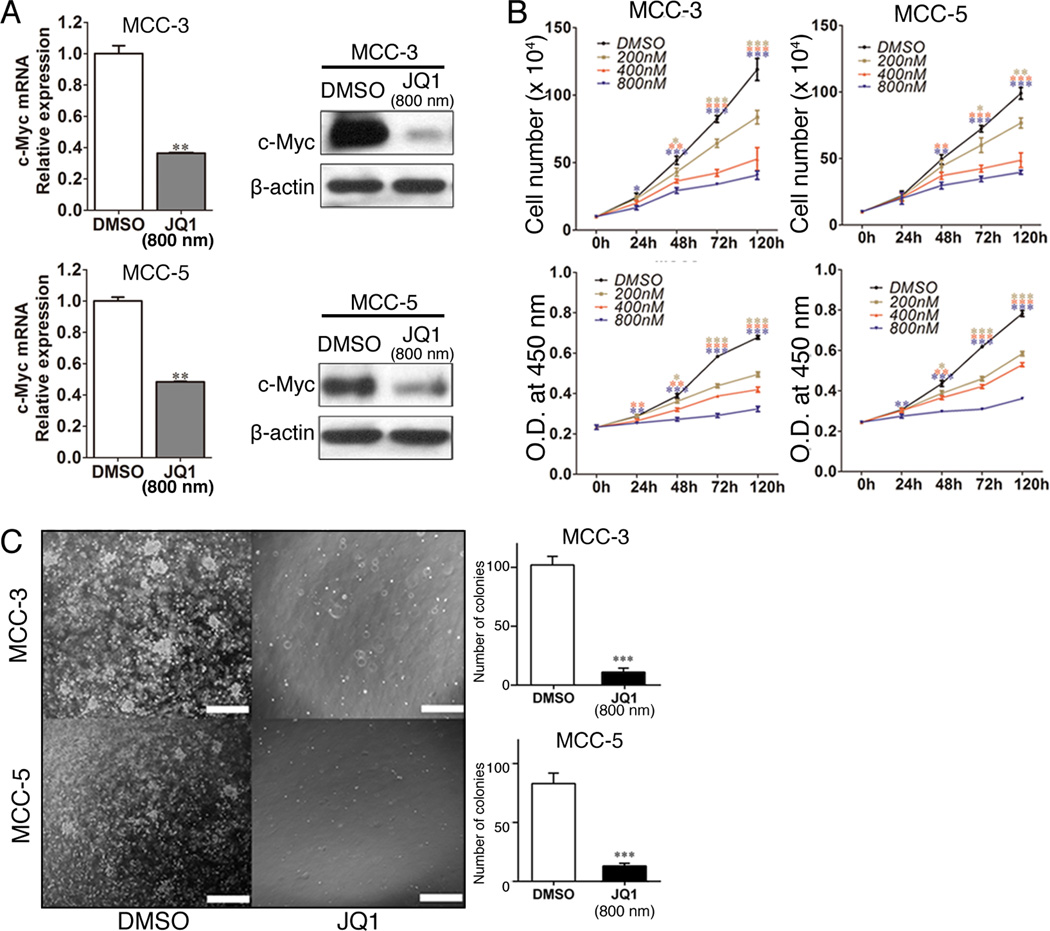
Targeting c-Myc by the BET inhibitor JQ1 has demonstrated efficient suppression of c-Myc expression as well as antitumor activity in many types of human cancer both in vitro and in vivo (28, 32). We therefore decided to examine the effects of growth inhibition by JQ1 in MCC cells. Based on published studies, there is a wide range of half maximal inhibitory concentration (IC50) of JQ1 used (200nM - 5µM) (25, 31). However, IC50 for most hematopoietic tumor cell lines are between 500nM -1000nM and the duration of treatment was between 3–5 days. Thus, we decided to test JQ1 at a series of concentration between 200nm and 800nM and at 24, 48 and 72 hours. In agreement with other published findings, a significant reduction of c-Myc expression was found after JQ1 treatment in both MCC-3 and MCC-5 cells (Figure 2A). A time- and dose-dependent inhibition of MCC cell proliferation was observed after JQ1 treatment as determined by CCK-8 assay and manual counting (Figure 2B). Consistent with our hypothesis, a greater inhibition was found in MCC-3 and MCC-5 cells with c-Myc overexpression. Interestingly a growth inhibition of MCC-2 cells was found with a higher concentration of JQ1 (800nm) (Figure S4A). After 72 hours treatment and at the lowest tested dosage of JQ1 (200nM), we observed ~20% of growth inhibition in MCC-3 and MCC-5, while MCC-2 cells were almost entirely unaffected. At the highest dosage of JQ1 (800nM) after 72 hours treatment, we observed >50% of growth inhibition in MCC-3 and MCC-5 as compared to 10% inhibition in MCC-2 cells (Figure 2B and Figure S4A). Although we observed suppression of cell growth in MCC cells without c-Myc overexpression by JQ1, in this study we decided to focus on the biological effects of c-Myc inhibition in MCC cell lines (MCC-3 and MCC-5) possessing c-Myc amplification. Since a fifty percent inhibition of growth was seen in both MCC cell lines at 800nM concentrations of JQ1 after 72 hours treatment, we chose JQ1 at the concentration of 800nM for all studies carried out in this manuscript. To evaluate the effect of long-term inhibition by JQ1, colony formation assay was performed. Similar to the results above, JQ1 (800nM) significantly decreased the number of colony formation in MCC cells compared with that of controls (Figure 2C). Our data suggest that BET inhibition has potent anti-proliferative effects in MCC cells with c-Myc overexpression in vitro.
Figure 2. BET protein Inhibitor JQ1 reduces c-Myc expression and attenuates primary MCC cell proliferation.
A. Decreased c-Myc expression in MCC-3 and MCC-5 treated with JQ1 (800nM) for 72 hours by qRT-PCR and immunoblotting. The mRNA expression of target genes was normalized to that of MRPS2 and a value of 1.0 was assigned to the mRNA expression of target genes in the control group (means ± SEM), (**P<0.01 vs control); β-actin was used as a loading control for immunoblotting. B. MCC-3 and MCC-5 cells are sensitive to JQ1 inhibition. MCC cell lines (MCC-3 and MCC-5) were cultured with JQ1 at different concentrations (200nM, 400nM and 800nM) for 24, 48, 72 and 120 hours respectively (means ± SEM), (*P<0.05, **P<0.01, ***P<0.001 vs control). C. Decreased colony formation in MCC-3 and MCC-5 cells treated with JQ1 (800nM) compared with that of DMSO control. A total of 3000 cells were mixed with methylcellulose medium with JQ1 (800nM) or DMSO and the mixture was plated in each 35-mm dish for 21 days at 37 °C (means ± SEM), (***P<0.001 vs control); scale bars = 200µm.
JQ1 induced cell cycle arrest is via upregulation of p21, p27 and p57 in MCC cell lines
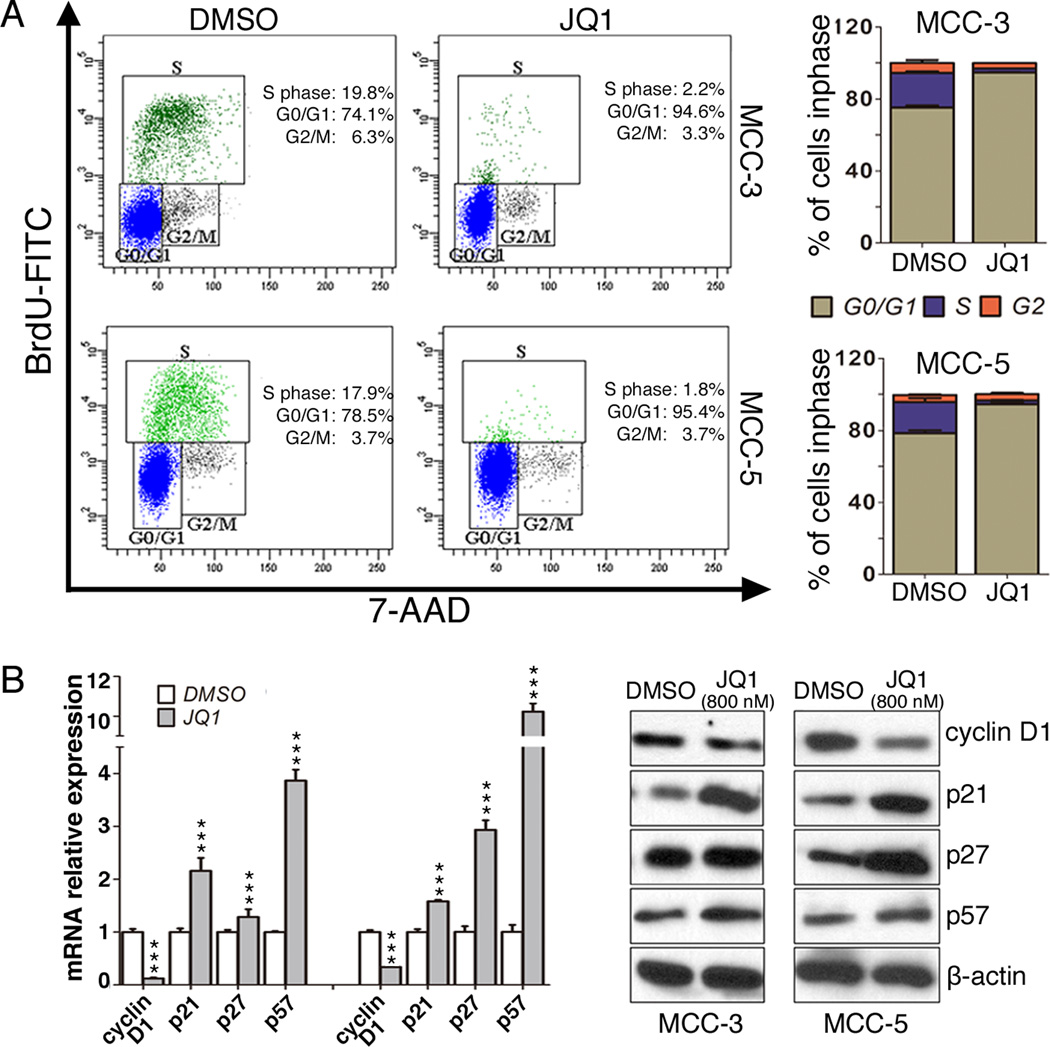
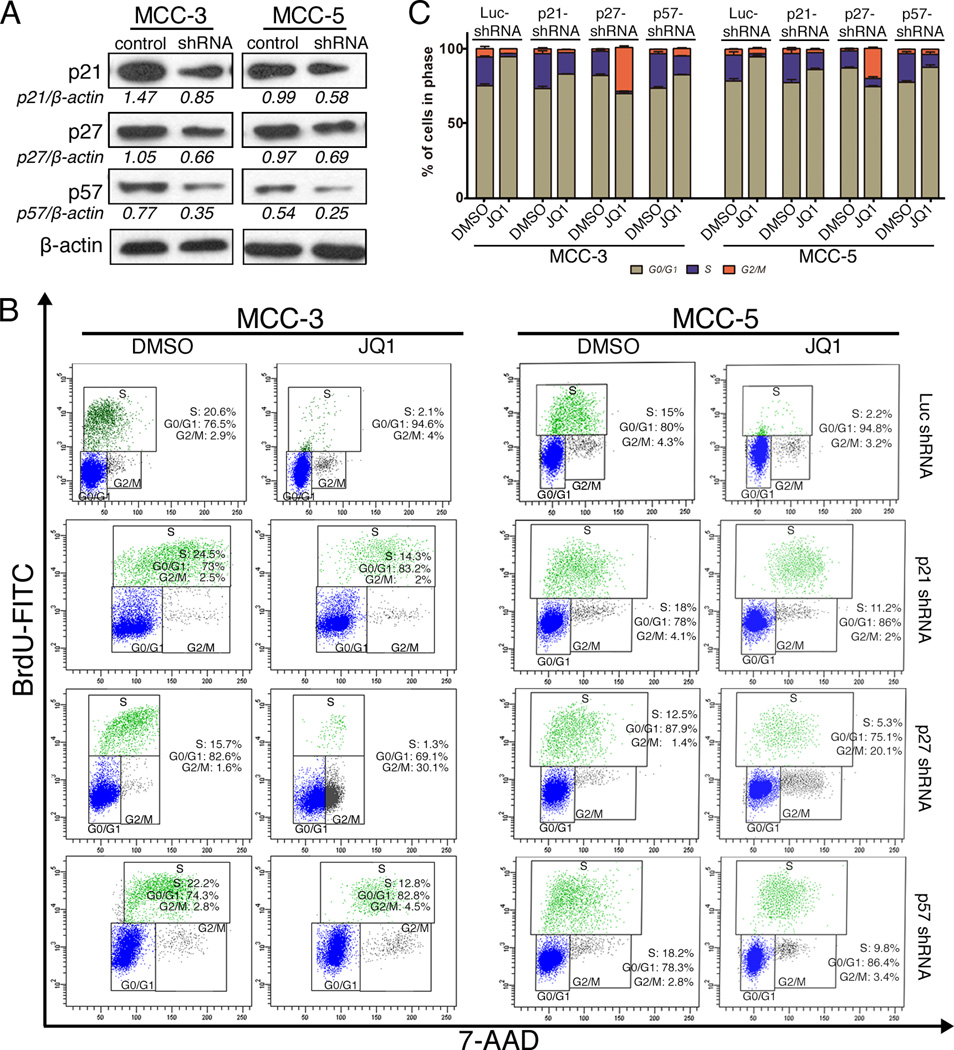
To further investigate the growth inhibitory mechanisms of JQ1 on MCC, we evaluated the effects of JQ1 on cell cycle progression and apoptosis by flow cytometry. Both MCC-3 and MCC-5 cells treated with JQ1 at 800nm for 72 hours were collected and subjected to BrdU cell cycle or Annexin V/PI analysis, respectively. Consistent with the antiproliferative effects of JQ1, a pronounced decrease of cells in S phase with a concomitant increase in cells in G0/G1 phase was observed in treated groups as compared to that in the control groups (Figure 3A), suggesting a cell cycle arrest at the G0/G1 phase. Further assessment of the involvement of the cell cycle target genes, downregulation of cell cycle promoter gene cyclin D1 and upregulation of negative cell cycle regulator genes p21, p27 and p57 was detected at the RNA and protein levels after JQ1 treatment in both MCC-3 and MCC-5 cells (Figure 3B). To further elucidate which cell cycle negative regulatory proteins contribute to cell cycle arrest by JQ1, we have successfully abolished expression of p21, p27 or p57 by shRNA in MCC cells, respectively (Figure 4A and Figure S4B). Interestingly, individual knockdown of p21, p27, or p57 expression partially rescued JQ1 induced cell cycle arrest to a similar degree, which suggested JQ1 induced suppressed proliferation is mainly due to upregulation of p21, p27 and p57 (Figures 4B and 4C). Of note, JQ1 failed to induce apoptosis in MCC cells, as demonstrated by Annexin V study (Supplementary Fig. S2A and S2B). MCC cells treated with staurosporine (1µM) for 4 hours served as positive controls.
Figure 3. JQ1 induced cell cycle arrest is via upregulation of p21, p27 and p57 in both MCC cell lines.
A. Representative FACS histograms show the distribution of MCC cells in cell cycle after JQ1 treatment. MCC-3 and MCC-5 cells were treated with JQ1 (800nM) for 72 hours and stained with BrdU and 7-AAD followed by flow cytometry analysis (means ± SEM). Bar graphs show the percentage of MCC cells in each cell cycle phase. Data presented as means ± SEM of triple experiments. B. Expressions of cell cycle associated genes in MCC cells (MCC-3 and MCC-5) treated with JQ1 (800nM) for 72 hours by qPCR and immunoblotting. The mRNA expression of target genes was normalized to that of MRPS2 and a value of 1.0 was assigned to the mRNA expression of target genes in the control group (means ± SEM), (***P<0.001 vs control); β-actin was used as a loading control.
Figure 4. JQ1 induced cell cycle arrest is depended on the expression of p21, p27 and p57 in MCC-3 and MCC-5 cells.
A. Immunoblotting demonstrates successful knockdown of p21, p27 and p57 expression by shRNA in both MCC-3 and MCC-5 transfected cells; β-actin was used as a loading control. B. Representative FACS histograms demonstrate the distribution of p21, p27, p57 knockdown cells in cell cycle after JQ1 (800nM) treatment for 72 hours (means ± SEM). C. Bar graphs show the percentage of MCC cells in each cell cycle phase. Data presented as means ± SEM of triple experiments.
c-Myc knockdown is sufficient to recapitulate the antitumor effect of JQ1 in MCC cells
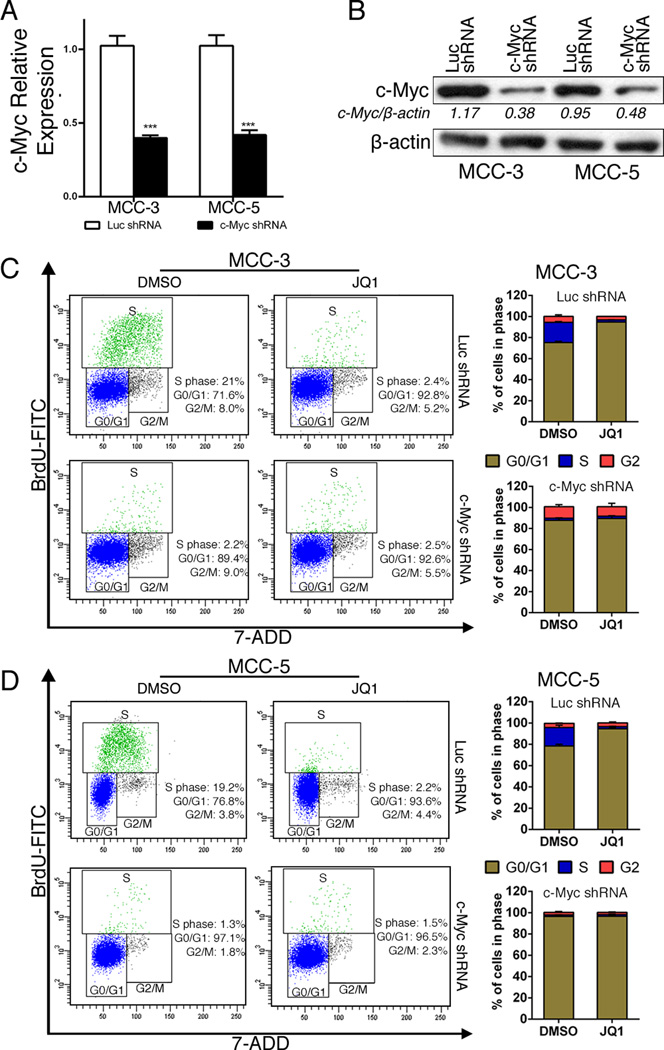
To confirm if c-Myc is the major effector of JQ1 inhibition in MCC cells, we next depleted c-Myc expression by shRNA followed by JQ1 treatment. Successful knockdown of c-Myc expression by shRNA in MCC cells is shown in Figures 5A and 5B. Similarly, c-Myc knockdown caused a significant reduction of cells in S phase that was associated with G1 arrest (Figures 5C and 5D). Cells in S phase reduced from 21% to 2.2% in MCC-3 cells, and from 19.2% to 1.3% in MCC-5 cells. Moreover, there was no additive effect by JQ1 treatment in c-Myc knockdown cells, suggesting that JQ1 induced cell cycle arrest was mediated by c-Myc. Although we cannot exclude the involvement of additional cellular targets, the strong concordance between phenotypes induced by c-Myc knockdown and JQ1 supports the notion that c-Myc is the major target of JQ1 in MCC with c-Myc amplification.
Figure 5. c-Myc knockdown recapitulates cell cycle arrest in MCC cells.
A–B. Successful knockdown of c-Myc expression in transfected cells (MCC-3 and MCC-5) by qRT-PCR and immunoblotting. The mRNA expression of target genes was normalized to that of MRPS2 and a value of 1.0 was assigned to the mRNA expression of target genes in the control group (means ± SEM), (***P<0.001 vs Luc shRNA); β-actin was used as a loading control for immunoblotting. C-D. Representative FACS histograms show the distribution of c-Myc knockdown or control MCC cells in cell cycle after JQ1 (800nM) treatment for 72 hours (means ± SEM).
In addition to c-Myc, we also examined other transcription factors, such as Notch and NFkB and c-Jun, which have been suggested to be regulated by BET brodomain proteins in other cell types (41, 42). c-Jun expression is extremely low in both MCC-3 and MCC-5 cells (data not show). In contrast to suppressed c-Myc expression upon JQ1 treatment, Notch and NFkB expressions are increased upon JQ1 treatment (Figure S4C), indicating that these molecules are less likely involved in JQ1 induced cell cycle arrest.
cDNA microarray combined with chromatin immunoprecipitation have revealed that Myc regulates all aspects of protein synthesis, including downstream of the mTOR pathway (12). Moreover, a recent study suggests mTOR dependent phosphorylation of the eukaryotic translation initiation factor 4E binding protein-1 (4E-BP1) is found to be required for Myc-driven hematological cancers (43). Therefore, we decided to examine if the mTOR pathway is downstream of c-Myc in MCC cells. Interestingly, both p-mTOR and p-4E-BP1 were unchanged after JQ1 treatment, suggesting that 4E-BP1 is not regulated by c-Myc in MCC-3 and MCC-5 cells (Figure S3A).
JQ1 impaired MCC xenograft tumor growth in vivo
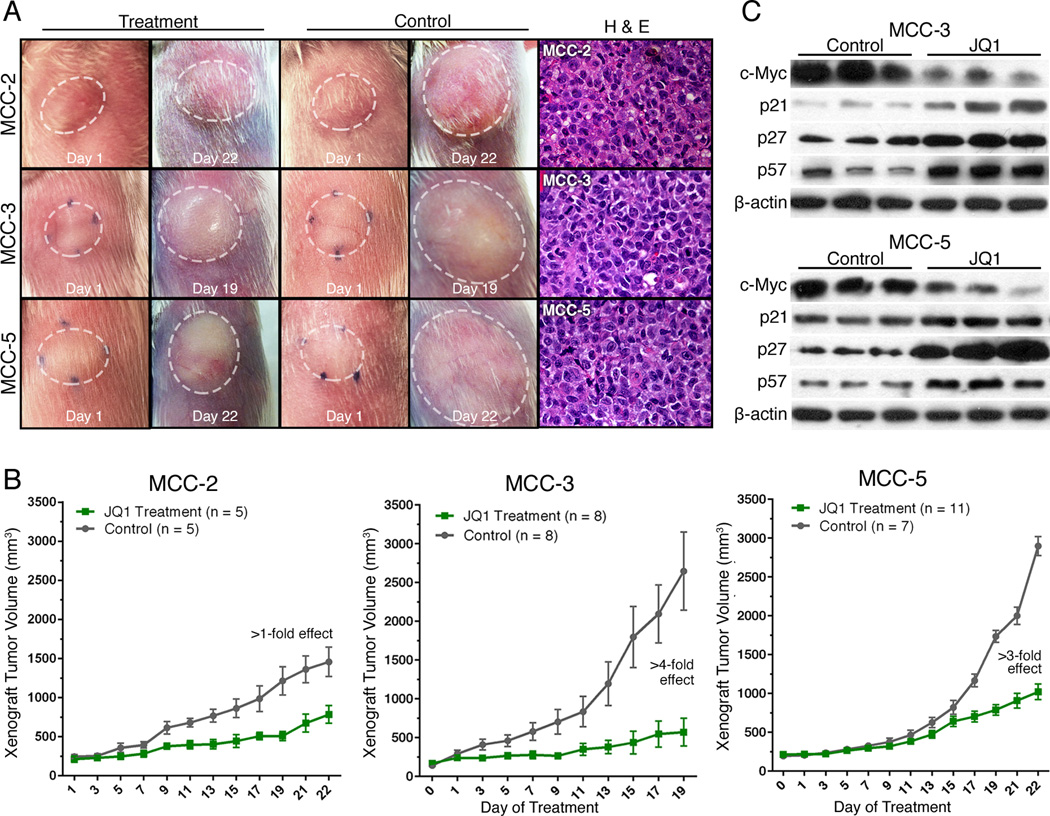
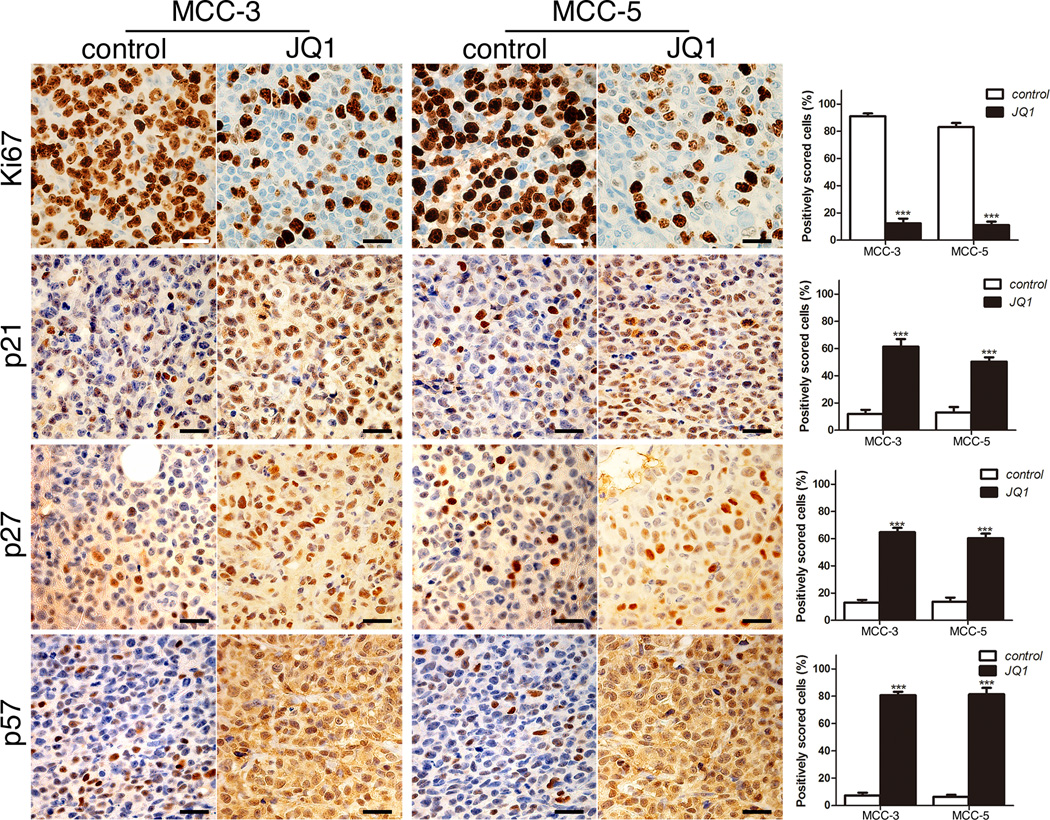
We proceeded to test the impact of JQ1 on MCC cells in our xenograft MCC mouse models. In order to strengthen our hypothesis that c-Myc was the major downstream effector of JQ1 in MCC cells, we included MCC-2 cells without c-Myc amplification as controls. MCC-2, MCC-3 and MCC-5 cells (2 × 107 cells from each cell line) were mixed with Matrigel (BD Biosciences) (80 µl of media + 120 µl of Matrigel), respectively. Tumor cells were inoculated subcutaneously into the rear flanks of NSG mice. When xenograft tumors approached ~100 mm3 in volume (or 7mm in diameter), treatment was started with as per standard treatment protocol as previously published (31). NSG mice bearing xenograft tumors began to receive intraperitoneal administration of 50 mg/kg/day JQ1 or vehicle for a 3-week duration. NSG mice bearing MCC-5 xenograft tumors were sacrificed after completion of 21-day treatment. However, NSG mice bearing MCC-3 tumors were terminated at Day 19 because control tumors were reaching 2cm in dimension. Mice treated with JQ1 had no obvious signs of toxicity (based on body weight, food and water intake, activity, and general exam). As shown in Figure 6A, xenograft tumors receiving JQ1 treatment showed great reductions of tumor volume as compared to xenograft tumors receiving vehicle. Histologically, xenograft tumor cells demonstrated large, round to oval, vesicular nuclei with scant cytoplasm, which is characteristic of MCC (Figure 6A). As measured by mean ± SEM, JQ1 significantly attenuated xenograft tumor growth (more than a 4-fold reduction in MCC-3 xenograft tumors and a 3-fold reduction in MCC-5 xenograft tumors) (Figure 6B). This was accompanied by prolonged event-free survival as tumor in the treatment group never attained 600mm3 and 1100mm3 in MCC-3 and MCC-5 tumors, respectively. Consistent with our central hypothesis, MCC-3 cells with highest c-Myc expression correlates with the greatest reduction (>4-fold) in MCC-3 xenograft tumor volume. Conversely, MCC-2 xenograft tumors lacking c-Myc amplification are less sensitive to JQ1 (>1-fold reduction). By RT-PCR analysis, xenograft tumors showed consistent expression pattern of MCC markers, such as cytokeratin 18, 19, 20, synaptophysin, neurospecific enolase and Merkel cell specific transcription factor Math-1 as expressed in MCC cells (Figure S3B). Immunoblotting analysis further confirmed suppressed c-Myc expression and upregulation of p21, p27 and p57 in the treatment groups (Figure 6C). As expected, decreased cell proliferation as determined by immunohistochemistry staining of Ki67, was found in the xenograft tumors in the treatment group as compared to those in the control group (Figure 7). Similarly, increased numbers of p21, p27 and p57 positive cells were found in xenograft tumors in the treatment group by immunohistochemistry (Figure 7). Although our in vivo data is promising, it is generated in NSG mice with profound immune deficiency. Additionally, JQ1 can potentially interact with several immune-related pathways. Nevertheless, our data provides initial evidence that the BET protein inhibitor JQ1 might be applicable in the clinic for MCC.
Figure 6. Effect of JQ1 on MCC xenograft growth in vivo.
A. Representative examples of xenograft tumors treated with JQ1 or vehicle and characteristic histology features of MCC in xenograft tumors. B. Comparison of tumor volumes ± SEM from tumor bearing NSG mice treated with JQ1 or vehicles. NSG mice bearing MCC-2, MCC-3 and MCC-5 xenograft tumors were treated with JQ1 at 50 mg/kg/day JQ1 by intraperitoneal injection between 18–21 days. C. Immunoblotting of MCC-3 and MCC-5 xenograft tumor tissues with the indicated antibodies; β-actin was used as a loading control.
Figure 7. Immunohistochemical staining of xenograft tumor tissues.
Immunohistochemical staining of xenograft tumor tissues with the indicated antibodies. p21, p27, p57 and Ki67 positive cells (brown staining) were quantified at ×400 magnification (means ± SEM), (**P < 0.01, ***P < 0.001 vs control). Scale bars = 10µm.
Discussion
MCC is an aggressive skin cancer. Standard treatment is surgery followed by radiation therapy for local and regional disease or chemotherapy for distant metastasis (44). Despite standard treatment, one third of patients will eventually develop distant metastases, for which currently there is no cure. Therefore, molecular events driving MCC pathogenesis need to be further defined in order to benefit heterogeneous patient populations. In search of receptor tyrosine kinase (RTK) involvement in MCC tumorigenesis (providing a rationale for the use of targeted molecular therapies), studies have found variable expression of c-kit, VEGFs, PDGF and PDGF in MCC compared to normal skin (45–47). Although a hyperactivated PI3K/Akt/mTOR pathway is reported in MCC (8, 9, 40), the etiology of this aberrant pathway activation is still elusive. Since c-Myc represents a unifying molecular feature in many hematopoietic malignancies (12), and amplification of L-Myc is reported in MCC (10), we were intrigued to study c-Myc. Interestingly, while we were conducting this study, a paper published by Moore’s group suggests that MCV small T antigen stabilizes c-Myc expression. However, the role of c-Myc overexpression in MCC pathogenesis still needs further elucidation (39). Consistent with the central role of Myc in the pathogenesis of human cancer, we are the first to report that c-Myc activation is common in MCC.
Although c-Myc is one of the most deregulated oncogenes in human cancers, a therapeutic approach to target c-Myc has remained elusive. Like many transcriptional targets, the lack of a ligand binding domain creates an obstacle towards direct inhibition (48). BETs are transcriptional regulators that epigenetically control gene expression, which are key in cell proliferation, cell cycle progression and apoptosis (22, 23). Dysfunction of BET proteins has been associated with the development of aggressive tumors, such as NUT midline carcinomas (26, 49). Recently developed selective BET protein inhibitors are attractive because of promising antitumor activity demonstrated in diverse preclinical models, such as multiple myeloma (31), hematologic malignancies (21, 24, 25, 27, 28, 32), glioblastoma (30), lung cancer (29, 33) and meduloblastoma (50). The efficacy of these compounds has been attributed mainly to their ability to suppress c-Myc expression as well as downstream targets. Based on these prior studies, BET inhibitors would be expected to have activity primarily against MCC possessing higher expression of c-Myc at the transcription level. In agreement with this notion, we have uncovered G1 cell cycle arrest by JQ1 in MCC cells with c-Myc amplification, particularly via suppressed c-Myc expression, downregulation of cyclin D1 and upregulation of cell-cycle inhibitors such as p21, p27 and p57 in JQ1 treated MCC cells. Moreover, knockdown of p21, p27 and p57 partially reverts JQ1 induced G0/G1 arrest in MCC cells. Interestingly, individual knockdown of p21, p27 and p57 exhibits the same extent of neutralization of JQ1 effect in MCC cells. Although JQ1 fails to confer cell death in MCC cells, this is unlikely due to the lower concentration used (800nM as compared to 1µM used in studies using models of hematopoietic malignancies). Thus, BET inhibition may have synergistic effects with targeted pro-apoptotic agents (e.g. ABT-737 and YM155) in the clinical setting.
Of note, several reports have demonstrated off-target effects on genes or signaling pathways by JQ1 in addition to c-Myc, such as TYRO3, pro-survival gene BIRC5/survivin, NFB target gene BIRC3 and JAK/STAT pathway gene IL-7R. Because of the strong concordance between the phenotypes by c-Myc knockdown and JQ1, it is most likely that c-Myc is the major gene involved in JQ1 induced cell cycle arrest in MCC cells with c-Myc overexpression. Since expression of Notch1 and NFB is increased in both MCC-3 and MCC-5 cells upon JQ1 treatment, they are less likely to be involved in JQ1-induced cell cycle arrest. Importantly, the greatest suppression of xenograft tumor by JQ1 is evident in MCC-3 cells with the highest c-Myc expression. Of note, MCC-2 cells without c-Myc amplification also confers JQ1 sensitivity at a higher concentration, suggesting alternative mechanisms in growth inhibition in MCC cells without c-Myc overexpression. Intriguingly, unlike previous publications demonstrating that the mTOR pathway and c-Myc converge at 4E-BP1, phosphorylation of 4E-BP1 is not regulated by JQ1 in our experimental system (43).
It is controversial as to whether BET protein levels are altered when cells are subjected to JQ1 (31). In our study, BRD4 expression at the protein level is unchanged in both MCC-3 and MCC-5 cells after JQ1 treatment, which is consistent with the theory that JQ1 competitively displaces BRD4 from chromatin without changing its level (data not shown). Interestingly, BRD4 expression is found to be higher in metastatic melanoma (26). It is debatable whether knockdown of BRD4 is able to phenocopy the antitumor effects of BET inhibitors in different cell types. Therefore, the concomitant displacement of other BET proteins may broaden the oncosuppression effects of these small molecule compounds despite the fact that BRD4 is the key BET protein which has been studied extensively so far. It would be compelling to explore the biological consequence of complete abrogation of BET proteins not only in MCC with c-Myc amplification but also in MCC lacking Myc amplification.
It is evidential that epigenetic modification impacts cancer initiation and progression. Of note, new small-molecule drugs modifying the epigenetic landscape of tumors has improved disease overall survival as well as potentiates the cytotoxic effects of chemotherapy (51). Here, we have demonstrated that epigenetic inhibition of c-Myc by JQ1 retards tumor growth in xenograft MCC mouse models with no obvious toxicity, which establishes the feasibility of c-Myc inhibition by BET protein inhibitors within an acceptable therapeutic window of tolerability as a single agent or in combination with other therapies in holding disease progression in check. However BET protein inhibitors may be most effective when employed in combination with agents possessing cytotoxicity in the clinical setting for MCC. Insights provided by our study identify BET protein inhibitors as rational therapeutic options and warrant further exploration in MCC with and without pathological c-Myc activation.
Supplementary Material
Acknowledgements
The project described was supported by the Translational Research Institute (TRI), grants UL1TR000039 and KL2TR000063 through the NIH National Center for Research Resources and the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
This study was also supported by funds from the Department of Dermatology and the Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences.
We thank Ms. Kimberly A. Hall for her excellent technical support.
Footnotes
Conflicts of Interest Statement: All authors have no financial disclosure.
REFERENCES
- 1.Paulson KG, Iyer JG, Byrd DR, Nghiem P. Pathologic nodal evaluation is increasingly commonly performed for patients with Merkel cell carcinoma. J Am Acad Dermatol. 2013;69:653–654. doi: 10.1016/j.jaad.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrama D, Becker JC. Merkel cell carcinoma--pathogenesis, clinical aspects and treatment. J Eur Acad Dermatol Venereol. 2011;25:1121–1129. doi: 10.1111/j.1468-3083.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 3.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afanasiev OK, Yelistratova L, Miller N, Nagase K, Paulson K, Iyer JG, et al. Merkel Polyomavirus-Specific T Cells Fluctuate with Merkel Cell Carcinoma Burden and Express Therapeutically Targetable PD-1 and Tim-3 Exhaustion Markers. Clin Cancer Res. 2013;19:5351–5360. doi: 10.1158/1078-0432.CCR-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. J Invest Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- 6.Weeraratna AT, Houben R, O'Connell MP, Becker JC. Lack of Wnt5A expression in Merkel cell carcinoma. Arch Dermatol. 2010;146:88–89. doi: 10.1001/archdermatol.2009.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houben R, Michel B, Vetter-Kauczok CS, Pfohler C, Laetsch B, Wolter MD, et al. Absence of classical MAP kinase pathway signalling in Merkel cell carcinoma. J Invest Dermatol. 2006;126:1135–1142. doi: 10.1038/sj.jid.5700170. [DOI] [PubMed] [Google Scholar]
- 8.Hafner C, Houben R, Baeurle A, Ritter C, Schrama D, Landthaler M, et al. Activation of the PI3K/AKT pathway in Merkel cell carcinoma. PLoS One. 2012;7:e31255. doi: 10.1371/journal.pone.0031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nardi V, Song Y, Santamaria-Barria JA, Cosper AK, Lam Q, Faber AC, et al. Activation of PI3K signaling in Merkel cell carcinoma. Clin Cancer Res. 2012;18:1227–1236. doi: 10.1158/1078-0432.CCR-11-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulson KG, Lemos BD, Feng B, Jaimes N, Penas PF, Bi X, et al. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J Invest Dermatol. 2009;129:1547–1555. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe. 2013;14:125–135. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494:145–160. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 15.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–437. doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 18.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 19.Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolani B, Gopalakrishnan R, Punj V, Matta H, Chaudhary PM. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene. 2013 doi: 10.1038/onc.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73:6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyspianska BS, Bannister AJ, Barbieri I, Nangalia J, Godfrey A, Calero-Nieto FJ, et al. BET protein inhibition shows efficacy against JAK2V617F-driven neoplasms. Leukemia. 2014;28:88–97. doi: 10.1038/leu.2013.234. [DOI] [PubMed] [Google Scholar]
- 29.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19:6183–6192. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Da Costa D, Agathanggelou A, Perry T, Weston V, Petermann E, Zlatanou A, et al. BET inhibition as a single or combined therapeutic approach in primary paediatric B-precursor acute lymphoblastic leukaemia. Blood Cancer J. 2013;3:e126. doi: 10.1038/bcj.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013;73:3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picaud S, Wells C, Felletar I, Brotherton D, Martin S, Savitsky P, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Z, McDermott A, Shao L, Kannan A, Morgan M, Stack BC, Jr, et al. Chronic mTOR activation promotes cell survival in Merkel cell carcinoma. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin ZKA, Shao Q, Stack B, Jr, James YS, Gao L. Dual mTOR inhibitor INK128 represses tumor growth in xenograft Merkel cell carcinoma mouse models. 2014 [Google Scholar]
- 38.Rothenberg ME, Clarke MF, Diehn M. The Myc connection: ES cells and cancer. Cell. 2010;143:184–186. doi: 10.1016/j.cell.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 39.Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. Merkel Cell Polyomavirus Small T Antigen Controls Viral Replication and Oncoprotein Expression by Targeting the Cellular Ubiquitin Ligase SCF(Fbw7.) Cell Host Microbe. 2013;14:125–135. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin ZMA, Shao LJ, Kannan A, Morgan M, Stack BC, Moreno M, Davis DA, Cornelius LA, Gao L. Chronic mTOR activation promotes cell survival in Merkel cell carcinoma. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, et al. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene. 2013 doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci U S A. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller NJ, Bhatia S, Parvathaneni U, Iyer JG, Nghiem P. Emerging and Mechanism-Based Therapies for Recurrent or Metastatic Merkel Cell Carcinoma. Curr Treat Options Oncol. 2013 doi: 10.1007/s11864-013-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krasagakis K, Fragiadaki I, Metaxari M, Kruger-Krasagakis S, Tzanakakis GN, Stathopoulos EN, et al. KIT receptor activation by autocrine and paracrine stem cell factor stimulates growth of merkel cell carcinoma in vitro. J Cell Physiol. 2011;226:1099–1109. doi: 10.1002/jcp.22431. [DOI] [PubMed] [Google Scholar]
- 46.Krasagakis K, Kruger-Krasagakis S, Eberle J, Tsatsakis A, Tosca AD, Stathopoulos EN. Co-expression of KIT receptor and its ligand stem cell factor in Merkel cell carcinoma. Dermatology. 2009;218:37–43. doi: 10.1159/000173704. [DOI] [PubMed] [Google Scholar]
- 47.Brunner M, Thurnher D, Pammer J, Geleff S, Heiduschka G, Reinisch CM, et al. Expression of VEGF-A/C, VEGF-R2, PDGF-alpha/beta, c-kit, EGFR, Her-2/Neu, Mcl-1 and Bmi-1 in Merkel cell carcinoma. Mod Pathol. 2008;21:876–884. doi: 10.1038/modpathol.2008.63. [DOI] [PubMed] [Google Scholar]
- 48.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 49.Evans AG, French CA, Cameron MJ, Fletcher CD, Jackman DM, Lathan CS, et al. Pathologic characteristics of NUT midline carcinoma arising in the mediastinum. Am J Surg Pathol. 2012;36:1222–1227. doi: 10.1097/PAS.0b013e318258f03b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET-bromodomain inhibition of MYC-amplified Medulloblastoma. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.