Abstract
Post-translational modifications (PTMs) are widely used by eukaryotes to control the enzymatic activity, localization or stability of their proteins. Traditionally, it was believed that the broad biochemical diversity of the PTMs is restricted to eukaryotic cells, which exploit it in extensive networks to fine-tune various and complex cellular functions. During the last decade, the advanced detection methods of PTMs and functional studies of the host–pathogen relationships highlight that bacteria have also developed a large arsenal of PTMs, particularly to subvert host cell pathways to their benefit. Legionella pneumophila, the etiological agent of the severe pneumonia legionellosis, is the paradigm of highly adapted intravacuolar pathogens that have set up sophisticated biochemical strategies. Among them, L. pneumophila has evolved eukaryotic-like and rare/novel PTMs to hijack host cell processes. Here, we review recent progress about the diversity of PTMs catalyzed by Legionella: ubiquitination, prenylation, phosphorylation, glycosylation, methylation, AMPylation, and de-AMPylation, phosphocholination, and de-phosphocholination. We focus on the host cell pathways targeted by the bacteria catalyzed PTMs and we stress the importance of the PTMs in the Legionella infection strategy. Finally, we highlight that the discovery of these PTMs undoubtedly made significant breakthroughs on the molecular basis of Legionella pathogenesis but also lead the way in improving our knowledge of the eukaryotic PTMs and complex cellular processes that are associated to.
Keywords: post-translational modification, Legionella pneumophila, Dot/Icm effectors, host cell pathways hijacking, Legionella virulence
INTRODUCTION
Post-translational modifications (PTMs) are widely used by eukaryotes to control quickly, locally and specifically the enzymatic activity, localization or stability of their proteins, and thus to fine-tune key factors of the cellular biology to environmental changes. Eukaryotic PTMs involve diverse modifications of specific residues of the protein by the covalent addition of simple or complex chemical groups; they include the addition of chemical group (e.g., phosphate, methyl, or acetate), more complex molecules (e.g., carbohydrates or lipids), the covalent linkage of small proteins (e.g., ubiquitin), and the irreversible hydrolysis of a specific peptide bond between two amino acids, or proteolysis (for review, see Walsh et al., 2005). PTMs are catalyzed by specific enzymes and most of them are reversed by antagonistic catalytic activities. Traditionally, it was believed that the broad biochemical diversity of the PTMs is restricted to complex eukaryotic cells, which exploit it in extensive networks to control various and complex cellular functions. During the last decade, the advanced detection methods of PTMs, including the modified peptides enrichment combined with high accuracy mass spectrometry, the pathogen genomes sequencing that predicts PTMs activities, and the functional studies of the host–pathogen relationships highlight that bacteria have also developed a large arsenal of PTMs, particularly to subvert host cell pathways to their benefit, to escape to the host defences, and finally to promote their replication (for review, see Ribet and Cossart, 2010a,b).
Legionella pneumophila, the etiological agent of the severe pneumonia legionellosis, is a paradigm of highly adapted intravacuolar pathogens that have set up sophisticated biochemical strategies to hijack host cell processes. Legionella pathogenic strains (i) emerge from the environment after intracellular multiplication in protozoans, especially in amoebae; (ii) are disseminated by contaminated aerosols; and (iii) can infect alveolar macrophages of its accidental human host. Within environmental phagocytic cells and human macrophages, L. pneumophila evades endocytic degradation (Horwitz and Maxfield, 1984; Clemens et al., 2000), controls the innate immune response, especially the NF-κB pathway (Schmeck et al., 2007; Shin et al., 2008), and triggers the biogenesis of a Legionella-containing vacuole (LCV), a rough endoplasmic reticulum-like compartment permissive for its intracellular replication (Horwitz, 1983; Kagan and Roy, 2002). Crucial for hijacking host cell vesicle trafficking necessary for LCV biogenesis, and subsequently for intracellular multiplication of L. pneumophila, is the Dot/Icm Type 4 Secretion System (T4SS; Marra et al., 1992; Andrews et al., 1998) that translocates into the host cell cytosol over 275 bacterial proteins, named effectors (Zhu et al., 2011). Many Dot/Icm effectors harbor eukaryotic domains (Cazalet et al., 2004), such as protein–protein interaction domains and enzymatic activity-associated domains, in particular for PTMs such as methylation, phosphorylation, ubiquitination, and glycosylation, which support that L. pneumophila has evolved eukaryotic-like PTMs to hijack host cell processes.
Here, we review recent progress about the diversity of PTMs catalyzed by Legionella. We focus on the host cell pathways targeted by the bacteria-catalyzed PTMs and we stress the importance of the PTMs in the Legionella infection strategy.
DIVERSITY OF PTMs CATALYZED BY L. pneumophila
A key finding of the L. pneumophila genome analysis was the identification of a large number of proteins similar to eukaryotic proteins. The wide variety of these proteins includes enzymatic activity–associated domains for various PTMs such as phosphorylation, glycosylation, methylation, prenylation, ubiquitination, reversible AMPylation, and phosphocholination of host cell proteins to modulate cellular functions (Table 1).
Table 1.
Diversity of PTMs catalyzed by Dot/Icm effectors of Legionella pneumophila.
| PTMs | Mechanism | Effector name | Motif | Target | Reference |
|---|---|---|---|---|---|
| Phosphorylation | 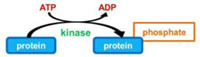 |
||||
| LegK1 | STPK | IκB | Ge et al. (2009); Hervet et al. (2011) | ||
| LegK2 | STPK | Hervet et al. (2011) | |||
| LegK3 | STPK | Hervet et al. (2011) | |||
| LegK4 | STPK | Hervet et al. (2011) | |||
| Methylation | 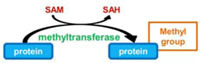 |
LegAS4/RomA | SET domain | H3 | Li et al. (2013), Rolando et al. (2013b) |
| Prenylation | 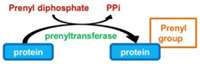 |
AnkB/LegAU13/Ceg27 | CAAX | Price et al. (2010a) | |
| Ubiquitination | 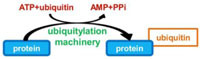 |
||||
| LubX/LegU2 | U-box | Clk1 /SidH | Kubori et al. (2008) | ||
| AnkB/LegAU13 /Ceg27 | F-box | Skp1/ParvB | Price et al. (2009), Lomma et al. (2010) | ||
| LegU1 | F-box | BAT3 | Ensminger and Isberg (2010) | ||
| SidC | Hsu et al. (2014) | ||||
| Glycosylation | 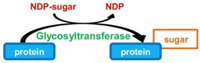 |
||||
| Lgt1 | Coiled- coil | eEF1A | Belyi et al. (2006) | ||
| Lgt2 | Coiled- coil | eEF1A | Belyi et al. (2008), Aktories (2011) | ||
| Lgt3/Legc5 | Coiled- coil | eEF1A | Belyi et al. (2008), Aktories (2011) | ||
| SetA | Heidtman et al. (2009) | ||||
| AMPylation | 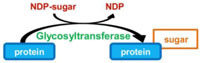 |
SidM/DrrA | Rab1 | Müller et al. (2010) | |
| DeAMPylation | 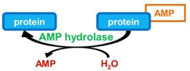 |
SidD | Rab1 | Neunuebel et al. (2011); Tan et al. (2011) | |
| Phosphocholination | 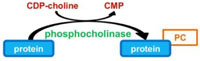 |
AnkX /AnkN /LegA8 | Ankyrin | Rab1 | Mukherjee et al. (2011) |
| Dephosphocholination |  |
Lem3 | Rab1 | Tan et al. (2011) |
STPK, Ser/Thr protein kinase.
PROTEIN PHOSPHORYLATION
Phosphorylation–dephosphorylation of proteins represents a powerful regulatory mechanism of cellular activity. Indeed, intensive research has revealed that eukaryotes contain numerous interconnected signal transduction networks in which protein phosphorylation plays a dominant role for controlling essential functions, such as growth, cell cycle and apoptosis, in response to extracellular stimuli and stresses. It consists in the reversible covalent addition of a phosphate group, from the phosphate donor ATP, to specific residues of a target protein, the most frequent being hydroxyl groups of serine, threonine or tyrosine residues. The phosphoester bond is catalyzed by eukaryotic protein kinases that share a common catalytic domain characterized by 11 conserved Hanks’s subdomains (Hanks, 2003). Conversely, phosphatases hydrolyze the phosphoester bond, thereby releasing the phosphate group and restoring the acceptor amino acid in its unphosphorylated form.
The genomes of the six sequenced L. pneumophila strains, Philadelphia, Lens, Paris, Corby, Alcoy, and 130b, have been reported to encode four putative eukaryotic-like serine/threonine kinases, named LegK1–LegK4 (Cazalet et al., 2004; de Felipe et al., 2005; D’Auria et al., 2010; Schroeder et al., 2010). Alignment with several eukaryotic protein kinases revealed residues that are highly conserved in the Hanks’ subdomains, including the glycine-rich loop and the invariant lysine in subdomains I and II, which are essential for binding and correct orientation of the phosphate donor ATP. In vitro phosphorylation assays confirmed that these kinases were functional for autophosphorylation and/or phosphorylation of the classical substrate for eukaryotic kinases Myelin-basic protein (Hervet et al., 2011; Table 1).
PROTEIN ALKYLATION
Protein alkylation consists in the addition of alkyl substituents on specific amino acids. The common alkyl groups transferred are the methyl (C1) or the C15 (farnesyl)/C20 (geranyl–geranyl) isoprenyl groups, leading to protein methylation and protein prenylation, respectively.
Protein methylation typically takes place on arginine or lysine residues in the protein sequence. Arginine can be methylated once or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylated arginine) or one on both nitrogens (symmetric dimethylated arginine) by peptidylarginine methyltransferases (PRMTs). Lysine can be methylated once, twice, or three times by lysine methyltransferases (Walsh et al., 2005). Protein methylation has been extensively studied in the histones. The transfer of methyl groups from S-adenosyl methionine (SAM) to histones is catalyzed by SET domain-containing proteins. This protein family is characterized by an ∼130 amino acid-long SET domain that possesses catalytic activity toward the ε-amino group of lysine residues. In vivo, lysine methylation can be dynamically regulated by the opposing actions of lysine methyltransferases and lysine demethylases (Herz et al., 2013). L. pneumophila genome analysis revealed that all the five strains Philadelphia, Lens, Paris, Corby, and Alcoy encode each an orthologous protein encoding a SET domain that show 95–100% sequence identity over the entire length (Cazalet et al., 2004). In vitro assays recently demonstrated that Lpp1683 in Paris strain and Lpg1718 in Philadelphia strain, display histone methyltransferase activity toward the histone H3 substrate (Li et al., 2013; Rolando et al., 2013b; Table 1).
Prenylation, i.e., addition of a farnesyl (C15) or a geranyl–geranyl (C20) group, is a PTM that covalently links a lipid moiety at the cysteine residue of the CAAX motif in the C-terminal region of proteins (where C represents cysteine and A an aliphatic amino acids). The Ras GTPases, Rab small GTPases, and protein kinases superfamilies have members that can be prenylated on cysteine thiolate side chains. The lipid anchors drive the modified proteins to partition more to membranes, thus controlling their subcellular localization (Walsh et al., 2005). Interestingly, bioinformatic approaches identified 11–12 (depending on the strains) different Legionella proteins containing a CAAX motif in the C terminus, which have been so called Pel proteins for Prenylated effectors of Legionella (Ivanov et al., 2010; Price et al., 2010a,b). Six of these proteins had highly conserved homologs across all Legionella stains, whereas four of the proteins were unique for either the Philadelphia or Lens strain. Host farnesyltransferase and class I geranylgeranyltransferase were both involved in the lipidation of the Legionella CAAX motif proteins, among which AnkB from L. pneumophila AA100 and Philadelphia Lp01 (Ivanov et al., 2010; Price et al., 2010a,b; Table 1).
PROTEIN UBIQUITINATION
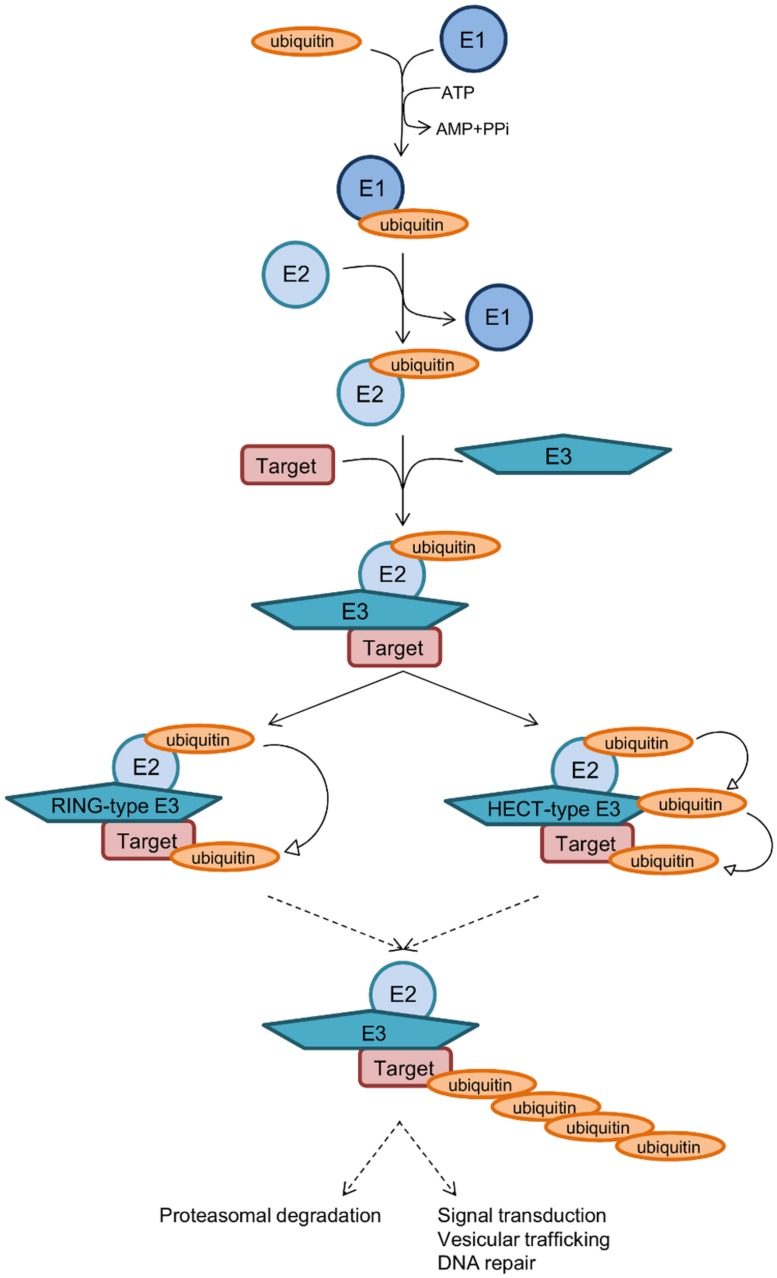
Ubiquitination consists in the addition of one or several ubiquitins on a target protein, most frequently on lysine residue, although linkages on cysteine, serine or threonine, or on the N-terminal amino group have also been reported. Ubiquitin is a small protein of 9 kDa, which contains itself seven lysines; all of these lysine residues can be used as a target for the addition of another ubiquitin moiety, thus leading to polyubiquitination. Polyubiquitin chains built up through Lys48 side chains are commonly associated with proteasome binding and degradation of the modified protein, whereas chains tethered through Lys63 participate in signal transduction, vesicular trafficking or DNA repair (Hochstrasser, 2009). The conjugation of ubiquitin requires different enzymes (Figure 1): E1 activating enzymes that bind ubiquitin in a ATP dependent manner; E2 conjugation enzymes that bind ubiquitin in a thioester bond; E3 ubiquitin ligases are then required to catalyze the efficient transfer of the activated ubiquityl protein tag to Lys side chains of target proteins. There are two different families of E3 ubiquitin ligases, the HECT family and the RING/U-Box family. In the RING family, some of the enzyme 3 are multicomponent catalysts, such as the SCF E3s that consist in four subunits: the invariable subunit Skp1, the central core component Cullin, the RING finger protein Rbx1/Roc1 and the variable F-Box protein that serves as a receptor for the target protein, such providing selectivity for a given protein (Lorick et al., 1999). There are several 100 isoforms of such E3 ubiquitin ligases in higher eukaryotes, which allow subtle discrimination among many target proteins selected for ubiquitination.
FIGURE 1.
Sequential steps of ubiquitination. The small protein ubiquitin is first activated by an E1 enzyme in an ATP-dependent manner, then transferred to an E2 conjugating enzyme. Two main classes of E3 enzymes, the RING, and HECT classes, transfer differently the ubiquitin to a specific protein substrate. Polyubiquitin chains are then built up through Lys48 side chains or through Lys63 of ubiquitin, which directs the protein to proteasome degradation or participate in signal transduction, vesicular trafficking, or DNA repair.
Genome analysis of the L. pneumophila strains Paris and Philadelphia revealed they encode a protein, named LubX, containing two U-box domains similar to eukaryotic E3 ubiquitin ligases (Cazalet et al., 2004). Indeed, in vitro and in host cells, LubX functions as an ubiquitin ligase (Kubori et al., 2008). Moreover, L. pneumophila encodes several proteins with predicted F-Box motifs (de Felipe et al., 2005). The best characterized is the AnkB protein, that is conserved in the five sequenced L. pneumophila strains (Table 1). Genome sequence of L. pneumophila Philadelphia reveals the presence of at least another four F-box containing proteins (Price and Kwaik, 2010). These include Lpg2224 (PpgA), Lpg2525 (MavK), LicA, and LegU1. Finally, the Dot/Icm effector SidC has been recently reported to define a unique family of ubiquitin ligase (Hsu et al., 2014). While the amino acid sequence of SidC does not exhibit significant homology with any known protein, the crystal structure of its N-terminal domain revealed a canonical catalytic triad C46-H444-D446 found in cystein-based proteases and deubiquitinases. Unexpectedly, in vitro assays demonstrated that SidC exhibits ubiquitin ligase rather than protease or deubiquitinase activity. More precisely, SidC catalyzes the formation of high–molecular-weight ubiquitinated conjugates in a manner that is dependent on the catalytic residue C46. Authors further showed that the SidC paralog SdcA has also ubiquitin ligase activity (Hsu et al., 2014; Table 1).
PROTEIN GLYCOSYLATION
O-glycosylation and N-glycosylation of proteins are very common in eukaryotes but only O-glycosylation has been described to date in the two major groups of bacterial toxins. O-glycosylation consists in the modification of serine or threonine residues. A 60 kDa protein that exihibited UDP-glycosyltransferase activity toward a 50 kDa protein from HeLa cell lysates was firstly purified from L. pneumophila (Belyi et al., 2003). This protein, named Lgt1 (for Legionella glycosyltransferase 1) contains a DXD motif, which is conserved in many prokaryotic and eukaryotic glucosyltransferases. In L. pneumophila strains Philadelphia, Lens, Paris, and Corby, two other proteins very similar to Lgt1 were then identified; they were thus called Lgt2 and Lgt3 and were shown to exhibit the same glycosylase activity (Belyi et al., 2008; Aktories, 2011). An additional protein, namely SetA, possesses a functional glycosyltransferase domain (Heidtman et al., 2009). However, its target in the host cell has not been yet identified (Table 1).
REVERSIBLE PROTEIN AMPylation
AMPylation or adenylylation is the addition of an adenosine monophosphate (AMP) group from ATP onto a threonine, tyrosine, or serine residue of a protein. This PTM was first and recently discovered on host cell proteins infected by Vibrio parahaemolyticus and Histophilus somni (Worby et al., 2009; Yarbrough et al., 2009). This activity involves a conserved domain, called Fic domain (for filamentation induced by cAMP domain), which was originally described in Escherichia coli as a stress response protein associated with filamentous bacterial growth in the presence of cAMP (Komano et al., 1991). The Fic domain is also found in eukaryotic proteins, and AMPylation has now been shown to be naturally occurring in eukaryotic cells (Kinch et al., 2009; Roy and Mukherjee, 2009; Worby et al., 2009; Yarbrough and Orth, 2009; Yarbrough et al., 2009). Despite the lack of a consensus Fic domain on its sequence and that its amino acid sequence did not suggest its function, the protein SidM from L. pneumophila has been recently shown to possess AMPylase activity (Müller et al., 2010). More precisely, its N-terminal domain exhibits structural similarities with the C-terminal domain of the glutamine synthase adenylyl transferase, which leads the authors to speculate that the N-terminal region of SidM might have AMPylase activity toward the small GTPase Rab1, the substrate of its GEF domain. Indeed, in vitro assays and mass spectrometry analysis demonstrates that SidM, more precisely its N-terminal domain, AMPylates Rab1 on the Tyr77 residue (Müller et al., 2010; Table 1).
AMPylation is a reversible process. Two independent groups simultaneously identified SidD from L. pneumophila as the first protein exhibiting a deAMPylase activity, by using two distinct approaches. Neunuebel et al. (2011) observed that a whole cell lysate from L. pneumophila but not from E. coli efficiently removes in vitro radiolabeled AMP from AMPylated Rab1. Given that genes of functionally linked proteins tend to be clustered on bacterial genomes, they deleted the immediately nearby sidM gene, namely sidD gene, and they observed that the whole cell lysate from the sidD mutant was not able anymore to deAMPylate SidM (Neunuebel et al., 2011). On the other hand, Tan and Luo identified SidD as a protein capable of suppressing the toxicity of the AMPylase SidM to yeast (Tan and Luo, 2011). Both groups demonstrated that SidD removes the AMP moiety from Tyr77 of Rab1, thus reversing the effect of SidM on this small GTPase activity (Table 1).
REVERSIBLE PHOSPHOCHOLINATION
As mentioned above, the Fic domain is associated to enzymes that trigger AMPylation of target proteins. In silico analysis revealed that another protein from L. pneumophila, namely AnkX, contains a Fic domain. However, mass spectrometry demonstrated that AnkX promotes a novel PTM, namely phosphocholination rather than AMPylation (Mukherjee et al., 2011). Phosphocholination consists in the covalent link of a phosphocholine group to a serine residue (Table 1). More precisely, AnkX catalyzes the phosphocholination of Ser76 of the small GTPase Rab1, immediately upstream the Tyr77 AMPylated by SidM (Mukherjee et al., 2011).
Like AMPylation, phosphocholination is a reversible PTM. Tan et al. (2011) recently identified the Dot/Icm effector Lem3 as a protein capable to rescue the growth of yeast transformed by AnkX expression vector, which suggested that Lem3 was able to antagonize the activity of phosphocholination of AnkX. Indeed, in vitro assays demonstrated that Lem3 reverses the AnkX-dependent phosphocholination of Rab1 by removing the phosphocholine moiety from Rab1 (Tan et al., 2011; Table 1).
PTMs FOR Legionella CONTAINING VACUOLE BIOGENESIS
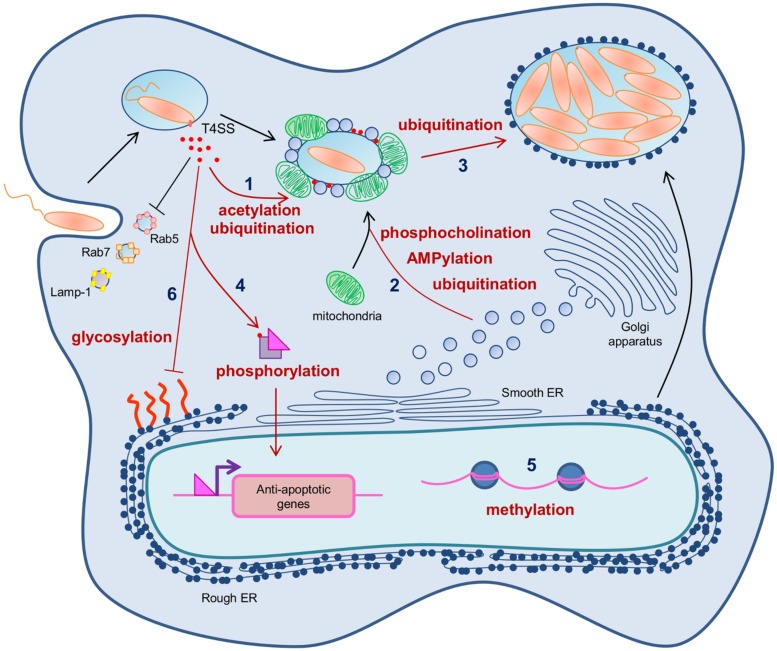
Legionella-containing vacuole biogenesis is a main trait of Legionella intracellular fate that allows the bacteria to generate a niche permissive for intracellular replication. Within 15 min of uptake, the LCV is surrounded and fused with ER-derived smooth vesicles and mitochondria (Horwitz, 1983), and 4 h post-contact it is decorated by host cell ribosomes (Horwitz and Silverstein, 1981; Roy and Tilney, 2002), thus resulting in a replication-permissive vacuole (Figure 2). Legionella-containing vacuole biogenesis mobilizes complex molecular mechanisms that are strictly dependent on the Dot/Icm T4SS and its exceptionally high number of effectors. PTMs of both host cell proteins and Dot/Icm effectors play a key role in the fine-tuned orchestration of this infection step.
FIGURE 2.
Post-translational modifications controlling various infectious cycle steps of Legionella pneumophila. Immediatly after uptake of the bacteria, L. pneumophila secretes a high number of Dot/Icm effectors into the host cell cytosol. (1) L. pneumophila exploits the host prenylation apparatus to alkylate some of these effectors and target them to the LCV surface. The effector LubX is able to ubiquitinate some effectors thus addressing them to proteasomal degradation and controlling their temporal presence on the LCV during infection. (2) Four Dot/Icm effectors reversibly AMPylate and phosphocholinate the host Rab1 small GTPase, thus controlling its activity to promote ER recruitment on the LCV, a prerequisite feature to make the phagosome a replicative niche. SidC and its paralog SdcA monoubiquitinate Rab1 and catalyze polyubiquitin chains formation necessary for ER recruitment on the LCV. (3) The Dot/Icm effector AnkB functions as a platform for the docking of polyubiquitinated proteins to the LCV membrane, thus promoting proteasome-mediated generation of free amino acids essential as energy and carbon sources for L. pneumophila intracellular proliferation. (4) The Dot/Icm effector LegK1 phosphorylates IκB thus mimicking the host IKKs, and triggering the activation of the NF-κB pathway and the transcription of NF-κB dependent genes. (5) The Dot/Icm effector LegAS4/RomA trigger the methylation of the histone H3, thus inducing epigenetic changes and subsequent transcriptional control of host genes. (6) Several Dot/Icm effectors exhibits glycosidase activity toward eEF1A, thus inhibiting the host cell translation.
Dot/Icm EFFECTORS ACETYLATION AND UBIQUITINATION SPATIO-TEMPORALLY CONTROL THEIR RECRUITMENT ON THE LCV
Given the high number of effectors, it could be assumed that both translocation into the host cell cytosol, organelles addressing, and degradation of each effector must be controlled such that it could sequentially participate to the LCV biogenesis. Many Dot/Icm effectors are targeted to the LCV surface. L. pneumophila uses [PI(4)P] to anchor some Dot/Icm substrates, such as SidC and SidM to the cytoplasmic face of LCV (Ragaz et al., 2008; Brombacher et al., 2009). Another way for L. pneumophila to address injected effectors to the LCV membrane, is the exploitation of the host cell prenylation apparatus (Ivanov et al., 2010; Price et al., 2010a). The Dot/Icm substrate AnkB, of strains L. pneumophila AA100 and Philadelphia Lp01, contains a CAAX motif. During infection, the CAAX motif of AnkB is modified by the host farnesylation machinery (Ivanov et al., 2010; Price et al., 2010a). Expression of a CAAX substituted-variant results in defective anchoring of AnkB to the LCV, severe defects in intracellular replication, and attenuation of intrapulmonary proliferation in a mouse model, thus demonstrating that the farnesyl-dependent vacuolar location of AnkB is essential to its role in the infectious cycle of L. pneumophila (Price et al., 2009).
In addition to the appropriate addressing of Dot/Icm effectors, a specific temporal control of their stability in the host cell is carried out during L. pneumophila infection. In that purpose, L. pneumophila interferes with the ubiquitin system to address some effectors to proteasomal degradation. LubX is a Dot/Icm effector containing two U-box domains and functions as a E3 ubiquitin ligase toward the cellular Clk1 protein. However, cellular consequences of ubiquitination of Clk1 remain unknown (Kubori et al., 2008). LubX was also shown to bind and polyubiquitinate in vitro SidH, another Dot/Icm effector. It mediates proteasomal degradation of SidH in infected cells. Thus, LubX is considered like a metaeffector that controls in space and time, the presence of another effector, by using ubiquitination PTM (Kubori et al., 2010).
AMPylation AND PHOSPHOCHOLINATION CONTROLS THE GTPase Rab1 ACTIVATION FOR ER RECRUITMENT ON THE LCV
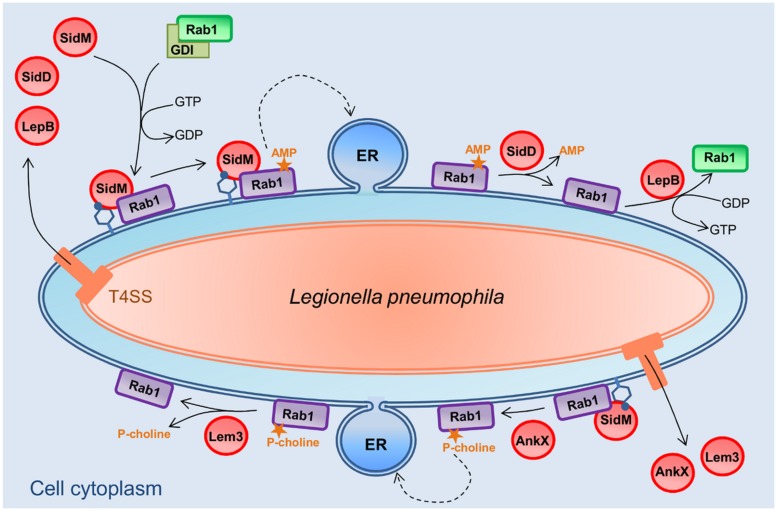
One main characteristic of the LCV is that it is fused with ER-derived vesicles. The manipulation of host cell vesicular trafficking by L. pneumophila is strictly dependent of the Dot/Icm T4SS. In particular, some of Dot/Icm substrates target host cell small GTPases. Among them, the effector SidM interacts with Rab1, and its GEF and GDF activities result in Rab1 release from GDI (Ingmundson et al., 2007), and in LCV membrane associated GTP-coupled Rab1 (Arasaki et al., 2012), respectively (Figure 3). An additional PTM-associated enzymatic activity of the multifunctional protein SidM has recently been revealed. The N-terminal domain of SidM, which exhibits similarities with the catalytic domain of glutamin synthetase adenylyl transferase, modifies the tyrosine 77 of Rab1 by AMPylation or adenylylation, i.e., the addition of a AMP moiety (Müller et al., 2010). This PTM inhibits GAP-stimulated GTP hydrolysis, thus locking Rab1 in the GTP-bound active state, and finally allows ER recruitment at the surface of the LCV (Figure 3).
FIGURE 3.
Post-translational modifications of small GTPase Rab1 by L. pneumophila for LCV biogenesis. The ER recruitment on the LCV is orchestrated by four Dot/Icm effectors-mediated PTMs. SidM releases Rab1 from GDI with its GEF activity. SidM then modifies Rab1 by AMPylation, i.e., the addition of a AMP moiety. This PTM locks Rab1 in the GTP-bound active state, and finally allows ER recruitment at the surface of the LCV. SidD removes AMP from Rab1, making it accessible for GAP activities, such as that exhibited by LepB. LepB promotes GTP hydrolysis of Rab1, removing it from the LCV. AnkX harbors a novel PTM activity, the phosphocholination, that transfers a phosphocholine moiety to Rab1, resulting in the same effect that the SidM-mediated AMPylation, i.e., locking Rab1 in the active form. The Dot/Icm effector Lem3 possesses an antagonistic activity to that of AnkX by removing the phosphocholine from Rab1.
The activation of Rab1 by SidM is counteracted by two others Dot/Icm effectors, SidD and LepB (Figure 3). SidD removes AMP from Tyr77 of Rab1 (Neunuebel et al., 2011; Tan and Luo, 2011) by a protein phosphatase-like catalytic mechanism, as suggested by structural analysis (Rigden, 2011). DeAMPylation of Rab1 makes it accessible for GAP activities, such as that exhibited by LepB. Despite any similarity with eukaryotic Rab-GAPs, LepB harbors a Rab1-specific GAP activity that promotes GTP hydrolysis and subsequent removal of Rab1 from the LCV (Ingmundson et al., 2007; Mihai Gazdag et al., 2013). Consistent with the SidD-dependent action of LepB, the phenotype of a lepB mutant is similar to that of a sidD mutant, i.e., a prolonged localization of Rab1 on the LCV (Neunuebel et al., 2011).
Two additional Dot/Icm effectors target the Rab1 GTPase for PTM and participate to the temporal control of its activation during Legionella infection cycle. AnkX harbors a novel PTM activity, namely phosphocholination, that transfers a phosphocholine moiety from CDP-choline to serine 76 of Rab1, preceding the SidM-modified tyrosine (Mukherjee et al., 2011; Figure 3). Although the biological effect of this PTM of Rab1 is not completely deciphered, it results in the same biochemical consequence as the SidM-mediated AMPylation, i.e., locking Rab1 in the active form. Like AMPylation, phosphocholination is reversible. The Dot/Icm effector Lem3 has been recently shown to possess an antagonistic activity to that of AnkX by removing the phosphocholine from the Ser76 of Rab1 (Tan et al., 2011; Goody et al., 2012). Thus, Rab1 is directly targeted and its activity is controlled by four different Dot/Icm effectors that catalyze different PTMs.
It is noteworthy that ubiquitination, mediated by the Dot/Icm effectors SidC and SdcA, could also participate to ER recruitment on the LCV. The Dot/Icm effector SidC and its paralog SdcA were proposed to function as vesicle fusion tethering factors involved in the recruitment of ER vesicles on the LCV (Luo and Isberg, 2004; Ragaz et al., 2008). Recently, infection by a WT L. pneumophila strain was shown to mediate the mono-ubiquitination of Rab1 on lysine 187 (Horenkamp et al., 2014). Given that cells infected with the double mutant sdcA-sidC did not exhibit this Rab1 PTM, it was assumed that Rab1 ubiquitination required the Dot/Icm effectors SidC and SdcA. However, ectopic expression of SidC or SdcA alone in HEK293 cells did not result in Rab1 ubiquitination, which suggests that neither SidC nor SdcA are E3 ubiquitin ligases. By contrast, another study demonstrated that the N-terminal domain of SidC exhibits ubiquitin ligase activity that catalyzes polyubiquitin chains formation and is necessary for ER recruitment on the LCV (Hsu et al., 2014). According to the authors, SidC does not seem to directly target Rab1 but more likely triggers a remodeling of proteins composition at the surface of the LCV. Although the mono-ubiquination of Rab1 would be mediated by an indirect unknown mechanism and that the substrates and impact of the SidC/SdcA-catalyzed polyubiquitination remains unclear, both these studies highlight the role of ubiquitination in ER recruitment on the LCV.
UBIQUITINATION OF BAT3 COULD MITIGATE THE EFFECTS OF DISRUPTING NORMAL VESICULAR TRAFFICKING
Legionella pneumophila co-opts host vesicular trafficking during infection, in particular to recruit ER on the LCV surface. It can be assumed that some Dot/Icm substrates are translocated to protect host cells against the cytotoxic stress generated by the ER traffic hijacking. Among them, the Dot/Icm effector LegU1 contains an F-box domain and interferes with ubiquitin signaling. It can be integrated into the functional SCF1 complex that confers E3 ubiquitin ligase activity. It specifically targets the host chaperone protein BAT3, a key regulator of the ER stress response. LegU1 associates with BAT3 and mediates its polyubiquitination in vitro (Ensminger and Isberg, 2010). Moreover, another translocated L. pneumophila protein, Lpg2160, plays a role in this complex by binding both the SCF complex and BAT3. These results suggest that this multicomplex formation leads to BAT3 ubiquitination, probably to modulate the ER stress response during infection (Ensminger and Isberg, 2010).
PTMs FOR SUSTAINING Legionella INTRACELLULAR REPLICATION
In the rough ER-like compartment of the LCV, L. pneumophila proliferates in a so-called replicative form until vacuolar nutrients become limiting. Polyubiquination of host cell proteins mediated by a Legionella effector has been recently proposed to be a bacterial strategy dedicated to generate sources of carbon and energy needed for microbial proliferation in vivo (Price et al., 2011). Indeed, in addition to the CAAX farnesylation motif described above, AnkB from L. pneumophila Philadelphia strain harbors two ankyrin (ANK) protein–protein interaction domains and a F-box domain. In both macrophages and protozoa, AnkB functions as a bona fide F-box protein where it recruits Skp1, thus subverting the host SCF1 complex and functionning as a platform for the docking of polyubiquitinated proteins to the LCV membrane. The polyubiquitinated proteins assembled by AnkB on the LCV are preferentially enriched for Lys48-linked polyubiquitinated proteins, which is a hallmark for proteasomal degradation, that generate 2–24 amino acid peptides (Price et al., 2011). Interestingly, substitution of Lys48 to Arg abolishes the decoration of the LCV with polyubiquitinated proteins and blocks intracellular proliferation. Moreover, inhibition of proteasome, or host amino- and oligo-peptidases that degrade the short peptides generated by proteasomal degradation, blocks intracellular proliferation (Price et al., 2011). However, both inhibitions are bypassed by excess amino acid supplementation. Together these data strongly support that AnkB promotes proteasome-mediated generation of free amino acids essential as energy and carbon sources for L. pneumophila intracellular proliferation (Figure 2).
It is noteworthy that in some L. pneumophila strains such as the strain Paris, AnkB does not contain the CAAX motif. Given they do not localize to the LCV, these AnkB homologues might not be key effectors of L. pneumophila that generate nutrients for intracellular growth. A yeast two-hybrid screen and co-immunoprecipitation analysis identified ParvB as one target of the L. pneumophila F-box protein AnkB encoded by strain Paris. ParvB, or affixin, is known to play important roles in focal adhesion, cell spreading and motility. Surprisingly, expression of AnkB led to a decrease of ubiquitination of ParvB. Thus, it was proposed that L. pneumophila modulates ubiquitination of ParvB by competing with eukaryotic E3 ligases for the specific protein–protein interaction site of ParvB. However, the role of AnkB in the infectious cycle of L. pneumophila strain Paris remains unknown (Lomma et al., 2010).
PTMs FOR CONTROLLING HOST CELL GENES EXPRESSION
PHOSPHORYLATION OF IκB FOR CONTROLLING THE NF-κB DEPENDENT GENES TRANSCRIPTION
After phagocytosis, L. pneumophila resides and replicates in the LCV within the host cytosol. Consequently, survival of the host cell is necessary for successful replication. To prevent cell death, some Dot/Icm translocated substrates interfere with pro-death pathways (Laguna et al., 2006; Banga et al., 2007). A second mechanism of preventing host cell death during infection is to stimulate the NF-κB pathway, which results in up-regulation of genes encoding anti-apoptotic proteins (Karin and Lin, 2002). NF-κB homo- and heterodimers are master transcription regulators of the mammalian innate immune response that control the expression of almost 400 genes (Karin and Lin, 2002; Ahn and Aggarwal, 2005; Hayden and Ghosh, 2008). NF-κB activation can result from sensing of pathogen associated molecular patterns (PAMPs) by the pattern recognition receptors (PRRs), which leads to activation of IκB kinases (IKKs). Once activated, IKKs phosphorylate IκB family members, inhibitory proteins that are bound to NF-κB subunits in the cell cytoplasm, thus triggering IκB ubiquitination, IκB degradation, and subsequent translocation of NF-κB into the nucleus (Hayden and Ghosh, 2008). L. pneumophila infection results in increased Dot/Icm-dependent transcription of NF-κB subunits as well as NF-κB regulated genes including pro-inflammatory cytokines and antagonists of apoptosis (Losick and Isberg, 2006; Abu-Zant et al., 2007; Shin et al., 2008). Besides the engagement of PRRs with PAMPs, direct targeting of the pathway by a Dot/Icm effector, namely LegK1, has been demonstrated (Ge et al., 2009; Figure 2). LegK1 efficiently phosphorylates IκB on Ser-32 and Ser-36 both in vitro and in cells, thus mimicking the host IKKs. Ectopic expression of the protein in mammalian cells results in activation of an NF-κB-dependent promoter. The kinase activity is necessary for this activation, as a point mutation in the ATP binding domain or a catalytic residue abolishes NF-κB activity (Ge et al., 2009; Losick et al., 2010), and cell-free reconstitution revealed that LegK1 stimulated NF-κB activation in the absence of IKKs (Ge et al., 2009).
METHYLATION OF HISTONES FOR CONTROLLING HOST CELL GENE TRANSCRIPTION
Legionella-containing vacuoles are studded with an increasing number of ribosomes during the first 8 h after bacterial internalization, after which the bacteria start to replicate in the vacuole. Besides, transcription of rRNA genes (rDNAs) in the nucleolus is known to be regulated by epigenetic chromatin modifications including histone H3 lysine (de)methylation. Recently, the Dot/Icm LegAS4 from L. pneumophila Philadelphia strain was shown to localize in the host nucleolus and promoted rDNA transcription (Li et al., 2013; Figure 2). LegAS4 contains an active SET-domain-sharing 35% sequence identity with eukaryotic NSD2/3 Lys Histone Methyltransferases of the SET2 family. In vitro studies on histone H3 substrate, using methylation-specific H3 antibodies, show that LegAS4 catalyses dimethylation of histone H3 on Lys4 (H3K4me2). Consistently, ectopic expression of LegAS4 in human cells is associated with increased levels of H3K4me2 at rDNA promoters and the activation of the transcription of these genes. LegAS4’s association with rDNA chromatin is mediated by interaction with host HP1a/c. Docking of LegAS4 to these regions through binding to HP1, and subsequent methylation of H3K4, might convert the epigenetically silent state of rDNA genes to an active state methylated H3. Stimulation of rDNA transcription might contribute to bacterial replication in two flavors. The enforced higher proliferation potential of infected cells, resulting from activation of rDNA transcription, could provide a better niche for bacterial replication. On the other hand, intracellular bacteria could exploit host ribosome activity for its own survival advantages (Li et al., 2013).
Interestingly, mass spectrometry analysis revealed that the equivalent effector of LegAS4 from the L. pneumophila strain Paris, named RomA (for regulator of methylation A) trimethylates in vitro Lys14 of H3 (H3K14me3), a histone mark not previously described in mammals (Rolando et al., 2013b). This epigenetic mark was confirmed by systematic site-directed mutagenesis of the lysine residues in the N-terminal tail of H3. It is noteworthy that while H3 methylation was almost completely decreased when H3 was mutated on K14, RomA enzymatic activity appeared to be also reduced on H3 carrying a mutated K4. However, no H3K4 methylation was revealed in western-blot probed with anti-H3K4me2 or H3K4me3 antibodies, thus suggesting that RomA only targets K14 of H3 and that H3K4 methylation could influence H3K14 methylation by being part of the motif required by RomA to bind to its substrate (Rolando et al., 2013b). By promoting a burst of H3K14me3, RomA decreases H3K14 acetylation, which is an activating mark, thus leading to repression of host gene expression. In addition, ChIP-seq analysis identified 4,870 H3K14 methylated promoter regions, including at innate immune genes, during Legionella infection.
Recently, the H3K14-specific methylation was shown to be conserved in cells infected by seven different strains of L. pneumophila, including the Philadelphia 1 (Lp02) strain (Rolando and Buchrieser, 2014). Thus, there is more likely no different specificity of the methylation activities of LegAS4 and RomA, and despite slight discrepancies about the biochemistry and the biological effect of these effectors, both these studies highlight the key role of histone PTMs during Legionella infection (Figure 2).
GLYCOSYLATION OF EF1A FOR INHIBITING THE HOST CELL TRANSLATION
In addition to controlling the host cell gene transcription, L. pneumophila is able to inhibit the overall host cell translation. L. pneumophila encodes three Dot/Icm effectors, namely Lgt1, Lgt2, and Lgt3, that monoglycosylate the serine residue Ser53 of the GTPase domain of the host translational factor eEF1A (eukaryotic Elongation Factor 1A), leading to the inhibition of protein synthesis, and consequently to the death of the host cell (Belyi et al., 2006). Although EF1A glycosylation seems to promote L. pneumophila pathogenesis, the biological role of this PTM remains to be addressed. Because their activities cause the host cell death, glysosyltransferases are usually considered like bacterial toxins rather than molecular tools that hijack host cell pathways to the benefit of the bacteria. However, it can be assumed that the inhibition of host cell protein synthesis leads to the overall decrease of the host metabolism, which promotes the ability of the bacteria to overcome the cellular response and consequently to replicate (Belyi et al., 2011). Moreover, it has been recently shown that Lgt1, Lgt2, Lgt3 plus two others Dot/Icm effectors, SidI and SidL that respectively, interacts with eEF1A and eEF1B (Shen et al., 2009) and inhibit protein synthesis by an unknown mechanism, are critical to control the host cell transcription response to Legionella infection (Fontana et al., 2011). In fact, these Dot/Icm effectors decrease the overall translation of host cell proteins, among which the NF-κB inhibitor IκB, thus promoting the activation of the NF-κB pathway. In that way, glycosylation of eEF1A by these effectors and thus inhibition of host cell translation could potentiate the activation of the NF-κB pathway, already controlled by the IκB phosphorylation by LegK1, as described above.
CONCLUSION
Given PTMs play key roles in the cellular biology, it is not surprising that interference with host PTMs is a strategy widely used by bacterial pathogens to not only escape from host cell defences but also to hijack host cell pathways to their benefit. However, recent technological progresses in the detection of PTMs and advanced functional studies of the host–bacteria relationship highlighted an unexpected diversity of the PTMs triggered by bacteria and the complexity of these processes in host–pathogen interactions, thus making studies of bacteria-mediated PTMs an emerging field of research.
Legionella pneumophila is a paradigm of a pathogenic bacteria that evolved sophisticated biochemical strategies to successfully infect and replicate into professional bacteria killer phagocytic cells. In fact, L. pneumophila is a unique example for the co-evolution of a bacterium with environmental hosts, namely amoeba, that results in the acquisition of many genes encoding proteins that can be secreted by the Dot/Icm T4SS and trigger diverse PTMs into the host cells. Indeed, the large repertoire of Dot/Icm effectors enables the bacteria to phosphorylate, alkylate, ubiquitinate, glycosylate, AMPylate, and phosphocholinate specific host cell proteins. Noteworthy, L. pneumophila also catalyze PTMs of its own proteins, namely some of Dot/Icm effectors, to control their localization and/or their stability in the host cell, and subsequently their activity during the infection. Importantly, despite PTMs are usually catalyzed by eukaryotic-like proteins, some of them are performed by enzymes that do not exhibit similarity with their eukaryotic counterparts. More interestingly, research on Dot/Icm effectors functional roles lead to the discovery of a new PTM, namely the reversible phosphocholination, that may also be used by eukaryotic cells to modulate cellular functions, as previously suggested by studies that detected phosphoryl-choline substituted peptides secreted by nematodes and from mammalian cells residing in the placenta (Lovell et al., 2007; Grabitzki et al., 2008). AMPylation had been also discovered by studying infections by V. parahaemolyticus and H. somni, a human pathogen and the causal agent of septicemia in cattle, respectively (Worby et al., 2009; Yarbrough et al., 2009). These discoveries reveal that studies of the relationship between pathogenic bacteria and their host cells could lead the way to improve our knowledge of the eukaryotic PTMs and complex cellular processes that are associated to.
Interestingly, L. pneumophila targets host proteins that have been already described to be preferential targeted for bacterial-induced PTMs. This is the case of regulators of the NF-κB pathway, which allows the bacteria to control both anti-apoptotic genes and host immune response, like previously demonstrated for Shigella flexneri (Kim et al., 2005), Salmonella typhimurium (Le Negrate et al., 2008), L. monocytogenes (Gouin et al., 2010), and Yersinia species (Mittal et al., 2006). Moreover, L. pneumophila joins those bacteria that secrete effectors manipulating PTMs at histones tails, allowing a fine-tuned regulation of host genes transcription (Hamon and Cossart, 2008; Bierne, 2013; Rolando et al., 2013a). These recent insights highlight the key role of both these processes and their control by PTMs in the pathogenic bacteria–host relationships.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Ph.D. grant of Céline Michard was provided by the Programme Avenir Lyon Saint-Etienne (ANR-11-IDEX-0007) of Université de Lyon, within the program “Investissements d’Avenir” operated by the French National Research Agency (ANR). We apologize to authors whose works cannot be cited here because of space limitations.
REFERENCES
- Abu-Zant A., Jones S., Asare R., Suttles J., Price C., Graham J., et al. (2007). Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol. 9 246–264 10.1111/j.1462-5822.2006.00785.x [DOI] [PubMed] [Google Scholar]
- Ahn K. S., Aggarwal B. B. (2005). Transcription factor NF-kappaB: a sensor for smoke and stress signals. Ann. N. Y. Acad. Sci. 1056 218–233 10.1196/annals.1352.026 [DOI] [PubMed] [Google Scholar]
- Aktories K. (2011). Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 9 487–498 10.1038/nrmicro2592 [DOI] [PubMed] [Google Scholar]
- Andrews H. L., Vogel J. P., Isberg R. R. (1998). Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K., Toomre D. K., Roy C. R. (2012). The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe 11 46–57 10.1016/j.chom.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S., Gao P., Shen X., Fiscus V., Zong W. X., Chen L., et al. (2007). Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. U.S.A. 104 5121–5126 10.1073/pnas.0611030104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi I., Popoff M. R., Cianciotto N. P. (2003). Purification and characterization of a UDP-glucosyltransferase produced by Legionella pneumophila. Infect. Immun. 71 181–186 10.1128/IAI.71.1.181-186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi Y., Jank T., Aktories K. (2011). Effector glycosyltransferases in Legionella. Front. Microbiol. 2:76 10.3389/fmicb.2011.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi Y., Niggeweg R., Opitz B., Vogelsgesang M., Hippenstiel S., Wilm M., et al. (2006). Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc. Natl. Acad. Sci. U.S.A. 103 16953–16958 10.1073/pnas.0601562103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi Y., Tabakova I., Stahl M., Aktories K. (2008). Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J. Bacteriol. 190 3026–3035 10.1128/JB.01798-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H. (2013). Nuclear microbiology–bacterial assault on the nucleolus. EMBO Rep. 14 663–664 10.1038/embor.2013.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E., Urwyler S., Ragaz C., Weber S. S., Kami K., Overduin M., et al. (2009). Rab1 guanine nucleotide exchange factor sidm is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284 4846–4856 10.1074/jbc.M807505200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C., Rusniok C., Bruggemann H., Zidane N., Magnier A., Ma L., et al. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36 1165–1173 10.1038/ng1447 [DOI] [PubMed] [Google Scholar]
- Clemens D. L., Lee B. Y., Horwitz M. A. (2000). Mycobacterium tuberculosis and Legionella pneumophila phagosomes exhibit arrested maturation despite acquisition of Rab7. Infect. Immun. 68 5154–5166 10.1128/IAI.68.9.5154-5166.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria G., Jiménez-Hernández N., Peris-Bondia F., Moya A., Latorre A. (2010). Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11:181 10.1186/1471-2164-11-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe K. S., Pampou S., Jovanovic O. S., Pericone C. D., Ye S. F., Kalachikov S., et al. (2005). Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187 7716–7726 10.1128/JB.187.22.7716-7726.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger A. W., Isberg R. R. (2010). E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect. Immun. 78 3905–3919 10.1128/IAI.00344-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M. F., Banga S., Barry K. C., Shen X., Tan Y., Luo Z. Q., et al. (2011). Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 7:e1001289 10.1371/journal.ppat.1001289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Xu H., Li T., Zhou Y., Zhang Z., Li S., et al. (2009). A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc. Natl. Acad. Sci. U.S.A. 106 13725–13730 10.1073/pnas.0907200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody P. R., Heller K., Oesterlin L. K., Müller M. P., Itzen A., Goody R. S. (2012). Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 31 1774–1784 10.1038/emboj.2012.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E., Adib-Conquy M., Balestrino D., Nahori M. A., Villiers V., Colland F., et al. (2010). The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the I{kappa}B kinase subunit IKK{alpha}. Proc. Natl. Acad. Sci. U.S.A. 107 17333–17338 10.1073/pnas.1007765107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabitzki J., Ahrend M., Schachter H., Geyer R., Lochnit G. (2008). The PCome of Caenorhabditis elegans as a prototypic model system for parasitic nematodes: identification of phosphorylcholine-substituted proteins. Mol. Biochem. Parasitol. 161 101–111 10.1016/j.molbiopara.2008.06.014 [DOI] [PubMed] [Google Scholar]
- Hamon M. A., Cossart P. (2008). Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4 100–109 10.1016/j.chom.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Hanks S. (2003). Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 4 111 10.1186/gb-2003-4-5-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M. S., Ghosh S. (2008). Shared principles in NF-kappaB signaling. Cell 132 344–362 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Heidtman M., Chen E. J., Moy M. Y., Isberg R. R. (2009). Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 11 230–248 10.1111/j.1462-5822.2008.01249.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervet E., Charpentier X., Vianney A., Lazzaroni J.-C., Gilbert C., Atlan D., et al. (2011). Protein kinase LegK2 Is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect. Immun. 79 1936–1950 10.1128/IAI.00805-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz H. M., Garruss A., Shilatifard A. (2013). SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38 621–639 10.1016/j.tibs.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (2009). Origin and function of ubiquitin-like proteins. Nature 458 422–429 10.1038/nature07958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horenkamp F. A., Mukherjee S., Alix E., Schauder C. M., Hubber A. M., Roy C. R., et al. (2014). Legionella pneumophila subversion of host vesicular transport by SidC effector proteins. Traffic 15 488–499 10.1111/tra.12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. A. (1983). Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158 1319–1331 10.1084/jem.158.4.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. A., Maxfield F. R. (1984). Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99 1936–1943 10.1083/jcb.99.6.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. A., Silverstein S. C. (1981). Interaction of the Legionnaires’ disease bacterium (Legionella pneumophila) with human phagocytes. I. L. pneumophila resists killing by polymorphonuclear leukocytes, antibody, and complement. J. Exp. Med. 153 386–397 10.1084/jem.153.2.386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., Luo X., Qiu J., Teng Y. B., Jin J., Smolka M. B., et al. (2014). The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. Proc. Natl. Acad. Sci. U.S.A. 111 10538–10543 10.1073/pnas.1402605111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A., Delprato A., Lambright D., Roy C. (2007). Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450 365–369 10.1038/nature06336 [DOI] [PubMed] [Google Scholar]
- Ivanov S. S., Charron G., Hang H. C., Roy C. R. (2010). Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem. 285 34686–34698 10.1074/jbc.M110.170746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J., Roy C. (2002). Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4 945–954 10.1038/ncb883 [DOI] [PubMed] [Google Scholar]
- Karin M., Lin A. (2002). NF-kappaB at the crossroads of life and death. Nat. Immunol. 3 221–227 10.1038/ni0302-221 [DOI] [PubMed] [Google Scholar]
- Kim D. W., Lenzen G., Page A. L., Legrain P., Sansonetti P. J., Parsot C. (2005). The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 102 14046–14051 10.1073/pnas.0504466102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch L. N., Yarbrough M. L., Orth K., Grishin N. V. (2009). Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS ONE 4:e5818 10.1371/journal.pone.0005818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Utsumi R., Kawamukai M. (1991). Functional analysis of the fic gene involved in regulation of cell division. Res. Microbiol. 142 269–277 10.1016/0923-2508(91)90040-H [DOI] [PubMed] [Google Scholar]
- Kubori T., Hyakutake A., Nagai H. (2008). Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67 1307–1319 10.1111/j.1365-2958.2008.06124.x [DOI] [PubMed] [Google Scholar]
- Kubori T., Shinzawa N., Kanuka H., Nagai H. (2010). Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 6:e1001216 10.1371/journal.ppat.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna R. K., Creasey E. A., Li Z., Valtz N., Isberg R. R. (2006). A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. U.S.A. 103 18745–18750 10.1073/pnas.0609012103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Negrate G., Faustin B., Welsh K., Loeffler M., Krajewska M., Hasegawa P., et al. (2008). Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J. Immunol. 180 5045–5056 10.4049/jimmunol.180.7.5045 [DOI] [PubMed] [Google Scholar]
- Li T., Lu Q., Wang G., Xu H., Huang H., Cai T., et al. (2013). SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO Rep. 14 733–740 10.1038/embor.2013.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomma M., Dervins-Ravault D., Rolando M., Nora T., Newton H. J., Sansom F. M., et al. (2010). The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 12 1272–1291 10.1111/j.1462-5822.2010.01467.x [DOI] [PubMed] [Google Scholar]
- Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 96 11364–11369 10.1073/pnas.96.20.11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V. P., Haenssler E., Moy M. Y., Isberg R. R. (2010). LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 12 1083–1097 10.1111/j.1462-5822.2010.01452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V. P., Isberg R. R. (2006). NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 203 2177–2189 10.1084/jem.20060766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell T. M., Woods R. J., Butlin D. J., Brayley K. J., Manyonda I. T., Jarvis J., et al. (2007). Identification of a novel mammalian post-translational modification, phosphocholine, on placental secretory polypeptides. J. Mol. Endocrinol. 39 189–198 10.1677/JME-07-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. Q., Isberg R. R. (2004). Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101 841–846 10.1073/pnas.0304916101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra A., Blander S. J., Horwitz M. A., Shuman H. A. (1992). Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. U.S.A. 89 9607–9611 10.1073/pnas.89.20.9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihai Gazdag E., Streller A., Haneburger I., Hilbi H., Vetter I. R., Goody R. S., et al. (2013). Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep. 14 199–205 10.1038/embor.2012.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R., Peak-Chew S. Y., Mcmahon H. T. (2006). Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc. Natl. Acad. Sci. U.S.A. 103 18574–18579 10.1073/pnas.0608995103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Liu X., Arasaki K., Mcdonough J., Galán J. E., Roy C. R. (2011). Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477 103–106 10.1038/nature10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. P., Peters H., Blümer J., Blankenfeldt W., Goody R. S., Itzen A. (2010). The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329 946–949 10.1126/science.1192276 [DOI] [PubMed] [Google Scholar]
- Neunuebel M. R., Chen Y., Gaspar A. H., Backlund P. S., Yergey A., Machner M. P. (2011). De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333 453–456 10.1126/science.1207193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T., Al-Khodor S., Al-Quadan T., Santic M., Habyarimana F., Kalia A., et al. (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5:e1000704 10.1371/journal.ppat.1000704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T., Al-Quadan T., Santic M., Jones S. C., Abu Kwaik Y. (2010a). Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med. 207 1713–1726 10.1084/jem.20100771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T., Jones S. C., Amundson K. E., Kwaik Y. A. (2010b). Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of legionella pneumophila. Front. Microbiol. 1:131 10.3389/fmicb.2010.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T., Al-Quadan T., Santic M., Rosenshine I., Abu Kwaik Y. (2011). Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334 1553–1557 10.1126/science.1212868 [DOI] [PubMed] [Google Scholar]
- Price C. T., Kwaik Y. A. (2010). Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front. Microbiol. 1:122 10.3389/fmicb.2010.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragaz C., Pietsch H., Urwyler S., Tiaden A., Weber S., Hilbi H. (2008). The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 10 2416–2433 10.1111/j.1462-5822.2008.01219.x [DOI] [PubMed] [Google Scholar]
- Ribet D., Cossart P. (2010a). Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 143 694–702 10.1016/j.cell.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D., Cossart P. (2010b). Post-translational modifications in host cells during bacterial infection. FEBS Lett. 584 2748–2758 10.1016/j.febslet.2010.05.012 [DOI] [PubMed] [Google Scholar]
- Rigden D. J. (2011). Identification and modelling of a PPM protein phosphatase fold in the Legionella pneumophila deAMPylase SidD. FEBS Lett. 585 2749–2754 10.1016/j.febslet.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Rolando M., Buchrieser C. (2014). Legionella pneumophila type IV effectors hijack the transcription and translation machinery of the host cell. Trends Cell Biol. 24 771–778 10.1016/j.tcb.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Rolando M., Rusniok C., Margueron R., Buchrieser C. (2013a). [Host epigenetic targeting by Legionella pneumophila]. Med. Sci. (Paris) 29 843–845 10.1051/medsci/20132910010 [DOI] [PubMed] [Google Scholar]
- Rolando M., Sanulli S., Rusniok C., Gomez-Valero L., Bertholet C., Sahr T., et al. (2013b). Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13 395–405 10.1016/j.chom.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Roy C. R., Mukherjee S. (2009). Bacterial FIC Proteins AMP Up Infection. Sci. Signal. 2 pe14. 10.1126/scisignal.262pe14 [DOI] [PubMed] [Google Scholar]
- Roy C. R., Tilney L. G. (2002). The road less traveled: transport of Legionella to the endoplasmic reticulum. J. Cell Biol. 158 415–419 10.1083/jcb.200205011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeck B., N’Guessan P. D., Ollomang M., Lorenz J., Zahlten J., Opitz B., et al. (2007). Legionella pneumophila-induced NF-kappaB- and MAPK-dependent cytokine release by lung epithelial cells. Eur. Respir. J. 29 25–33 10.1183/09031936.00141005 [DOI] [PubMed] [Google Scholar]
- Schroeder G. N., Petty N. K., Mousnier A., Harding C. R., Vogrin A. J., Wee B., et al. (2010). Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J. Bacteriol. 192 6001–6016 10.1128/JB.00778-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Banga S., Liu Y., Xu L., Gao P., Shamovsky I., et al. (2009). Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 11 911–926 10.1111/j.1462-5822.2009.01301.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Case C. L., Archer K. A., Nogueira C. V., Kobayashi K. S., Flavell R. A., et al. (2008). Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 4:e1000220 10.1371/journal.ppat.1000220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Arnold R. J., Luo Z. Q. (2011). Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U.S.A. 108 21212–21217 10.1073/pnas.1114023109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Luo Z. Q. (2011). Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475 506–509 10.1038/nature10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T., Garneau-Tsodikova S., Gatto G. J. (2005). Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 44 7342–7372 10.1002/anie.200501023 [DOI] [PubMed] [Google Scholar]
- Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., et al. (2009). The fic domain: regulation of cell signaling by adenylylation. Mol. Cell. 34 93–103 10.1016/j.molcel.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., Orth K. (2009). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323 269–272 10.1126/science.1166382 [DOI] [PubMed] [Google Scholar]
- Yarbrough M. L., Orth K. (2009). AMPylation is a new post-translational modiFICation. Nat. Chem. Biol. 5 378–379 10.1038/nchembio0609-378 [DOI] [PubMed] [Google Scholar]
- Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., et al. (2011). Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6:e17638 10.1371/journal.pone.0017638 [DOI] [PMC free article] [PubMed] [Google Scholar]