Abstract
Invadopodia are actin-driven membrane protrusions that show oscillatory assembly and disassembly causing matrix degradation to support invasion and dissemination of cancer cells in vitro and in vivo. Profilin1, an actin and phosphoinositide binding protein, is downregulated in several adenocarcinomas and it's been shown that its depletion enhances invasiveness and motility of breast cancer cells by increasing PI(3,4)P2 levels at the leading edge. In this study we show for the first time that depletion of profilin1 leads to an increase in the number of mature invadopodia and these assemble and disassemble more rapidly than in control cells. Previous work by Sharma et al. 2013a, has shown that the binding of the protein Tks5 with PI(3,4)P2 confers stability to the invadopodium precursor causing it to mature into a degradation-competent structure. We found that loss of profilin1 expression increases the levels of PI(3,4)P2 at the invadopodium and as a result, enhances recruitment of the interacting adaptor Tks5. The increased PI(3,4)P2 - Tks5 interaction accelerates the rate of invadopodium anchorage, maturation, and turnover. Our results indicate that profilin1 acts as a molecular regulator of the levels of PI(3,4)P2 and Tks5 recruitment in invadopodia to control the invasion efficiency of invadopodia.
Keywords: profilin1; PI(3,4)P2; Tks5; matrix degradation
INTRODUCTION
Through the years, there has been an improvement in the treatment of breast cancer patients. However, metastasis remains as the principal cause of death for these patients and improvements in prediction and treatment have been lacking. There is a need for more research to understand the basic mechanisms of metastasis.
To achieve the invasive phenotype and escape from the primary tumor, cancer cells form actin-based protrusions called invadopodia (Linder, 2007; Murphy and Courtneidge, 2011; Yamaguchi and Condeelis, 2007). These structures can degrade the basal membrane and the extracellular matrix by delivering matrix metalloproteinases (MMPs) to the membrane, a process that facilitates tumor cells entry into the blood stream and dissemination to distant organs (Gligorijevic et al., 2014, 2012; Yamaguchi and Condeelis, 2007). Invadopodia contain an actin-rich core and actin regulatory molecules including, cortactin, cofilin, Tks5, MMPs, neural Wiskott-Aldrich syndrome protein (N-WASP) and Arp2/3 complex (Clark et al., 2007; Gimona and Buccione, 2006; Magalhaes et al., 2011; Murphy and Courtneidge, 2011; Oser et al., 2009; Sakurai-Yageta et al., 2008; Yamaguchi et al., 2005). Invadopodium formation is a highly regulated multistep process that starts with the formation of an invadopodium precursor which is a structure enriched in all the proteins that constitute the invadopodium core but hasn't yet acquired the capacity to degrade the extracellular matrix. The invadopodium precursor will mature into a structure with proteolytic activity (mature invadopodium) upon integrin/Arg-mediated cofilin activation (Beaty et al., 2014, 2013; Mader et al., 2011; Oser et al., 2009). Several reports have shown that any dysregulation of the actin cytoskeleton leads to impaired invadopodium formation and matrix degradation (Beaty et al., 2013; Clark et al., 2007; Desmarais et al., 2009; Diaz et al., 2010; Liu et al., 2009; Magalhaes et al., 2011; Sakurai-Yageta et al., 2008; Sharma et al., 2013a; Yamaguchi et al., 2005).
Profilin1, a small actin-binding protein (12-15 KDa), is downregulated in several adenocarcinomas such as, breast (Janke et al., 2000), pancreatic (Grønborg et al., 2006) and hepatic (Wu et al., 2006) cancers but the signaling pathways affected by this down-regulation remain unclear. It has been previously reported that loss of expression of profilin1 increases motility and invasiveness of breast cancer cells (Bae et al., 2010, 2009; Ding et al., 2013; Zou et al., 2007). Profilin2, another isoform of profilin, binds to actin but with less affinity than profilin1 (Witke, 2004). Recently, Mouneimne et al. (2012) reported that profilin2 has an active role in motility and invasion of breast cancer cells. Profilins can regulate actin polymerization in various ways: they can inhibit actin polymerization by sequestering G-actin in a 1:1 complex, or they can promote actin polymerization by catalyzing the exchange of ADP- to ATP-G-actin and this complex (ATP-G-actin) binds to the free barbed end of the actin filament and profilin is dissociated from the complex (Goldschmidt-Clermont et al., 1992; Gutsche-Perelroizen, 1999; Kang et al., 1999; Pollard and Borisy, 2003; Witke, 2004). Besides actin, profilins can bind to other classes of ligands: poly-L-proline rich sequences and phosphoinositides. Through the binding to poly-L-proline rich sequences profilin can interact with many proteins involved in actin polymerization, including Enabled/vasodilator stimulated phosphoprotein (Ena/VASP), N-WASP, WASP-associated verprolin homology protein (WAVE), Diaphanous, Mena, among others (Ding et al., 2012; Rawe et al., 2006; Witke, 2004; Wittenmayer et al., 2004; Yang et al., 2000). Moreover, profilin1 binds to several membrane phosphoinositides with high affinity (PI(4,5)P2, PI(3,4)P2, PI(3,4,5)P3) in vitro (Lu et al., 1996). In a previous study, it was reported that the effect of profilin1 on breast cancer cell motility is mediated by its phosphoinositide interaction with PI(3,4)P2 (Bae et al., 2010, 2009).
Profilin1 has been shown to play an important role in actin regulation as well as in cell migration and invasion, but its role on invadopodium formation and function has not yet been investigated. In the present study, we investigated the role of profilin1 in invadopodium formation. We show that profilin1 down-regulation enhances invadopodium formation, maturation and matrix degradation through phosphoinositide PI(3,4)P2 binding and Tks5 recruitment. We propose that this mechanism explains how profilin1 can act as a negative regulator of breast cancer cell invasion.
RESULTS
Profilin1 but not profilin2 regulates invadopodium formation, maturation and matrix degradation in breast cancer cells
To determine whether profilin1 is involved in invadopodium formation and function in invasive breast cancer cells, we first transiently depleted profilin1 expression in MDA-MB-231 cells by a single oligo siRNA transfection. Western blot analysis of whole cell lysates confirmed that profilin1 levels were efficiently reduced by 85%, while the expression levels of profilin2 remained unaffected by profilin1 siRNA (Figure 1A).
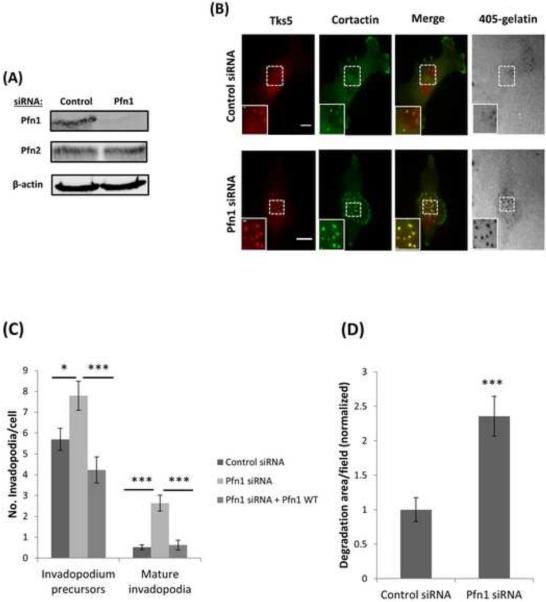
Figure 1. Profilin1 depletion increases the number of invadopodia and matrix degradation.
(A) MDA-MB-231 cells were transfected with control or Pfn1 siRNA and whole lysates were immunoblotted after 96 h of transfection with Pfn1, Pfn2 and β-actin antibodies. Pfn1 levels in Pfn1 siRNA sample was reduced by 85 %. (B) Representative images of MDA-MB-231 cells treated with control or Pfn1 siRNA and plated on 405-labeled gelatin and stained for Tks5 and cortactin as invadopodia markers. Bar, 10 μm. Inset shows magnified image of invadopodia in the box, bar, 1 μm. (C) Quantification of the number of invadopodium precursors and mature invadopodia per cell. n>800 invadopodia, n>130 cells; three independent experiments. The knockdown-rescue experiment was performed using a RNAi resistant profilin1-WT-GFP construct. n>300 invadopodia, n>65 cells; three independent experiments. *, p=0.017 (invadopodium precursors, Control siRNA vs Pfn1 siRNA), ***, p=0.00018 (invadopodium precursors, Pfn1 siRNA vs Pfn1siRNA + WT), ***p=4.35E-07 (mature invadopodia, Control siRNA vs Pfn1 siRNA), ***, p=1.29E-05 (mature invadopodia, Pfn1 siRNA vs Pfn1siRNA + WT). (D) Quantification of invadopodial matrix degradation area per field normalized to control. n>60 fields; three independent experiments. ***, p=0.000112 compared with control cells. Error bars indicate SEM.
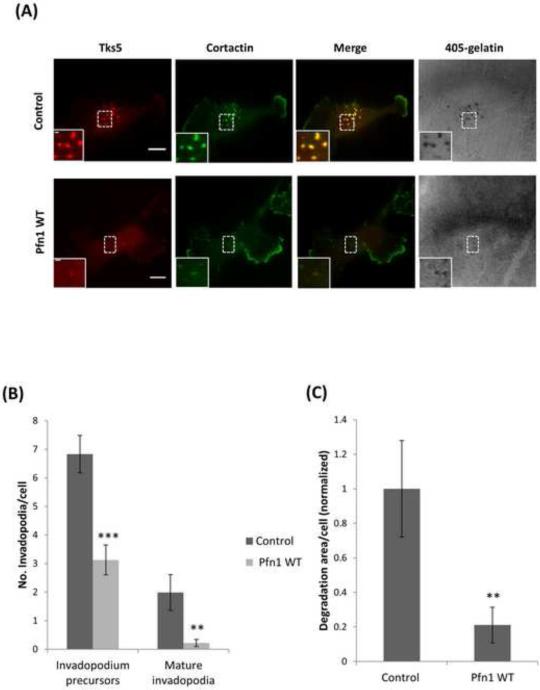
Next, we assayed profilin1 siRNA cells for invadopodia formation after overnight plating, and found that upon depletion of profilin1 the number of invadopodia were increased (Figure 1B). Then, we explored which invadopodium maturation stage was affected by profilin1 depletion. Our analyses showed that in both cases the number of mature invadopodia and invadopodium precursors was significantly increased as compared with control siRNA (Figure 1C). Consistent with these results, the matrix degradation area was also increased due to the increased number of mature invadopodia (Figure 1D). These results were reproduced by using a pool of siRNAs targeting profilin1 (Figure S1A-D). Then to confirm the specificity of our findings, we next performed a knockdown-rescue experiment. Cells were rescued by using a RNAi resistant profilin1-WT-GFP construct (Bae et al., 2010; Ding et al., 2009) (RNAi resistance of the rescue construct is demonstrated by immunoblot data as shown in Figure S1E). Rescuing knockdown cells restored the number of invadopodium precursors and mature invadopodia to the same level as in control cells (Figure 1C). Consistent with these phenotypes of profilin1-knockdown cells, we found that overexpression of profilin1-WT-GFP (Das et al., 2009; Zou et al., 2007) led to reduction in the numbers of invadopodia, mature invadopodia and invadopodium precursors, and the amount of matrix degradation when compared to control (Figure 2A-C). Overall, these results demonstrate that profilin1 negatively regulates invadopodium formation and maturation, and matrix degradation capacity of human breast cancer cells.
Figure 2. Overexpression of profilin1 decreases the number of invadopodia and matrix degradation.
(A) Representative images of MDA-MB-231 control cells and MDA-MB-231 cells transfected with profilin1-WT-GFP, plated on 405-labeled gelatin and stained for Tks5 and cortactin as invadopodia markers. Bar, 10 μm. Inset shows magnified image of invadopodia in the box, bar, 1 μm. (B) Quantification of the number of invadopodium precursors and mature invadopodia per cell. n>200 invadopodia, n>65 cells; three independent experiments. ***, p=1.84E-05 (invadopodium precursors), **, p=0.007 (mature invadopodia). (C) Quantification of invadopodial matrix degradation area per cell normalized to control. n>60 cells; three independent experiments. **, p=0.009 compared with control cells. Error bars indicate SEM.
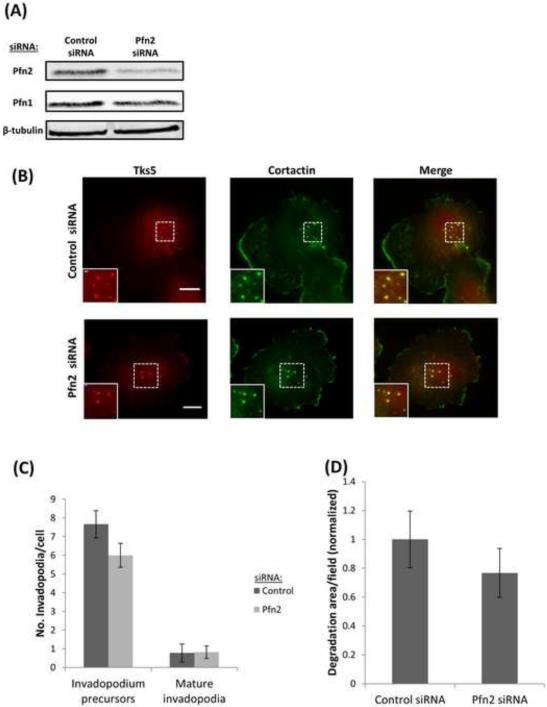
Interestingly, when profilin2 expression was selectively downregulated (72%) by using a pool of siRNA targeting profilin2 (Figure 3A), it did not affect the number of invadopodia, the numbers of mature invadopodia and invadopodium precursors, or the matrix degradation area (Figure 3B-D), further suggesting that profilin2 activity is not essential for invadopodium formation and function.
Figure 3. Profilin2 is not necessary for invadopodium formation.
(A) Western blot of cell lysates from MDA-MB-231 cells after 96 h of transfection with control or Pfn2 siRNA and blotted for Pfn2, Pfn1 and β-tubulin antibodies. Pfn2 levels in Pfn2 siRNA sample was reduced by 72 %. (B) Representative images of MDA-MB-231 cells plated on 405-labeled gelatin and stained for Tks5 and cortactin as invadopodia markers. Bar, 10 μm. Inset shows magnified image of invadopodia in the box, bar, 1 μm. (C) Quantification of the number of invadopodium precursors and mature invadopodia per cell. n>500 invadopodia, n>60 cells; three independent experiments. (D) Quantification of invadopodial matrix degradation area per field normalized to control. n>30 fields; three independent experiments.
Profilin1 depletion enhances local actin polymerization
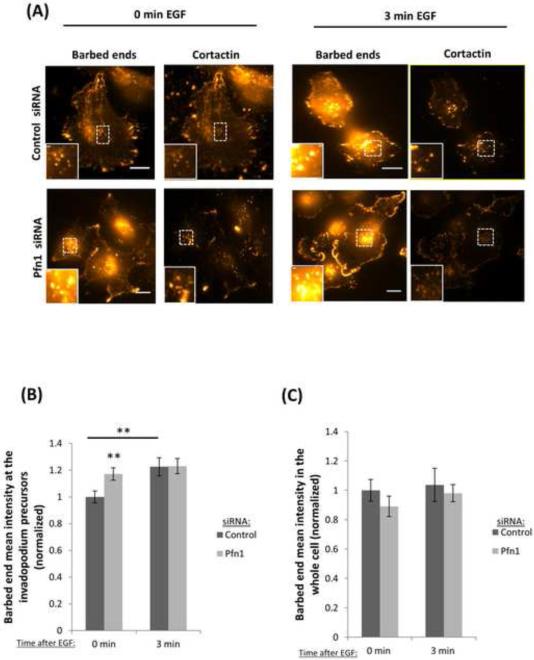
It has been shown that actin polymerization is a critical step in invadopodia formation (Bravo-Cordero et al., 2012; Liu et al., 2009; Magalhaes et al., 2011; Yamaguchi et al., 2005). Invadopodium formation starts with the assembly of a precursor, which must initiate actin polymerization within its structure. Previous work showed that carcinoma cells respond to EGF stimulation by increasing barbed ends at invadopodium precursors and lamellipodia. Based on the role of profilin1 on actin dynamics, we asked whether depletion of profilin1 has any effect on local actin polymerization (barbed end formation) during the early stages of invadopodium formation. In order to answer this question, we performed a barbed end assay using MDA-MB-231 cells serum-starved. Next, cells were stimulated with EGF to induce invadopodium precursors formation. Surprisingly, we found that the number of barbed ends in profilin1-knockdown cells was higher than control cells at baseline, reaching levels seen in the control cells only after EGF stimulation (Figure 4A-B). Quantification of barbed ends in the whole cell showed no differences between control and profilin1-depleted cells (Figure 4C), indicating that the change in barbed ends is specific to the invadopodium precursors. Next, we investigated if the early stages of invadopodium formation were affected by EGF stimulation. Interestingly, the number of invadopodium precursors formed in response to EGF stimulation, an event that does not require barbed end formation, was the same in control and profilin1-depleted cells (Figure 4D). Together these results show that profilin1 regulates actin polymerization at invadopodia.
Figure 4. Profilin1 depletion increases barbed end formation.
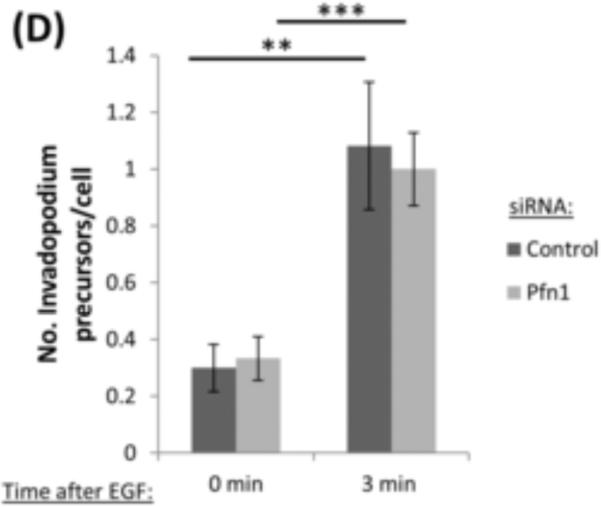
(A) Representative images of a barbed end assay in MDA-MB-231 cells transfected with control or Pfn1 siRNA and stimulated with EGF for 0 (untreated) or 3 minutes and stained for anti-biotin (barbed ends), cortactin and Arp2. Bar, 10 μm. Inset shows magnified image of barbed ends at invadopodium precursors, bar, 1μm. (B) Quantification of barbed ends intensity in response to EGF stimulation. Results are normalized to control at time 0 min. n>170 invadopodia; three independent experiments. **, p=0.0053 (Control siRNA 0 min vs Control siRNA 3 min) **, p=0.008 (Control siRNA 0 min vs Pfn1 siRNA 0 min). (C) Quantification of the mean barbed ends intensity in the whole cell in response to EGF stimulation. Results are normalized to control time 0 minutes. n>30 cells; three independent experiments. (D) Quantification of the number of invadopodium precursors in response to EGF stimulation. n>150 invadopodia, n>53 cells; three independent experiments. **, p=0.0016 (Control siRNA 0 min vs Control siRNA 3 min), ***, p= 2.8E-05 (Pfn1 siRNA 0 min vs Pfn1siRNA 3 min). Error bars indicate SEM.
Profilin1 regulates PI(3,4)P2 and PI(3,4)P2-dependent recruitment of Tks5 at the invadopodium
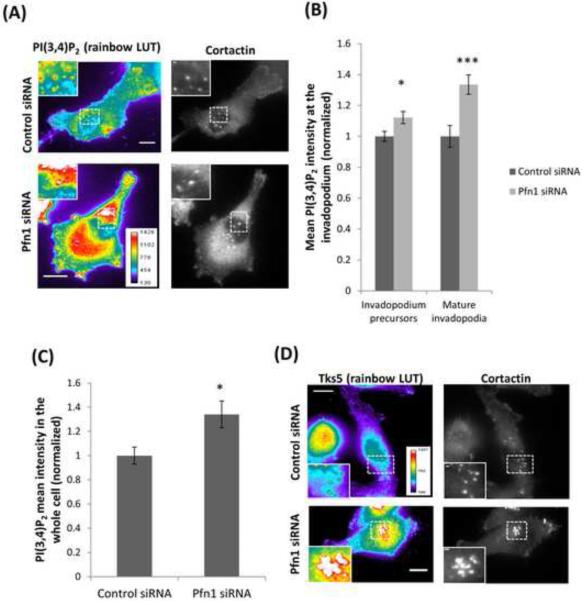
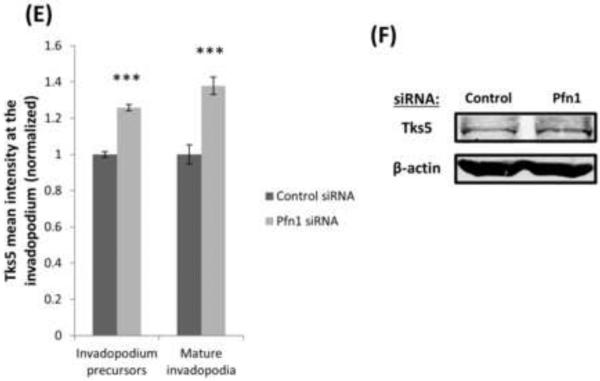
Profilin1 has affinity for various phosphoinositides. Previous work have been shown that profilin1 depletion induces a hypermotile phenotype in breast cancer cells via increasing membrane presentation of PI(3,4)P2 thereby promoting PI(3,4)P2- dependent recruitment of pro-migratory protein complexes to the leading edge (Bae et al., 2010; Ding et al., 2013, 2012). It was also reported that the binding of PI(3,4)P2 with the scaffold protein Tks5 stabilizes invadopodium precursors, allowing them to grow into a degradation-competent mature invadopodia (Sharma et al., 2013a). These observations prompted us to further examine whether profilin1 depletion affects PI(3,4)P2 accumulation and in turn, Tks5 recruitment at the invadopodium. Immunostaining data showed that profilin1 depletion enhanced total levels of PI(3,4)P2, specifically in mature invadopodia and invadopodium precursors (Figure 5A-C). Likewise, after profilin1 depletion Tks5 recruitment is also increased in mature invadopodia and invadopodium precursors (Figures 5D-E), but the total Tks5 protein levels remain unchanged (Figure 5F). Therefore this enrichment of Tks5 is specific to invadopodia and due to recruitment of Tks5. Together, these observations suggest that profilin1 plays an important role in regulating PI(3,4)P2-dependent recruitment of Tks5 to the invadopodium further leading to its maturation.
Figure 5. Profilin1 depletion enhances the levels of PI(3,4)P2 and Tks5.
(A) Images of MDA-MB-231 cells transfected with control or Pfn1 siRNA, plated on 405-labeled gelatin and stained for PI(3,4)P2 and cortactin. Bar, 10 μm. Insets show magnified image of invadopodia in the box, bar, 1μm. (B) Quantification of PI(3,4)P2 mean intensity at the invadopodium precursor and mature invadopodia and normalized to control. n>330 invadopodia; three independent experiments.*, p=0.017 (invadopodium precursors), ***, p=0.0005 (mature invadopodia). (C) PI(3,4)P2 mean intensity quantification in the whole cell and normalized to control. n>30 cells; three independent experiments.*, p=0.012. (D) Representative images of MDA-MB-231 cells transfected with control or Pfn1 siRNA, plated on 405-labeled gelatin and stained for Tks5 and cortactin as invadopodia markers. Bar, 10 μm. Insets show magnified image of invadopodia in the box, bar, 1μm. Rainbow lookup table (LUT) is used to highlight the difference in the expression levels of the proteins at invadopodia. (E) Quantification of Tks5 mean intensity at the invadopodium precursor and mature invadopodia and normalized to control. n>800 invadopodia; three independent experiments. ***, p=4.39E-30 (invadopodium precursors), ***, p=2.50E-07 (mature invadopodia). Error bars indicate SEM. (F) Western blot of whole cell lysates from MDA-MB-231 cells transfected with control or Pfn1 siRNA and blotted for Tks5 and β-actin antibodies.
Profilin1 regulates invadopodium dynamics and lifetime
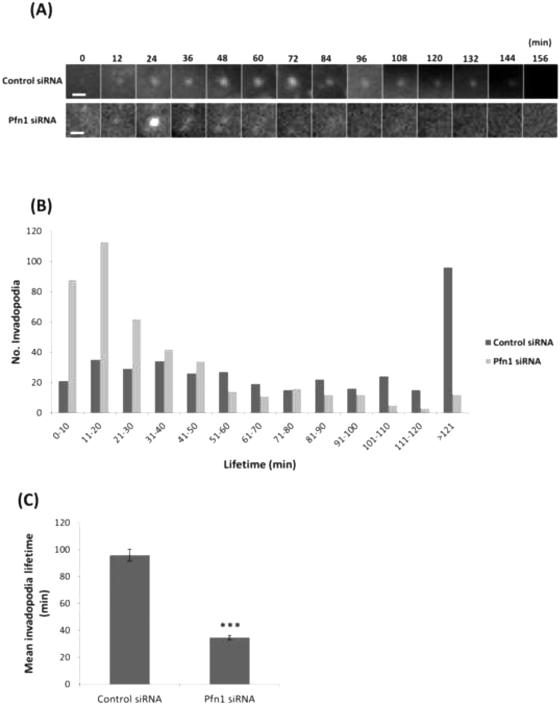
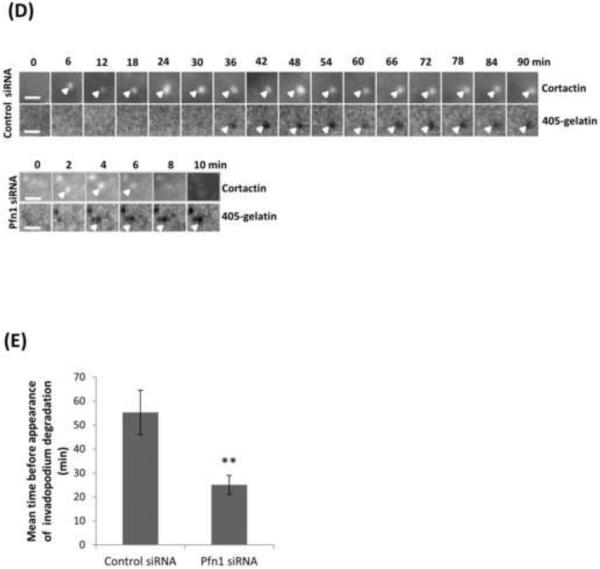
To directly visualize invadopodium dynamics, we performed time-lapse imaging of MDA-MB-231 cells treated with either control or profilin1 siRNA, followed by transfection with TagRFP-cortactin and GFP-Tks5 as invadopodia markers, and plated on 405-labeled gelatin. We found that upon profilin1 knockdown, cells form highly dynamic invadopodia that assemble and disassemble more rapidly than control cells (Videos 1-2). While control cells form long-lived invadopodia, profilin1-depleted cells form invadopodia that are unstable and disassemble shortly after formation (Figure 6A). Quantification of the lifetime confirms that profilin1-depleted cells form invadopodia with an average lifetime of 30 minutes, while the average lifetime of control cells is 90 minutes (Figure 6B-C). Consistent with these results, profilin1-depleted invadopodia started to degrade the matrix after 25 minutes of assembly, while for control invadopodia this process was longer, starting only after 55 minutes of assembly (Figure 6D-E and Videos 3-4). Thus, the rate of invadopodium turnover is faster in profilin1-knockdown cells than in control cells.
Figure 6. Profilin1-knockdown cells have short-lived invadopodia.
(A) Time-lapse montage during invadopodium assembly and disassembly of MDA-MB-231 cells treated with control or Pfn1 siRNA and transfected with TagRFP-cortactin and GFP-Tks5 as invadopodia markers. Cortactin channel is shown, bar, 5μm (see Videos 1-2). (B) Invadopodia lifetime quantification (minutes) for control and Pfn1 siRNA. n>380 invadopodia; two independent experiments. (C) Mean invadopodia lifetime quantification (minutes) for control and Pfn1 siRNA. ***, p=3.53E-33 compared with control cells. (D) Time-lapse montage during invadopodium assembly and disassembly and matrix degradation capacity of MDA-MB-231 cells treated with control or Pfn1 siRNA and transfected with TagRFP-cortactin and GFP-Tks5 as invadopodia markers. Cortactin and gelatin channels are shown, bar, 5 μm (see Videos 3-4). (E) Mean time before appearance of invadopodium degradation (minutes) by control and Pfn1 siRNA. n>21 invadopodia. **, p= 0.005. Error bars indicate SEM.
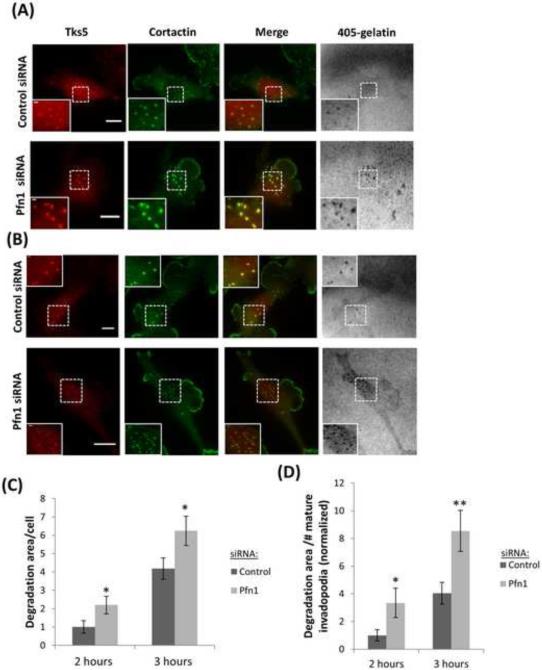
Profilin1-depleted cells degrade the ECM at earlier stages of assembly
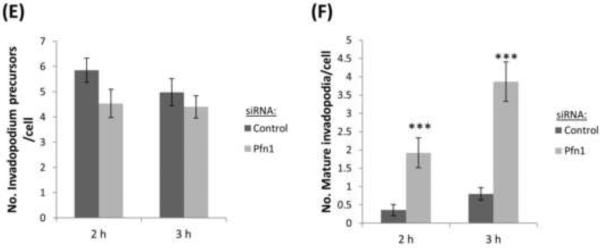
Previous work correlated long-lived invadopodia with higher matrix degradation activity (Yamaguchi et al., 2005), but in this study the lifetime of invadopodia showed a decrease upon profilin1 depletion, while the appearance of matrix degradation was shortly after invadopodia formation, suggesting that invadopodia in these cells begin to degrade the matrix earlier resulting from an increase in invadopodium maturation rate. To confirm this, we knocked down profilin1 and evaluated the degradation activity by invadopodia over time (2 h and 3 h post-plating) in fixed cells. Quantification of the matrix degradation area per cell as a function of time showed that the degradation area in profilin1-depleted cells was larger than in control cells (Figure 7A-C). Degradation area per number of mature invadopodia confirmed that invadopodia in profilin1-depleted cells degrade the matrix at earlier time points (Figure 7D). In agreement with these data, the number of mature invadopodia found in profilin1-depleted cells as a function of time was also increased, while the number of invadopodium precursors remained unaffected (Figure 7E-F). These data, together with the short-lifetime of profilin1-depleted cells and the short time in which these cells degrade the matrix, suggest that profilin1 depletion accelerates the maturation of invadopodium precursors resulting in a larger matrix degradation area.
Figure 7. Profilin1-depleted cells have a higher degradation activity by invadopodia.
(A and B) Representative images of MDA-MB-231 cells transfected with control or Pfn1 siRNA and plated on 405-labeled gelatin for 2 h (A) and 3 h (B). Cells were stained for Tks5 and cortactin as invadopodia markers. Bar, 10 μm. Inset shows magnified image of invadopodia in the box, bar, 1μm (C) Quantification of degradation area per cell at 2 h and 3 h after plating and normalized to control at 2 h. n>60 cells; three independent experiments. *, p=0.04 (2 h), *, p=0.03 (3 h). (D) Quantification of invadopodial matrix degradation area per number of invadopodia after 2 h and 3 h plating and normalized to control at 2 h. n>60 fields; three independent experiments. *, p=0.042 (2 h), **, p=0.008 (3 h). (E-F) Quantification of invadopodium precursors and mature invadopodia after 2 h and 3 h plating. n>500 invadopodia, n>60 cells; three independent experiments. ***, p=0.0004 (mature invadopodia at 2 h fix), ***, p=3.43E-07 (mature invadopodia at 3 h fix). Error bars indicate SEM.
DISCUSSION
Invasive cancer cells use invadopodia to invade surrounding tissue and intravasate into blood vessels to disseminate to distant organs (Bravo-Cordero et al., 2012; Condeelis and Segall, 2003; Yamaguchi and Condeelis, 2007). Regulation of the actin cytoskeleton during invadopodium formation is a critical step in the maturation process of these structures. In this study, we investigated the role of profilin1 in regulating invadopodium formation and function. Profilin1 has been reported to be downregulated in several types of adenocarcinomas, and its depletion enhances motility and invasion in cancer cells. Because of these findings, we wanted to explore whether the invasion was increased due to enhanced invadopodium formation in breast cancer cells. The data presented here, shows for the first time that profilin1 has a role in the regulation of invadopodium formation and function.
Our study has shown that loss of profilin1 expression leads to an increase in the number of invadopodia. Although there was a small, but significant increase in the number of invadopodium precursors, the number of mature invadopodia in profilin1-depleted cells plated overnight was substantially increased, resulting in an increase in matrix degradation. Overexpression of profilin1 had the opposite effect. A similar trend of higher number of mature invadopodia and matrix degradation was noted in synchronized cell populations of control and profilin1-depleted cells plated for only 2-3 h. The number of precursors in this experiment remained unaffected, leading us to infer that longer cell incubation time (eg. overnight) causes differences to emerge in precursor numbers between control and profilin1-depleted cells, which is not seen with short incubation times. This is further confirmed by the experiment in figure 4D, where EGF-induced precursors -which are formed within minutes of EGF stimulation-, showed no difference in numbers between control and profilin1-knockdown cells. This defect seems to be mediated by the changes in actin polymerization dynamics after profilin1 knockdown.
Invadopodium formation is a multistep process where actin polymerization plays a critical role (Bravo-Cordero et al., 2012; Liu et al., 2009; Magalhaes et al., 2011). It has been reported that an invadopodium precursor that is unable to polymerize actin within its core structure cannot mature into an invadopodium with matrix degradation activity (Stylli et al., 2009; Yamaguchi et al., 2005). In our study we found that barbed ends formed by profilin1-depleted cells are higher at baseline, reaching levels seen in the control cells only after EGF stimulation. Surprisingly 3 minutes after EGF stimulation the number of barbed ends formed in control and profilin1-depleted cells is similar. We can hypothesize that profilin1 depletion affects the action of other actin-binding proteins that also regulate barbed ends such as cofilin, which may explain the higher numbers of barbed ends in profilin1-depleted cells in the serum-starved state. In particular, the higher levels of PI(3,4)P2 in profilin1-deficient cells will lead to recruitment of Tks5 to stabilize the invadopodium core allowing recruitment of NHE1 and cofilin activation (Beaty et al., 2014).
In this study, we found that after loss of profilin1 expression, there is an increase in the levels of PI(3,4)P2 and Tks5 in the invadopodium. Previous work reported a role for profilin1 in cell migration (Bae et al., 2010), where the binding of profilin1 and PI(3,4)P2 reduced the availability of PI(3,4)P2 at the membrane, therefore reducing the recruitment of the PI(3,4)P2-binding protein lamellipodin and Ena/VASP to the leading edge inhibiting in this way the motility of MDA-MB-231 cells. Consistent with this, previous work has shown that the invasiveness of profilin1-depleted cells involves a phosphatidylinositol 3-kinase-PI(3,4)P2 signaling axis (Ding et al., 2013). It was previously demonstrated that binding of PI(3,4)P2 and Tks5 stabilizes the invadopodium precursor, allowing it to mature into a degradation-competent structure (Sharma et al., 2013a). Our findings indicate that loss of profilin1 expression enhances the levels of PI(3,4)P2 and as a result, the recruitment of PI(3,4)P2 interacting adaptor protein Tks5 to the invadopodium, resulting in a higher number of mature invadopodia. It has been established that the Ena/VASP protein family regulates actin filament elongation by its anti-capping activity at free barbed ends (Pasic et al., 2008). Previous work has shown that this characteristic of Ena/VASP can promote invadopodium maturation in MTLn3 cells (Philippar et al., 2009). Therefore we hypothesize that increased PI(3,4)P2 levels at invadopodia resulting from profilin1 depletion will also increase the recruitment of Ena/VASP proteins through recruitment of lamellipodin the PI(3,4)P2 binding Ena/VASP interacting protein as previously shown for the leading edge (Krause et al., 2004).
The other profilin isoform, profilin2 suppress protrusive activity of tumor cells inhibiting invasion. Profilin2-induced contractile actin bundles are generated by the Ena/VASP protein EVL, which suppresses protrusive activity, migration and invasion, and depletion of profilin2 increases protrusive activity as well as migration and invasion (Mouneimne et al., 2012). In our study we found that depletion of profilin2 has no effect in invadopodium formation and/or matrix degradation. Based on these results we can speculate that profilin2 may play a role at different stages of tumor cell invasion and metastasis. It is known that, while migration is important for tumor cell invasion, the invadopodium is essential for tumor cell intravasation and dissemination ultimately leading to metastasis (Gligorijevic et al., 2014). Therefore, profilin2 may regulate protrusion formation during cell migration steps but not degradation of basement membranes and extracellular matrix, which are mediated by invadopodia and regulated by profilin1. Although it is not clear at this point why profilin2 silencing does not affect invadopodia, we speculate two reasons. First, abundance of profilin2 is negligible compared to profilin1 in MDA-MB-231 cells (Mouneimne et al., 2012). Second, profilin2 has a weaker affinity for phosphoinositides than profilin1 (Lambrechts et al., 2000). Therefore it is possible that perturbing profilin2 expression may have negligible effects on PI(3,4)P2 distribution and Tks5 localization. These reasons may explain our observations of profilin2 depletion having no functional consequence on invadopodial dynamics in MDA-MB-231 cells.
Invadopodia oscillate their assembly and disassembly to enhance invasion (Magalhaes et al., 2011). The mature invadopodia formed after loss of profilin1 expression are more dynamic and short-lived than control cells. In addition, the degradation activity is higher and starts earlier than control cells, suggesting that profilin1 acts as a break in controlling invadopodium turnover by modulating the levels of PI(3,4)P2 to control Tks5 recruitment. Depletion of profilin1 removes the break thereby accelerating the rate of actin polymerization, maturation, acquisition of matrix degradation activity, and invadopodium turnover, thus providing an explanation for the enhanced invasiveness seen in these cells.
In summary, the present study for the first time unveils the role of profilin1 in the regulation of invadopodial dynamics in carcinoma cells.
MATERIALS AND METHODS
Cell culture
MDA-MB-231 cells were cultured in DMEM media supplemented with 10 % FBS and antibiotics. For EGF stimulation experiments, MDA-MB-231 cells were serum starved in 0.5 % FBS and 0.8 % BSA in DMEM media for 12-16 h. Next, cells were starved in 0.345 % BSA in L-15 media for 10 minutes immediately prior to stimulation with 2.5 nM EGF. Live cell imaging experiments were performed at 37° C using L-15 media supplemented with 10 % FBS.
Antibodies, DNA constructs and transfection
The profilin1 (3237) antibody was from Cell Signaling. The Tks5 (sc-30122), cortactin (sc-30771) and Arp2 (sc-H-84) antibodies were from Santa Cruz Biotechnology, Inc. The cortactin (ab33333), cortactin (ab81208) and profilin2 (ab55611) antibodies were from Abcam. The PI(3,4)P2 (D143-3) antibody was from MBL International. The FITC-anti-biotin (200-092-211) antibody was from Jackson Immuno Research Laboratories. The β-tubulin (ATN02) antibody was from Cytoskeleton. The wild type profilin1 construct has been previously described (Bae et al., 2010). The TagRFP-cortactin has been previously described by Oser et al., 2009. The GFP-Tks5 was from Dr. Sara Courtneidge (Sanford-Burnham Institute, La Jolla, CA). The β-actin (AC-15) antibody was from Sigma-Aldrich.
For cell imaging, 1×106 MDA-MB-231 cells were transfected with 2 μg of TagRFP-cortactin and 2 μg of GFPTks5 using the Amaxa V kit (Lonza) nucleofection system 24 h before each experiment. For overexpression experiments, cells were transfected with 0.5 μg of wild type profilin1.
siRNA
Control siRNA (1027281) was from Qiagen. The sense-strand sequence of a single target specific profilin1 siRNA is 5’-AGAAGGUGUCCACGGUGGUUU-3’ and it was purchased from Dharmacon Inc. The sense strand sequences of SMARTpool profilin1 siRNA (also available for Dharmacon Inc.) are 5’-GGCCAGAAAUGUUCGGUGAUU-3’, 5’-GUGGUUUGAUCAACAAGAAUU-3’, 5’-CAAUGUCACUGUCACCAAGUU-3’ and 5’-GGUGGAACGCCUACAUCGAUU-3’. The siGENOME SMARTpool profilin2 siRNA (M-011750-01-0005) was from Dharmacon, Inc. The sense strand sequences are 5’-GAGCAUUACGCCAAUAGA-3’, 5’-GCUACUGCGACGCCAAAU-3’, 5’-GUCUAUACGUCGAUGGUG-3’ AND 5’-ACAAUGGACAUCCGGACA-3’.
For siRNA transfection, 1×106 MDA-MB-231 cells were transfected with 2 μM siRNA using the Amaxa V kit (Lonza) nucleofection system 96 h before each experiment.
Invadopodial matrix degradation assay
15×105 MDA-MB-231 cells were plated on 405-labeled gelatin overnight. The next day, cells were fixed in 3.7% paraformaldehyde for 20 min, permeabilized in 0.1% Triton-X-100 for 5 min and stained with appropriate antibodies. Ten different fields were acquired and each independent experiment was performed three times. Invadopodium precursors were identified as a cortactin and Tks5 positive punctate structures, while mature invadopodia were identified as a cortactin and Tks5 positive punctate structures colocalizing with a degradation hole. Invadopodia were manually counted from images and reported as invadopodia per cell. Degradation area was calculated as the total area covered by degradation holes/field in thresholded images using the Analyze Particles command in ImageJ, and normalized to the number of cells in each field to give degradation area/cell, or normalized to the number of total fields to give degradation area/field.
Imaging was performed on a wide-field microscope (Inverted Olympus IX70) and images were acquired with a cooled CCD camera (Sensicam QE cooled CCD camera) with a 60X NA=1.4 oil immersion objective using IP Laboratory 4.0 software.
Immunofluorescence
For cortactin and Tks5 antibodies, cells were fixed in 3.7% PFA for 20 min and permeabilized in 0.1% Triton-X-100 for 5 min. Cells were blocked in 1% BSA + 1% FBS for 1 h, and incubated in primary and secondary antibodies for 1 h each at RT. For PI(3,4)P2 antibody, cells were fixed following Eddy fix protocol (Song et al., 2006). Cells were blocked in 2% BSA for 1 h and incubated with primary and secondary antibodies for 1 h each at RT.
Invadopodia lifetime analysis
Cells treated with control or profilin1 siRNA were transfected with 2 μg of TagRFP-cortactin and 2 μg of GFP-Tks5 as invadopodia markers and incubated overnight. Then, cells were plated on 405-labeled gelatin for 4 h prior to imaging. Cell media was changed to L-15 supplemented with 10% FBS. Multiple-position imaging was performed on a Delta Vision microscope (Applied Precision). Images were acquired at 37° C, one frame every 2 min for RFP and GFP channels and every 10 min for 405 channel for up to 6 h. Invadopodia lifetimes were calculated using “Invadopodia Tracker” plugin (Sharma et al., 2013b).
Barbed end assay
The barbed end assay was performed as described previously (Beaty et al., 2013; Bravo-Cordero et al., 2011; Chan et al., 1998; Oser et al., 2009). In brief, cells were starved, stimulated with 2.5 nM EGF and permeabilized using a permeabilization buffer (20 mM Hepes, pH 7.5, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.2 mg/mL saponin, 1 mM ATP, and 1% BSA) containing 0.4 μM biotin-actin (Cytoskeleton, Inc.) for 1 min at 37°C and then fixed with 3.7% PFA for 5 min, blocked in 1% FBS/1% BSA/PBS containing 3 μM phalloidin, and stained with FITC anti-biotin to visualize barbed ends, as well as cortactin and Arp2 to identify invadopodium precursors. Ten different fields were acquired in each condition and each independent experiment was performed three times. The barbed end intensity at invadopodium precursors was quantified by measuring the mean gray value at invadopodium precursors after subtracting the mean grey value of the background. The data were normalized to the control condition for each experiment.
Imaging was performed on a wide-field microscope (Inverted Olympus IX70) and images were acquired with a cooled CCD camera (Sensicam QE cooled CCD camera) with a 60X NA=1.4 oil immersion objective using IP Laboratory 4.0 software.
Western blotting
Whole cell lysates were obtained by using a SDS-PAGE sample buffer. Next, cells were sonicated for 10 s and boiled at 95° C for 5 min. Samples were loaded in a polyacrylamide 15% gels and run at 120V for 1.5 h. Protein was transferred to nitrocellulose membrane, blocked in Odyssey blocking buffer (LI-COR Bioscience), incubated with primary antibodies at 4° C overnight. Secondary antibodies (mouse 680, mouse 800, and rabbit 680 [LI-COR Bioscience]) were incubated for 1 h at RT and scanned using a high sensitivity Odyssey Infrared Imaging System (LI-CORBiosciences). β-actin and β-tubulin were used as a loading controls. For quantification, the median gray value of the band was calculated after subtraction of the background.
Statistical analysis
Statistical analysis was performed using an unpaired, two-tailed Student's t-test. Statistical significance was defined as: * p < 0.05, ** p<0.01 and *** p< 0.001. Error bars represent the standard error of the mean (SEM). All graphs are displayed as mean ± SEM.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of the Condeelis, Cox and Hodgson labs for their helpful discussions and the Analytical Imaging Facility of the Gruss Lipper Biophotonics Center at Albert Einstein College of Medicine. This work was funded by CA150344 (J.C, J.J.B.C., B.T.B), KG111405 (V.P.S), 2R01 CA108607 (Z.D, P.R,), MSTP training grant T32-GM007288 (B.T.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bae YH, Ding Z, Das T, Wells A, Gertler F, Roy P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc. Natl. Acad. Sci. 2010;107:21547–21552. doi: 10.1073/pnas.1002309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YHO, Ding Z, Zou LI, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol. 2009;219:354–364. doi: 10.1002/jcp.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty BT, Wang Y, Bravo-Cordero JJ, Sharma VP, Miskolci V, Hodgson L, Condeelis J. Talin regulates moesin–NHE-1 recruitment to invadopodia and promotes mammary tumor metastasis. J. Cell Biol. 2014;205:737–751. doi: 10.1083/jcb.201312046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty BT, Sharma VP, Bravo-Cordero JJ, Simpson MA, Eddy RJ, Koleske AJ, Condeelis J. β1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol. Biol. Cell. 2013;24:1661–1675. doi: 10.1091/mbc.E12-12-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Oser M, Chen X, Eddy R, Hodgson L, Condeelis J. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr. Biol. 2011;21:635–644. doi: 10.1016/j.cub.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 1998;111:199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 2007;67:4227–4235. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- Das T, Bae YH, Wells A, Roy P. Profilin-1 over-expression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218:436–443. doi: 10.1002/jcp.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais V, Yamaguchi H, Oser M, Soon L, Mouneimne G, Sarmiento C, Eddy R, Condeelis J. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil. Cytoskelet. 2009;66:303–316. doi: 10.1002/cm.20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz B, Shani G, Pass I, Anderson D, Quintavalle M, Sara A. Tks5-dependent, Nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci Signal. 2010;2:1–30. doi: 10.1126/scisignal.2000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Joy M, Bhargava R, Gunsaulus M, Lakshman N, Miron-Mendoza M, Petroll M, Condeelis J, Wells a, Roy P. Profilin-1 downregulation has contrasting effects on early vs late steps of breast cancer metastasis. Oncogene. 2013:1–10. doi: 10.1038/onc.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Bae YH, Roy P. Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adh. Migr. 2012;6:442–449. doi: 10.4161/cam.21832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Gau D, Deasy B, Wells A, Roy P. Both actin and polyproline interactions of profilin-1 are required for migration, invasion and capillary morphogenesis of vascular endothelial cells. Exp Cell Res. 2009;315:2963–2973. doi: 10.1016/j.yexcr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Buccione R. Adhesions that mediate invasion. Int. J. Biochem. Cell Biol. 2006;38:1875–1892. doi: 10.1016/j.biocel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Gligorijevic B, Bergman A, Condeelis J. Multiparametric classification links tumor microenvironments with tumor cell phenotype. Plos Biology. 2014 doi: 10.1371/journal.pbio.1001995. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B, Wyckoff J, Yamaguchi H, Wang Y, Roussos ET, Condeelis J. N-WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. Journal of Cell Science. 2012;125:724–734. doi: 10.1242/jcs.092726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD. The control of actin nucleotide exchange by thymosinβ4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell. Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- Gutsche-Perelroizen I. Filament assembly from profilin-actin. J. Biol. Chem. 1999;274:6234–6243. doi: 10.1074/jbc.274.10.6234. [DOI] [PubMed] [Google Scholar]
- Janke J, Schlüter K, Jandrig B, Theile M, Kölble K, Arnold W, Grinstein E, Schwartz A, Estevéz-Schwarz L, Schlag PM, Jockusch BM, Scherneck S. Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J. Exp. Med. 2000;191:1675–1686. doi: 10.1084/jem.191.10.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, Yaffe MB, Boussiotis VA, Gertler FB. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Braun A, Jonckheere V, Aszodi A, Lanier LM, Robbens J, Van I, Vandekerckhove J, Fässler R, Ampe C. Profilin II is alternatively spliced , resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol. Cell. Biol. 2000;20:8209–8219. doi: 10.1128/mcb.20.21.8209-8219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue P, Artym VV, Mueller SC, Guo W. The role of the exocyst in matrix metalloproteinase secretion and actin dynamics during tumor cell invadopodia formation. 2009;20:3763–3771. doi: 10.1091/mbc.E08-09-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Shieh WR, Rhee SG, Yin HL, Chen CS. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry. 1996;35:14027–14034. doi: 10.1021/bi961878z. [DOI] [PubMed] [Google Scholar]
- Mader CC, Oser M, Magalhaes M, Bravo-Cordero JJ, Condeelis J, Koleske AJ, Gil-Henn H. An EGFR–Src–Arg–Cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes MAO, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, Chen X, Koleske AJ, Condeelis J. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 2011;195:903–920. doi: 10.1083/jcb.201103045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, Mullins RD, Brugge JS. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–630. doi: 10.1016/j.ccr.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J, Koleske AJ, Condeelis J. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA. Ena/VASP proteins capture actin filament barbed ends. J. Biol. Chem. 2008;283:9814–9819. doi: 10.1074/jbc.M710475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim D, Giampieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, Sahai E, Condeelis JS, Frank B. A mena invasion isoform potentiates EGF-induced carcinoma. Dev Cell. 2009;15:813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Rawe VY, Payne C, Schatten G. Profilin and actin-related proteins regulate microfilament dynamics during early mammalian embryogenesis. Hum. Reprod. 2006;21:1143–1153. doi: 10.1093/humrep/dei480. [DOI] [PubMed] [Google Scholar]
- Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita J-B, Daviet L, Camonis J, D'Souza-Schorey C, Chavrier P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VP, Eddy R, Entenberg D, Kai M, Gertler FB, Condeelis J. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr. Biol. 2013a;23:2079–2089. doi: 10.1016/j.cub.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VP, Entenberg D, Condeelis J. High-resolution live-cell imaging and time-lapse microscopy of invadopodium dynamics and tracking analysis. Methods Mol Biol. 2013b;1046:343–357. doi: 10.1007/978-1-62703-538-5_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Chen X, Yamaguchi H, Mouneimne G, Condeelis JS, Eddy RJ. Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 2006;119:2871–2881. doi: 10.1242/jcs.03017. [DOI] [PubMed] [Google Scholar]
- Stylli SS, Stacey TTI, Verhagen AM, Xu SS, Pass I, Courtneidge S, Lock P. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 2009;122:2727–2740. doi: 10.1242/jcs.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wittenmayer N, Jandrig B, Rothkegel M, Arnold W, Haensch W, Scherneck S, Jockusch BM, Ro R. Tumor suppressor activity of profilin requires a functional actin binding site. Molecular Biolofy of the Cell. 2004;15:1600–1608. doi: 10.1091/mbc.E03-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Zhang W, Yang Y, Liang YL, Wang LY, Jin JW,N. Profilin1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics. 2006;6:6095–6106. doi: 10.1002/pmic.200500321. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Huang M, DeBiasio J, Pring M, Joyce M, Miki H, Takenawa T, Zigmond SH. Profilin enhances Cdc42-induced nucleation of actin polymerization. J. Cell Biol. 2000;150:1001–1012. doi: 10.1083/jcb.150.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, Roy P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br. J. Cancer. 2007;97:1361–1371. doi: 10.1038/sj.bjc.6604038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.