Abstract
Y-box protein 1 (YB-1) is a multifunctional DNA/RNA-binding protein that regulates transcription and translation. The specificity of YB-1’s RNA binding and its consequences are unknown. Because expression and subcellular localization of YB-1 have been reported to be important in breast cancer, we determined the specificity and functional impact of YB-1 mRNA-binding in MCF7 breast cancer cells. We used YB-1 antibodies to immunoprecipitate YB-1 and microarray profiling to compare YB-1-bound and total poly(A) RNA. We demonstrated that YB-1 mRNA-binding was preferential. Transcript sequences significantly associated with this binding had high GC content. Selected YB-1 mRNA-binding targets were confirmed by QRT-PCR. However, downregulation of YB-1 levels by siRNA did not affect their RNA or protein expression. Thus, YB-1 has RNA-binding specificity; however, YB-1 binding does not necessarily regulate the stability or translation of its mRNA targets. Further study is needed to determine the functional consequences of selective YB-1 mRNA binding.
Keywords: YB-1, translation elongation, RNA-binding, poly(A) RNA, microarray, breast cancer
Introduction
Y-box protein 1 (YB-1) is a multifunctional protein that binds DNA and RNA, and plays an important role in transcriptional and translational regulation. By binding to specific DNA sequences, particularly the inverted CCAAT box (Y-box) sequence motif, YB-1 regulates the transcription of several cellular genes. In addition, YB-1 associates with RNA for almost every step of RNA biogenesis. YB-1 not only stabilizes mRNA in vitro and in vivo 1, but also selectively regulates the synthesis of certain proteins, even its own expression. 2 YB-1 possesses specific and non-specific mRNA-binding capabilities. Studies have shown that YB-1 has a preference for poly(G) nucleotides in RNA and DNA binding. 3, 4 Using RNA footprinting to localize specific YB-1 binding sites 2, we identified a sequence motif of UCCAG/ACAA as such a binding site. Using in vitro selection to select nucleotide sequences specifically bound by the Xenopus Y-box binding proteins FRGY1 and FRGY2 5, showed that the hexanucleotide AACAUC was the FRGY recognized sequence. Nevertheless, the generalizability of binding preferences and functional impact remain unclear.
The purpose of our study was to better define RNA-binding specificity of YB-1 and functional consequences of this binding. We show that mRNA-binding is preferential and directly correlated with GC content. However, YB-1 binding does not necessarily regulate stability or translation of its mRNA targets. Thus, the functional impact of YB-1 binding on its RNA targets, if any, remains to be determined.
Results
Isolation of YB-1-bound poly(A) RNA
To determine YB-1’s RNA-binding ability, MCF7 cells were chosen for study as YB-1 has been shown to be primarily cytoplasmic in this cell line, and to regulate its growth and chemosensitivity. 6, 7 YB-1 bound RNA was immunoprecipitated using anti-YB-1 antibody. To identify the YB-1-preferentially-bound-poly(A) RNA, we used the GeneChip Human Genome U133A Plus 2.0 Array (Affymetrix, Santa Clara, CA) to hybridize the samples with the immunoprecipitated poly(A) RNA using YB-1 specific antibody (referred to as “IP RNA”) and the total poly(A) RNA (referred to as “No IP RNA”) for comparison. Two independent experiments were performed. We focused on 1,339 genes present in the No IP RNA samples and that had enrichment in IP RNA (low No IP RNA/IP RNA ratio consistent with YB1-bound transcripts) or in No IP RNA (high No IP RNA/IP RNA ratio consistent with YB-1-unbound transcripts). The top unique 100 genes with No IP RNA/IP RNA ratio < 0.376 (genes preferentially enriched in IP RNA samples by 2.7-fold or greater) and the bottom unique 100 genes with No IP RNA/IP RNA ratio > 5.06 (genes not enriched in IP RNA samples) are listed in Supplementary Tables 1 and 2, respectively.
Sequence characteristics YB-1-bound mRNA
We next asked what transcript features contributed to YB-1 binding. First, we found that there was no statistical difference in the length of the YB-1-bound and YB-1-unbound mRNA. By analyzing the 5′ untranslated region (UTR), coding sequence (CDS), and 3′ UTR separately, we found the length of the 5′ UTRs of YB-1-bound and YB-1-unbound transcripts also did not differ. Although the CDSs of the IP-enriched transcripts were slightly longer than IP-non-enriched transcripts (2280+/−1878 vs. 1778+/−1727), and the 3′ UTRs of the IP-enriched transcripts were slightly shorter than IP-non-enriched transcripts (966+/−918 vs. 1578+/−1176), these differences did not achieve statistical significance.
We next evaluated the following sequences that have been proposed to confer greater YB-1 binding affinity: the original Y-box sequence; the AACAUC sequence for the Xenopus Y-box binding proteins FRGY1 and FRGY2; and the UCCAACAA and UCCAGCAA YB-1 binding sites identified by RNA footprinting. 2 Comparing the sequences of the 100 genes most preferentially enriched in IP RNA samples with the sequences of the 100 IP-non-enriched RNA, we found no significant difference in the presence of these motifs in the IP-enriched genes.
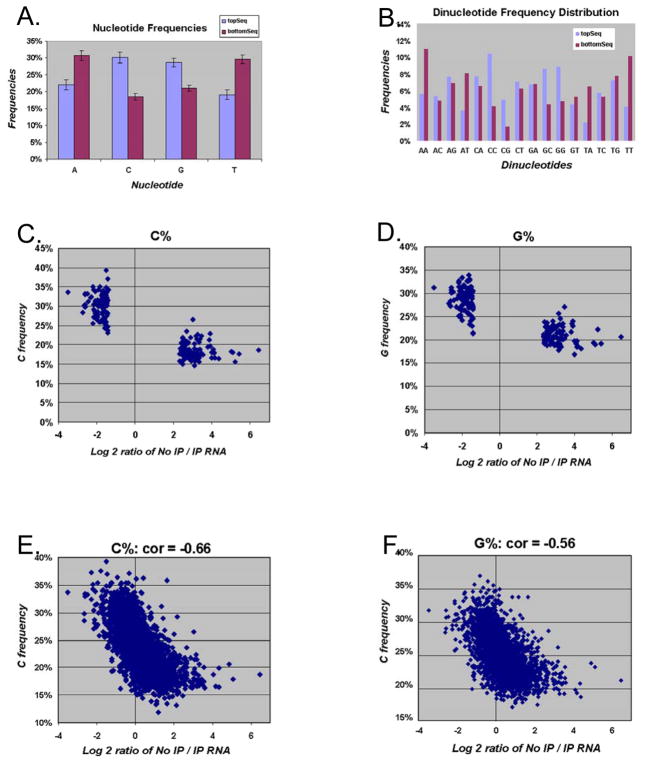
Nucleotide and dinucleotide frequency were calculated for each sequence from the top and the bottom 100 genes. The average nucleotide and dinucleotide frequencies for the top and bottom gene lists are shown in Figures 1A and 1B respectively. IP-enriched genes had more C and G components (C, G, CC, CG, GC, GG), while IP-non-enriched genes had more A and T components (A, T, AA, AT, TA, TT). Nucleotide/dinucleotide frequency distribution for CDS, 5′ UTR and 3′ UTR showed a similar difference between IP-enriched and IP-non-enriched genes (data not shown). Two sample t-tests were used to assess the difference between the two groups of genes (Fig. 1C, D). GC content was found to be significantly different (p = 6.68E-60 for %G; p = 5.14E-78 for %C). To further explore the relationship between GC content and IP enrichment, we randomly extracted 2,500 unique genes expressed at the mRNA level. Figure 1C and D show a scatter plot of the log2 ratio of IP/No IP and GC content for the 100 top and bottom genes. The scatter plot of the log2 ratio of IP/No IP and GC content for randomly extracted 2500 unique genes are shown in Figure 1E and 1F. These findings confirmed the general trend of higher GC ratios corresponding to lower No IP/IP ratios (%G correlation = −0.56; %C correlation = −0.66).
FIGURE 1. Average nucleotide/dinucleotide frequency distribution.
(A) Nucleotide frequency distribution of the top 100 IP-enriched genes / bottom 100 IP-non-enriched genes are compared. Error bars represent standard deviation. Dinucleotide frequency distribution of the top 100 IP-enriched genes and the bottom 100 IP-non-enriched genes are compared. Error bars represent standard deviation (C, D). Percentage of C nucleotide content (C) and percentage of G nucleotide content (D) compared to the log2 ratio of No IP /IP to for the top 100 IP enriched and 100 bottom non-enriched genes. (E and F) Percentage of C nucleotide content (E) and percentage of G nucleotide content (F) compared to the log2 ratio of No IP /IP to for 2500 randomly extracted unique genes.
Functional activities of YB-1-bound mRNA
To determine whether there were functional differences between proteins encoded by IP-enriched mRNA and those encoded by IP-non-enriched mRNA, we analyzed the 100 top IP-enriched genes using all human genes as a reference. The biological process/molecular functions enriched (p < 0.01) by gene ontology analysis are listed in Supplementary Table 3. Proteins encoded by YB-1-bound mRNA are involved in a variety of biological processes, including cell metabolism, translation elongation, regulation of the IκB kinase cascade, and DNA repair, in contrast to proteins encoded by IP-non-enriched mRNA.
Validation of YB-1-bound mRNA
To validate YB-1-bound mRNA, we selected genes that were preferentially bound to YB-1 by microarray analysis (EEF1D, EEF1A2, EEF2, Mark4, MTA1, p53, SMARCA, PRPF1 and AKT1) and S6K from the preferentially unbound group of genes (Table 1). We performed QRT-PCR to compare the relative ratios of these mRNA, with cyclophilin A as a control. We used cyclophilin A as the control because microarray analysis revealed that its transcript was not preferentially bound by YB-1. QRT-PCR results confirmed that the mRNA for EEF1D, EEF1A2, EEF2, Mark4, MTA1, AKT1, and p53 were enriched in the IP RNA samples compared to the No IP RNA samples (Fig. 2). In contrast, mRNA levels of the negative control S6K were higher in the No IP RNA than in the IP RNA, as was expected.
FIGURE 2. QRT-PCR validation of YB-1 mRNA-binding targets.
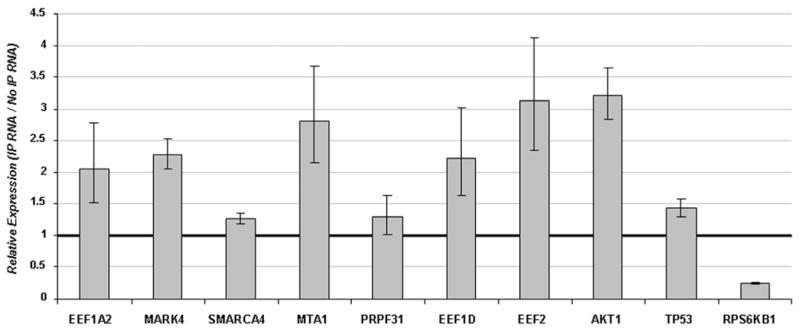
Relative levels of mRNA expression of EEF1A2, MARK4, SMARCA4, MTA1, PRPF31, EEF1D, EEF2, AKT1, p53, and S6K1 (negative control for YB-1 mRNA binding) in IP RNA compared to in No IP RNA, using cyclophilin A as endogenous control. Values >1 correspond to enrichment in IP RNA, while values <1 depict low levels of RNA in immunocomplexes.
Evaluation of effect of YB-1 downregulation on its targets
We hypothesized that YB-1 binding affects gene expression of its mRNA targets by altering their mRNA levels, affecting mRNA stability. To test this hypothesis, we suppressed YB-1 protein expression in MCF7 cells by siRNA and then assessed mRNA levels of EEF1D, EEF1A2, EEF2, Mark4, MTA1, AKT1, and p53 relative to that of cyclophilin A using QRT-PCR. YB-1 protein knockdown was confirmed by western blotting analysis (data not shown). We found no differences in relative expression of these mRNA in the YB-1-expressing control siRNA-transfected MCF7 cells compared to the YB-1 siRNA-transfected cells (data not shown). Thus, we concluded that it is unlikely that YB-1 binding alters the mRNA levels of its binding targets.
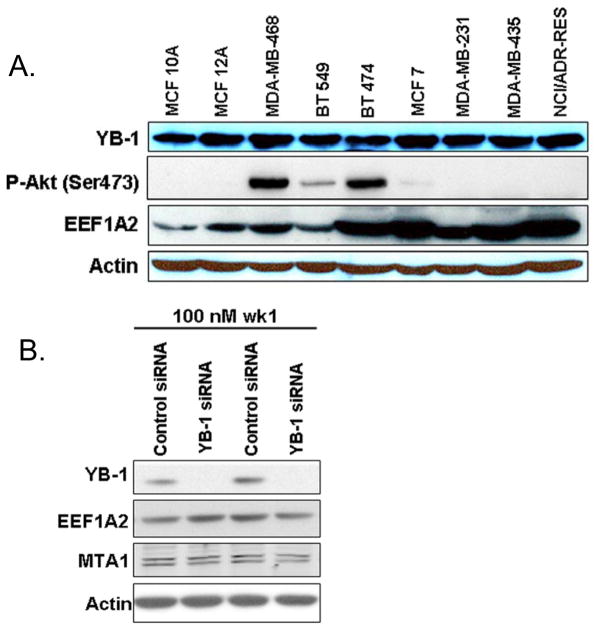
Next, we hypothesized that YB-1 binding affects protein expression of its mRNA targets by altering translation of YB-1-bound mRNA. We focused on EEF1A2 because it has been reported to be overexpressed in breast cancer and also a regulator of cell signaling and cancer progression 8–10. First, we wanted to determine if YB-1 protein levels varied in breast cancer cell lines, and if the expression EEF1A2 varied with YB-1 levels (Fig. 3A). We found that although EEF1A2 levels were variable, YB-1 was ubiquitously expressed. Although YB-1 mRNA expression has been reported to be regulated by PI3K signaling 11, we found that YB-1 expression was not affected by varying levels of phosphorylated Akt (P-Akt Ser473), a marker of activity of the PI3K pathway.
FIGURE 3. YB-1 and EEF1A2 expression.
(A) Cell lysates from normal-like cell lines MCF10A, MCF12A, MDA-MB-468, BT549, BT474, MCF7, MDA-MB-231, MDA-MB-435, and NCI/ADR-RES were separated with SDS-PAGE and western blotting was performed for antibodies to YB-1, EEF1A2, and p-Akt (S473) and actin. (B) Effect of YB-1 knockdown on EEF1A2 and MTA1 levels. MCF7 breast cancer cells were transfected with YB-1 or control siRNA. After 7 days cells were harvested and western blotting was performed for YB-1, EEF1A2, MTA1, and actin.
Next, we wanted to determine if altering YB-1 levels affected EEF1A2 levels. We transfected MCF7 cells with YB-1 siRNA or control siRNA and harvested the cells after 7 days to assess EEF1A2 levels (Fig. 3B). Although YB-1 knockdown was evident, we did not observe a change in the protein expression of EEF1A2 and MTA1. We also did not observe any changes in protein levels of these YB-1 targets when the levels were assessed earlier (day 3 or 4) or when YB-1 knockdown was continued for 2 weeks with repeated siRNA transfections (data not shown). In all experiments the cells remained healthy after YB-1 knockdown, suggesting that YB-1 is not essential for MCF7 cell viability. Also, no change was observed in the protein expression of EEF1D and EEF2 (data not shown). Thus, we conclude that at least in MCF7 breast cancer cells in which YB-1 has been shown to preferentially bind the mRNA of EEF1A2, EEF1D, EEF2, and MTA1, modulating YB-1 expression did not affect protein expression of its targets.
Discussion
YB-1 is a multifunctional DNA/RNA-binding protein that plays an important role in transcriptional and translational regulation. In this study, we found that YB-1 mRNA binding is preferential and that YB-1 has a significantly greater affinity to messages with a high GC content. However, YB-1 binding does not necessarily regulate stability or translation of its mRNA targets.
To date, little is known about the specific mRNA bound by YB-1 and functional consequences of this mRNA binding. Consistent with previous reports that YB-1 binds its own mRNA, the mRNA for YB-1 itself (YBX1), was also on the top 100 YB-1 bound mRNA in our study (Supplemental Table 1). 2 In a recent study, Evdokimova et al. (2006) used an approach similar to ours and found that mRNA bound to ectopically expressed YB-1 in K-Ras-transfected NIH3T3 cells. 12 Comparing their results with ours, we found that only two genes were common to both sets of results, one being EEF1A2. This finding suggests that YB-1’s RNA-binding specificity may be dynamic and context dependent. In our work, we elected to use microarray profiling of precipitated mRNA due to the extensive coverage of the transcriptome with this approach. However, other approaches, such as deep sequencing of precipitated RNAs may have revealed additional important mRNAs that might be missed because they are not on the array.
In our study, we confirmed that YB-1 mRNA-binding targets EEF1A2, EEF1D, EEF2, MTA1, and AKT1 were preferentially enriched by YB-1 IP, suggesting binding specificity for YB-1. We hypothesized that YB-1-RNA binding may alter the expression of the putative oncogene EF1A2. 8, 9 However, we did not observe a change in EEF1A2 expression at the RNA or protein level when we modulated YB-1 levels by siRNA. Similarly, we found that levels of EEF1D and EEF2 were not altered by YB-1 knockdown. Our results are consistent with those of Evdokimova et al. (2006), who found that YB-1 depletion did not affect translation elongation. 12 However, Y-box proteins’ effect may be dependent on the YB-1: RNA ratio. 13 Thus, although YB-1 did not modulate the expression of its RNA targets in our experiments, we cannot rule out the possibility that YB-1 regulates expression of its targets at other YB-1: mRNA molar ratios. YB-1’s potential role in oncogenesis has been an area of active study. YB-1 overexpression disrupts apoptosis and enhances chemoresistance 6, 14; YB-1 downregulation suppresses cell growth. 14 In a transgenic model, YB-1 overexpression promoted malignant mammary tumor development, demonstrating that YB-1 is a bona fide oncogene. 15 High YB-1 levels also correlate with poor clinical outcome. 6, 16 Even though YB-1 is mainly cytoplasmic, its nuclear localization, induced by chemotherapy and Akt activity, has been associated with the development of multi-drug resistance and poor prognosis. 7, 16–19 While many studies have implicated YB-1 in oncogenesis, some studies have found that YB-1 has an anti-oncogenic function, specifically by interference with PI3K-induced transformation. 11 In spite of growing interest in YB-1’s role in cancer biology, its exact impact on oncogenesis and how it accomplishes these effects remains poorly understood.
YB1 protein is known to have RNA chaperone activity20. YB1 displays RNA annealing, melting and strand exchange activities4, 21. RNA chaperones have non-specific RNA binding affinities, and the property to destabilize RNA structures and to resolve RNA-RNA interactions20. As there are many other proteins with RNA chaperone activity (e.g. hnRNPs, histone-like proteins, and ribosomal proteins), thus it is conceivable that other proteins may take over the role of YB1 when YB-1 levels decline20. This may, in part, explain the lack of modulation of the expression of YB1’s RNA-binding targets with YB-1 siRNA knockdown. It is also possible that YB-1’s RNA binding effects may be more relevant under conditions such as cold shock or other stress conditions that have not been tested in our study.
We had hypothesized that YB-1 modulates the malignant phenotype by altering the target mRNA’s stability (i.e., by stabilizing oncogenic mRNA) or by modulating its translation (i.e., by inhibiting translation at steady state but releasing oncogenic mRNA for translation upon YB-1’s nuclear translocation). In our study YB-1 did not play a major role in regulation of its RNA-binding targets. It is possible that YB-1 binding may not be a critical regulator of expression of its RNA targets in all systems and that its effect on translation varies with the expression and activation of other translational regulators. Alternatively, YB-1’s oncogenic effect may be primarily attributable to its nuclear function as a transcription factor.22–24 Thus, it is possible that YB-1’s RNA binding partly serves as a storage mechanism for YB-1 and with stimuli inducing YB-1 nuclear translocation leading to the activation of its oncogenic effects. Further investigation is needed to better understand YB-1’s multiple functions, especially those in carcinogenesis.
Materials and methods
Cell cultures, western blotting, and siRNA transfection
MCF10A, MCF12A, BT474, BT549, MCF7 MDA-MB-231, and MDA-MB-468 cells were purchased from the American Type Culture Collection (Manassas, VA). MDA-MB-435 and NCI/ADR-RES cell lines were from NCI (Frederick, MD). The following antibodies were used for western blotting : EEF2, AKT1, P-Akt (Ser473), p53, and YB-1 (Cell Signaling Technology, Danvers, MA), EEF1D and YB-1 (Abcam Inc., Cambridge, MA); beta actin (Sigma-Aldrich, St. Louis, MO); and MTA1 (Santa Cruz Biotechnology, Santa Cruz, CA). The EEF1A2 antibody was a gift from Dr. Cathy Abbott (University of Edinburgh, United Kingdom) and the YB-1 antibody used for immunoprecipitation was a gift from Dr. Thomas A. Cooper (Baylor College of Medicine, Houston, TX). Pooled siRNA to YB-1 and control siRNA were obtained from Dharmacon, Inc. (Lafayette, CO). Transfection was performed using the Dharma FECT 1 Transfection Reagent (Dharmacon, Inc.).
IP, Poly (A) isolation and microarray analysis
Immunoprecipitation and recovery of YB-1-RNA immunocomplexes was done as described before. 25 The poly(A) RNA samples were prepared from recovered immunocomplexes using the MicroPoly(A)Purist(tm) Kit (Ambion, Austin, TX). RNA was reverse transcribed into first-strand cDNA in a reaction containing 200 U SuperScript II RT (Invitrogen) and 50 pmol of T7-oligo(dT) primer. The reaction was incubated at 42°C for 1 h. The second strand of cDNA was synthesized in the presence of 40 U E. coli DNA polymerase I, 10 U E. coli DNA ligase, and 2 U E. coli RNase H. The reaction was incubated at 16°C for 2 h. Then 10 U T4 DNA ligase was added and after incubating at 16°C for 5 min, the reaction mixture was purified by phenol:chloroform extraction. cDNA was transcribed into cRNA using the Bioarray High Yield RNA Transcript Labeling Kit (Enzo Life Sciences, Inc., Farmingdale, NY) by following manufacturer’s recommendations. Biotin-labeled cRNA was purified using the RNeasy kit, quantified, and fragmented by fragmentation buffer (Affymetrix, Santa Clara, CA). The fragmented cRNA was then hybridized to GeneChip U133A Plus 2.0 Arrays (Affymetrix, Santa Clara, CA) at 45°C overnight. Washing and staining of chips were done according to the Affymetrix technical manual.
QRT-PCR analysis
RNA was reverse transcribed from a 25-μl sample of the reaction mixture. Random hexamers were added (1 μg of total RNA to 0.4 μg of random hexamers) to mixture and heated at 70°C for 10 min. The tubes were incubated at room temperature for 10 min, and the following added: 1X Superscript II RT Buffer (Invitrogen Corporation, Carlsbad, CA), 10 mM DTT (Invitrogen Corporation), 0.5 mM dNTP’s (ISC Bioexpress, Kaysville, UT), 20U RNase Inhibitor (Ambion, Austin, TX), and 200U Superscript II Reverse Transcriptase (Invitrogen Corporation). The mixture was incubated at room temperature for 10 min for primer annealing and then at 37°C for 1 h. The mixture was spiked with 200U Superscript II Reverse Transcriptase (Invitrogen Corporation) and then incubated at 42°C for 1.5 h then at 50°C for 30 min. Quantitative real-time PCR (QRT-PCR) was performed on the ABI Prism 7900HT Sequence Detection System using the cyclophilin Vic-labeled Pre-Developed Assay Reagent (4326316E; Applied Biosystems, Foster City, CA) endogenous control, without multiplexing, as an endogenous control. A 25-μl final reaction volume containing 1X TaqMan Universal PCR Master Mix (Applied Biosystems) and 1X Assays-on-Demand was used to amplify ~5 ng of cDNA with the following cycling conditions: 10 min at 95°C followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min. The detection system’s software automatically determined fold changes for the target RNAs in the IP samples relative to the No IP samples, or in the YB-1 siRNA samples relative to the control siRNA samples, using the comparative Ct method with a 95% confidence interval.
Statistical analysis
We performed a standard two-sample, unequal variance t-test between the total poly(A) RNA (No IP RNA) samples and YB-1-immunoprecipitated poly(A) RNA (IP RNA) samples for each probe.
Supplementary Material
Table 1 – Top 100 IP-enriched genes
Table 2 – Bottom 100 IP-enriched genes (or Top 100 IP non-enriched genes)
Table 3- Gene ontology functional categories that have been enriched in top 100 gene list using all human genes as the reference group
Acknowledgments
We thank Toi Clayton Soh for assistance with manuscript preparation, Haixia Zhang for technical assistance, and David Cogdell for assistance in QRT-PCR. Grant support: This work was supported by a grant from NIH1 grant R01 CA112199 (to FM-B), the Gillson-Longenbaugh Foundation (to FM-B), and the M. D. Anderson Cancer Center Core Grant CA16672.
References
- 1.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes & development. 2000;14:1236–48. [PMC free article] [PubMed] [Google Scholar]
- 2.Skabkina OV, Lyabin DN, Skabkin MA, Ovchinnikov LP. YB-1 autoregulates translation of its own mRNA at or prior to the step of 40S ribosomal subunit joining. Molecular and cellular biology. 2005;25:3317–23. doi: 10.1128/MCB.25.8.3317-3323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minich WB, Maidebura IP, Ovchinnikov LP. Purification and characterization of the major 50-kDa repressor protein from cytoplasmic mRNP of rabbit reticulocytes. European journal of biochemistry / FEBS. 1993;212:633–8. doi: 10.1111/j.1432-1033.1993.tb17701.x. [DOI] [PubMed] [Google Scholar]
- 4.Zasedateleva OA, Krylov AS, Prokopenko DV, Skabkin MA, Ovchinnikov LP, Kolchinsky A, Mirzabekov AD. Specificity of mammalian Y-box binding protein p50 in interaction with ss and ds DNA analyzed with generic oligonucleotide microchip. Journal of molecular biology. 2002;324:73–87. doi: 10.1016/s0022-2836(02)00937-3. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet P, Matsumoto K, Wolffe AP. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. The Journal of biological chemistry. 1995;270:28297–303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 6.Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dorken B, Royer HD. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature medicine. 1997;3:447–50. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 7.Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, Leighton Grimes H, Miller K, Badve S, Huntsman D, Blake-Gilks C, Chen M, Pallen CJ, Dunn SE. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–92. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 8.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, Lee JM. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nature genetics. 2002;31:301–5. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson VA, Newbery HJ, Wray NR, Jackson J, Larionov A, Miller WR, Dixon JM, Abbott CM. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC cancer. 2005;5:113. doi: 10.1186/1471-2407-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amiri A, Noei F, Jeganathan S, Kulkarni G, Pinke DE, Lee JM. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene. 2007;26:3027–40. doi: 10.1038/sj.onc.1210101. [DOI] [PubMed] [Google Scholar]
- 11.Bader AG, Felts KA, Jiang N, Chang HW, Vogt PK. Y box-binding protein 1 induces resistance to oncogenic transformation by the phosphatidylinositol 3-kinase pathway. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12384–9. doi: 10.1073/pnas.2135336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Molecular and cellular biology. 2006;26:277–92. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto K, Meric F, Wolffe AP. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. The Journal of biological chemistry. 1996;271:22706–12. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 14.Schittek B, Psenner K, Sauer B, Meier F, Iftner T, Garbe C. The increased expression of Y box-binding protein 1 in melanoma stimulates proliferation and tumor invasion, antagonizes apoptosis and enhances chemoresistance. International journal of cancer. 2007;120:2110–8. doi: 10.1002/ijc.22512. [DOI] [PubMed] [Google Scholar]
- 15.Bergmann S, Royer-Pokora B, Fietze E, Jurchott K, Hildebrandt B, Trost D, Leenders F, Claude JC, Theuring F, Bargou R, Dietel M, Royer HD. YB-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer research. 2005;65:4078–87. doi: 10.1158/0008-5472.CAN-04-4056. [DOI] [PubMed] [Google Scholar]
- 16.Janz M, Harbeck N, Dettmar P, Berger U, Schmidt A, Jurchott K, Schmitt M, Royer HD. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. International journal of cancer. 2002;97:278–82. doi: 10.1002/ijc.1610. [DOI] [PubMed] [Google Scholar]
- 17.Fujita T, Ito K, Izumi H, Kimura M, Sano M, Nakagomi H, Maeno K, Hama Y, Shingu K, Tsuchiya S, Kohno K, Fujimori M. Increased nuclear localization of transcription factor Y-box binding protein 1 accompanied by up-regulation of P-glycoprotein in breast cancer pretreated with paclitaxel. Clin Cancer Res. 2005;11:8837–44. doi: 10.1158/1078-0432.CCR-05-0945. [DOI] [PubMed] [Google Scholar]
- 18.Basaki Y, Hosoi F, Oda Y, Fotovati A, Maruyama Y, Oie S, Ono M, Izumi H, Kohno K, Sakai K, Shimoyama T, Nishio K, Kuwano M. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene. 2007;26:2736–46. doi: 10.1038/sj.onc.1210084. [DOI] [PubMed] [Google Scholar]
- 19.Shibahara K, Sugio K, Osaki T, Uchiumi T, Maehara Y, Kohno K, Yasumoto K, Sugimachi K, Kuwano M. Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin Cancer Res. 2001;7:3151–5. [PubMed] [Google Scholar]
- 20.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA biology. 2007;4:118–30. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 21.Skabkin MA, Evdokimova V, Thomas AA, Ovchinnikov LP. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. The Journal of biological chemistry. 2001;276:44841–7. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- 22.Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. The Journal of biological chemistry. 2003;278:27988–96. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- 23.Wu J, Lee C, Yokom D, Jiang H, Cheang MC, Yorida E, Turbin D, Berquin IM, Mertens PR, Iftner T, Gilks CB, Dunn SE. Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2. Cancer research. 2006;66:4872–9. doi: 10.1158/0008-5472.CAN-05-3561. [DOI] [PubMed] [Google Scholar]
- 24.Stratford AL, Habibi G, Astanehe A, Jiang H, Hu K, Park E, Shadeo A, Buys TP, Lam W, Pugh T, Marra M, Nielsen TO, Klinge U, Mertens PR, Aparicio S, Dunn SE. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res. 2007;9:R61. doi: 10.1186/bcr1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong J, Peng J, Zhang H, Mondesire WH, Jian W, Mills GB, Hung MC, Meric-Bernstam F. Role of glycogen synthase kinase 3beta in rapamycin-mediated cell cycle regulation and chemosensitivity. Cancer research. 2005;65:1961–72. doi: 10.1158/0008-5472.CAN-04-2501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1 – Top 100 IP-enriched genes
Table 2 – Bottom 100 IP-enriched genes (or Top 100 IP non-enriched genes)
Table 3- Gene ontology functional categories that have been enriched in top 100 gene list using all human genes as the reference group