Abstract
Corticosteroids (CS) such as methylprednisolone (MPL) affect almost all liver functions through multiple mechanisms of action, and long-term use results in dysregulation causing diverse side effects. The complexity of involved molecular mechanisms necessitates a systems approach. Integration of information from the transcriptomic and proteomic responses has potential to provide deeper insights into CS actions. The present report describes the tandem analysis of rich time-series transcriptomic and proteomic data in rat liver after a single dose of MPL. Hierarchical clustering of the common genes represented in both mRNA and protein datasets displayed two dominant patterns. One of these patterns exhibited complementary mRNA and protein expression profiles indicating that MPL affected the regulation of these genes at the transcriptional level. Some of the classic pharmacodynamic markers for CS actions, including tyrosine aminotransferase (TAT), were among this group, together with genes encoding urea cycle enzymes and ribosomal proteins. The other pattern was rather unexpected. For this group of genes, MPL had distinctly observable effects at the protein expression level, although a change in the reverse direction occurred at the transcriptional level. These genes were functionally associated with metabolic processes that might be essential to elucidate side effects of MPL on liver, most importantly including modulation of oxidative stress, fatty acid oxidation, and bile acid biosynthesis. Furthermore, profiling of gene and protein expression data was also done independently of one another by a two-way sequential approach. Prominent temporal shifts in expression and relevant cellular functions were described together with the assessment of changes in the complementary side.
Introduction
Glucocorticoids are steroid hormones produced by the adrenal cortex that have diverse effects on a variety of physiological processes including carbohydrate, lipid, and protein metabolism, immune-regulation, bone homeostasis, and developmental processes (Barnes, 1998; Vegiopoulos and Herzig, 2007). The properties of glucocorticoids in regulating the immune system are exploited clinically with numerous synthetic corticosteroids (CS) that are used as anti-inflammatory and immunosuppressive agents (Barnes, 1998; Swartz and Dluhy, 1978). The CS including dexamethasone, prednisolone, fluticasone, and methylprednisolone are extensively used in the treatment of a variety of conditions including organ transplantation, rheumatoid arthritis, lupus erythematosus, asthma, and allergic rhinitis (Barnes, 1998; Swartz and Dluhy, 1978). However, because of their strong effects on systemic metabolism, long-term usage of CS can cause many side effects, including metabolic syndrome, dyslipidemia, steroid-induced diabetes, atherosclerosis, and muscle atrophy (Bialas and Routledge, 1998; Schacke et al., 2002).
Because of their diverse physiological effects, CS can influence the functioning of many tissues. Liver is one of the primary targets of CS action and plays a central role in maintaining systemic energy balance (Andrews and Walker, 1999; Morand and Leech, 1999). The liver stores glucose in the form of glycogen, which can be released in response to hormonal signals to maintain systemic glucose concentrations. In addition, de novo synthesis of glucose through gluconeogenesis involving utilization of carbon backbones from free amino acids (released by protein breakdown in muscle) and glycerol (released from adipose tissue) occurs in the liver (Andrews and Walker, 1999). Furthermore, the liver plays a critical role in lipid metabolism and affects synthesis, storage, and utilization of different lipid molecules (Hazra et al., 2008b). CS have been shown to affect almost all of these functions in liver, and long-term use of CS results in dysregulation of these processes causing diverse side-effects (Schacke et al., 2002).
The well-established molecular mechanisms of action for CS include the passive diffusion of the highly lipophilic CS molecule through the cell membrane and binding to the cytosolic glucocorticoid receptor, which is held inactive through the association with heat shock proteins (Schaaf and Cidlowski, 2002). Binding of the drug to the receptor causes conformational changes, phosphorylation, and activation of receptor, resulting in the formation of a homodimer of the drug receptor complex (Oakley and Cidlowski, 2011; Schaaf and Cidlowski, 2002). This activated complex translocates into the nucleus and binds to regulator sites, glucocorticoid regulatory elements (GREs) in the DNA, resulting in the regulation of transcription rate. However, in addition to direct binding, the activated complex can regulate gene expression by other mechanisms including tethering and composite binding to other transcription factors, activators, or repressors (Barnes, 1998; Schaaf and Cidlowski, 2002). Some of the critical transcription factors that are affected by CS include NF-κB, AP-1, and STAT. In addition to these genomic mechanisms, studies have shown that CS can regulate pathways by signaling through its receptor in a transcription-independent manner, although the exact mechanisms for the non-genomic effects are still unclear (Schaaf and Cidlowski, 2002).
Because of the diverse effects of CS and different molecular mechanisms potentially involved in these actions, an ‘omics’ approach can be effective in gaining better understanding of the effects of CS on different pathways and functions (Nguyen et al., 2010). Previously we used Affymetrix gene chips to profile temporal changes in mRNA expression in multiple tissues following CS administration in rats (Almon et al., 2003; 2005; 2007a; 2007b). These studies characterized global dynamics of the system that are regulated by CS at the transcriptional level. Although this information is useful and highly relevant, direct profiling of the protein expression changes and integrating the information from the genomic and proteomic response will provide deeper insights into CS actions, given their diverse and complex mechanisms. Recently, we were able to develop high-throughput methodology to perform comprehensive and accurate profiling of the tissue proteome using an ion current-based liquid chromatography/mass spectrometry (LC/MS) strategy (Tu et al., 2012). Using this methodology, we characterized the temporal changes in the expression of thousands of proteins in rat liver after methylprednisolone (MPL) administration (Nouri-Nigjeh et al., 2014).
The present report describes the combined analysis of rich time-series transcriptomic and proteomic data in rat liver after a single dose of MPL. We identified two distinct groups of genes that are significantly affected by MPL. Impact of MPL administration on one group of genes was at the transcriptional level, which was complemented by subsequent alterations in protein translation. For the other group, although there was an observable change at the proteomic level, this change was not accompanied by transcriptional signatures of the genes. Furthermore, we performed proteomic and transcriptomic profiling independent of one another and defined functional characteristics of hepatic proteins affected by MPL administration.
Methods
Experiments
All animal protocols adhered to “Principles of Laboratory Animal Care” (NIH publication 85-23, revised in 1985) and were approved by the University at Buffalo IACUC committee.
Proteomics
Sixty adrenalectomized (ADX) Wistar rats were injected with 50 mg/kg methylprednisolone (MPL) intramuscularly and sacrificed at 12 different time points between 0.5 and 66 h post-dosing (5 animals/time point). Five animals, injected with saline and sacrificed at random time points in the same time window, served as controls. In order to remove the high concentrations of blood protein, it was necessary to use perfused tissue for proteomic analyses, which precluded the use of the same tissues employed for transcriptomics (below). Proteins from perfused and flash frozen livers were extracted, digested, and analyzed using a nano-LC /LTQ/ Orbitrap instrument. The Nano Flow Ultra-high Pressure LC system (nano-UPLC) consisted of a Spark Endurance autosampler (Emmen, Holland) and an ultra-high pressure Eksigent (Dublin, CA) Nano-2D Ultra capillary/nano-LC system, with a LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) used for detection. Protein quantification was based on the area under the curve (AUC) of the ion-current peaks. A more extensive description of the experimental setup and the analytical methodology can be found in our published report (Nouri-Nigjeh et al., 2014).
Transcriptomics
Forty-three ADX Wistar rats were given a bolus dose of 50 mg/kg MPL intravenously. Animals were sacrificed at 16 different time points between 0.25 and 72 h post-dosing. Four untreated animals sacrificed at 0 h served as controls. The mRNA expression profiles of the liver were arrayed via Affymetrix GeneChips Rat Genome U34A (Affymetrix, Inc.), which contained 8800 full-length sequences and approximately 1000 expressed sequence tag clusters (Jin et al., 2003). This dataset was previously submitted to the GEO (GSE490).
Computational analysis
Hierarchical clustering of concatenated datasets
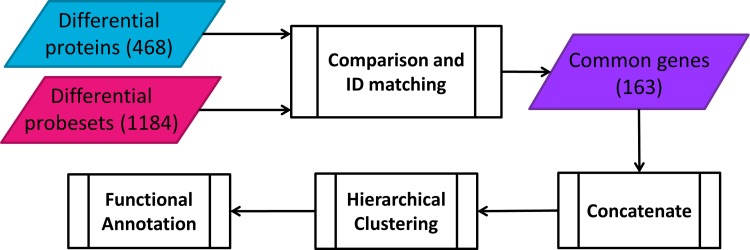
Data analysis for both proteomic and transcriptomic datasets started first by filtering for differential expression over time. Proteins and transcripts with differential temporal profiles were determined by using software for the extraction and analysis of gene expression (EDGE). We employed within-class differential expression to extract profiles that have a differential expression over time (Leek et al., 2006; Storey et al., 2005; 2007). Integration of these two datasets for any further analysis required matching the object identifiers, which was achieved through running a comparison between two filtered datasets in Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, www.ingenuity.com). This analysis helped us identify the genes that were differentially expressed both at the transcriptional and translational levels. In order to find potential co-regulatory relationships at these two levels, hierarchical clustering was used for first-pass analysis. For this purpose, temporal transcriptomic and proteomic data for the common genes were first concatenated and then clustered using the clustergram function in the Bioinformatics toolbox of MATLAB (Mathworks, Natick MA). The two clusters were obtained by using correlation as the distance metric. One cluster contained genes that were regulated in the same general pattern, while the second cluster contained genes for which the direction of regulation differed in the protein and transcript datasets. Biological implications of the genes populated in each cluster were determined through investigating the enriched canonical pathways and predicted upstream regulators obtained in IPA.
Two-way sequential clustering of individual proteomic and transcriptomic datasets
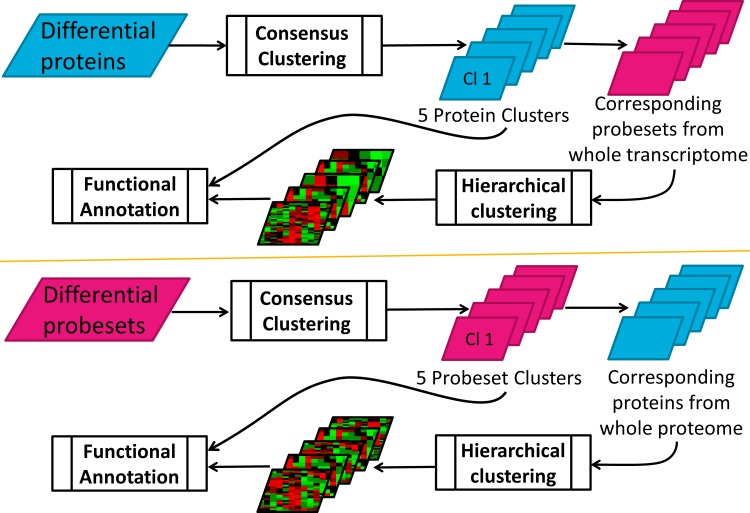
While hierarchical clustering analysis described above identifies the potential co-regulatory schemes for the genes in the intersection of transcriptomic and proteomic datasets; it fails to capture the dynamics in the rest of the genes that may also show differences in expression over time, although they may not co-exist in both datasets. In order to evaluate the overall dynamic patterns and extract the most useful information integrating these two datasets, a consensus clustering (Nguyen et al., 2009) method was applied to these two “-omic” datasets separately. First, proteins with differential temporal profiles were clustered using p values of 0.05 for significant clusters and an agreement level of 0.70 for the genes in each cluster. Then, probe sets corresponding to the proteins in each cluster were identified through the comparison function in IPA as before. Temporal profiles of these probe sets corresponding to the proteins were compiled and separately subclustered through the less stringent hierarchical clustering method, again using the clustergram function in MATLAB.
The reverse of the same procedure was also performed—starting from transcriptional analysis and continuing with the corresponding proteomic analysis. Here, differential transcriptional profiles were first determined and then clustered using the same procedures described above. As with the previous analysis, proteins that were coded by the probesets within each of these clusters were then identified and subclustered.
Biological interpretation
Functional annotations of proteins and transcripts at each level of analysis were conducted in IPA by running a core analysis for each cluster and evaluating the enriched canonical pathways (at p value threshold of 0.05) and predicted upstream regulators obtained in IPA.
Results
Studies focused on understanding the relationship between global mRNA transcription and protein translation have produced mixed results, many of which concluded that the transcriptomic and proteomic data is far from being easily described as complementary (Greenbaum et al., 2003; Haider and Pal, 2013; Hegde et al., 2003; Nicholson et al., 2004; Waters et al., 2006). This study aimed to compare and contrast the transcriptional and translational changes in liver induced by the exposure to a synthetic CS at a pharmacological dose. Although high-throughput omics analyses have been obtained from samples collected from two independent studies, the strain of experimental animals, dose and type of pharmacologic agent, sampled tissue, sampling procedures, and most of the time points for sample collection were the same for these studies. These conditions allowed us to assume that the experiments are similar enough to conduct individual and integrated bioinformatics analyses. The preprocessing before performing the first-pass analysis involved identifying the significant genes whose both transcripts and proteins existed in the individual datasets. The followed procedure is schematically shown in Figure 1. Differential expression analysis through EDGE, which utilizes an optimal discovery procedure in order to test each element in the dataset for differential expression (Leek et al., 2006), identified that 475 out of 959 proteins and 1624 out of around 8800 transcripts had temporal profiles that significantly varied over time (meeting p value<0.05 and q-value <0.01 cut-offs). After this filtering step, both datasets were fed into IPA in order to match distinct identifiers used (Swiss-Prot IDs for proteins and Affymetrix IDs for transcripts). A comparison between two datasets indicated that 163 genes were found in both transcriptomic and proteomic datasets (i.e., both mRNAs and proteins corresponding to these genes were differentially expressed over time).
FIG. 1.
Workflow for hierarchical clustering of the concatenated transcriptomic-proteomic dataset.
Hierarchical clustering of concatenated datasets
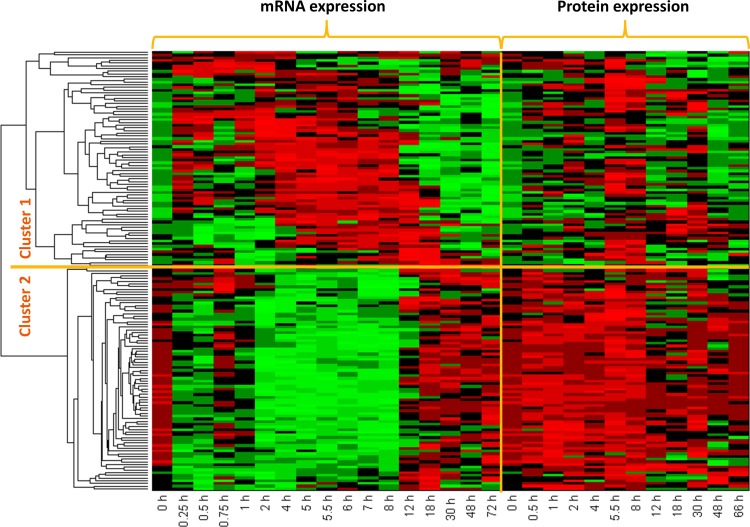
Temporal transcriptional and protein expression data for 163 common genes were concatenated and clustered through hierarchical clustering by using correlation as the distance metric. Overall, this analysis identified two dominant patterns as shown in Figure 2. Cluster 1 was populated with 80 genes for which corresponding mRNA and protein expression profiles were essentially parallel in direction, while for 83 genes in Cluster 2 the directionality was reversed. For both clusters, the first 8 hours is seemingly the most critical time period during which mRNA and protein expression profiles change direction. Genes in Cluster 1 display upregulation for both mRNA and protein expression profiles in the first 8 hours, after which downregulation predominates, most markedly in the transcriptional profiles. In the second cluster, early downregulation predominates for transcriptional profiles; however corresponding protein expression profiles are not complementary. While downregulation is observed in these transcripts most notably in the first 8 hours, expression of the same proteins seems to be upregulated in the same time frame. After the 8th hour, both transcriptional and protein expression profiles approach basal levels, though from opposite directions; elevated mRNA levels start to be downregulated and reduced protein levels start to be upregulated.
FIG. 2.
Heat map of clustered concatenated dataset. Red color indicates increase in expression while green indicates decrease.
Functional annotation of the genes in these two clusters, obtained through enrichment analysis in IPA, is shown in Table 1. Cluster 1 (where direction of regulation is similar) included a number of genes coding for heat shock proteins, which take part in the negative regulation of CS signaling through direct protein–protein interaction with glucocorticoid receptor to prevent its translocation to nucleus (Chrousos and Kino, 2005). Complementary transcriptional and proteomic profiles of these genes indicated that this is a negative feed-back control induced by MPL delivery, which is regulated at the transcriptional level. Proteins functioning in the regulation of protein degradation and translation machinery were also among the genes in Cluster 1 (including PSMCs, HSPs, EIFs, RPLs, and RPSs), implying that these processes are also controlled at the transcriptional level after CS exposure. In contrast, functions enriched by the genes in Cluster 2 appear to be regulated at post-transcriptional levels, likely through control of mRNA processing, initiation of protein translation or protein stability, since the transcriptional profiles are not emulated by protein expression, (Waters et al., 2006). Among these functions most notable are the modulation of oxidative stress, lipid metabolism, and bile acid biosynthesis.
Table 1.
Functional Annotation of Differentially Expressed Genes in Both Transcriptional and Translational Levels
| Cluster 1 | Cluster 2 |
|---|---|
| mRNA expression is essentially parallel with protein expression | mRNA expression and protein expression moves in opposite directions |
| Functional annotation | |
|---|---|
| CS signaling | Xenobiotic metabolism, oxidative stress modulation, hormone degradation Cytochrome p450 family Sulfo-transferases Glutathione S-transferases Aldehyde dehydrogenases Catalase |
| Protein ubiquitination Proteasome Heat shock proteins | |
| Protein translation Eukaryotic translation factors and ribosomal proteins | |
| Urea cycle enzymes | Fatty acid oxidation, valine-tryptophan degradation, ketolysis |
| Hormone degradation Sulfo- and glucuronyl-transferases |
Bile acid biosynthesis |
Functional annotation through IPA core analysis also allows the identification of upstream regulators that can explain the observed changes in gene/protein expression based on the prior knowledge of expected effects between the upstream regulators and target genes/proteins in the dataset. Definition of upstream regulator, however, is used rather loosely here, as almost any type of molecule that affects the expression of other molecules can be upstream regulators whether they are transcription factors, kinases, or hormones. The analysis first examines how many known target genes of each candidate upstream regulator are present in the dataset. It then also compares the direction of change in those targets with what is expected from the literature in order to predict relevant upstream regulators. If the observed direction of change is consistent with a particular activation state (i.e., activated or inhibited) of that candidate regulator then a prediction is made about the activation state. Using the genes coexisting in both transcriptomic and proteomic datasets, a number of upstream regulators were identified for each time point; predicted results are shown in Table 2. Two clusters obtained through hierarchical clustering were examined separately; however there are common predicted regulators of the elements in different clusters. Reliability of a predicted element increases as the number of times it is identified from consecutive datasets and also if there is a coherent pattern in its predicted state (i.e., whether it is activated or inhibited). Based on this, nuclear factor erythroid-derived 2-like 2 (NFE2L2) seems to contribute reliably to the observed gene/protein expression patterns by inducing early upregulation and late downregulation. Two other important genes are solute carrier family 13 (sodium/sulfate symporter), member 1 (SLC13A1), and leptin (LEP), both of which seem to have an inhibitory effect on gene/protein expression throughout the time course of the study.
Table 2.
Predicted Upstream Regulators and their Activation States* Based on Gene Groups Obtained by Hierarchical Clustering
| 0.5 h | 1 h | 2 h | 4 h | 5.5 h | 8 h | 12 h | 18 h | 30 h | 48 h | 66 h | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | NFE2L2 | SLC13A1 | NFE2L2 | NR1I3 | NR1I3 | SLC13A1 | NFE2L2 | LEP | |||
| SLC13A1 | SLC13A1 | LEP | LEP | ||||||||
| LEP | SLC13A1 | ||||||||||
| Cluster 2 | ZBTB20 | NFE2L2 | NR1I3 | ZBTB20 | ACOX1 | ZBTB20 | ZBTB20 | ||||
| NR1I3 | NFE2L2 | ||||||||||
| NR1I2 | |||||||||||
| NFE2L2 |
Green: activated; Red: inhibited.
Two-way sequential clustering of individual proteomic and transcriptomic datasets
The approach followed for the first part of the analysis described above imposes the stringency that a gene has to be differentially expressed both in the transcriptional and translational levels to be included in the final hierarchically clustered dataset. To fully characterize the temporary patterns of protein translation induced by MPL and to get additional insights into how that process is connected with the transcriptional events during the corresponding time frame, we also followed a sequential approach as schematically shown in Figure 3. What is distinct about the sequential approach is that it allows focusing on each dataset independently of the other (i.e., regardless of the complementary dataset being also differentially expressed). This increases the number of elements in clustering analysis. However, consensus clustering, being an inherently more stringent approach compared to hierarchical clustering, elicits the most coherent expression patterns in the dataset that it is applied to. Therefore, this second approach with stringency in coherency at one level of expression rather than commonality at both levels aims to identify the dominant patterns in one level of regulation (transcription or translation) and to check how closely it is associated with the patterns in the other level.
FIG. 3.
Workflow for sequential clustering analysis carried out in both forward and reverse directions between proteomic and transcriptomic datasets.
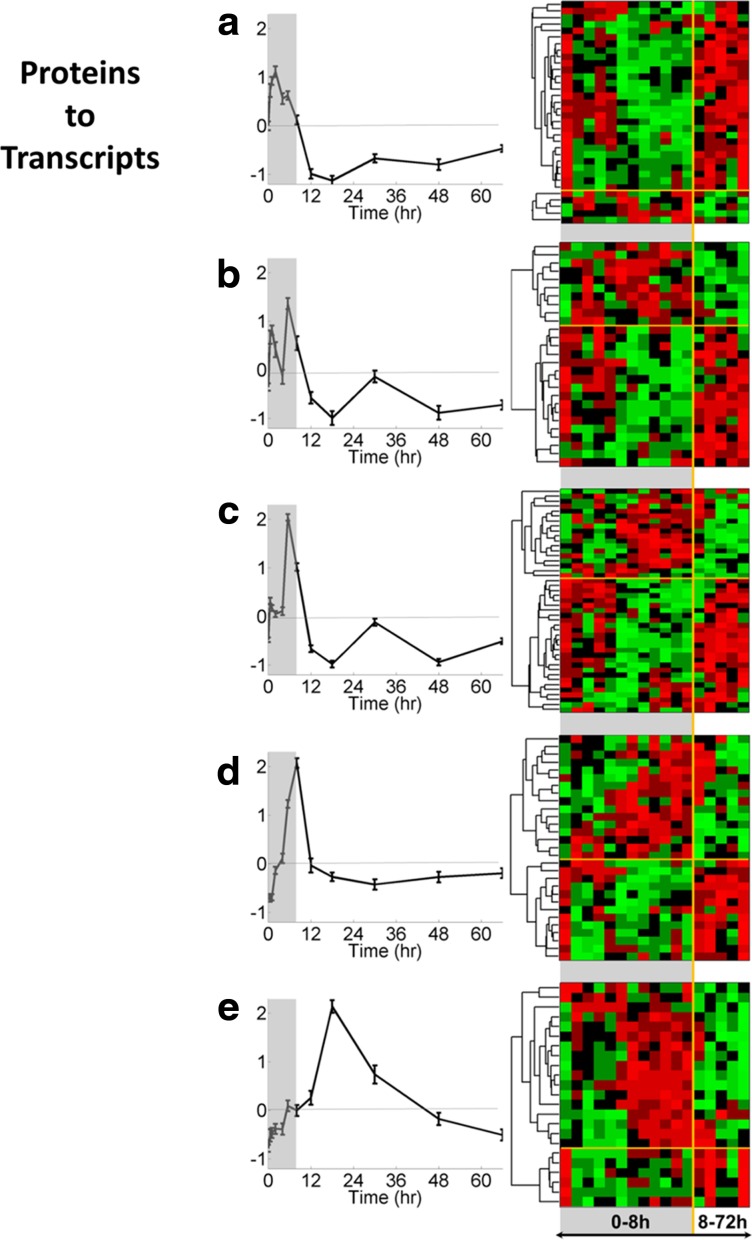
First, an in-depth analysis of the proteomic dataset was done in order to capture the dynamics of protein expression in liver following MPL dosing. EDGE identified 475 out of 959 proteins to be differentially expressed over time. Consensus clustering revealed five coherent temporal profiles containing 217 of the 475 regulated proteins. The detailed distribution of these five clusters is given in Table 3. The first two clusters (Fig. 4a, b) show early upregulation within the first 5.5 hours after MPL, followed by a recovery period. Proteins in Clusters 3 through 5 (Fig. 4c–e) display later increase in expression, peaking at 5.5, 8, and 18 hours after the MPL dose. Hierarchical clustering of the corresponding probesets for each protein cluster was used to investigate the dependency of protein translation (or lack of it) on transcription. Of the 217 clustered proteins, 158 showed regulation of its mRNA as well. Interestingly, this analysis showed that, while a small number of the transcripts roughly correlate with expression of corresponding proteins, in most of the clusters a greater number of transcripts displayed the opposite pattern. This is consistent with the observations in hierarchical clustering of the concatenated dataset and emphasizes the prominent role of post-transcriptional regulation in establishing the pharmacologic effects of MPL in liver.
Table 3.
Distribution of Elements after Clustering and Subclustering of Data in Two-Way Analysis
| Proteomics → Transcriptomics | ||
|---|---|---|
| # of proteins | Corresponding # of probesets in the transcriptomic dataset | |
| Cluster 1 | 44 | 34 |
| Cluster 2 | 30 | 27 |
| Cluster 3 | 72 | 45 |
| Cluster 4 | 42 | 29 |
| Cluster 5 | 29 | 23 |
| Transcriptomics → Proteomics | ||
|---|---|---|
| # of probesets | Corresponding # of proteins in the proteomic dataset | |
| Cluster 1 | 413 | 66 |
| Cluster 2 | 155 | 30 |
| Cluster 3 | 92 | 25 |
| Cluster 4 | 334 | 85 |
| Cluster 5 | 138 | 11 |
FIG. 4.
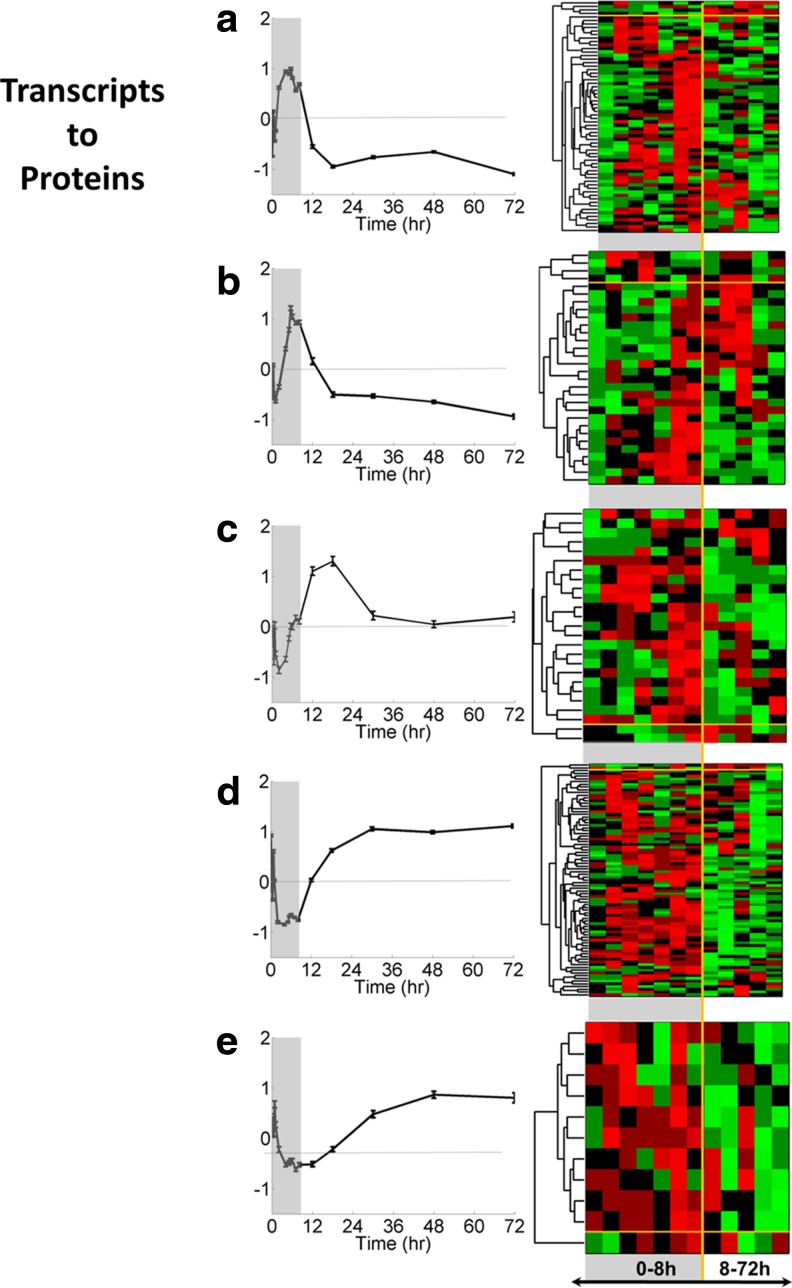
Five clusters of proteins obtained by consensus clustering (a–e, left side), and heat maps of corresponding hierarchically clustered probesets (a–e, right side). Canonical pathways enriched by the proteins in these clusters are given in Table 6.
In addition, this analysis was repeated in the reverse direction (i.e., starting from the transcriptomic dataset and progressing to the proteomic dataset). 1624 of the probe sets were differentially expressed, 1132 of those were in five clusters obtained by consensus clustering. Only 217 of these 1132 probe sets had corresponding proteins in the proteomic dataset. Distribution of these 217 proteins to their correspondent probe set clusters is shown in Table 3. Two of the clusters were considerably more densely populated than the others. The transcriptional profiles of these clusters are shown in Figure 5a and 5d (left side), and they can be considered as the most dominant early-up/late-downregulation and early-down/late-upregulation patterns. Heat maps on the right display the expression patterns of corresponding proteins for each cluster. Compared to the first part of sequential clustering analysis, considerably fewer proteins actually correlate with the transcriptional profile of their respective clusters. Especially for the most densely populated cluster, Figure 5d, only a couple of proteins show early-down/late-upregulation pattern in parallel with the corresponding temporal pattern of the first cluster. The only cluster in which expression of the majority of proteins go more or less in parallel with the temporal profile of the corresponding transcriptional cluster is the first cluster (Fig. 5a).
FIG. 5.
Five clusters of probesets obtained by consensus clustering (a–e, left side), and heat maps of corresponding hierarchically clustered protein datasets (a–e, right side).
Considering that protein expression is a more reliable predictor of function, the annotation analysis was based on the proteomic data in this part of the analysis. The proteins included in the clusters shown in Figure 4 were functionally annotated through core analysis in IPA, pathways with an enrichment score higher than 0.05 were considered significant and obtained results are shown in Table 4. The protein clusters were numbered according to the time at which the peak activity was observed. Four clusters displayed increasing activity sequentially within the first 8 hours, and one showed a relatively delayed activity, around 18 h after MPL dosing.
Table 4.
Functional Annotation of Groups of Proteins with Distinct Expression Patterns in Response to MPL as Identified in Clustering Analysis*
| Canonical pathway | Genes |
|---|---|
| Cluster 1 | |
| L-Cysteine degradation II | CTH |
| Citrulline degradation | OTC |
| Formaldehyde oxidation II (glutathione-dependent) | ADH5 |
| Fatty acid β-oxidation I | ACAA1, ACAA2 |
| LPS/IL-1 mediated inhibition of RXR function | GSTM2, Cyp2a2, SULT1E1, FABP1 |
| Methylglyoxal degradation I | HAGH |
| Cysteine biosynthesis/Homocysteine degradation | CTH |
| Estrogen biosynthesis | CYP2C18, HSD17B2 |
| Proline biosynthesis II (from arginine) | OTC |
| Phenylalanine degradation I (aerobic) | QDPR |
| CMP-N-acetylneuraminate biosynthesis I (eukaryotes) | GNE |
| PPARα/RXRα activation | ACAA1, CYP2C18, Cyp2c44 |
| Arginine biosynthesis IV | OTC |
| Urea cycle | OTC |
| Citrulline biosynthesis | OTC |
| Acyl-CoA hydrolysis | Ces1e |
| Glutathione redox reactions I | GPX1 |
| Cluster 2 | |
| Ketogenesis | HADHB, BDH1, HMGCL |
| Ketolysis | HADHB, BDH1 |
| Serotonin degradation | UGT2B17, ADH1C, UGT2B15 |
| Xenobiotic metabolism signaling | ALDH1L2, UGT2B17, UGT2B15, Gsta3, HSP90AA1 |
| Isoleucine degradation I | HADHB, ACADSB |
| Valine degradation I | HADHB, ACADSB |
| Thyroid hormone metabolism II (via conjugation and/or degradation) | UGT2B17, UGT2B15 |
| Sulfate activation for sulfonation | PAPSS2 |
| Thiosulfate disproportionation III (rhodanese) | TST |
| Sorbitol degradation I | SORD |
| Aryl hydrocarbon receptor signaling | ALDH1L2, Gsta3, HSP90AA1 |
| Nicotine degradation | UGT2B17, UGT2B15 |
| Glutamate degradation II | GOT2 |
| Aspartate biosynthesis | GOT2 |
| Superpathway of melatonin degradation | UGT2B17, UGT2B15 |
| Glycogen biosynthesis II (from UDP-D-Glucose) | UGP2 |
| L-Cysteine degradation I | GOT2 |
| Aspartate degradation II | GOT2 |
| LPS/IL-1 mediated inhibition of RXR function | ALDH1L2, Gsta3, PAPSS2 |
| Sucrose degradation V | KHK |
| Leucine degradation I | HMGCL |
| Colanic acid building blocks biosynthesis | UGP2 |
| Mevalonate pathway I | HADHB |
| Phenylalanine degradation IV (mammalian, via side chain) | GOT2 |
| Superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | HADHB |
| Glutaryl-CoA degradation | HADHB |
| Cluster 3 | |
| Mitochondrial dysfunction | HSD17B10, SDHA, NDUFA9, XDH, CYB5R3, COX5A, NDUFS3, MAOA, AIFM1 |
| TCA cycle II | SDHA, SUCLA2, ACO2, ACO1 |
| Serotonin degradation | HSD17B10, ALDH3A2, PECR, SULT1C3, MAOA |
| Ethanol degradation II | HSD17B10, ALDH3A2, PECR, ACSL1 |
| Noradrenaline and adrenaline degradation | HSD17B10, ALDH3A2, PECR, MAOA |
| Tryptophan degradation X (mammalian, via tryptamine) | ALDH3A2, DDC, MAOA |
| Dopamine degradation | ALDH3A2, SULT1C3, MAOA |
| LPS/IL-1 mediated inhibition of RXR function | ALDH3A2, ACOX1, ALDH8A1, Cyp2a12/Cyp2a22, SULT1C3, ACSL1, MAOA |
| Adenosine nucleotides degradation II | XDH, AOX1 |
| Urate biosynthesis/Inosine 5′-phosphate degradation | XDH, AOX1 |
| Guanosine nucleotides degradation III | XDH, AOX1 |
| Purine nucleotides degradation II (aerobic) | XDH, AOX1 |
| Oxidative ethanol degradation III | ALDH3A2, ACSL1 |
| Phenylalanine degradation IV (mammalian, via side chain) | ALDH3A2, MAOA |
| Serotonin receptor signaling | DDC, MAOA |
| Putrescine degradation III | ALDH3A2, MAOA |
| Ethanol degradation IV | ALDH3A2, ACSL1 |
| γ-Linolenate biosynthesis II (animals) | CYB5R3, ACSL1 |
| tRNA charging | DARS, TARS |
| Glutamine biosynthesis I | Glul |
| Thyroid hormone biosynthesis | CTSD |
| GDP-glucose biosynthesis | PGM1 |
| 4-Aminobutyrate degradation I | SUCLG2 |
| Acetate conversion to acetyl-CoA | ACSL1 |
| Fatty acid β-oxidation I | HSD17B10, ACSL1 |
| Glucose and glucose-1-phosphate degradation | PGM1 |
| Pentose phosphate pathway (oxidative branch) | PGLS |
| Heme degradation | BLVRB |
| Serotonin and melatonin biosynthesis | DDC |
| Catecholamine biosynthesis | DDC |
| Phenylethylamine degradation I | ALDH3A2 |
| Melatonin degradation II | MAOA |
| Glutamate degradation III (via 4-aminobutyrate) | SUCLG2 |
| Glycogen degradation II | PGM1 |
| NAD phosphorylation and dephosphorylation | NADK2 |
| Pentose phosphate pathway | PGLS |
| Glycogen degradation III | PGM1 |
| Leucine degradation I | MCCC2 |
| Purine nucleotides de novo biosynthesis II | ADSS |
| Cluster 4 | |
| EIF2 signaling | RPL3, EIF4A2, EIF4G1, RPLP0 |
| Acetyl-CoA biosynthesis III (from citrate) | ACLY |
| 4-Hydroxybenzoate biosynthesis | TAT |
| 4-Hydroxyphenylpyruvate biosynthesis | TAT |
| Remodeling of epithelial adherens junctions | TUBB3, ARF6 |
| Methylmalonyl pathway | MUT |
| Histidine degradation III | FTCD |
| 2-Oxobutanoate degradation I | MUT |
| Tyrosine degradation I | TAT |
| Aspartate degradation II | MDH1 |
| Tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde | TDO2 |
| NAD biosynthesis II (from tryptophan) | TDO2 |
| Regulation of eIF4 and p70S6K signaling | EIF4A2, EIF4G1 |
| Lipid antigen presentation by CD1 | ARF6 |
| Bile acid biosynthesis, neutral pathway | CYP27A1 |
| Gluconeogenesis I | MDH1 |
| Cluster 5 | |
| Superpathway of methionine degradation | CBS, MAT1A, GOT1, MAT2A, BHMT2 |
| Cysteine biosynthesis III (mammalia) | CBS, MAT1A, MAT2A |
| S-Adenosyl-L-methionine biosynthesis | MAT1A, MAT2A |
| Acute phase response signaling | HPX, HP, C3, TF, FGB |
| LXR/RXR activation | HPX, C3, TF, ACACA |
| Methionine degradation I (to homocysteine) | MAT1A, MAT2A |
| L-Cysteine degradation III | GOT1 |
| Cysteine biosynthesis/Homocysteine degradation | CBS |
| Tyrosine biosynthesis IV | PAH |
| Melatonin degradation I | POR, Sult1a1 |
| 1, 25-Dihydroxyvitamin D3 biosynthesis | POR |
| Methionine salvage II (mammalian) | BHMT2 |
| Phenylalanine degradation I (aerobic) | PAH |
| Glutamate degradation II | GOT1 |
| Aspartate biosynthesis | GOT1 |
| Superpathway of melatonin degradation | POR, Sult1a1 |
| Biotin-carboxyl carrier protein assembly | ACACA |
| L-Cysteine degradation I | GOT1 |
| Aspartate degradation II | GOT1 |
| TR/RXR activation | HP, ACACA |
| Glycine betaine degradation | BHMT2 |
| Lipid antigen presentation by CD1 | CANX |
| Phenylalanine degradation IV (mammalian, via side chain) | GOT1 |
Cluster profiles are shown in Figure 4.
Discussion
Characterization and analysis of global gene expression changes has become an integral part in studying mechanisms of actions of various pharmacological agents (Butte, 2002). Developments in high-throughput methodologies such as microarrays allow for relatively affordable and faster characterization of the transcriptomics. These genomic approaches offer a powerful tool in understanding drug effects at the molecular level and aid in target and biomarker discoveries and in gaining insights into modulation of relevant pathways. For a long time, our laboratories have utilized Affymetrix gene chips to understand the tissue-specific effects of CS actions in a variety of peripheral tissues (Almon et al., 2003; 2005; 2007a). These studies helped in understanding of various pathways modulated by CS at the transcriptional level that are either common across tissues or unique to certain tissue types (Nguyen et al., 2010; Yang et al., 2009). In addition, through clustering and other bioinformatic analyses of time series, we were able to identify genes that share similar expression patterns across different dosing regimens and relate drug effects/side effects to functional pathways and gene clusters showing distinct temporal profiles (Nguyen et al., 2010).
Although information on mRNA expression changes helps in understanding mechanisms of drug action, some studies show that message expression changes may not correlate well with protein changes and hence might not accurately reflect drug effects (as the majority of pathway modulators and drug targets are proteins) (Nishizuka et al., 2003; Shankavaram et al., 2007). Hence characterization of changes in protein expression at the proteome level (along with gene expression profiling) will not only reveal the dynamic and temporal features of drug-induced protein changes, but will also provide rich biological information that may lead to improved understanding of diverse drug effects at both transcriptional and translational levels. However, comprehensive, accurate, and reliable profiling of protein expression remains highly challenging because of the extreme diversity of the chemical and physical properties of proteins, the large dynamic ranges in concentrations in most proteomes, and the fact that drug-responsive proteins are often in low abundance. We recently developed a robust and highly sensitive label-free quantification strategy for accurate expression profiling of complex tissue proteomes, with the capacity for analyzing large numbers of biological samples (Tu et al., 2012). This strategy was utilized to characterize the temporal changes in expression of thousands of proteins after a single dose of MPL (Nouri-Nigjeh et al., 2014).
Availability of rich time-series datasets for changes in both mRNA and protein expression after single-dose administration of MPL allowed us to integrate both transcriptional and translational states and enabled us to perform a unique analysis to compare and contrast the two. With this analysis we developed an algorithm to compare temporal changes in both gene and protein expression. This allowed us to examine the relationship between the two and to differentiate the transcriptional and translational effects of CS dosing. These drugs affect a wide range of pathways that are involved in metabolism (carbohydrates, lipids, and proteins), immune-regulation and other critical cellular functions (Bialas and Routledge, 1998; Swartz and Dluhy, 1978). Because CS regulate diverse sets of genes and proteins, the dynamic effects of these drugs provide a relevant system to compare, contrast, and integrate both the genomic and proteomic data. Although our data were obtained from different but very similar animal studies, it is reasonable to integrate the two for the following reasons: both studies were performed in the same strain of rats (Wistar) that were adrenalectomized and maintained under similar conditions; MPL was given in both studies and identical doses of 50 mg/kg were used. The only major difference between the two studies is that the gene expression analysis were performed in animals that were given an intravenous dose of the drug and the proteomic measurements were performed in rats given an intramuscular injection of MPL. However, our previous studies comparing the two routes of MPL dosing indicated that, though there are some early differences in the pharmacokinetic profiles of MPL, the pharmacodynamics of an important biomarker tyrosine aminotransferase (TAT) expressed in liver were comparable (Hazra et al., 2007).
With the hierarchical clustering of the concatenated genomic and proteomic data we identified two dominant patterns, one of which showed upregulation of expression at both mRNA and protein levels (Fig. 2). Most of the genes and proteins in this cluster show similar temporal patterns (with peak expression occurring at similar times) or with a slight delay in the peak expression time of the protein compared to the gene profiles. For example, some of the classic pharmacodynamics markers for CS actions, including tyrosine aminotransferase (TAT) and aspartate aminotransferase (GOT1), fall into this category with upregulation of both the mRNA and protein expression after MPL dosing. Similarly, this group includes genes/proteins involved in glucocorticoid signaling, confirming the direct pharmacological action of MPL at both the genomic and proteomic levels (Hazra et al., 2008a). One of the primary effects of CS in liver is to stimulate gluconeogenesis (production of glucose) from amino acids released by protein breakdown in muscle (because of CS action) through deamination and utilization of the carbon backbone for glucose production in the liver. As a result, the excess amines are removed through the urea cycle to maintain proper homeostasis (Bialas and Routledge, 1998; Hazra et al., 2008a). As illustrated in Table 1, Cluster 1 includes genes and proteins involved in the urea cycle that are upregulated after MPL dosing and both gene and protein expression share similar temporal profiles. The other functional sets that are enriched in Cluster 1 include the ones involved in protein translation and processing (Table 1). Ribosomal proteins and translational factors that play critical roles in translation of mRNA to proteins and heat shock proteins that help in proper chaperoning of newly formed proteins show upregulation at both gene and protein levels (Warner and McIntosh, 2009). All the genes/proteins in Cluster 1 represent direct transcriptional effects of CS, resulting in temporal expression changes in mRNA that directly translate to concurrent or slightly delayed protein expression changes. Since mRNA profiling is more straightforward, well established, and cheaper than the protein counterpart, an important point to note here is that mRNA expression markers can be representative of their corresponding proteins in assessing the effects of CS actions for genes/proteins populated in Cluster 1.
Genes and protein expression profiles in Cluster 2 are more intriguing, and we have very limited understanding of the biology and mechanisms behind this observation. Although previous studies have been conducted in identifying differences in the changes at the transcriptional and translational levels for one or selected few proteins, global assessment through high throughput methodologies have never tried to address this issue (Nouri-Nigjeh et al., 2014; Qu et al., 2006; Sukumaran et al., 2011). Studies that have characterized and compared genomic and proteomic data have almost always been single time-point studies that do not provide the relevant temporal information necessary to identify and characterize transcriptional and translational differences (de Godoy et al., 2008; Gry et al., 2009). These single time-point high throughput studies showed poor correlation between mRNA and protein expression data (Gry et al., 2009). Analysis from our study shows that some of the functional pathways involved in lipid, protein, and xenobiotic metabolism that are important pharmacological targets of CS showed different temporal expression patterns for mRNA and proteins. This suggests that, in addition to the direct transcriptional effects of CS, there could be additional translational or post-translational effects that result in different protein expression temporal patterns compared to the mRNA profiles. Factors including regulation of microRNAs (which can alter protein translation) and direct or indirect translational controls could produce the different mRNA and protein profiles (He and Hannon, 2004). Whatever the mechanisms that control the difference between gene and protein expression are, mRNA profiles in Cluster 2 cannot be directly used for deciphering the regulation of the functional pathways they represent.
Conclusions
The results of the present study elicited both expected and unexpected relationships between mRNA transcription and protein translation in liver after a single CS dose. Roughly half of the genes commonly found in both transcriptomic and proteomic datasets had complementary temporal profiles, indicating regulation at the transcriptional level. Some of the functions with which these genes are associated were regulation of corticosteroid signaling, protein degradation, and translation machinery. The lack of complementarity between message and protein expression profiles in the other half of genes was intriguing. Although our understanding of the involved mechanisms is limited at this point, this result suggested additional translational or post-translational impacts of CS in addition to their direct transcriptional effects.
Independent from the corresponding profiles, we also examined the rich time-series data through stringent two-way clustering and subclustering approach. We used proteomic data to portray the cellular landscape after CS dose due to its higher priority in representing the actual phenotype. This allowed us to define the prominent temporal shifts in protein expression and to determine associated cellular functions.
Acknowledgments
KK and IPA gratefully acknowledge the financial support from NIH Grant GM082974. This work was also supported, in part, by NIH Grants GM24211 (WJJ), U54HD071594 (WJJ and JQ), DA027528 (JQ), AI060260 (JQ), HD075363 (JQ), and HL103411 (JQ), by the Center for Protein Therapeutics (JQ), and by American Heart Association (AHA) Award 12SDG9450036 (JQ).
Author Disclosure Statement
The authors declare there are no conflicting financial interests.
References
- Almon RR, DuBois DC, and Jusko WJ. (2007a). A microarray analysis of the temporal response of liver to methylprednisolone: A comparative analysis of two dosing regimens. Endocrinology 148, 2209–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almon RR, DuBois DC, Pearson KE, Stephan DA, and Jusko WJ. (2003). Gene arrays and temporal patterns of drug response: Corticosteroid effects on rat liver. Funct Integrat Genomics 3, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almon RR, DuBois DC, Yao Z, Hoffman EP, Ghimbovschi S, and Jusko WJ. (2007b). Microarray analysis of the temporal response of skeletal muscle to methylprednisolone: Comparative analysis of two dosing regimens. Physiol Genomics 30, 282–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almon RR, Lai W, DuBois DC, and Jusko WJ. (2005). Corticosteroid-regulated genes in rat kidney: Mining time series array data. Am J Physiol Endocrinol Metabol 289, E870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RC, and Walker BR. (1999). Glucocorticoids and insulin resistance: Old hormones, new targets. Clin Sci (Lond) 96, 513–523 [DOI] [PubMed] [Google Scholar]
- Barnes PJ. (1998). Anti-inflammatory actions of glucocorticoids: Molecular mechanisms. Clin Sci (Lond) 94, 557–572 [DOI] [PubMed] [Google Scholar]
- Bialas MC, and Routledge PA. (1998). Adverse effects of corticosteroids. Adverse Drug Reactions Toxicol Rev 17, 227–235 [PubMed] [Google Scholar]
- Butte A. (2002). The use and analysis of microarray data. Nature Rev Drug Discovery 1, 951–960 [DOI] [PubMed] [Google Scholar]
- Chrousos GP, and Kino T. (2005). Intracellular glucocorticoid signaling: A formerly simple system turns stochastic. Science's STKE 2005, pe48. [DOI] [PubMed] [Google Scholar]
- De Godoy LM, Olsen JV, Cox J, et al. (2008). Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, and Gerstein M. (2003). Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gry M, Rimini R, Stromberg S, et al. (2009). Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics 10, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, and Pal R. (2013). Integrated analysis of transcriptomic and proteomic data. Curr Genomics 14, 91–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A, DuBois DC, Almon RR, Snyder GH, and Jusko WJ. (2008a). Pharmacodynamic modeling of acute and chronic effects of methylprednisolone on hepatic urea cycle genes in rats. Gene Reg Systems Biol 2, 1–19 [PMC free article] [PubMed] [Google Scholar]
- Hazra A, Pyszczynski N, DuBois DC, Almon RR, and Jusko WJ. (2007). Pharmacokinetics of methylprednisolone after intravenous and intramuscular administration in rats. Biopharmaceut Drug Disposition 28, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A, Pyszczynski NA, DuBois DC, Almon RR, and Jusko WJ. (2008b). Modeling of corticosteroid effects on hepatic low-density lipoprotein receptors and plasma lipid dynamics in rats. Pharmaceut Res 25, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, and Hannon GJ. (2004). MicroRNAs: Small RNAs with a big role in gene regulation. Nature Rev Genetics 5, 522–531 [DOI] [PubMed] [Google Scholar]
- Hegde PS, White IR, and Debouck C. (2003). Interplay of transcriptomics and proteomics. Curr Opin Biotechnol 14, 647–651 [DOI] [PubMed] [Google Scholar]
- Jin JY, Almon RR, DuBois DC, and Jusko WJ. (2003). Modeling of corticosteroid pharmacogenomics in rat liver using gene microarrays. J Pharmacol Exp Therapeut 307, 93–109 [DOI] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, and Storey JD. (2006). EDGE: Extraction and analysis of differential gene expression. Bioinformatics 22, 507–508 [DOI] [PubMed] [Google Scholar]
- Morand EF, and Leech M. (1999). Glucocorticoid regulation of inflammation: The plot thickens. Inflam Res 48, 557–560 [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Almon RR, DuBois DC, Jusko WJ, and Androulakis IP. (2010). Comparative analysis of acute and chronic corticosteroid pharmacogenomic effects in rat liver: Transcriptional dynamics and regulatory structures. BMC Bioinformat 11, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Nowakowski RS, and Androulakis IP. (2009). Unsupervised selection of highly coexpressed and noncoexpressed genes using a consensus Clustering approach. OMICS 13, 219–237 [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Lindon JC, and Wilson ID. (2004). The challenges of modeling mammalian biocomplexity. Nat Biotechnol 22, 1268–1274 [DOI] [PubMed] [Google Scholar]
- Nishizuka S, Chen ST, Gwadry FG, et al. (2003). Diagnostic markers that distinguish colon and ovarian adenocarcinomas: Identification by genomic, proteomic, and tissue array profiling. Cancer Res 63, 5243–5250 [PubMed] [Google Scholar]
- Nouri-Nigjeh E, Sukumaran S, Tu C, et al. (2014). Highly multiplexed and reproducible ion current-based strategy for large-scale quantitative proteomics and the application to protein expression dynamics induced by methylprednisolone in 60 rats. Anal Chem 86, 8149–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley RH, and Cidlowski JA. (2011). Cellular processing of the glucocorticoid receptor gene and protein: New mechanisms for generating tissue-specific actions of glucocorticoids. J Biol Chem 286, 3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Jusko WJ, and Straubinger RM. (2006). Utility of cleavable isotope-coded affinity-tagged reagents for quantification of low-copy proteins induced by methylprednisolone using liquid chromatography/tandem mass spectrometry. Anal Chem 78, 4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf MJ, and Cidlowski JA. (2002). Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem Mol Biol 83, 37–48 [DOI] [PubMed] [Google Scholar]
- Schacke H, Docke WD, and Asadullah K. (2002). Mechanisms involved in the side effects of glucocorticoids. Pharmacol Therapeut 96, 23–43 [DOI] [PubMed] [Google Scholar]
- Shankavaram UT, Reinhold WC, Nishizuka S, et al. (2007). Transcript and protein expression profiles of the NCI-60 cancer cell panel: An integromic microarray study. Mol Cancer Therapeut 6, 820–832 [DOI] [PubMed] [Google Scholar]
- Storey JD, Dai JY, and Leek JT. (2007). The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics 8, 414–432 [DOI] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, and Davis RW. (2005). Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA 102, 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Jusko WJ, DuBois DC, and Almon RR. (2011). Mechanistic modeling of the effects of glucocorticoids and circadian rhythms on adipokine expression. J Pharmacol Exp Therapeut 337, 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz SL, and Dluhy RG. (1978). Corticosteroids: Clinical pharmacology and therapeutic use. Drugs 16, 238–255 [DOI] [PubMed] [Google Scholar]
- Tu C, Li J, Bu Y, Hangauer D, and Qu J. (2012). An ion-current-based, comprehensive and reproducible proteomic strategy for comparative characterization of the cellular responses to novel anti-cancer agents in a prostate cell model. J Proteomics 77, 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegiopoulos A, and Herzig S. (2007). Glucocorticoids, metabolism and metabolic diseases. Mol Cell Endocrinol 275, 43–61 [DOI] [PubMed] [Google Scholar]
- Warner JR, and Mcintosh KB. (2009). How common are extraribosomal functions of ribosomal proteins? Mol Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters KM, Pounds JG, and Thrall BD. (2006). Data merging for integrated microarray and proteomic analysis. Brief Funct Genomic Proteomic 5, 261–272 [DOI] [PubMed] [Google Scholar]
- Yang EH, Almon RR, DuBois DC, Jusko WJ, and Androulakis IP. (2009). Identification of global transcriptional dynamics. PLoS ONE 4, e5992. [DOI] [PMC free article] [PubMed] [Google Scholar]