Abstract
Reversible protein phosphorylation, regulated by protein kinases and phosphatases, mediates a switch between protein activity and cellular pathways that contribute to a large number of cellular processes. The Mycobacterium tuberculosis genome encodes 11 Serine/Threonine kinases (STPKs) which show close homology to eukaryotic kinases. This study aimed to elucidate the phosphoproteomic landscape of a clinical isolate of M. tuberculosis. We performed a high throughput mass spectrometric analysis of proteins extracted from an early-logarithmic phase culture. Whole cell lysate proteins were processed using the filter-aided sample preparation method, followed by phosphopeptide enrichment of tryptic peptides by strong cation exchange (SCX) and Titanium dioxide (TiO2) chromatography. The MaxQuant quantitative proteomics software package was used for protein identification. Our analysis identified 414 serine/threonine/tyrosine phosphorylated sites, with a distribution of S/T/Y sites; 38% on serine, 59% on threonine and 3% on tyrosine; present on 303 unique peptides mapping to 214 M. tuberculosis proteins. Only 45 of the S/T/Y phosphorylated proteins identified in our study had been previously described in the laboratory strain H37Rv, confirming previous reports. The remaining 169 phosphorylated proteins were newly identified in this clinical M. tuberculosis Beijing strain. We identified 5 novel tyrosine phosphorylated proteins. These findings not only expand upon our current understanding of the protein phosphorylation network in clinical M. tuberculosis but the data set also further extends and complements previous knowledge regarding phosphorylated peptides and phosphorylation sites in M. tuberculosis.
Keywords: M. tuberculosis, phosphoproteomics, tyrosine phosphorylation, serine phosphorylation, threonine phosphorylation
Introduction
According to the World Health Organization (WHO), tuberculosis (TB) ranks as the second leading cause of death from an infectious disease worldwide, after HIV (WHO|Global tuberculosis report 2014, 2014). It is estimated that one third of the world's population is infected with Mycobacterium tuberculosis, the causative agent of TB and 8.6 million new TB cases were reported in 2012 alone (WHO|Global tuberculosis report 2013, 2013). In order to control this epidemic there is a critical need for the development of effective and affordable anti-TB therapy and diagnostic tools.
Harnessing the power of the field of proteomics provides a unique opportunity to identify novel protein candidates for diagnosis and drug targets of pathogenic bacteria. Of particular interest is the identification of proteins with post-translational modifications (PTMs) as these modifications are often critical to protein functions, such as regulating protein-protein interactions, subcellular localization or modification of catalytic sites (Seo and Lee, 2004; Gupta et al., 2007). Protein phosphorylation is an important reversible PTM that directly or indirectly regulates signal transduction cascades linking the intracellular and extracellular environments. In bacteria, protein phosphorylation plays a fundamental role in the regulation of key processes ranging from metabolism and cellular homeostasis to cellular signaling which can be mediated by two classes of phosphorylation events (Cozzone, 1998). The underlying molecular mechanisms regulating protein phosphorylation and dephosphorylation is of great physiological importance due to its ability to ultimately affect protein activity, function, half-life or subcellular localization (McConnell and Wadzinski, 2009). Until recently it was thought that histidine/aspartate phosphorylation was the main mediator of signal transduction in bacteria (Frasch and Dworkin, 1996). However, with the advancement of mass spectrometry-based analyses serine/threonine and tyrosine kinases have been identified in a number of different bacteria (Macek et al., 2007; Macek and Mijakovic, 2011; Mijakovic and Macek, 2012).
The M. tuberculosis genome encodes 11 Serine/Threonine kinases (STPK's) (PknA, PknB, PknD, PknE, PknF, PknG, PknH, PknI, PknJ, PknK, PknL), two tyrosine phosphatases (PtpA, PtpB) and 11 two-component systems, highlighting the complexity of signaling network mediated by protein phosphorylation and thereby their potential as drug targets (Chopra et al., 2003; Koul et al., 2004; Sharma et al., 2004; Sala and Hartkoorn, 2011). Prisic et al. described the Serine/Threonine (S/T) phosphorylation profiles of the laboratory strain M. tuberculosis H37Rv under 6 different culture conditions (Prisic et al., 2010). This study identified 301 phosphorylated proteins after combining data from six different culture conditions (Prisic et al., 2010) and identified four phosphorylated STPKs, ribosomal and ribosome-associated proteins as well as phosphorylated substrates which suggest that protein phosphorylation provides a mechanism for regulating key physiological process during infection. A more recent study of H37Rv further expanded the knowledge of the phosphoproteome by identifying novel tyrosine (Y) phosphorylated proteins in M. tuberculosis further supporting the broad regulation of its physiology by phosphorylation (Kusebauch et al., 2014).
In this study we report the phosphoproteome of a previously described clinical Beijing genotype M. tuberculosis isolate at early-logarithmic growth phase in liquid culture to provide further insight the influence of S/T/Y phosphorylation events on bacterial growth and virulence (de Souza et al., 2010). We used a combination of strong cation exchange (SCX) with Titanium dioxide (TiO2) enrichment in a mass spectrometry-based phosphoproteomic analysis of a hyper-virulent clinical M. tuberculosis isolate (de Souza et al., 2010). We confirmed the presence of previously described M. tuberculosis phosphorylated proteins and also identified novel phosphorylated proteins and sites. In addition, this dataset identified novel tyrosine phosphorylation events, and thereby confirmed that there are multiple tyrosine kinase targets in this clinically relevant M. tuberculosis strain.
Materials and methods
Cell culture and lysate preparation
A previously described hyper-virulent clinical Beijing genotype Mycobacterium tuberculosis isolate, SAW5527, isolated from a TB patient attending a primary health care clinic in the Western Cape province, South Africa was used for this phosphoproteomics analysis (de Souza et al., 2010). Secondary cultures were inoculated into 50 ml 7H9 Middlebrooks medium supplemented with Dextrose and Catalase and incubated at 37°C until early-logarithmic phase (OD600 between 0.6 and 0.7). Mycobacterial cells were collected by centrifugation (2000 × g for 10 min at 4°C) and washed two times with cold lysis buffer containing 10 mM Tris-HCl (pH 7.4), 0.1% Tween-80, Complete Protease inhibitor cocktail (Roche, Mannheim Germany) and Phosphatase inhibitor cocktail (Roche, Mannheim Germany). An equal amount of 0.1 mm glass beads (Biospec Products Inc., Bartlesville, OK) was added to the cell pellet after centrifugation together with cold 300 μl lysis buffer and 10 μl DNaseI (2U/ml) (NEB, New England Laboratories). Lysis was achieved by mechanical bead-beating in a Rybolyser (Bio101 SAVANT, Vista, CA) for 6 cycles of 20 s at a speed of 4.0 m.s−1, with 1 min cooling periods on ice. The whole cell lysates were filter-sterilized with a sterile 0.22 μm pore acrodisc 25 mm PF syringe filter (Pall Life Sciences, Pall Corporation, Ann Arbour, MI) and stored at −80°C. The protein concentration of the whole cell lysate was determined using the RC DC Protein assay according to manufacturer's instructions (BioRad). A single biological replicate was analyzed in triplicate for downstream phosphoproteomic analysis.
Filter aided sample preparation and trypsin digestion
Four milligrams of concentrated whole cell lysate proteins was heated in 4% SDS buffer and 0.1 M dithiothreitol (DTT) in 100 mM Tris/HCl pH 7.5. The samples were processed using Filter Aided Sample Preparation (FASP) (Wiśniewski et al., 2009). In brief, 4 mg dried whole cell lysate protein was resuspended in 250 μl of urea (UA) and loaded onto a 15 ml Amicon filtration device (30 kDa MWCO) and centrifuged at 2000 × g for 40 min at 25°C. After centrifugation, the flow-through was collected in a clean falcon tube and discarded. The concentrated whole cell lysate proteins in the filter unit were diluted in 2 ml 8 M Urea in 0.1 M Tris/HCl (pH 8.5) and re-centrifuged to remove the SDS. The flow-through was discarded and the remaining proteins in the filter unit were alkylated by mixing with 1.5 ml 50 mM iodacetamide (IAA) and incubated in the dark for 20 min to irreversibly modify cysteine. The alkylated proteins were equilibrated with 2 ml 50 mM ammonium bicarbonate (ABC) and digested with trypsin (Promega) in a protein to enzyme ratio of 100:1 at 37°C overnight. After trypsin digestion the filter unit is transferred to a clean collection tube and the peptides were eluted by centrifuged at 14 000 × g for 10 min. The eluted peptides were diluted in 50 μl water to avoid desalting for further processing of the peptide and acidified with trifluoroacetic acid (TFA).
Fractionation of peptides by strong cation exchange (SCX)
Extracted trypsin digested peptides were diluted to a volume of 7 ml in Solvent A (5 mM monopotassium phosphate (KH2PO4) 30% acetonitrile (ACN), pH 2.7). The pH of the diluted peptide samples was adjusted to 2.7 and made up to a volume of 10 ml with 100% ACN. The respective peptide samples were then separated at pH 2.7 by strong cation exchange (SCX ) by loading each peptide mixture onto a cation exchanger column (3.0 mm × 20 cm) (Poly LC, Columbia, MD) containing 5 μm polysulfoethyl aspartamine beads with a 200 Å pore size and a flow rate of 350 μl/min−1 equilibrated with SCX solvent A. The flow-through was collected and the bound peptides were eluted from the columns using an increasing salt gradient (0–30%) containing 5 mM KH2PO4 pH and 150 mM KCl. A total of 9 fractions were collected; 5 fractions generated by SCX based on UV absorbance (220 and 280), 3 from the flow-through and 1 salvage fraction (containing washes from the cation exchanger column) from the SCX column as an additional fraction.
Enrichment of phosphopeptides with TiO2 beads
All nine fractions (5 SCX, 3 SCX flow-through, 1 salvage fraction) were subjected to Titanium dioxide (TiO2) phosphopeptide enrichment. The TiO2-beads,10 μm in size, (GL Sciences, Inc., Japan) was resuspended 30 mg/ml dihydrobenzoic acid (DHB) (Sigma) to prevent non-specific binding. Each of the 9 fractions was incubated 4 times with TiO2 at a peptide to bead ratio of 1:2–1:8. Each fraction was rotated for 30 min, and then briefly centrifuged (14,000 g × 30 s). The supernatants were aspirated and transferred to a new labeled tube and the phosphopeptides bound to the TiO2beads were washed twice with 30% ACN and 3% TFA followed by washing twice with 75% ACN and 0.3% TFA. The enriched phosphorylated peptides were eluted with elution buffer containing 25% ammonia and ACN pH10. The eluted phosphopeptides were desalted using in house prepared C18 Stage tips.
LC-MS/MS analysis
The peptides were separated on a column packed in-house with C18 beads (reprosil-AQ Pur, Rd Maisch) on an Proxeon Easy-nLC system (Proxeon Biosystems, Odense, Denmark) using a binary gradient with buffer A (0.5% acetic acid in water) and buffer B (0.5% acetic acid and 80% ACN). The 4 μl of the enriched phosphopeptides from each of the 9 fractions were injected 3 times and were loaded directly without any trapping column with buffer A at a flow rate of 500 nl/min. Elution was carried out at a flow rate of 250 nl/min, with a linear gradient from 10 to 35% buffer B over a period of 95 min followed by 50% buffer B for 15 min. At the end of the gradient the column was washed with 90% buffer B and equilibrated with 5% buffer B for 10 min. The LC system was directly coupled in-line with the LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific) via the Proxeon Biosystems nanoelectrospray source. The mass spectrometer was programmed to acquire in a data-dependant mode with a resolution of 30,000 at 400 m/z with lock mass option enabled for the 445.120025. However, the target lock mass abundance was set to 0% in order to save the injection time for lock mass. For full scans 1E6 ions were accumulated within a maximum injection time of 250 ms in the C trap and detected in the Orbitrap mass analyser. The 10 most intense ions with charge states ≥2 were sequentially isolated and fragmented by high-energy collision dissociation (HCD) mode in the collision cell with normalized collision energy of 40% and detected in the Orbitrap analyser at 75,000 resolution. For HCD based method, the activation time option in the Xcalibur file was set to 0.1 ms. For the high-low strategy, full scans were acquired in the Orbitrap analyser at 60,000 resolution as parallel acquisition is enabled in the high-low mode. Up to the 10 most intense peaks with charge states ≥2 were selected for sequencing to a target value of 5000 with a maximum injection time of 25 ms and fragmented in the ion trap by HCD with normalized collision energy of 35%, activation of 0.25 and activation time of 10 ms.
Database search
The raw data acquired were processed using MaxQuant software version (1.4.1.2) and processed as per default workflow. Since HCD spectra were acquired in profile mode, deisotoping was performed similar to survey MS scans to obtain singly charged peak lists and searched against the M. tuberculosis H37Rv protein database (version R11 tuberculist.epfl.ch). The search included cysteine carbamidomethylation as a fixed modification while N-acetylation, oxidation of methionine and phosphorylation at serine, threonine and tyrosine were set as variable modifications. Up to two missed cleavages were allowed for protease digestion and a peptide had to be fully tryptic. Identifications were filtered at 1% FDR at three levels namely; site, peptide and protein using reversed sequences. As such there is no fixed cut-off score threshold but instead spectra were accepted until the 1% false discovery rate (FDR) is reached. Only peptides with a minimum length of 7 amino acids were considered for identification and detected in at least one or more of the replicates. All phosphopeptide spectra were manually validated by applying stringent acceptance criteria: only phosphorylation events on S/T/Y with a localization probability of ≥0.75 and PEP ≤ 0.01 were used for further analysis and reported as high confidence localized phosphosites.
Gene ontology analysis
The categorization of identified phosphorylated proteins in terms of functional categorization, molecular function, biological processes and cellular components was carried out using TubercuList-Mycobacterium tuberculosis Database.
Results
In this study we set out to analyse the phosphoproteome of a hyper-virulent Beijing genotype M. tuberculosis isolate by extracting whole cell lysate proteins at early-logarithmic growth (OD600 of 0.8) (Figure 1) which resulted in the identification of 619 MS/MS spectra. The 274 LC-MS/MS spectra that fulfilled the criteria for high confidence phosphosites identified a total of 414 (38:59:3%) S/T/Y phosphorylation sites present in 214 M. tuberculosis H37Rv proteins (Supplementary Table S2; Supplementary Figure S1).
Figure 1.
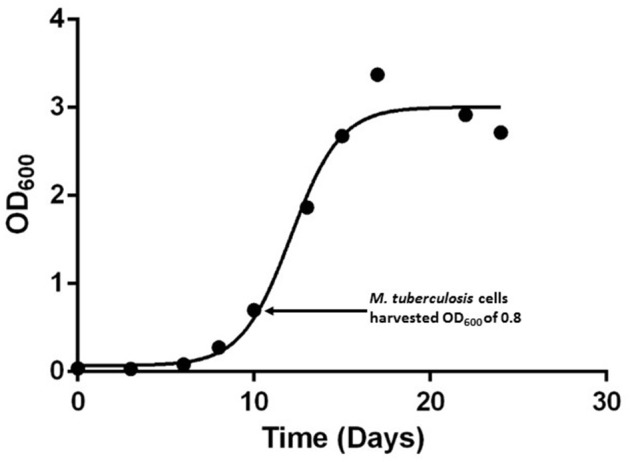
Growth curves of hyper-virulent M. tuberculosis Beijing strain. Growth monitored over a 24 day period using OD600 measurements in 7H9 Middlebrooks liquid media supplemented with dextrose and catalase.
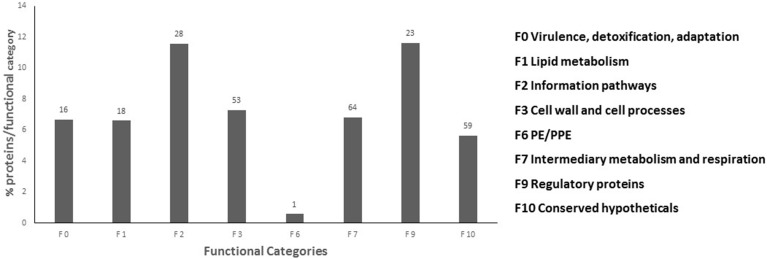
Of the 401 serine/threonine phosphorylation sites (pS/T), only 156 had been previously described for M. tuberculosis (Table 1). Only 6 of 13 tyrosine phosphorylation sites (pY) has been previously described for M. tuberculosis (Kusebauch et al., 2014). The remaining 245 pS/T and 7 pY were uniquely identified in this study (Supplemental Table S2). To determine whether phosphorylated proteins were differentially represented in any particular cellular process, we grouped the phosphorylated proteins based on their functional category according to Tuberculist (Lew et al., 2011) (Figure 2). One hundred and seventy (79.4%) of the phosphorylated proteins had an annotated function, while the remaining 59 phosphorylated proteins (20.5%) were assigned as hypothetical. The biological function of the annotated proteins varied from transcription, translation, protein biosynthesis, fatty acid metabolism to phosphorylation. Our analysis identified phosphorylated forms of the 9 of the 11 M. tuberculosis STPK's: PknA, PknB, PknD, PknF, PknG PknE, PknH, PknJ, and PknL (Table 2). Of these, phosphorylated forms of PknE, PknH, PknJ, and PknL had not been previously described in M. tuberculosis.
Table 1.
List of phosphopeptides identified in this and previous studies of M. tuberculosis.
| Rv numbers | Protein name | Phosphopeptides | Phospho-residue | References |
|---|---|---|---|---|
| Rv0007 | Rv0007 | FISGASAPVTGPAAAVR | Not known | Prisic et al., 2010 |
| FIS*GAS*APVT*GPAAAVR | S; T | This study | ||
| FIS*GASAPVT*GPAAAVR | S; T | This study | ||
| Rv0014c | PknB | AIADSGNSVTQTAAVIGTAQYLSPEQAR | Not known | Prisic et al., 2010 |
| AIADSGNSVT*QT*AAVIGTAQYLSPEQAR | T | This study | ||
| AIADSGNS*VT*QTAAVIGTAQYLSPEQAR | S; T | This study | ||
| TSLLSSAAGNLSGPRTDPLPR | Not known | Prisic et al., 2010 | ||
| TSLLSSAAGNLS*GPRTDPLPR | S | This study | ||
| TSLLSSAAGNLSGPRT*DPLPR | T | This study | ||
| Rv0015c | PknA | RPFAGDGALT$VAMK | T | Prisic et al., 2010; This study |
| Rv0020c | FhaA | FEQSSNLHTGQFR | Not known | Prisic et al., 2010 |
| FEQS*SNLHT*GQFR | S; T | This study | ||
| HPDQGDY$PEQIGYPDQGGYPEQR | Y | Kusebauch et al., 2014; This study | ||
| QDYGGGADY$TR | Y | Kusebauch et al., 2014; This study | ||
| VPGY$APQGGGYAEPAGR | Y | Kusebauch et al., 2014; This study | ||
| Rv0175 | Rv0175 | AADSAESDAGADQTGPQVK | Not known | Prisic et al., 2010 |
| AADSAESDAGADQT*GPQVK | T | This study | ||
| Rv0204c | Rv0204c | DPPT$DPNLR | Prisic et al., 2010; This study | |
| Rv0227c | Rv0227c | GGFEEPVPGAEAETEKLPTQRPDFPR | Not known | Prisic et al., 2010 |
| GGFEEPVPGAEAET$EK | T | Prisic et al., 2010; This study | ||
| Rv0351 | GrpE | RIDPET#GEVR | T | Prisic et al., 2010 |
| IDPET*GEVR | T | This study | ||
| Rv0389 | PurT | AAGHQVQPQT$GGVSPR | T | Prisic et al., 2010; This study |
| Rv0410c | PknG | SGPGTQPADAQTAT#SATVRPL | T | O'Hare et al., 2008 |
| S*GPGT*QPADAQTATSATVR | S; T | This study | ||
| SGPGTQPADAQTAT*S*ATVRPLSTQAVFR | S; T | This study | ||
| SGPGTQPADAQTAT*SAT*VR | T | This study | ||
| PLST#QAVFRPDFGDEDNFPHPTLGPDTEPQDR | T | O'Hare et al., 2008 | ||
| PLS*T*QAVFR | S; T | This study | ||
| Rv0421c | Rv0421c | GLAEGPLIAGGHS#YGGR | S | Prisic et al., 2010 |
| GLAEGPLIAGGHS*Y*GGR | S; Y | This study | ||
| Rv0440 | GroEL2 | KWGAPTIT$NDGVSIAK | T | Molle et al., 2006 |
| WGAPTIT*NDGVSIAK | T | This study | ||
| AVEKVT#ETLLK | T | Molle et al., 2010 | ||
| VTET*LLK | T | This study | ||
| Rv0497 | Rv0497 | RGDSDAITVAELT$GEIPIIR | T | Prisic et al., 2010; This study |
| T*GPHPETESSGNR | T | This study | ||
| Rv0685 | Tuf | PDLNET$KAFDQ | T | Sajid et al., 2011 |
| VLHDKFPDLNET*K | T | This study | ||
| Rv0733 | Adk | LGIPQISTGELFR | Not known | Prisic et al., 2010 |
| LGIPQIS*TGELFR | S | This study | ||
| Rv0822c | Rv0822c | VHDDADDQQDTEAIAIPAHSLEFLSELPDLR | Not Known | Prisic et al., 2010 |
| VHDDADDQQDT*EAIAIPAHSLEFLSELPDLR | T | This study | ||
| Rv0896 | GltA2 | ADTDDT$ATLR | T | Prisic et al., 2010; This study |
| Rv0931c | PknD | PGLTQT$GTAVG | T | Durán et al., 2005; This study |
| AASDPGLT*QT*GTAVGTYNYMAPER | T | This study | ||
| WSPGDS*AT*VAGPLAADSR | S; T | This study | ||
| WSPGDSATVAGPLAADSR | Not known | Prisic et al., 2010 | ||
| WSPGDS*AT*VAGPLAADSR | S; T | This study | ||
| WSPGDS*ATVAGPLAADS*R | S | This study | ||
| Rv1388 | MihF | AQEIMTELEIAPT$RR | T | Prisic et al., 2010; This study |
| Rv1719 | Rv1719 | S$GGIQVIAR | S | Prisic et al., 2010This study |
| Rv1746 | PknF | DDTRVS$QPVAV | S | Durán et al., 2005; This study |
| Rv1747 | Rv1747 | YPTGGQQLWPPSGPQR | T | Prisic et al., 2010 |
| YPT*GGQQLWPPS*GPQR | S; T | This study | ||
| IPAAPPSGPQPR | Not known | Prisic et al., 2010 | ||
| IPAAPPS*GPQPR | S | This study | ||
| Rv1820 | IlvG | STDTAPAQTMHAGR | Not Known | Prisic et al., 2010 |
| STDT*APAQTMHAGR | T | This study | ||
| Rv1827 | GarA | DQTSDEVTVETTSVFR | Not known | Prisic et al., 2010 |
| DQTSDEVT*VET*T*SVFR | T | This study | ||
| VTVETT$SVFRA | T | Prisic et al., 2010; This study | ||
| DQTSDEVTVET$TSVFR | T | Villarino et al., 2005; This study | ||
| DQT*SDEVTVET*TSVFR | T | This study | ||
| DQTSDEVT*VET*TSVFR | T | This study | ||
| DQTSDEVTVET*T*SVFR | T | This study | ||
| DQTSDEVT*VET*T*SVFR | T | This study | ||
| Rv2094c | TatA | VDPSAASGQDS*T*EARPA | S; T | This study |
| AEASIETPTPVQSQR | Not known | Prisic et al., 2010 | ||
| AEAS*IETPTPVQSQR | S | This study | ||
| AEASIETPT*PVQSQR | T | Prisic et al., 2010; This study | ||
| VDPSAASGQDSTEARPA | Not known | Chou et al., 2012 | ||
| VDPSAASGQDST*EARPA | T | This study | ||
| Rv2127 | AnsP1 | ERLGHT$GPFPAVANPPVR | T | Prisic et al., 2010; This study |
| Rv2151c | FtsQ | VADDAADEEAVT#EPLATESK | T | Prisic et al., 2010; This study |
| Rv2197c | Rv2197c | MAEAEPATRPT#GASVR | T | Prisic et al., 2010; This study |
| MAEAEPAT#RPTGASVR | T | Prisic et al., 2010 | ||
| MAEAEPAT*RPT*GASVR | T | This study | ||
| Rv2198c | MmpS3 | ASGNHLPPVAGGGDKLPSDQTGETDAYSR | Not known | Prisic et al., 2010 |
| ASGNHLPPVAGGGDKLPSDQT*GETDAY$SR | T; Y | Kusebauch et al., 2014; This study | ||
| Rv2536 | Rv2536 | ADDSPTGEMQVAQPEAQTAAVATVER | Not known | Prisic et al., 2010 |
| AADTDVFSAVR | Not known | Prisic et al., 2010 | ||
| AADT*DVFS*AVR | T | This study | ||
| Rv2536 | Rv2536 | EAPT$EVIR | T | Prisic et al., 2010; This study |
| ADDSPT*GEMQVAQPEAQTAAVAT*VER | T | This study | ||
| ADDS*PTGEMQVAQPEAQTAAVATVER | S | This study | ||
| Rv2606c | SnzP | MDPAGNPATGT$AR | T | Prisic et al., 2010; This study |
| MDPAGNPAT*GTAR | T | This study | ||
| Rv2694c | Rv2694c | RIPGIDT$GR | T | Prisic et al., 2010; This study |
| Rv2696c | Rv2696c | EAAAAQADT#QRQAAAGVAR | T | Prisic et al., 2010 |
| EAAAAQADT*QR | T | This study | ||
| Rv2921 | FtsY | IDTSGLPAVGDDATVPR | Not known | Prisic et al., 2010 |
| IDTS*GLPAVGDDATVPR | S | This study | ||
| IDT*SGLPAVGDDAT*VPR | T | This study | ||
| IDT*SGLPAVGDDAT*VPR | T | This study | ||
| Rv2940c | Mas | ALAQYLADTLAEEQAAAPAAS$ | S | Prisic et al., 2010; This study |
| Rv2996c | SerA1 | SATTVDAEVLAAAPK | Not known | Prisic et al., 2010 |
| SAT*TVDAEVLAAAPK | T | This study | ||
| S*ATTVDAEVLAAAPK | S | This study | ||
| Rv3181c | Rv3181c | LAALDST#DTLER | T | Prisic et al., 2010 |
| LAALDST*DT*LER | T | This study | ||
| Rv3197 | Rv3197 | S#KDEVTAELMEK | S | Prisic et al., 2010 |
| Rv3200c | Rv3200c | QSGADTVVVSS$ETAGR | S | Prisic et al., 2010; This study |
| QSGADTVVVS*SETAGR | S | This study | ||
| Rv3248c | SahH | GVTEETTTGVLR | Not known | Prisic et al., 2010 |
| GVTEETT*T*GVLR | T | This study | ||
| GVTEET*TTGVLR | T | This study | ||
| Rv3604c | Rv3604c | TADTPPDDSGGLHAR | Not known | Prisic et al., 2010 |
| TADTPPDDS*GGLHAR | S | This study | ||
| DPLTGGQSVADLMAR | Not known | Prisic et al., 2010 | ||
| DPLT$GGQSVADLMAR | T | Prisic et al., 2010; This study | ||
| Rv3801c | FadD32 | FDPEDTSEQLVIVGER | Not known | Prisic et al., 2010 |
| FDPEDT*SEQLVIVGER | T | This study | ||
| Rv3817 | Rv3817 | LWQAEDDS*S*R | S | This study |
| RLWQAEDDSSR | Not known | Prisic et al., 2010 | ||
| Rv3868 | EccA1 | LAQVLDIDT$LDEDRLR | T | Prisic et al., 2010; This study |
Novel phosphorylated amino acid identified in this study.
Phosphorylated residue identified in previous studies.
Phosphorylated residue identified in current and previous studies.
Figure 2.
Functional classification of phosphorylated proteins. Percentage phosphorylated proteins per functional category were classified according to Tuberculist. Number of phosphorylated proteins identified in each functional category depicted above the black bars.
Table 2.
Serine/threonine kinases and their phosphorylation sites identified in this study.
| STPK | Phosphorylated residue (position in protein) | Phosphopeptides | References |
|---|---|---|---|
| PknA | T8, S10, T224, S299, T301, T302 | AAPAAIPS*GT*T*AR | This study |
| VGVT*LS*GR | This study | ||
| RPFAGDGALT#VAMK | Prisic et al., 2010 | ||
| PknB | S169, T171, T173, S305, T309 | AIADSGNS*VT*QTAAVIGTAQYLSPEQAR | This study |
| TSLLSSAAGNLS*GPRTDPLPR | This study | ||
| AIADSGNSVT*QT*AAVIGTAQYLSPEQAR | This study | ||
| TSLLSSAAGNLSGPRT*DPLPR | This study | ||
| PknD | T169, T171, S332,T334,S343,S350 | GGNWPS*QTGHSPAVPNALQASLGHAVPPAGNK | This study |
| WSPGDS*AT*VAGPLAADSR | This study | ||
| WSPGDS*ATVAGPLAADS*R | This study | ||
| AASDPGLT*QT*GTAVGTYNYMAPER | This study | ||
| PknE | S304 | LPVPSTHPVS*PGTR | This study |
| PknF | T289, S290 | LGGAGDPDDT*RVS*QPVAVAAPAK | This study |
| PknG | S10, T14, T23, S24, T26, S31, T32, T55 | PLS*T*QAVFR | This study |
| S*GPGT*QPADAQTATSATVR | This study | ||
| SGPGTQPADAQTAT*S*ATVRPLSTQAVFR | This study | ||
| PDFGDEDNFPHPTLGPDT*EPQDR | This study | ||
| SGPGTQPADAQTAT*SAT*VR | This study | ||
| PknH | T174 | LTQLGT*AVGTWK | This study |
| PknJ | S498 | HLADLAS*IWRR | This study |
| PknL | S306, T309 | S*RIT*QQGQLGAK | This study |
Novel phosphorylation sites identified in this study.
Phosphorylated residue identified in previous studies.
We detected 13 Y phosphorylation sites located on 10 proteins in M. tuberculosis during early-logarithmic growth. Six of the 13 Y phosphorylation sites (Table 3) were located on two proteins, FhaA and MmpS3 (Kusebauch et al., 2014) and have recently been described in a M. tuberculosis H37Rv at stationary growth phase. The remaining 7 Y phosphorylated sites were uniquely identified in this study. Amongst these with known annotations were 2 virulence factors (GroS and GroEL2) and Ppa involved in macromolecule biosynthesis.
Table 3.
Tyrosine phosphorylation sites identified.
| Rv number | Protein name | Tyrosine phosphopeptides | Biological Function | References |
|---|---|---|---|---|
| Rv0020c | FhaA | GGQGQGRPDEY*YDDR | Signal transduction | This study |
| GGYPPETGGYPPQPGY*PRPR | This study | |||
| HEEGSY*VPSGPPGPPEQR | This study | |||
| HPDQGDY$PEQIGYPDQGGYPEQR | Kusebauch et al., 2014; This study | |||
| QDYGGGADY$TR | Kusebauch et al., 2014; This study | |||
| VPGY$APQGGGYAEPAGR | Kusebauch et al., 2014; This study | |||
| Rv0421c | Rv0421c | GLAEGPLIAGGHS*Y*GGR | Hypothetical | This study |
| Rv0440 | GroEL2 | QEIENSDSDY$DREK | Protein refolding | Kusebauch et al., 2014; This study |
| Rv0613c | RecC | IVLAGY*DEELLER | Exonuclease V gamma chain | This study |
| Rv1513 | Rv1513 | HQDAFPPANY*VGAQR | Hypothetical | This study |
| Rv2198c | MmpS3 | ASGNHLPPVAGGGDKLPSDQT*GETDAY*SR | Integral Membrane protein | This study |
| AYS*APESEHVTGGPY$VPADLR | Kusebauch et al., 2014 | |||
| AYS*APESEHVT*GGPY*VPADLR | This study | |||
| Rv3418c | GroS | Y*GGTEIK | Response to stress | This study |
| Rv3628 | Ppa | HFFVHY*K | Phosphate metabolic process | This study |
#Previously identified pY site.
Novel pY site identified in this study.
pY site identified in current and previous studies.
A large number of proteins involved in intermediary metabolism and respiration processes such as lipid and fatty acid metabolism (KasB, FadD32, AccD4, and MmaA3) were found to be phosphorylated in this study (Supplemental Table S2). In addition, several proteins from the ESX-1 type seven secretion system (T7SS) in M. tuberculosis including EspR, EccA, CFP10, EspI, EspL, EspB were amongst the identified phosphorylated proteins (Supplemental Table S1). We also identified virulence factors such Pks15, AceA5, FadD5, EsxB, KatG, GlpX, Rv2032, and PtbB that were phosphorylated in this hyper-virulent M. tuberculosis strain (Supplemental Table S2).
Of the 21 phosphorylated proteins involved in information pathways, we identified 6 phosphorylated ribosomal proteins; two phosphorylated small subunit (30S) ribosomal proteins (S3, S19), and four large subunit (50S) ribosomal proteins (L3, L24, L29, L31) with a total of 8 S/T phosphorylation sites. In addition, phosphorylated sites on the ribosomal proteins RpsS and RplX were also identified (Table 4).
Table 4.
List of phosphorylated ribosomal proteins identified in this study and other bacteria.
| Protein name | Protein name | Phosphopeptides | Phospho-residue | References |
|---|---|---|---|---|
| rpsC | Streptomyces coelicolor | Not known | Not known | Mikulík et al., 2011 |
| M. tuberculosis | NPES*QAQLVAQGVAEQLSNR | S | This study | |
| AAGGEEAAPDAAAPVEAQSTES* | S | This study | ||
| rpsS | M. tuberculosis | HVPVFVTES*MVGHK | S | This study |
| rplC | Streptomyces coelicolor | Not known | Not known | Mikulík et al., 2011 |
| Streptococcus pneumonia | Not known | Not known | Zhang et al., 2000 | |
| Halobacterium salinarum | Not known | Not known | Aivaliotis et al., 2009 | |
| M. tuberculosis | IVVEVCSQCHPFYT*GK | T | This study | |
| rplX | M. tuberculosis | S*GGIVTQEAPIHVSNVMVVDSDGKPTR | S | This study |
| rpmC | Streptococcus agalatiae | FQAAAGQLEKT*AR | T | Burnside et al., 2011 |
| Streptomyces coelicolor | RERELGIET*VESA | T | Manteca et al., 2011 | |
| Listeria monocytogenes | FQLATGQLENT*AR | T | Misra et al., 2011 | |
| DLSTTEIQDQEK | Not known | Misra et al., 2011 | ||
| Lactococcus lactis | MKLSETK | Not known | Soufi et al., 2008 | |
| M. tuberculosis | ELGLATGPDGKES* | S | This study | |
| rpmE | Klebsiella pneumonia | S*TVGHDLNLDVCGK | S | Lin et al., 2009 |
| Halobacterium salinarum | ASSEFDDRFVTVPLRDVTK | Not known | Aivaliotis et al., 2009 | |
| M. tuberculosis | T*GGLVMVR | T | This study |
Novel pY site identified in this study.
Discussion
Here we report 214 phosphorylated proteins extracted during early-logarithmic growth phase from a hyper-virulent clinical Beijing genotype Mycobacterium tuberculosis isolate. These proteins can be categorized into different biological functions according to Tuberculist (Lew et al., 2011). The impact of phosphorylation on these Hank's type Ser/Thr kinases (STPKs), phosphatases and their substrates, and the functional role of phosphorylated residues still remains to be elucidated. However, as in previous studies, the identification of phosphorylated residues clearly suggests functional importance.
Regulatory proteins
In recent years, bacterial S/T/Y kinases and phosphatases have been extensively investigated, with indications that they might play a crucial role in pathogenicity. These Hank's type kinases have the ability to short-circuit the host defense mechanism since they are mostly involved in key biological processes. We identified phosphopeptides from 9 of the 11 STPKs encoded by the M. tuberculosis genome. This included both previously described S/T phosphorylation sites (Boitel et al., 2003; Young et al., 2003; Durán et al., 2005; Villarino et al., 2005; Molle et al., 2006; O'Hare et al., 2008; Prisic et al., 2010; Sajid et al., 2011; Chou et al., 2012; Kusebauch et al., 2014) and novel sites on these STPKs thereby highlighting the complexity of the signal transduction mechanism of this pathogen. Phosphorylated peptides were not detected for PknI and PknK. However MS/MS spectra for these peptides for these proteins were detected with mass spectrometry thereby confirming the presence of these proteins (data not shown).
Of the phosphorylated STPKs, PknA, PknB, and PknG have been shown to be essential for in vitro growth (Sassetti et al., 2003) and to regulate cell growth and cell division and interfere with host signaling pathways (Fernandez et al., 2006). PknE, PknH, PknJ, and PknL have been implicated in the adaptation to the extracellular environment or intracellular survival of M. tuberculosis (Sharma et al., 2006; Lakshminarayan, 2009; Arora et al., 2010; Parandhaman et al., 2014) which is in agreement with reports that during early growth the bacilli undergo a period of adaptation to their external environment (Stock et al., 1989; Soares et al., 2013). PknE is involved in the suppression of apoptosis during nitrate stress (Kumar and Narayanan, 2012) and intracellular survival and adaptation to hostile environments (Parandhaman et al., 2014). In M. tuberculosis, PknH controls the expression of a variety of cell wall related enzymes and regulates in vivo growth in mice (Zheng et al., 2007). PknJ undergoes autophosphorylation and phosphorylates the Thr168, Thr171, and Thr173 residues of Embr (a transcriptional regulator), MmaA4/Hma (a methyltransferase involved in mycolic acid biosynthesis) and PepE (a peptidase located adjacent to the pknJ gene in the M. tuberculosis genome), respectively (Jang et al., 2010). Lastly, PknL is involved in an adaptive response to nutrient starvation. This kinase regulates transcription which allows the bacilli to maintain metabolic activity without sourcing energy from elsewhere (Lakshminarayan, 2009). Furthermore, we identified a number of STPK substrates that were phosphorylated in clinical hyper-virulent M. tuberculosis strain (list not shown) thereby highlighting the complexity of the phosphorylation regulatory network in M. tuberculosis. Even though the role of STPKs in bacterial physiology is not yet fully understood the data presented here could underpin a targeted approach to improving our understanding of STPK-mediated signal transduction mechanisms in M. tuberculosis.
Tyrosine phosphorylation
The M. tuberculosis genome encodes for two putative tyrosine phosphatases (PtpA and PtpB) but is not predicted to encode tyrosine kinases (Cole et al., 1998; Bach et al., 2009). Most bacterial phosphorylation sites are on serine and threonine; a survey of 11 bacterial phosphoproteomes revealed that S/T phosphorylation accounted for an average of 48 and 40% of phosphorylated sites, respectively, while tyrosine phosphorylation events account for less than 10% of the overall phosphoproteome (Ge and Shan, 2011). Tyrosine phosphorylated proteins have been previously shown to play important regulatory roles through their involvement in biological functions such as exopolysaccharide production, DNA metabolism, stress responses (Ge et al., 2011; Whitmore and Lamont, 2012). Recently, Kusebauch et al. identified tyrosine phosphorylated proteins in M. tuberculosis and demonstrated that a number of STPKs can phosphorylate tyrosine in either cis or trans (Kusebauch et al., 2014). This suggests that STPKs have the ability to phosphorylate S/T/Y. In this study we identified 13 tyrosine phosphorylation sites in 8 proteins (Table 3). An overlap of three proteins (FhaA, MmpS3, and GroES) and 6 tyrosine phosphorylated sites we similar between this and the previous study (Kusebauch et al., 2014). Five of the tyrosine phosphorylated proteins (FhaA, GroEL2, MmpS3, GroES, and Ppa) identified in this study are essential for in vitro growth (Sassetti et al., 2003; Griffin et al., 2011) and involved in a variety of functions.
This study confirmed and expanded work by Kusebauch et al., where multiple tyrosine phosphopeptides were identified for FhaA. FhaA is a regulatory protein which has been implicated in cell wall biosynthesis (Fernandez et al., 2006) and has a strong association with PknA and PknB (Pallen et al., 2002; Roumestand et al., 2011). We found that the highly S/T/Y phosphorylated FHA-domain contained 6 tyrosine phosphopeptides of which 4 were previously been identified in H37Rv (Kusebauch et al., 2014). FhaA is a major substrate of PknB and has been implicated in the formation of a regulatory complex with MviN required for peptidoglycan biosynthesis (Warner and Mizrahi, 2012). In our dataset, we found that all three of the proteins (PknB, FhaA, and MviN) in the regulator complex were phosphorylated.
We also confirm the presence of a previously reported Y phosphopeptide of MmpS3 and identified a second phosphopeptide. MmpS3 forms part of the mycobacterial membrane protein small family and is an essential protein for mycobacterial growth and cholesterol metabolism (Griffin et al., 2012). The role of phosphorylation of this protein has yet to be determined.
The two proteins GroEL2 and GroES have been identified as potential candidates for antituberculosis treatment (Al-Attiyah et al., 2006). We confirmed the presence of the Y phosphopeptide in GroEL2 identified in H37Rv (Kusebauch et al., 2014). Recently it has been shown that the antigen GroES is sufficient to protect BALB/c mice against challenge infection (Lima et al., 2003) and up-regulated in kanamycin and amikacin resistant isolates (Kumar et al., 2013). Ppa is an inorganic pyrophosphate and is involved in macromolecule biosynthesis. The M. tuberculosis Ppa is highly similar to a well conserved homolog of Legionella. pneumophila PPase which is induced in macrophages, although the M. tuberculosis PPa promotor is not responsive to any specific intracellular triggers (Triccas and Gicquel, 2001).
Virulence factors
The identification of virulence factors is crucial in order to improve our understanding of the mechanisms involved in pathogenesis of M. tuberculosis. Several of the phosphorylated virulence factors identified in this study were found to be involved in basic metabolic pathways such a lipid and fatty acid metabolism, secretion systems and response and adaptation to environmental changes. The virulence factor KasB and key enzymes (FadD32, AccD4, and MmaA3) in the mycolic acids biosynthesis pathway were phosphorylated in this hyper-virulent M. tuberculosis strain. The kasB gene is not essential for growth, however, the deletion mutant, ΔkasB, resulted in an alteration in growth morphology and loss of acid-fast staining (Bhatt et al., 2007). This suggests that modification of this protein could influence the synthesis of mycolic acids and thereby the pathogenicity of the bacilli.
The specialized ESX-1 Type VII secretion system (T7SS), unique to pathogenic mycobacteria is responsible for the secretion of two culture filtrate proteins EsxA and EsxB (ESAT-6 and CFP-10). These secretion systems have been shown to be involved in virulence and are critical for intracellular survival (Bitter and Kuijl, 2014) due to their ability to secrete proteins that lack classical signal peptides across the complex cell envelope to host cells during infection (Houben et al., 2014, p. 5). M. tuberculosis have several different ESX regions (ESX-1 to ESX-5) (Daleke et al., 2012) with varying gene numbers and size for each of these secretion machinery. In this study we found 6 T7SS proteins to be phosphorylated in the hyper-virulent strain. In a previous study, proteomics of whole cell extracts of this hyper-virulent M. tuberculosis strain revealed an under-representation of virulence factors such as ESAT-6 and Esx-like proteins (de Souza et al., 2010). The authors showed the abundance of ESAT-6 gene expression was reduced in the hyper-virulent M. tuberculosis suggesting that the low levels of this protein might be as a result of its ability to export these proteins more efficiently into the extracellular environment (de Souza et al., 2010).
Protein synthesis and interactions
The impact of phosphorylation on the functionality of ribosomal proteins is not fully understood. Mikulik et al. hypothesized that phosphorylation of ribosomal proteins induces or stabilizes conformational changes during proteins synthesis which could allow modification of subunit association or changes in interactions with proteins and RNAs (Mikulík et al., 2011). According to the protein phosphorylation database, phosphopeptides of RpmC have been identified in four different bacteria (Soufi et al., 2008; Burnside et al., 2011; Manteca et al., 2011; Misra et al., 2011). The implication of phosphorylation on RpmC has not been investigated. However, in E.coli, RpmC, RplW and Trigger factor are located at the exit tunnel in the ribosome, suggesting that phosphorylation may impact on multiple stages of transcription (Kramer et al., 2002). In our study we identified phosphopeptides for 7 ribosomal proteins. We also identified unique phosphopeptides on ribosomal proteins RpsS and RplX (Table 3).
Overlap of phosphorylated proteins with other bacteria
Twenty-five of phosphorylated proteins identified in our study were also identified in phosphoproteomics studies of other bacteria such as Klebsiella pneumonia (Lin et al., 2009), Helicobacter pylori (Ge et al., 2011), Steptococcus pneumonia (Sun et al., 2010), Bacillus subtillis (Macek et al., 2007), Halobacterium salinarum (Aivaliotis et al., 2009), etc. (Table 4). In our dataset the distribution of S/T/Y seem to be bias toward pT and is in accordance with previously described phosphoproteomes of M. tuberculosis (Prisic et al., 2010; Kusebauch et al., 2014). Manual evaluation of the genome found an over-representation of Threonine relative to Serine (52:48%). This compared to other bacteria such as Acinetobacter baumannii (Soares et al., 2014), Bacillus subtilis (Macek et al., 2007), Escherichia coli (Macek et al., 2008; Soares et al., 2013) and Halobacterium salinarum (Aivaliotis et al., 2009), Pseudomonas aeruginosa (Ravichandran et al., 2009), and Streptomyces coelicolor (Parker et al., 2010) which demonstrate a bias toward pS.
Forty-five of the phosphorylated proteins identified in our study were previously described for M. tuberculosis H37Rv (Boitel et al., 2003; Young et al., 2003; Molle et al., 2004, 2006; Durán et al., 2005; Kang et al., 2005; Villarino et al., 2005; O'Hare et al., 2008; Thakur et al., 2008; Prisic et al., 2010; Sajid et al., 2011; Gee et al., 2012). The reason for not identifying all of the previously identified phosphorylated proteins in the protein phosphorylation database could be ascribed to different genetic backgrounds of the analyzed M. tuberculosis strains, culture conditions, sample preparation and different MS-based proteomics approaches used in each of the studies. Our analysis was performed on a hyper-virulent clinical isolate of M. tuberculosis and a member of the Beijing genotype which is genetically distinct from the laboratory strain M. tuberculosis H37Rv analyzed by Prisic et al. and Kusebauch et al. In addition, the Prisic et al. study reported on the combined phosphoproteome from 6 different conditions (5 different culture conditions and 2 different growth phases) (Prisic et al., 2010) while Kusebauch et al. reported on the phosphoproteome of late-logarithmic phase cultures (Kusebauch et al., 2014), whereas our study analyzed early-logarithmic phase cultures. Even though the overlap between our study of clinical M. tuberculosis and that of the previously described laboratory M. tuberculosis H37Rv is low this work substantially extends our knowledge of the M. tuberculosis phosphoproteome. During logarithmic growth phase of bacterial growth the cells are adapting to the environment of the growth media and biological process such as RNA synthesis, DNA replication and synthesis of micro- and macromolecules are up-regulated. It is important to note that in this study the whole cell lysate proteins were enriched for phosphopeptides and we detected a number of phosphorylated proteins involved in these biological processes such as fatty acid- and lipid biosynthetic metabolism; RNA modification and translation; DNA repair, replication and modification. It is believed that environmental conditions, cell density and growth phase influence the expression of virulence factors by a pathogen (McIver et al., 1995). This is consistent other bacterial phosphoproteomes, thereby emphasizing that S/T/Y phosphorylation is an important process required for the regulation of numerous cellular processes.
Conclusion
Recent developments in the methodology and mass spectrometry technology for phosphoproteomics have highlighted the need to explore the involvement of phosphorylation in disease development and progression. However, the impact of the protein phosphorylation cascade on the physiology of pathogenic bacteria such as M. tuberculosis has yet to be fully elucidated. Improved preparative techniques and more sensitive instrumentation are required to fully appreciate the complexity of protein modification. This can only be achieved if concomitant methods are developed to elucidate the impact of phosphorylation on protein function. Although this qualitative study was done in clinical hyper-virulent M. tuberculosis, without any follow-up validation studies it still provides a valuable resource for further investigating and understanding the impact of protein phosphorylation regulation in M. tuberculosis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was sponsored by the National Research Foundation Norway/RSA research cooperation programme, the Medical research Council of South Africa, DST/NRF Centre of Excellence for Biomedical Tuberculosis Research Stellenbosch University (Professor Paul van Helden). We further want to acknowledge Prof. M. Mann who permitted us to use the proteomics and mass spectrometry facilities at the Max Planck Institute, Munich, Germany.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00006/abstract
References
- Aivaliotis M., Macek B., Gnad F., Reichelt P., Mann M., Oesterhelt D. (2009). Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum—a representative of the third domain of life. PLoS ONE 4:e4777. 10.1371/journal.pone.0004777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Attiyah R., Madi N. M., El-Shamy A. M., Wiker H. G., Andersen P., Mustafa A. S. (2006). Cytokine profiles in tuberculosis patients and healthy subjects in response to complex and single antigens of Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 47, 254–261. 10.1111/j.1574-695X.2006.00110.x [DOI] [PubMed] [Google Scholar]
- Arora G., Sajid A., Gupta M., Bhaduri A., Kumar P., Basu-Modak S., et al. (2010). Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS ONE 5:10772. 10.1371/journal.pone.0010772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach H., Wong D., Av-Gay Y. (2009). Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem. J. 420, 155–160. 10.1042/BJ20090478 [DOI] [PubMed] [Google Scholar]
- Bhatt A., Fujiwara N., Bhatt K., Gurcha S. S., Kremer L., Chen B., et al. (2007). Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 104, 5157–5162. 10.1073/pnas.0608654104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W., Kuijl C. (2014). Targeting bacterial virulence: the coming out of type VII secretion inhibitors. Cell Host Microbe 16, 430–432. 10.1016/j.chom.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Boitel B., Ortiz-Lombardía M., Durán R., Pompeo F., Cole S. T., Cerveñansky C., et al. (2003). PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 49, 1493–1508. 10.1046/j.1365-2958.2003.03657.x [DOI] [PubMed] [Google Scholar]
- Burnside K., Lembo A., Harrell M. I., Gurney M., Xue L., BinhTran N.-T., et al. (2011). Serine/threonine phosphatase Stp1 mediates post-transcriptional regulation of hemolysin, autolysis, and virulence of group B Streptococcus. J. Biol. Chem. 286, 44197–44210. 10.1074/jbc.M111.313486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra P., Meena L. S., Singh Y. (2003). New drug targets for Mycobacterium tuberculosis. Indian J. Med. Res. 117, 1–9. [PubMed] [Google Scholar]
- Chou M. F., Prisic S., Lubner J. M., Church G. M., Husson R. N., Schwartz D. (2012). Using bacteria to determine protein kinase specificity and predict target substrates. PLoS ONE 7:e52747. 10.1371/journal.pone.0052747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- Cozzone A. J. (1998). Post-translational modification of proteins by reversible phosphorylation in prokaryotes. Biochimie 80, 43–48. [DOI] [PubMed] [Google Scholar]
- Daleke M. H., Ummels R., Bawono P., Heringa J., Vandenbroucke-Grauls C. M. J. E., Luirink J., et al. (2012). General secretion signal for the mycobacterial type VII secretion pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 11342–11347. 10.1073/pnas.1119453109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza G. A., Fortuin S., Aguilar D., Pando R. H., McEvoy C. R. E., van Helden P. D., et al. (2010). Using a label-free proteomics method to identify differentially abundant proteins in closely related hypo- and hypervirulent clinical Mycobacterium tuberculosis Beijing isolates. Mol. Cell. Proteomics 9, 2414–2423. 10.1074/mcp.M900422-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán R., Villarino A., Bellinzoni M., Wehenkel A., Fernandez P., Boitel B., et al. (2005). Conserved autophosphorylation pattern in activation loops and juxtamembrane regions of Mycobacterium tuberculosis Ser/Thr protein kinases. Biochem. Biophys. Res. Commun. 333, 858–867. 10.1016/j.bbrc.2005.05.173 [DOI] [PubMed] [Google Scholar]
- Fernandez P., Saint-Joanis B., Barilone N., Jackson M., Gicquel B., Cole S. T., et al. (2006). The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188, 7778–7784. 10.1128/JB.00963-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch S. C., Dworkin M. (1996). Tyrosine phosphorylation in Myxococcus xanthus, a multicellular prokaryote. J. Bacteriol. 178, 4084–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R., Shan W. (2011). Bacterial phosphoproteomic analysis reveals the correlation between protein phosphorylation and bacterial pathogenicity. Genomics Proteomics Bioinformatics 9, 119–127. 10.1016/S1672-0229(11)60015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R., Sun X., Xiao C., Yin X., Shan W., Chen Z., et al. (2011). Phosphoproteome analysis of the pathogenic bacterium Helicobacter pylori reveals over-representation of tyrosine phosphorylation and multiply phosphorylated proteins. Proteomics 11, 1449–1461. 10.1002/pmic.201000649 [DOI] [PubMed] [Google Scholar]
- Gee C. L., Papavinasasundaram K. G., Blair S. R., Baer C. E., Falick A. M., King D. S., et al. (2012). A phosphorylated pseudokinase complex controls cell wall synthesis in Mycobacteria. Sci. Signal. 5, ra7. 10.1126/scisignal.2002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011). High-resolution phenotypic profiling defines genes essential for Mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E., Pandey A. K., Gilmore S. A., Mizrahi V., McKinney J. D., Bertozzi C. R., et al. (2012). Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227. 10.1016/j.chembiol.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Tanner S., Jaitly N., Adkins J. N., Lipton M., Edwards R., et al. (2007). Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 17, 1362–1377. 10.1101/gr.6427907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben E. N. G., Korotkov K. V., Bitter W. (2014). Take five—Type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta 1843, 1707–1716. 10.1016/j.bbamcr.2013.11.003 [DOI] [PubMed] [Google Scholar]
- Jang J., Stella A., Boudou F., Levillain F., Darthuy E., Vaubourgeix J., et al. (2010). Functional characterization of the Mycobacterium tuberculosis serine/threonine kinase PknJ. Microbiol. Read. Engl. 156, 1619–1631. 10.1099/mic.0.038133-0 [DOI] [PubMed] [Google Scholar]
- Kang C.-M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005). The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704. 10.1101/gad.1311105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A., Herget T., Klebl B., Ullrich A. (2004). Interplay between mycobacteria and host signalling pathways. Nat. Rev. Microbiol. 2, 189–202. 10.1038/nrmicro840 [DOI] [PubMed] [Google Scholar]
- Kramer G., Rauch T., Rist W., Vorderwülbecke S., Patzelt H., Schulze-Specking A., et al. (2002). L23 protein functions as a chaperone docking site on the ribosome. Nature 419, 171–174. 10.1038/nature01047 [DOI] [PubMed] [Google Scholar]
- Kumar B., Sharma D., Sharma P., Katoch V. M., Venkatesan K., Bisht D. (2013). Proteomic analysis of Mycobacterium tuberculosis isolates resistant to kanamycin and amikacin. J. Proteomics 94, 68–77. 10.1016/j.jprot.2013.08.025 [DOI] [PubMed] [Google Scholar]
- Kumar D., Narayanan S. (2012). pknE, a serine/threonine kinase of Mycobacterium tuberculosis modulates multiple apoptotic paradigms. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 12, 737–747. 10.1016/j.meegid.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Kusebauch U., Ortega C., Ollodart A., Rogers R. S., Sherman D. R., Moritz R. L., et al. (2014). Mycobacterium tuberculosis supports protein tyrosine phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 111, 9265–9270. 10.1073/pnas.1323894111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayan H. (2009). Involvement of serine threonine protein kinase PknL, from Mycobacterium tuberculosis H37Rv in starvation response of Mycobacteria. J. Microb. Biochem. Technol. 1, 30–36. [Google Scholar]
- Lew J. M., Kapopoulou A., Jones L. M., Cole S. T. (2011). TubercuList—10 years after. Tuberc. Edinb. Scotl. 91, 1–7. 10.1016/j.tube.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Lima K. M., Santos S. A., Lima V. M. F., Coelho-Castelo A. A. M., Rodrigues J. M., Silva C. L. (2003). Single dose of a vaccine based on DNA encoding mycobacterial hsp65 protein plus TDM-loaded PLGA microspheres protects mice against a virulent strain of Mycobacterium tuberculosis. Gene Ther. 10, 678–685. 10.1038/sj.gt.3301908 [DOI] [PubMed] [Google Scholar]
- Lin M.-H., Hsu T.-L., Lin S.-Y., Pan Y.-J., Jan J.-T., Wang J.-T., et al. (2009). Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol. Cell. Proteomics 8, 2613–2623. 10.1074/mcp.M900276-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., et al. (2008). Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307. 10.1074/mcp.M700311-MCP200 [DOI] [PubMed] [Google Scholar]
- Macek B., Mijakovic I. (2011). Site-specific analysis of bacterial phosphoproteomes. Proteomics 11, 3002–3011. 10.1002/pmic.201100012 [DOI] [PubMed] [Google Scholar]
- Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., et al. (2007). The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707. 10.1074/mcp.M600464-MCP200 [DOI] [PubMed] [Google Scholar]
- Manteca A., Ye J., Sánchez J., Jensen O. N. (2011). Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res. 10, 5481–5492. 10.1021/pr200762y [DOI] [PubMed] [Google Scholar]
- McConnell J. L., Wadzinski B. E. (2009). Targeting protein serine/threonine phosphatases for drug development. Mol. Pharmacol. 75, 1249–1261. 10.1124/mol.108.053140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver K. S., Heath A. S., Scott J. R. (1995). Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63, 4540–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I., Macek B. (2012). Impact of phosphoproteomics on studies of bacterial physiology. FEMS Microbiol. Rev. 36, 877–892. 10.1111/j.1574-6976.2011.00314.x [DOI] [PubMed] [Google Scholar]
- Mikulík K., Bobek J., Ziková A., Smětáková M., Bezoušková S. (2011). Phosphorylation of ribosomal proteins influences subunit association and translation of poly (U) in Streptomyces coelicolor. Mol. Biosyst. 7, 817–823. 10.1039/c0mb00174k [DOI] [PubMed] [Google Scholar]
- Misra S. K., Milohanic E., Aké F., Mijakovic I., Deutscher J., Monnet V., et al. (2011). Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165. 10.1002/pmic.201100259 [DOI] [PubMed] [Google Scholar]
- Molle V., Brown A. K., Besra G. S., Cozzone A. J., Kremer L. (2006). The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J. Biol. Chem. 281, 30094–30103. 10.1074/jbc.M601691200 [DOI] [PubMed] [Google Scholar]
- Molle V., Leiba J., Zanella-Cléon I., Becchi M., Kremer L. (2010). An improved method to unravel phosphoacceptors in Ser/Thr protein kinase-phosphorylated substrates. Proteomics 10, 3910–3915. 10.1002/pmic.201000316 [DOI] [PubMed] [Google Scholar]
- Molle V., Soulat D., Jault J.-M., Grangeasse C., Cozzone A. J., Prost J.-F. (2004). Two FHA domains on an ABC transporter, Rv1747, mediate its phosphorylation by PknF, a Ser/Thr protein kinase from Mycobacterium tuberculosis. FEMS Microbiol. Lett. 234, 215–223. 10.1016/j.femsle.2004.03.033 [DOI] [PubMed] [Google Scholar]
- O'Hare H. M., Durán R., Cerveñansky C., Bellinzoni M., Wehenkel A. M., Pritsch O., et al. (2008). Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70, 1408–1423. 10.1111/j.1365-2958.2008.06489.x [DOI] [PubMed] [Google Scholar]
- Pallen M., Chaudhuri R., Khan A. (2002). Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends Microbiol. 10, 556–563. 10.1016/S0966-842X(02)02476-9 [DOI] [PubMed] [Google Scholar]
- Parandhaman D. K., Hanna L. E., Narayanan S. (2014). PknE, a serine/threonine protein kinase of Mycobacterium tuberculosis initiates survival crosstalk that also impacts HIV coinfection. PLoS ONE 9:e83541. 10.1371/journal.pone.0083541 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Parker J. L., Jones A. M. E., Serazetdinova L., Saalbach G., Bibb M. J., Naldrett M. J. (2010). Analysis of the phosphoproteome of the multicellular bacterium Streptomyces coelicolor A3(2) by protein/peptide fractionation, phosphopeptide enrichment and high-accuracy mass spectrometry. Proteomics 10, 2486–2497. 10.1002/pmic.201000090 [DOI] [PubMed] [Google Scholar]
- Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C.-M., et al. (2010). Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526. 10.1073/pnas.0913482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran A., Sugiyama N., Tomita M., Swarup S., Ishihama Y. (2009). Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9, 2764–2775. 10.1002/pmic.200800655 [DOI] [PubMed] [Google Scholar]
- Roumestand C., Leiba J., Galophe N., Margeat E., Padilla A., Bessin Y., et al. (2011). Structural insight into the Mycobacterium tuberculosis Rv0020c protein and its interaction with the PknB kinase. Structure 19, 1525–1534. 10.1016/j.str.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Sajid A., Arora G., Gupta M., Singhal A., Chakraborty K., Nandicoori V. K., et al. (2011). Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation. J. Bacteriol. 193, 5347–5358. 10.1128/JB.05469-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C., Hartkoorn R. C. (2011). Tuberculosis drugs: new candidates and how to find more. Future Microbiol. 6, 617–633. 10.2217/fmb.11.46 [DOI] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H., Rubin E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- Seo J., Lee K.-J. (2004). Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 37, 35–44. 10.5483/BMBRep.2004.37.1.035 [DOI] [PubMed] [Google Scholar]
- Sharma K., Chopra P., Singh Y. (2004). Recent advances towards identification of new drug targets for Mycobacterium tuberculosis. Expert Opin. Ther. Targets 8, 79–93. 10.1517/14728222.8.2.79 [DOI] [PubMed] [Google Scholar]
- Sharma K., Gupta M., Pathak M., Gupta N., Koul A., Sarangi S., et al. (2006). Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 188, 2936–2944. 10.1128/JB.188.8.2936-2944.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares N. C., Spät P., Krug K., Macek B. (2013). Global dynamics of the Escherichia coli proteome and phosphoproteome during growth in minimal medium. J. Proteome Res. 12, 2611–2621. 10.1021/pr3011843 [DOI] [PubMed] [Google Scholar]
- Soares N. C., Spät P., Méndez J. A., Nakedi K., Aranda J., Bou G. (2014). Ser/Thr/Tyr phosphoproteome characterization of Acinetobacter baumannii: comparison between a reference strain and a highly invasive multidrug-resistant clinical isolate. J. Proteomics 102, 113–124. 10.1016/j.jprot.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., et al. (2008). The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8, 3486–3493. 10.1002/pmic.200800069 [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. (1989). Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53, 450–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ge F., Xiao C.-L., Yin X.-F., Ge R., Zhang L.-H., et al. (2010). Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J. Proteome Res. 9, 275–282. 10.1021/pr900612v [DOI] [PubMed] [Google Scholar]
- Thakur M., Chaba R., Mondal A. K., Chakraborti P. K. (2008). Interdomain interaction reconstitutes the functionality of PknA, a eukaryotic type Ser/Thr kinase from Mycobacterium tuberculosis. J. Biol. Chem. 283, 8023–8033. 10.1074/jbc.M707535200 [DOI] [PubMed] [Google Scholar]
- Triccas J. A., Gicquel B. (2001). Analysis of stress- and host cell-induced expression of the Mycobacterium tuberculosis inorganic pyrophosphatase. BMC Microbiol. 1:3. 10.1186/1471-2180-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A., Duran R., Wehenkel A., Fernandez P., England P., Brodin P., et al. (2005). Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J. Mol. Biol. 350, 953–963. 10.1016/j.jmb.2005.05.049 [DOI] [PubMed] [Google Scholar]
- Warner D. F., Mizrahi V. (2012). A pseudokinase debut at the mycobacterial cell wall. Sci. Signal. 5, pe3–pe3. 10.1126/scisignal.2002785 [DOI] [PubMed] [Google Scholar]
- Whitmore S. E., Lamont R. J. (2012). Tyrosine phosphorylation and bacterial virulence. Int. J. Oral Sci. 4, 1–6. 10.1038/ijos.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO|Global tuberculosis report 2013. (2013). WHO. Available online at: http://www.who.int/tb/publications/global_report/en/ [Accessed October 1, 2014].
- WHO|Global tuberculosis report 2014. (2014). WHO|Global Tuberculosis Report 2014, Geneva. [Google Scholar]
- Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Young T. A., Delagoutte B., Endrizzi J. A., Falick A. M., Alber T. (2003). Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat. Struct. Mol. Biol. 10, 168–174. 10.1038/nsb897 [DOI] [PubMed] [Google Scholar]
- Zhang W., Li L., Jiang W., Zhao G., Yang Y., Chiao J. (2000). A novel transmembrane serine/threonine protein kinase gene from a rifamycin SV-producing amycolatopsis mediterranei U32. Eur. J. Biochem. FEBS 267, 3744–3752. 10.1046/j.1432-1327.2000.01410.x [DOI] [PubMed] [Google Scholar]
- Zheng X., Papavinasasundaram K. G., Av-Gay Y. (2007). Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 355, 162–168. 10.1016/j.bbrc.2007.01.122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.