Abstract
Background
Obesity and diabetes mellitus are complex metabolic problems of pandemic proportion, contributing to significant cardiovascular mortality. Recent studies have shown altered mitochondrial function in the hearts of diabetic animals. We hypothesized that regulatory events involved in the control of mitochondrial function are activated in the prediabetic, insulin-resistant stage.
Methods and Results
Morphometric analyses demonstrated that cardiac myocyte mitochondrial volume density was increased in insulin-resistant uncoupling protein-diptheria toxin A (UCP-DTA) transgenic mice, a murine model of metabolic syndrome, compared with littermate controls. Mitochondrial DNA content and expression of genes involved in multiple mitochondrial pathways were also increased in insulin-resistant UCP-DTA hearts. The nuclear receptor, peroxisome proliferator-activated receptor-α (PPARα), is known to activate metabolic genes in the diabetic heart. Therefore, we evaluated the role of PPARα in the observed mitochondrial biogenesis response in the insulin-resistant heart. Insulin-resistant UCP-DTA mice crossed into a PPARα-null background did not exhibit evidence of mitochondrial biogenesis or induction of mitochondrial gene expression. Conversely, transgenic mice with cardiac-specific overexpression of PPARα exhibited signatures of cardiac mitochondrial biogenesis. A screen for candidate mediators of the PPARα-driven mitochondrial biogenic response revealed that expression of PPARγ coactivator-1α (PGC-1α), a known regulator of mitochondrial biogenesis, was activated in wild-type UCP-DTA mice but not in PPARα-deficient UCP-DTA mice.
Conclusions
These results demonstrate that mitochondrial biogenesis occurs early in the development of diabetic cardiac dysfunction through a transcriptional regulatory circuit that involves activation of PGC-1α gene expression by the fatty acid–activated nuclear receptor PPARα.
Keywords: diabetes mellitus, cardiomyopathy, mitochondria, metabolism
We are witnessing an emerging pandemic of obesity fueling a dramatic increase in the prevalence of type 2 diabetes mellitus.1 Cardiovascular disease is the leading cause of death in the diabetic population.2,3 Cardiac dysfunction is common in diabetics regardless of risk factors for coronary artery disease and hypertension.4–6 Emerging evidence suggests that diabetic cardiomyopathy is linked to alterations in myocardial fuel and energy metabolism.7–9 The normal mammalian heart is capable of generating ATP from multiple substrates. Although fatty acids (FAs) serve as the chief energy substrate for the heart, dynamic shifts in the proportion of ATP generated from oxidation of FAs and glucose provide the heart with an efficient and constant fuel source in diverse physiological and nutritional circumstances.7 In insulin-resistant and insulin-deficient forms of diabetes, cardiac energy substrate flexibility becomes constrained. Altered insulin signaling in the diabetic state reduces myocardial glucose uptake and utilization concordant with increased uptake and utilization of FAs to meet cardiac energy demands.8,10,11 Although this shift in myocardial fuel preference is initially adaptive, long-term reliance on mitochondrial FA oxidation (FAO) imposes a stress on the cardiac myocyte that includes accumulation of toxic lipid intermediates and reactive oxygen species and increased myocardial oxygen consumption, which predisposes to cardiac dysfunction.10,11
In addition to derangements in fuel metabolism, increasing evidence supports a role for mitochondrial dysfunction in the end-organ damage of diabetic striated muscle. Mitochondrial structural and functional derangements have been shown in skeletal muscle of insulin-resistant and diabetic animal models and humans.12–18 Studies of mitochondria in the diabetic heart are more limited, but several recent investigations with animal models have identified diabetes-related mitochondrial abnormalities. Studies performed with a model of chronic type 1 diabetes mellitus (OVE26 mice19) demonstrated evidence of mitochondrial biogenesis, focal mitochondrial damage, and altered mitochondrial function. Other investigators have found derangements in mitochondrial ultrastructure and reduced respiratory capacity in hearts of obese or diabetic animals.20–23 These results indicate that diabetes is associated with mitochondrial abnormalities in both skeletal muscle and heart, but that in the latter, a mitochondrial biogenic response is mounted. The role of the mitochondrial abnormalities as causal or secondary in diabetic cardiac dysfunction is unknown, however. Temporal pattern and mechanisms controlling mitochondrial biogenesis and dysfunction in the diabetic heart have not been delineated. Moreover, the role of the biogenic response as adaptive or maladaptive for cardiac energetics and function is unknown.
The present study was designed to test the hypothesis that the regulatory pathway involved in triggering mitochondrial biogenesis in the diabetic heart is activated in the prediabetic, insulin-resistant stage, possibly as an adaptive response to support increased flux of FAs through the mitochondrial β-oxidation pathway. In addition, we sought to identify the specific molecular regulatory pathway involved in this cardiac mitochondrial biogenic response. We found that the expression of nuclear and mitochondrial genes encoding enzymes involved in multiple mitochondrial pathways is activated in the hearts of insulin-resistant uncoupling protein-diptheria toxin A (UCP-DTA) transgenic mice, concomitant with a robust cardiac mitochondrial biogenic response. The results of the present study provide evidence for a regulatory loop that requires the FA-activated nuclear receptor peroxisome proliferator-activated receptor-α (PPARα), possibly together with the transcriptional coactivator PGC-1α (PPARγ coactivator-1α), a known regulator of mitochondrial biogenesis.
Methods
Animal Models
UCP-DTA transgenic mice24 (The Jackson Laboratory, Bar Harbor, Me) were crossed with PPARα-null (PPARα–/–)25,26 animals, both in an FVB/N background. Three-month-old male nontransgenic (NTG), UCP-DTA, PPARα–/–, and UCP-DTA×PPARα–/– mice were studied. In all experiments, mice were compared directly with strain-matched littermate NTG mice. Mice were fasted for 4 hours in the morning, during the early part of the light cycle, for evaluation of tail blood glucose concentration (HemoCue Blood Glucose Analyzer, HemoCue AB, Ängelholm, Sweden) and then euthanized to obtain plasma for insulin and triglyceride measurements (Tables 1 and 2). These measurements were performed by the Diabetes Research and Training Core (DRTC) Core at Washington University. The insulin resistance index was calculated by multiplying the blood glucose concentration by the concentration of plasma insulin.
TABLE 1.
Parameters of UCP-DTA Mice
| NTG | UCP-DTA | |
|---|---|---|
| Body weight, g | 30.8±1.5 | 35.6±2.9* |
| Glucose, mg/dL | 107.0±7.8 | 124.9±11 |
| Insulin, ng/mL | 0.7±0.32 | 2.78±0.62* |
| Insulin resistance index | 75.9±10.8 | 347.2±25.3* |
| Triacylglycerol, mg/dL | 48.8±5.4 | 132.8±9.0* |
Values are mean±SE; n≥6 for each group.
P<0.05 vs NTG mice.
TABLE 2.
Parameters of UCP-DTA×PPARα−/− Mice
| NTG | UCP-DTA | |
|---|---|---|
| Body weight, g | 30.7±1.1 | 35.3±3.9* |
| Glucose, mg/dL | 89.3±18.6 | 107.0±15.8 |
| Insulin, ng/mL | 0.37±0.06† | 0.93±0.21*† |
| Insulin resistance index | 33.0±5.4† | 99.5±16.4*† |
| Triacylglycerol, mg/dL | 55.0±3.2 | 156.8±11.3* |
Values are mean±SE; n≥6 for each group.
P<0.05 vs NTG mice of the same PPARα genotype.
P<0.05 vs PPARα+/+ mice of the same UCP-DTA genotype (values in Table 1).
MHC-PPARα mice (line 404-3)27 were backcrossed 6 times into the C57BL/6J background. Two-month-old MHC-PPARα mice and their NTG littermates were studied. All experiments and protocols were conducted in strict accordance with the National Institutes of Health guidelines for humane treatment of animals and were reviewed and approved by the Washington University Animal Studies Committee.
Electron Microscopy
Papillary muscle was dissected from the left ventricle of the heart, fixed, and sectioned as described previously.28 Cardiac mitochondrial and myofibrillar volume densities were determined from electron micrographs as described previously.28,29 For each animal, 3 different fields were quantified at the magnification 10 000× in a blinded fashion. Data are expressed as mean volume density (volume of mitochondria or myofibrils [μm3] per cytoplasmic volume [μm3]) in each field.
Quantitative Real-Time Polymerase Chain Reaction for Mitochondrial DNA
Mitochondrial DNA (mtDNA) content was quantified by real-time reverse-transcription polymerase chain reaction (rtPCR) with cardiac DNA as described previously.19 Briefly, DNA was extracted from frozen heart tissue of NTG and insulin-resistant UCP-DTA mice by proteinase K digestion followed by phenol-chloroform extraction. DNA was precipitated with ammonium acetate and ethanol. Total DNA concentration was determined with a fluorometer. Five nanograms of genomic DNA was assayed in triplicate with Sybrgreen core reagents (Applied Biosystems, Foster City, Calif) and cytochrome b (mitochondrial) or β-actin (nuclear) and a Prism 7500 Sequence Detector (Applied Biosystems). mtDNA per nuclear genome was calculated as the ratio of cytochrome b DNA to β-actin DNA quantity. Primer sequences for cytochrome b and β-actin are listed in Table I in the Data Supplement.
RNA and Protein Analyses
Total RNA was isolated from hearts by the RNAzol method (Tel-Test, Friendswood, Tex) as described previously.30 First-strand cDNA was generated, and real-time rtPCR was performed with triplicate reactions as described previously.28 Arbitrary units of target mRNA were corrected by measuring the levels of 36B4 RNA. The mouse-specific primer-probe sets used to detect gene expression can be found in supplemental Table I.
Western blotting was performed with whole-cell lysates as described previously31 with antibodies directed against medium-chain acyl-CoA dehydrogenase (MCAD),32 cytochrome oxidase complex II (COX II; Santa Cruz Biotechnology, Santa Cruz, Calif) and IV (COX IV; Molecular Probes, Eugene, Ore), and mitochondrial transcription factor A (mtTFA; Santa Cruz Biotechnology). Ponceau S stain (Sigma, St. Louis, Mo) was used as a control for loading.
Mitochondrial Respiration and ATP Measurements
Mitochondrial respiration was assessed in saponin-permeabilized cardiac fibers as described previously.21,28,33 Oxygen consumption (V̇o) was measured at 25°C with an optical probe (Oxygen FOXY probe, Ocean Optics, Dunedin, Fla) in the presence of palmitoyl-l-carnitine (0.02 mmol/L) and malate (2 mmol/L). After measurement of basal respiration, maximal (ADP-stimulated) state 3 respiration was determined by exposing the fibers to 1 mmol/L ADP, and then respiration in the absence of ADP phosphorylation was determined in the presence of 1 mg/mL oligomycin. Respiration rates were expressed as nanomoles of O2 per minute per milligram of dry fiber weight. ATP synthesis was evaluated with aliquots taken from the respiration chamber over a 2-minute period after the addition of ADP. ATP was quantified with a luciferase based assay (ENLITEN ATP assay, Promega, Madison, Wis). ATP/O ratio was calculated with the state 3 respiratory rate for each sample.
Statistical Analysis
For quantitative data, statistical comparisons were made with Student t test, assuming unequal variances. All data are presented as mean±SE, with a statistically significant difference defined as P<0.05.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Insulin-Resistant UCP-DTA Mice Exhibit Cardiac Mitochondrial Biogenesis
We first sought to examine the mitochondrial ultrastructure in hearts of obese, insulin-resistant mice. To this end, we used UCP-DTA mice, a transgenic model that exhibits many features of metabolic syndrome caused by partial ablation of brown adipose tissue via tissue-specific targeting of a diphtheria toxigene.24,34 By 3 months of age on standard chow diet, mean body weight was significantly greater in UCP-DTA mice than in NTG littermates (Table 1). The UCP-DTA mice were normoglycemic but hyperinsulinemic and hypertriglyceridemic at this age (Table 1). There was no change in cardiac function or left ventricular mass by echocardiographic assessment in the UCP-DTA mice (data not shown), as described previously.34 Furthermore, at this age, there was no difference in heart weight to body weight ratio and no evidence of hypertension in the UCP-DTA mice compared with NTG controls (data not shown).
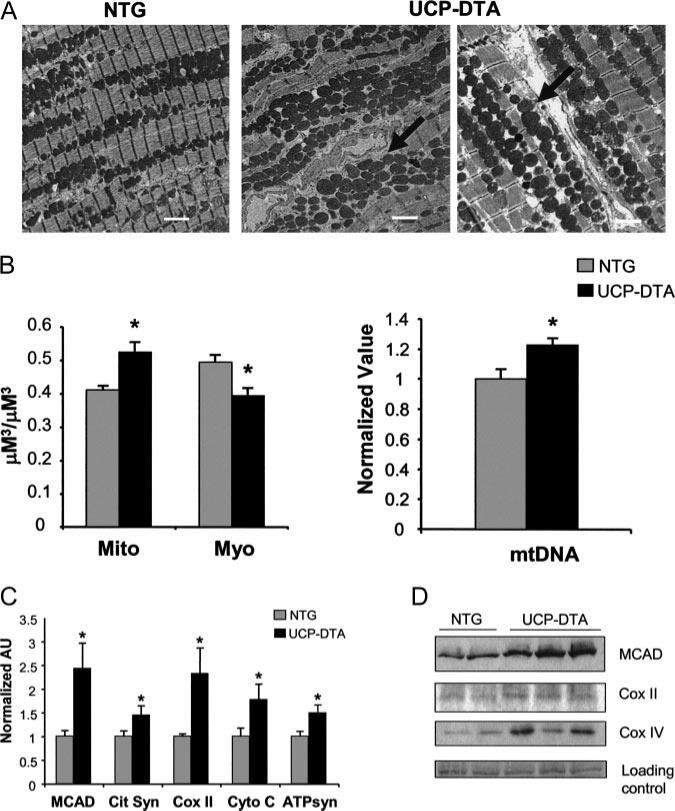
Transmission electron microscopy was performed on cardiac ventricular samples prepared from insulin-resistant UCP-DTA and NTG littermate control mice. The mitochondria in the hearts of UCP-DTA mice were larger in size and number than those in controls (Figure 1A). Quantitative morphometric analysis confirmed that mitochondrial volume density was significantly greater in the UCP-DTA samples (Figure 1B, left). Furthermore, myofibril volume density was significantly decreased (Figure 1B, left). Consistent with the observed increase in mitochondrial volume density, levels of mtDNA were greater in the insulin-resistant UCP-DTA mouse hearts than in NTG controls (Figure 1B, right).
Figure 1.
Mitochondrial biogenesis in insulin-resistant hearts. A, Representative electron micrographs of papillary muscles from NTG and insulin-resistant UCP-DTA hearts. White bar=2 μm. Arrows indicate areas of increased mitochondria. B, Quantitative morphometric measurement (left) of mitochondrial (Mito) or myofibril (Myo) cellular volume density (μm3/μm3) based on analysis of electron micrographs (n=6 animals/group). Mean cardiac mtDNA levels (right) determined by real-time PCR analysis shown as arbitrary units (AU; n=5 to 7 hearts/group) normalized to the NTG value (=1.0). C, Results of real-time rtPCR analysis of mitochondrial metabolic genes in insulin-resistant UCP-DTA and NTG control hearts. Cit Syn indicates citrate synthase; Cyto C, cytochrome c; and ATPsyn, ATP synthase-β; other abbreviations defined in text. n=6 to 9 mice/group. Bars represent mean (±SE) AU normalized to the corresponding NTG value. *P<0.05 compared with NTG control. D, Representative immunoblots performed with protein prepared from NTG and insulin-resistant UCP-DTA hearts with medium-chain acyl-CoA dehydrogenase, COX II, and COX IV antibodies. Representative ponceau-stained bands are shown as a loading control. MCAD indicates medium-chain acyl-CoA dehydrogenase.
We next determined expression levels of nuclear and mitochondrial genes involved in multiple mitochondrial energy metabolic pathways. The levels of transcripts encoding proteins involved in mitochondrial FAO (medium-chain acyl-CoA dehydrogenase), the tricarboxylic acid cycle (citrate synthase), electron transport (cytochrome C and COX II), and oxidative phosphorylation (β-subunit of ATP synthase) were all significantly increased in insulin-resistant hearts compared with controls (Figure 1C). Protein levels of medium-chain acyl-CoA dehydrogenase, COX II, and COX IV were also increased in the hearts of the UCP-DTA mice (Figure 1D). Taken together, these data demonstrate a cardiac mitochondrial biogenic response in the hearts of insulin-resistant UCP-DTA mice.
Insulin-Resistant Mitochondria Exhibit Increased Respiratory Uncoupling
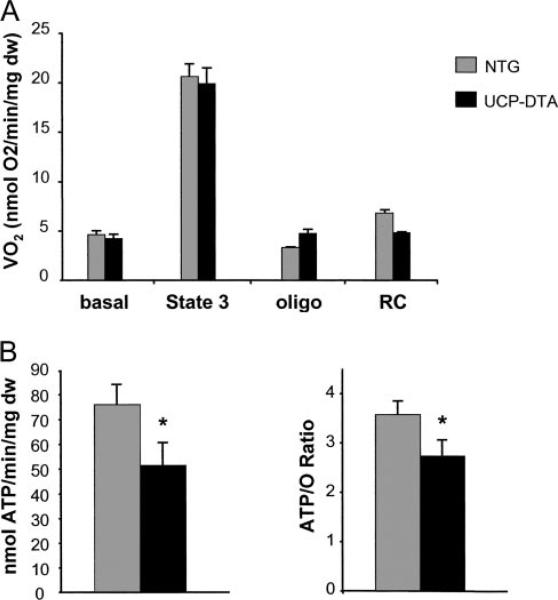
Mitochondrial respiratory rates were assessed in permeabilized muscle strips from insulin-resistant UCP-DTA mice and their littermates. There was no change in baseline or maximal ADP-stimulated (state 3) respiration in the insulin-resistant hearts (Figure 2A). However, there was a tendency toward increased oligomycin-inhibited respiration and reduced respiratory control ratio, suggestive of uncoupling. In addition, ATP synthesis rates and the ATP/O2 consumption ratio in muscle strips from UCP-DTA mice were significantly lower than in controls (Figure 2B). These data indicate that the respiration of cardiac mitochondria in the insulin-resistant animals was inefficient, likely owing to an increase in uncoupled respiration. Interestingly, rtPCR studies demonstrated that expression of the genes encoding UCP-2 and -3 was not increased in the UCP-DTA animals (data not shown).
Figure 2.
Mitochondrial respiration is inefficient in insulin-resistant UCP-DTA hearts. A, Mitochondrial respiration rates (V̇o) in saponin-permeabilized muscle strips prepared from NTG and UCP-DTA hearts in the presence of palmitoyl-l-carnitine/malate. Mean values (±SE) are shown for basal, state 3 (ADP-stimulated), and oligomycin-inhibited (oligo) respiration. The respiratory control (RC) ratio represents the ratio of oligo to state 3 respiration; n=6 animals/group. B, ATP synthesis rates (left) and ratio of ATP synthesis to maximal V̇o (ATP/O; right) obtained from the same muscle strips shown in Figure 2A. *P<0.05 compared with NTG value. mg dw indicate milligrams of dry weight.
PPARα Is Required for Activation of the Cardiac Mitochondrial Biogenic Program in Insulin-Resistant Mice
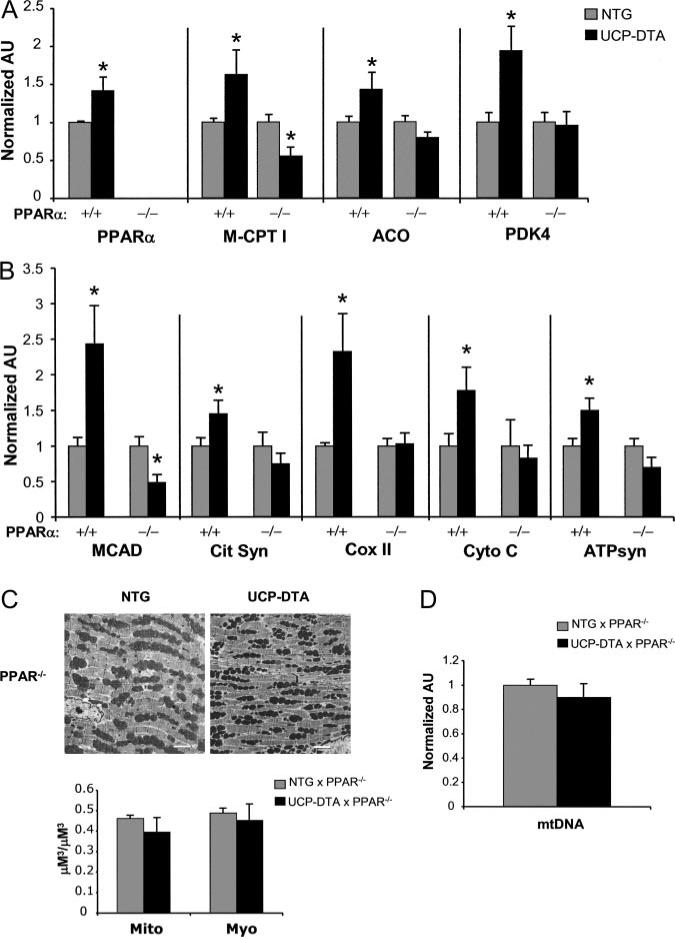
Previous studies have shown that the expression of genes involved in mitochondrial FAO is increased in the hearts of insulin-resistant and diabetic rodents and that this is driven, at least in part, by the nuclear receptor PPARα.26,35,36 As expected, we found that the expression of the PPARα gene and several of its target genes, including muscle carnitine palmitoyltransferase I, acyl-CoA oxidase, and pyruvate dehydrogenase kinase, was increased in insulin-resistant UCP-DTA mouse hearts compared with controls (Figure 3A).
Figure 3.
PPARα gene expression is induced in insulin-resistant hearts and is required for activation of genes involved in mitochondrial metabolism and biogenesis. A, Quantitative real-time rtPCR analysis of cardiac transcripts encoding PPARα and several of its target genes as denoted in insulin-resistant UCP-DTA and NTG control mice in wild-type (+/+) and PPARα-null (–/–) backgrounds (n=6 to 7 animals/group). M-CPT I indicates muscle carnitine palmitoyltransferase I; ACO, acyl-CoA oxidase; and PDK4, pyruvate dehydrogenase kinase. B, Results of real-time rtPCR analysis of the mitochondrial genes shown in Figure 1 in wild-type PPARα or PPARα-null backgrounds (n=6 to 9 animals/group). Bars represent mean (±SE) arbitrary units (AU) normalized to the NTG value (=1.0) in each case. *P<0.05 compared with NTG control for each PPARα background. C, Representative electron micrographs performed on papillary muscle from NTG and UCP-DTA hearts in PPARα-null (PPAR–/–) background. White bars=2 μm. Quantitative morphometric measurement (bottom) of mitochondrial (Mito) or myofibril (Myo) cellular volume density (μm3/μm3) based on analysis of electron micrographs (n=4 animals/group). D, Mean cardiac mtDNA levels determined by real-time PCR analysis shown as mean AU (n=7 to 8 hearts/group) normalized to the NTG value (=1.0).
To investigate the role of PPARα in the observed mitochondrial biogenic response in insulin-resistant hearts, the UCP-DTA mice were backcrossed into a PPARα-null background25,35 (UCP-DTA×PPARα–/–). The UCP-DTA×PPARα–/– mice exhibited higher insulin and triacylglycerol levels than corresponding PPARα–/– mice, albeit to a lesser degree than wild-type UCP-DTA mice. As we have shown previously in streptozotocin-treated diabetic mice,37 the upregulation of FAO enzyme genes was not observed in the UCP-DTA×PPARα–/– mice (Figure 3A). In addition, the observed induction of genes encoding other mitochondrial enzymes was not activated in the UCP-DTA×PPARα–/– animals (Figure 3B). The mitochondrial biogenic response was also absent in the UCP-DTA×PPARα–/– mice, as determined by morphometry (Figure 3C) and mtDNA quantification (Figure 3D). These results indicate that PPARα is necessary for the mitochondrial biogenic response related to the insulin-resistant state.
PPARα Is Sufficient to Drive the Cardiac Mitochondrial Biogenic Response
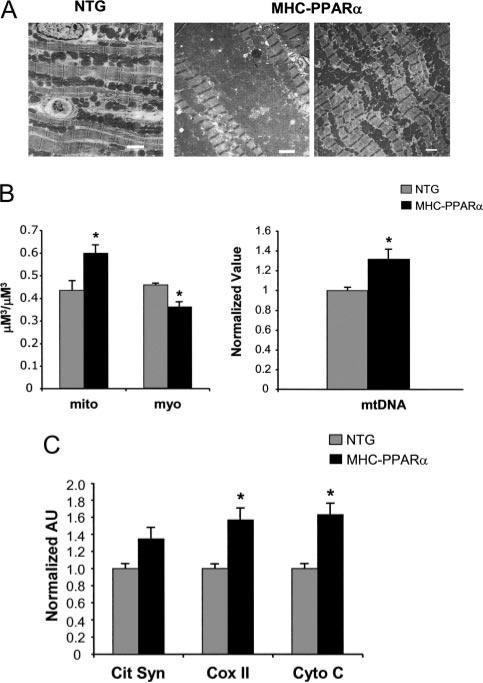
To determine whether activation of PPARα, such as occurs in the insulin-resistant heart, is sufficient to activate mitochondrial biogenesis in the absence of insulin resistance, we examined cardiac mitochondria of transgenic mice with cardiac-specific overexpression of PPARα (MHC-PPARα mice).27 Compared with NTG littermate controls, the hearts of MHC-PPARα mice exhibited a significant increase in mitochondrial volume density (Figure 4A and 4B, left) and mtDNA levels (Figure 4B, right). Quantitative rtPCR demonstrated a 50% to 60% increase in mean levels of transcripts encoding cytochrome C and COX II and a tendency toward increased citrate synthase (P=0.06; Figure 4C), similar to the UCP-DTA mice. Taken together with the results of the UCP-DTA×PPARα–/– mice, these results indicate that PPARα is both necessary and sufficient for the mitochondrial biogenic response of the insulin-resistant heart.
Figure 4.
Overexpression of PPARα induces cardiac mitochondrial biogenesis. A, Representative electron micrographs performed on cardiac papillary muscle of NTG and MHC-PPARα littermates. White bar=2 μm. B, Quantitative electron micrograph morphometric measurements (left) and mtDNA quantification (right) from MHC-PPARα mice. For morphometry, bars represent mean (±SE) volume density (μm3/μm3). For mtDNA, bars represent mean (±SE) arbitrary units (AU) normalized to the NTG value (=1.0). *P<0.05 compared with NTG mice. C, Results of real-time rtPCR analysis of mitochondrial metabolic genes (abbreviations as in Figure 1 legend) in hearts of MHC-PPARα mice (n=8 to 10 littermates/group). Values were normalized to the NTG value (=1.0) in each case. Bars represent mean (±SE) AU for each gene. *P<0.05 compared with corresponding NTG mice.
Evidence That PPARα-Mediated Mitochondrial Biogenic Response Involves the Transcriptional Coactivator PGC-1α
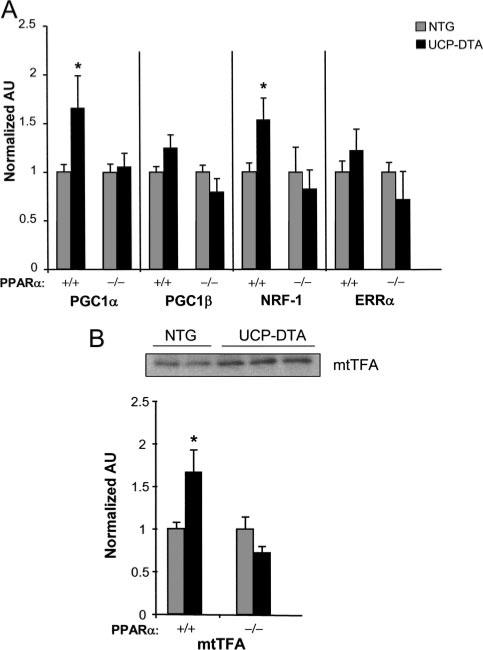
Whereas PPARα is known to activate expression of many genes involved in cellular FA utilization, it has not been shown to regulate genes involved in mitochondrial respiratory function or biogenesis. Therefore, it seemed likely that the observed effects of PPARα on the mitochondrial biogenic response in UCP-DTA and MHC-PPARα mice occurred indirectly, perhaps via regulatory pathways known to control postnatal mitochondrial function and biogenesis. Recently, a transcriptional regulatory cascade involved in the control of mitochondrial biogenesis has been defined (reviewed in Kelly and Scarpulla38). The PGC-1 family of transcriptional coactivators serve as the drivers of this cascade.39–41 The expression of genes encoding PGC-1α and PGC-1β and downstream components of the PGC-1 circuit, including the transcription factors nuclear respiratory factor 1 (NRF-1) and the estrogen-related receptor-α (ERRα), was evaluated. Real-time rtPCR studies demonstrated that levels of transcripts encoding both PGC-1α and NRF-1 were increased in the UCP-DTA hearts but not in UCP-DTA×PPARα–/– hearts (Figure 5A). Levels of mitochondrial transcription factor A (mtTFA), a critical regulator of mtDNA replication downstream of NRF-1, were also increased in the UCP-DTA samples in wild-type but not in the PPARα–/– background (Figure 5B). ERRα mRNA was not increased in either group (Figure 5A). However, ERRα is capable of cooperating with the NRFs and PGC-1α and thus may “amplify” the PGC-1α cascade even without being induced at the gene expression level.39 PGC-1β has been reported to have overlapping functions with PGC-1α,42 but it was not significantly increased in the insulin-resistant heart. These results suggest that PPARα stimulates the mitochondrial gene regulatory response by activation of the PGC-1α/NRF-1 regulatory pathway.
Figure 5.
Induction of PGC-1α and NRF-1 in insulin-resistant UCP-DTA hearts is dependent on PPARα. A, Results of real-time rtPCR analysis of candidate transcription factors (abbreviations defined in text) involved in mitochondrial biogenesis in insulin-resistant UCP-DTA hearts in wild-type (+/+) and PPARα-null (–/–) backgrounds (n=4 to 8 mice/group). AU indicates arbitrary units. B, Representative immunoblot (top) and real-time rtPCR analysis of mtTFA expression in the same mice as in (A). Bars represent mean (±SE) AU normalized to NTG control (=1.0) in each group. *P<0.05 compared with corresponding NTG mice.
Discussion
The role of mitochondrial dysfunction as a causal or secondary phenomenon in the development of diabetic cardiomyopathy is unknown. As an initial step to address this question, we sought to determine whether signatures of mitochondrial abnormalities were present in the insulin-resistant heart, before the development of overt diabetes mellitus. Herein, we demonstrate that a cardiac mitochondrial biogenic response occurs in the insulin-resistant, prediabetic stage in UCP-DTA mice. The present results implicate a PPARα-dependent, PGC-1α–mediated regulatory mechanism as a key driver in this mitochondrial response. We found that hearts of insulin-resistant UCP-DTA mice exhibit a robust mitochondrial biogenic response characterized by a coordinate increase in mitochondrial volume density, mtDNA content, and expression of nuclear and mitochondrial genes involved in energy transducing and ATP synthetic pathways. These results are consistent with observations by others showing cardiac mitochondrial proliferation in insulin-deficient diabetic mice.19 Given that this mitochondrial biogenic response occurs at an early stage, before the onset of overt diabetes, the signals involved in triggering this response are likely related to the insulin-resistant state. It is possible that alterations in cellular insulin signaling or increased cellular import of FAs, both of which are known to occur in the insulin-resistant heart, lead to alterations in bioenergetics, activating a mitochondrial biogenic response aimed at increasing capacity for ATP generation.
Previous studies have shown that mitochondrial respiratory function is altered in the hearts of animal models of diabetes.19,21,23,43 We found that cardiac mitochondria of insulin-resistant UCP-DTA mice exhibit normal state 3 respiration rates. However, the present results suggest early mitochondrial dysfunction. Specifically, despite the observed biogenic response, respiratory capacity was not augmented beyond levels in the normal controls. In addition, the mitochondria of the insulin-resistant UCP-DTA mice were inefficient, as evidenced by a reduced ATP synthesis/oxygen consumption ratio. These latter results are consistent with the findings of Boudina and coworkers21 indicating that mitochondrial respiratory uncoupling is increased in the hearts of mouse models of diabetes. Interestingly, UCP-2 and -3 gene expression was not increased in the UCP-DTA hearts. Consistent with this finding, Buchanan et al43 found reduced cardiac efficiency, suggestive of uncoupling, in isolated hearts from obese mice, before the induction of UCP gene expression. It is possible that other mechanisms, such as FA toxicity or accumulation of superoxide44 could lead directly to respiratory uncoupling. Taken together with the findings of others, the present results suggest that the mitochondrial biogenic response in the insulin-resistant and diabetic heart may be triggered by reduced mitochondrial efficiency, a response that is not adequate to significantly increase respiratory capacity.
Several lines of evidence support the conclusion that the PPARα gene regulatory pathway is involved in the cardiac mitochondrial biogenic response observed in insulin-resistant UCP-DTA mice. First, insulin-resistant UCP-DTA mice in a PPARα-null background did not exhibit mitochondrial proliferation or gene expression signatures of mitochondrial biogenesis. Second, we found that a significant cardiac mitochondrial biogenic response occurs in mice overexpressing PPARα in heart (MHC-PPARα mice), which indicates that PPARα is sufficient to activate mitochondrial biogenesis in the absence of insulin resistance. Interestingly, although the related nuclear receptor PPARβ/δ has been implicated as a factor involved in the regulation of mitochondrial respiratory functional capacity in skeletal muscle,45 PPARβ/δ expression was unchanged in the insulin-resistant UCP-DTA mice (data not shown), which suggests that PPARα is the predominant PPAR isotype influencing the biogenic response in this disease state. It is also of interest that the cardiac mitochondrial ultrastructure is not abnormal in PPARα–/– mice, which indicates that PPARα is not essential for normal biogenesis. This is not surprising given that PGC-1α remains active with mitochondrial biogenic factors such as NRF-1 in PPARα–/– mice.38 Although we found an increase in PPARα gene expression in the insulin-resistant heart, it is likely that PPARα is also activated via increased delivery of FA ligand to the heart. Indeed, we found that the insulin-resistant UCP-DTA mice had significantly increased triacylglycerol levels. However, UCP-DTA×PPARα–/– mice exhibit normal cardiac mitochondrial volume density in association with increased triacylglycerol levels, which indicates the requirement for PPARα.
The present results implicate the PGC-1α gene regulatory circuit in the PPARα-dependent mitochondrial biogenic response of the insulin-resistant heart. Although PPARα is known to regulate the expression of genes involved in mitochondrial FAO, it has not been shown to directly regulate the expression of genes involved in mitochondrial respiratory function and biogenesis. Recently, the regulatory cascade that controls postnatal mitochondrial biogenesis and functional capacity has been defined.38,41,46 Specifically, the inducible factors PGC-1α and PGC-1β coordinately regulate mitochondrial gene expression and biogenesis by serving as coactivators of multiple transcription factors involved in the control of specific mitochondrial pathways, including the PPAR (FAO) and ERR (FAO, electron transport, and oxidative phosphorylation) families of nuclear receptors, NRF-1 and NRF-2 (electron transport and oxidative phosphorylation), and mtTFA (mtDNA transcription and replication). Myocardial PGC-1α gene expression has been shown to be increased in insulin-resistant db/db mice.43 We found that expression of the genes encoding PGC-1α and its downstream regulators, NRF-1 and mtTFA, was significantly increased in the hearts of insulin-resistant UCP-DTA mice. In striking contrast, expression of the regulators was not induced in insulin-resistant UCP-DTA mice in a PPARα-deficient background. These latter results suggest the existence of a regulatory loop in which PPARα activates PGC-1α gene expression. Consistent with this notion, the human PGC-1α promoter sequence contains several putative PPAR recognition elements (data not shown), including a site 1982 base pairs upstream of the hPGC-1α gene transcription start site (5′-TGACCTTTGTCCT-3′) that exhibits 100% nucleotide identity with a PPARγ response element identified very recently in the mouse PGC-1α promoter.47
In summary, we propose the following model for regulation of mitochondrial biogenesis in the insulin-resistant heart: (1) Caloric excess drives insulin resistance and increased circulating FA, which activates PPARα; (2) PPARα is coactivated by PGC-1α to induce FA uptake and oxidation; (3) in addition, PPARα exerts reciprocal activation of PGC-1α gene expression, which amplifies coactivation of NRF-1 and mtTFA (and likely other factors) to orchestrate mitochondrial biogenesis. This series of events is likely triggered as an adaptive response to increase capacity for mitochondrial FAO and cope with mitochondrial inefficiency, which is probably a very early event. Over the long term, we speculate that this response is inadequate or becomes maladaptive, contributing to diabetic cardiac dysfunction.
Supplementary Material
CLINICAL PERSPECTIVE.
We are currently witnessing a marked increase in the prevalence of metabolic syndrome and obesity-related diabetes mellitus. Cardiovascular disease is the leading cause of death in the diabetic population. Diabetic patients develop a unique form of cardiac dysfunction that often leads to heart failure. The basis for diabetic cardiac dysfunction is unknown. Evidence is emerging that abnormalities in energy metabolism contribute to diabetic myocardial disease. In the present study, using mouse models that mimic the stages of progression from insulin resistance to diabetes, we demonstrate that at the early insulin-resistant stage, the heart exhibits abnormalities in mitochondrial structure and function. Specifically, signatures of increased mitochondrial number and gene expression are observed. Using genetically modified mice, we demonstrate that this early mitochondrial biogenic response is mediated by the nuclear receptor peroxisome proliferator-activated receptor-α and its coactivator, PGC-1α. In the absence of peroxisome proliferator-activated receptor-α, the insulin-resistant heart is incapable of increasing mitochondrial number. These results are important because they demonstrate that abnormalities in mitochondrial structure and regulation occur at an early stage in the development of diabetic cardiac dysfunction. It is likely that these changes are initially adaptive; evidence is presented, however, that the mitochondrial regulatory response is inadequate and leads to an inefficient form of energy production. The observation that the peroxisome proliferator-activated receptor-α α/PGC-1α regulatory circuit is involved in this response identifies a potential future therapeutic target for the prevention or treatment of diabetic cardiac dysfunction.
Acknowledgments
The authors thank Michelle Croce for assistance with mouse experimentation, Jacqueline Siljee and Michael Brown for assistance with the PGC-1α studies, Bill Kraft for technical assistance with electron microscopy, and Mary Wingate for her excellent assistance in manuscript preparation.
Sources of Funding
Dr Duncan was supported by National Institutes of Health grant T32 HD043010. This work was also supported by the Diabetes Research and Training Center (P60 DK20579) and National Institutes of Health grants P50 HL077113 and P01 HL057278.
Footnotes
The online-only Data Supplement, consisting of a table, is available with this article at http://circ.ahajournals.org/cgi/content/full/CIRCULATIONAHA.106.662296/DC1.
Disclosures
Dr Kelly is a scientific consultant for Bristol-Myers Squibb, GlaxoSmithKline, and Phrixus, Inc. The remaining authors report no conflicts.
References
- 1.Mokdad AH, Bowman BA, Ford ES, Vinicor F, Marks JS, Koplan JP. The continuing epidemics of obesity and diabetes in the United States. JAMA. 2001;286:1195–1200. doi: 10.1001/jama.286.10.1195. [DOI] [PubMed] [Google Scholar]
- 2.Koskinen P, Manttari M, Manninen V, Huttunen JK, Heinonen OP, Frick MH. Coronary heart disease incidence in NIDDM patients in the Helsinki Heart Study. Diabetes Care. 1992;15:820–825. doi: 10.2337/diacare.15.7.820. [DOI] [PubMed] [Google Scholar]
- 3.Stamler J, Vaccaro O, Neaton JD, Wentworth D. Diabetes, other risk factors, and 12-yr cardiovascular mortality for men screened in the Multiple Risk Factor Intervention Trail. Diabetes Care. 1993;16:434–444. doi: 10.2337/diacare.16.2.434. [DOI] [PubMed] [Google Scholar]
- 4.Rubler S, Dlugash J, Yuceoglu YZ, Kumral T, Branwood AW, Grishman A. New type of cardiomyopathy associated with glomerulosclerosis. Am J Cardiol. 1972;30:595–602. doi: 10.1016/0002-9149(72)90595-4. [DOI] [PubMed] [Google Scholar]
- 5.Regan TJ, Lyons MM, Ahmed SS, Levinson GE, Oldewurtel HA, Ahmed MR, Haider B. Evidence for cardiomyopathy in familial diabetes mellitus. J Clin Invest. 1977;60:885–899. doi: 10.1172/JCI108843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fein FS, Sonnenblick EH. Diabetic cardiomyopathy. Prog Cardiovasc Dis. 1985;4:255–270. doi: 10.1016/0033-0620(85)90009-x. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues B, Cam MC, McNeill JH. Myocardial substrate metabolism: implications for diabetic cardiomyopathy. J Mol Cell Cardiol. 1995;27:169–179. doi: 10.1016/s0022-2828(08)80016-8. [DOI] [PubMed] [Google Scholar]
- 8.Stanley WC, Lopaschuk GD, McCormack JG. Regulation of energy substrate metabolism in the diabetic heart. Cardiovasc Res. 1997;34:25–33. doi: 10.1016/s0008-6363(97)00047-3. [DOI] [PubMed] [Google Scholar]
- 9.Taegtmeyer H, McNulty P, Young ME. Adaptation and maladaptation of the heart in diabetes: part I: general concepts. Circulation. 2002;105:1727–1733. doi: 10.1161/01.cir.0000012466.50373.e8. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues B, McNeill JH. The diabetic heart: metabolic causes for the development of cardiomyopathy. Cardiovasc Res. 1992;26:913–922. doi: 10.1093/cvr/26.10.913. [DOI] [PubMed] [Google Scholar]
- 11.Belke DD, Larsen TS, Gibbs EM, Severson DL. Altered metabolism causes cardiac dysfunction in perfused hearts from diabetic (db/db) mice. Am J Physiol Endocrinol Metabol. 2000;279:E1104–E1113. doi: 10.1152/ajpendo.2000.279.5.E1104. [DOI] [PubMed] [Google Scholar]
- 12.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 13.Yechoor VK, Patti ME, Saccone R, Kahn CR. Coordinated patterns of gene expression for substrate and energy metabolism in skeletal muscle of diabetic mice. Proc Natl Acad Sci U S A. 2002;99:10587–10592. doi: 10.1073/pnas.142301999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman BM, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, Shatzkes N, Neschen S, White MF, Bilz S, Sono S, Pypaert M, Shulman GI. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest. 2005;115:3587–3593. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 19.Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type I diabetes. Am J Physiol Endocrinol Metabol. 2004;287:E896–E905. doi: 10.1152/ajpendo.00047.2004. [DOI] [PubMed] [Google Scholar]
- 20.Flarsheim CE, Grupp IL, Matlib MA. Mitochondrial dysfunction accompanies diastolic dysfunction in diabetic rat heart. Am J Phys. 1996;271:H192–H202. doi: 10.1152/ajpheart.1996.271.1.H192. [DOI] [PubMed] [Google Scholar]
- 21.Boudina S, Sena S, O'Neill BT, Tathireddy P, Young ME, Abel ED. Reduced mitochondrial oxidative capacity and increased mitochondrial uncoupling impair myocardial energetics in obesity. Circulation. 2005;112:2686–2695. doi: 10.1161/CIRCULATIONAHA.105.554360. [DOI] [PubMed] [Google Scholar]
- 22.Boudina S, Abel ED. Mitochondrial uncoupling: a key contributor to reduced cardiac efficiency in diabetes. Physiology. 2006;21:250–258. doi: 10.1152/physiol.00008.2006. [DOI] [PubMed] [Google Scholar]
- 23.How OJ, Aasum E, Severson DL, Chan WY, Essop MF, Larsen TS. Increased myocardial oxygen consumption reduces cardiac efficiency in diabetic mice. Diabetes. 2006;55:466–473. doi: 10.2337/diabetes.55.02.06.db05-1164. [DOI] [PubMed] [Google Scholar]
- 24.Lowell BB, Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee SST, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finck B, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, Han X, Gross RW, Kozak R, Lopaschuk GD, Kelly DP. The cardiac pheno-type induced by PPARα overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1α deficient mice exhibit multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control, and hepatic steatosis. PLoS Biol. 2005;3:672–687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros D, Kelly DP. PPARγ coactivator-1 (PGC-1) promotes cardiac mitochondrial bio-genesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly DP, Gordon JI, Alpers R, Strauss AW. The tissue-specific expression and developmental regulation of the two nuclear genes encoding rat mitochondrial proteins: medium-chain acyl-CoA dehydrogenase and mitochondrial malate dehydrogenase. J Biol Chem. 1989;264:18921–18925. [PubMed] [Google Scholar]
- 31.Cresci S, Wright LD, Spratt JA, Briggs FN, Kelly DP. Activation of a novel metabolic gene regulatory pathway by chronic stimulation of skeletal muscle. Am J Physiol. 1996;270(pt 1):C1413–C1420. doi: 10.1152/ajpcell.1996.270.5.C1413. [DOI] [PubMed] [Google Scholar]
- 32.Kelly DP, Kim JJ, Billadello JJ, Hainline BE, Chu TW, Strauss AW. Nucleotide sequence of medium-chain acyl-CoA dehydrogenase mRNA and its expression in enzyme-deficient human tissue. Proc Natl Acad Sci U S A. 1987;84:4068–4072. doi: 10.1073/pnas.84.12.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saks VA, Veksler VI, Kuznetsov AV, Kay L, Sikk P, Tiivel T, Tranqui L, Olivares J, Winkler K, Wiedemann F, Kunz WS. Permeabilized cell and skinned fiber techniques in studies of mitochondrial function in vivo. Mol Cell Biochem. 1998;184:81–100. [PubMed] [Google Scholar]
- 34.Cittadini A, Mantzoros CS, Hampton TG, Travers KE, Katz SE, Morgan JP, Flier JS, Douglas PS. Cardiovascular abnormalities in transgenic mice with reduced brown fat: an animal model of human obesity. Circulation. 1999;100:2177–2183. doi: 10.1161/01.cir.100.21.2177. [DOI] [PubMed] [Google Scholar]
- 35.Djouadi F, Weinheimer CJ, Saffitz JE, Pitchford C, Bastin J, Gonzalez FJ, Kelly DP. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator-activated receptor α-deficient mice. J Clin Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finck B, Han X, Courtois M, Aimond F, Nerbonne JM, Kovacs A, Gross RW, Kelly DP. A critical role for PPARα-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation of phenotype by dietary fat content. Proc Natl Acad Sci U S A. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finck BN, Bernal-Mizrachi C, Han DH, Coleman T, Sambandam N, LaRiviere LL, Holloszy JO, Semenkovich CF, Kelly DP. A potential link between muscle peroxisome proliferator-activated receptor α signaling and obesity-related diabetes. Cell Metabolism. 2005;1:133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 41.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 42.St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 43.Buchanan J, Mazumder PK, Hu P, Chakrabarti G, Roberts MW, Jeong Yun U, Cooksey RC, Litwin SE, Abel ED. Reduced cardiac efficiency and altered substrate metabolism precedes the onset of hyperglycemia and contractile dysfunction in two mouse models of insulin resistance and obesity. Endocrinology. 2005;146:5341–5349. doi: 10.1210/en.2005-0938. [DOI] [PubMed] [Google Scholar]
- 44.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, Clapham JC, Brand MD. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y-X, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:E294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hondares E, Mora O, Yubero P, de la Concepcion MR, Iglesias R, Giralt M, Villarroya F. Thiazolidinediones and rexinoids induce PGC-1α gene transcription: an auto-regulatory loop controls PGC-1α expression in adipocytes via PPARγ co-activation. Endocrinology. 2006;147:2829–2838. doi: 10.1210/en.2006-0070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.