Abstract
Nuclear receptors (NRs) are widely targeted to treat a range of human diseases. Feed-forward loops are an ancient mechanism through which single cell organisms organize transcriptional programming and modulate gene expression dynamics, but they have not been systematically studied as a regulatory paradigm for NR-mediated transcriptional responses. Here, we provide an overview of the basic properties of feed-forward loops as predicted by mathematical models and validated experimentally in single cell organisms. We review existing evidence implicating feed-forward loops as important in controlling clinically relevant transcriptional responses to estrogens, progestins, and glucocorticoids, among other NR ligands. We propose that feed-forward transcriptional circuits are a major mechanism through which NRs integrate signals, exert temporal control over gene regulation, and compartmentalize client transcriptomes into discrete subunits. Implications for the design and function of novel selective NR ligands are discussed.
Keywords: Nuclear receptor, Feed forward, Selective ligand, Glucocorticoid, Estrogen
1. Introduction
The nuclear receptor (NR) superfamily forms one of the most abundant classes of metazoan transcription factors (Mangelsdorf et al., 1995). Many NRs function as ligand-dependent, DNA-binding transcription factors that transduce specific molecular signals into adaptive responses at the level of gene expression. Their intrinsic abilities to bind and respond to small drug-like molecules, coupled with the regulatory control they exert over myriad physiologic and pathologic processes, have led to effective pharmacological targeting of a number of NRs (Huang, Chandra, & Rastinejad, 2010). However, the impact of therapeutic exploitation of NRs is substantially reduced by treatment resistance that variably occurs across a spectrum of diseases treated with NR modulators, and also by frequently severe side effects that are associated with systemic use of various NR ligands (Kremoser et al., 2007; McMaster & Ray, 2007; Gross et al., 2009; Higgins & Depaoli, 2010; Ryan & Tindall, 2011; Green et al., 2012; Rosenson et al., 2012; Ahmadian et al., 2013). Efforts to design new NR-based therapies that improve on these drawbacks have been complicated by several underlying features of NR biology (Aranda & Pascual, 2001; Deblois & Giguere, 2008; Bagamasbad & Denver, 2011; Jagannathan & Robinson-Rechavi, 2011), including: 1) a single NR can impact the expression of thousands of genes, including those encoding other NRs and DNA-binding transcription factors, 2) many downstream genes are regulatory targets of multiple NRs and other NR-regulated transcription factors, and 3) transcription of NR-regulated genes is a highly complex and tightly regulated event, involving other transcription factors and co-factors, the basal transcription machinery, and covalent modifications of both regulatory factors and chromatin. Thus, as highly intricate networks of interactions between target genes, proteins, and regulatory pathways influence NR signaling, developing compounds that precisely control a limited subset of downstream responses subject to regulation by a given NR remains a significant challenge.
Complex transcription regulatory networks have been intensely studied in lower model organisms, and these detailed theoretical, mathematical, and experimental analyses have clearly revealed that their structural organization is not random (see (Alon, 2007) for review). Rather, these networks were found to contain a common set of simple connectivity patterns called network motifs, each of which is predicted to carry out specific information processing functions (Milo et al., 2002). One of the most commonly recurring network motifs identified initially within the transcription networks of the bacterium Escherichia coli (Shen-Orr et al., 2002) and the yeast Saccharomyces cerevisiae (Lee et al., 2002) is the feed-forward loop (FFL). The FFL appears in hundreds of gene networks in bacteria and yeast, and has since been recognized to be prevalent in regulatory hierarchies of plant (Saddic et al., 2006), animal (Duggan et al., 1998; Davidson et al., 2002; Iranfar et al., 2006), and even human (Moroni et al., 2001; Mullen et al., 2002; Boyer et al., 2005; Swiers et al., 2006; Krejci et al., 2009) cells, suggesting an important role for this highly conserved motif in controlling metazoan gene expression. Here, we will briefly review the architecture of the FFL, as well as its predicted functional properties based on the different structural configurations it can assume. We will present evidence suggesting that NRs participate in canonical FFLs to regulate subsets of downstream target genes, and then examine how this organization may confer specific timing and signal integration properties to client gene expression in mammalian cells. We will lastly consider how feed-forward logic could potentially explain some of the pharmacologic outcomes of NR targeting that remain poorly understood.
2. Overview of feed-forward loop (FFL) structure and function
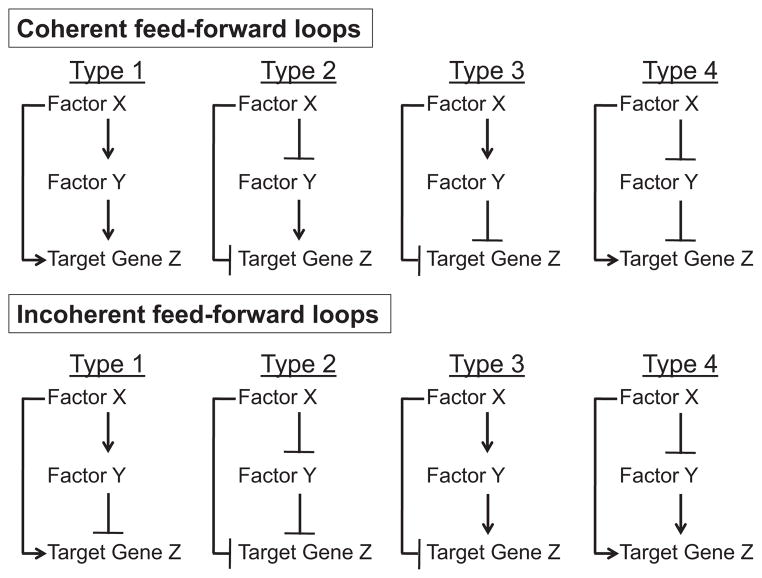
In contrast to a basic positive feedback (autoregulatory) transcriptional loop, which consists of a single transcription factor X that directly or indirectly enhances its own rate of production (Alon, 2007), the feed-forward loop is represented by a three-node directional structure (Mangan & Alon, 2003) that is driven by a primary, inducible transcription factor X. In the FFL, the regulatory effect of factor X on target gene Z is the combinatorial result of 1) a direct path from factor X to target gene Z, where X binds to and directly regulates Z expression, and 2) an indirect path from factor X to gene Z via a secondary, inducible transcription factor Y, in which X binds to and directly regulates Y expression, and then Y binds to and directly regulates Z expression. Thus, there are three separate, obligate regulatory events within an FFL (X to Z, X to Y, and Y to Z), and each can result in either positive (induction) or negative (repression) effects on transcription, providing 8 possible structural configurations of the circuit (Fig. 1). If the regulatory effect of the direct regulation path (X to Z) is the same as the overall effect of the indirect regulation path (X through Y to Z), the FFL has a coherent configuration (Fig. 1, top); if these effects are opposing, the FFL has an incoherent configuration (Fig. 1, bottom). The combinatorial effect of X and Y on Z expression will further depend on the type of logic that integrates the two inputs (X and Y), for example, AND-like logic is often utilized in biological systems, in which both X and Y are required to regulate Z. As will be explored in greater detail below, these distinct connectivity patterns can be modeled mathematically and validated experimentally to bestow unique response characteristics to feed-forward target genes. This suggests the intriguing possibility that a given target gene’s response to a specific signal or set of signals could be predicted if the structural configuration of its regulatory FFL were known.
Fig. 1.
Structural configurations of the transcriptional feed-forward loop (FFL). The FFL consists of a primary transcription factor X, which regulates a secondary transcription factor Y, such that X and Y both individually and combinatorially regulate expression of target gene Z. Each of the three regulatory events can be positive or negative, providing for the 8 possible structural configurations of the FFL presented here. In this schematic, lines heading in arrowheads represent induction, and lines ending with a perpendicular dash indicate repression. The two most common types of transcriptional feed-forward networks of E. coli and yeast are the coherent-type 1 and incoherent-type 1 FFLs, depicted in the top left and bottom left panels, respectively. Each distinct connectivity pattern has been found/is predicted to confer unique response profiles to feed-forward target genes.
In E. coli and S. cerevisiae, two of the possible eight structural configurations of transcriptional FFLs have been found to occur significantly more frequently than any other (Mangan & Alon, 2003; Alon, 2007). The most abundant FFL configuration in both species was the coherent-type 1, in which both X and Y are activators of target gene Z (Fig. 1, top left). In the coherent-type 1 FFL with AND-like logic (both X and Y are required), theoretical and synthetic circuit analyses in isolated systems predict a temporal delay in the response of gene Z to activation of primary factor X, as Z production would not begin until secondary factor Y had accumulated to a sufficient amount to cross the activation threshold for Z (Shen-Orr et al., 2002; Mangan & Alon, 2003). Such a response delay could provide a useful mechanism by which only persistent signals would be sufficient to induce Z, as any background noise generating brief pulses of signal would be filtered out. This anticipated function was experimentally validated in the context of the living cell using the well-characterized L-arabinose utilization system of E. coli (Mangan et al., 2003). In this FFL with demonstrated coherent-type 1 connectivity and AND-like logic, addition of primary input signal (cAMP, a cellular indicator of glucose deprivation) was followed by a nearly 20 minute delay before significant changes in target gene expression were detectable, indicating that a temporal delay function can be fulfilled by coherent-type 1 FFL architecture in vivo. The second most frequently recurring circuit structure in E. coli and yeast transcriptional FFLs was the incoherent-type 1, in which X is an activator of Y and Z, but Y represses Z (Fig. 1, bottom left). Although only one of the three regulatory events is different in this configuration (Y repressing Z) as compared to the coherent-type 1 FFL, the predicted effects on the response of target gene Z are fundamentally different. For example, modeling analyses in isolated incoherent-type 1 FFL systems predict an accelerated response time (to reach steady-state) of target gene Z following activation of primary factor X, as production of Z (driven by a strong promoter) would achieve rapid initial induction/overshoot followed by a delayed reduction to desired steady-state levels as the concentration of repressor Y accumulates to threshold levels (Mangan & Alon, 2003). This behavior was observed in studies of the galactose utilization system of living E. coli (Mangan et al., 2006), an FFL exhibiting incoherent-type 1 connectivity that showed a nearly threefold faster response time (to reach steady-state) of its target gene (galETK) compared to a reference gene under simple (not feed-forward) regulatory control.
Experimental validation of the predicted dynamic properties of the eight possible FFL configurations has been, to our knowledge, limited to type 1 coherent and incoherent circuitries, likely reflecting their vast overrepresentation among FFLs identified in lower model organisms. However, functions are still predictable for the other less common configurations. According to Alon and colleagues (Mangan & Alon, 2003), all four coherent FFL types could provide a delay function, and all four incoherent FFLs could facilitate response acceleration, but the specific properties of these responses would be dictated by the interactions among the major regulatory players. For example, a type 3-coherent FFL (see Fig. 1) is predicted to confer a delayed return to baseline expression of the downstream target Z if accumulated repressor Y maintains active repression of Z even after the activity of factor X has terminated. FFL regulatory systems thus appear to confer distinct timing properties to target gene expression responses that depend on the underlying circuit architecture.
In addition to providing a mechanism to control expression dynamics, some feed-forward configurations enable the effects of two signals to be integrated at the transcriptional level (Mangan & Alon, 2003). For example, in a coherent-type 1 or incoherent-type 1 FFL (Fig. 1), a signal that influences the expression of factor Y would be expected to alter the timing and magnitude of gene Z expression in response to a primary signal that induces factor X activity. Moreover, if the level of factor Y that is required for regulation of Z cannot be achieved simply through factor X-mediated induction of factor Y, a second signal that induces factor Y could serve as a gating event required for downstream regulation of gene Z in response to factor X activation. Under this regime, clients of factor X:Y FFLs would only be induced in the presence of signals activating both factors X and Y. Thus, pharmacologic manipulation of factor X would be predicted to variably alter gene Z expression depending on the presence or absence of signals regulating factor Y activity.
3. Evidence for NR-controlled FFLs
NRs are subject to a wide range of regulatory mechanisms including cell-type specific combinations of co-regulator expression, tissue-restricted chromatin organization, and post-translational modifications (George et al., 2011; Yokoyama et al., 2011; King et al., 2012; Magnani et al., 2012; Knutson & Lange, 2013; Oakley & Cidlowski, 2013; Sever & Glass, 2013; Abdel-Hafiz & Horwitz, 2014). In addition to these, a growing number of reports directly or indirectly establish that feed-forward circuitry contributes to the hierarchical organization and control of NR-regulated gene expression networks (Zhang et al., 2002, 2003; Laganiere et al., 2005; Carroll et al., 2006; Villanueva et al., 2011; Grunewald et al., 2012; Ross-Innes et al., 2012; Tan et al., 2012; Sasse et al., 2013). Our working definition of an NR-controlled FFL is congruent with that of Mangan and Alon (2003), in that 1) it would be driven by the NR as the primary transcription factor (factor X) that would directly regulate the expression of a secondary transcription factor (factor Y), and 2) the NR and factor Y would both bind the regulatory region of target gene Z and jointly modulate its transcription. While few studies have systematically demonstrated fulfillment of all of the above defining criteria for a single putative NR-driven FFL (e.g. Villanueva et al., 2011, Sasse et al., 2013), we identified examples within existing publications focusing on progesterone receptor (PR; Zhang et al., 2002, 2003; Velarde et al., 2006; Pabona et al., 2012; Rubel et al., 2012), estrogen receptor-α (ERα; Robyr et al., 2000; Laganiere et al., 2005; Carroll et al., 2006; Fullwood et al., 2009; Al Saleh et al., 2011; Ross-Innes et al., 2012; Mohammed et al., 2013), androgen receptor (AR; Thomas et al., 2010; Tan et al., 2012), peroxisome proliferator-activated receptor gamma (PPARγ; Villanueva et al., 2011), and glucocorticoid receptor (GR; Sasse et al., 2013) signaling that are consistent with these NRs forming multiple feed-forward circuits that regulate target gene expression. Examples will be briefly reviewed below and are summarized in Table 1.
Table 1.
Selected nuclear receptors, their feed-forward regulatory partners, and examples of (putative) feed-forward gene targets highlighted in this review.
| Primary transcription factor (nuclear receptor)
|
Secondary transcription factor/alternative name
|
Feed-forward target gene(s)
|
|---|---|---|
| [Factor X] | [Factor Y] | [Gene Z] |
| PR | KLF9/BTEB1 | SLP1, DKK1, PRC1, UHRF1, IGFBP1, FAM167A |
| PR | SOX17/VUR3 | Ihh, Areg, SCGB1A1 |
| ERα | FOXA1/HNF3A | TFF1 |
| ERα | NRIP1/RIP140 | BCAS4, IRX4, GUSB, MUC1 |
| ERα | GREB1 | CXCL12, TRIM5 |
| AR | NKX3-1/NKX3A | RAB3B, RAB36, RAB20 |
| PPARγ | TLE3/GRG3 | Ap2, Cd36, Aqp7, Agpat2, Lpl |
| GR | KLF15/KKLF | AASS, PRODH, MT2A |
3.1. PR forms FFLs with KLF9 and SOX17
Induction of PR signaling is a cornerstone in the treatment of endometriosis, a painful disorder defined by ectopic growth of endometrial tissue that afflicts roughly 5% of Caucasian females between the ages of 18 and 49 (Johnson & Hummelshoj, 2013; Rogers et al., 2013). Thus, mechanisms of PR action have important implications for therapeutics. With that backdrop, it was intriguing to find a series of studies suggesting that PR and the transcription factor Kruppel-like factor 9 (KLF9) meet many of the basic requirements for factors X and Y of a feed-forward circuit. For example, progesterone treatment induced KLF9 expression in endometrial epithelial cells that was abrogated by PR knockdown (Velarde et al., 2006), and several PR-occupied genomic sites were discovered in close proximity to the KLF9 locus (Rubel et al., 2012), indicating that KLF9 is a direct transcriptional target of PR. Velarde et al. (2006) subsequently identified secretory leukocyte peptidase inhibitor (SLPI) as a PR target gene whose induction by progesterone was reduced following KLF9 knockdown. Further, SLPI expression was dose-dependently increased by KLF9 overexpression, and the SLPI promoter was co-occupied by PR and KLF9, suggestive of coherent-type 1 feed-forward regulation of SLPI by PR and KLF9 (see Fig. 1). PR and KLF9 were also shown to physically interact in endometrial epithelial cells (Zhang et al., 2002), where they co-regulated the expression of several progesterone-responsive genes, including dickkopf WNT signaling pathway inhibitor 1 (DKK1), whose promoter was bound by both PR and KLF9 (Pabona et al., 2012).
Like many transcription factors (Latchman, 2001), KLF9 can act as both transcriptional activator and repressor, depending on cell and promoter context (Zhang et al., 2003). This implies that the same two factors (PR and KLF9 in this case) are capable of regulating downstream target genes through either type I coherent or incoherent feed-forward logic. Accordingly, KLF9 knockdown in endometrial stromal cells was shown to decrease the expression of some direct PR targets (e.g. protein regulator of cytokinesis 1 (PRC1), ubiquitin-like with PHD and ring finger domains 1 (UHRF1)) while simultaneously increasing the expression of others (e.g. insulin-like growth factor binding protein 1 (IGFBP1), family with sequence similarity 167, member A (FAM167A)), consistent with both coherent-type 1 (in the former case) and incoherent-type 1 (in the latter case) feed-forward regulation by PR:KLF9 cross-talk (Pabona et al., 2012). Thus, while additional work is required to establish direct binding and regulation of these putative target genes by PR and KLF9, these findings strongly suggest that subsets of the transcriptional response to progesterone in the endometrium are differentially regulated by distinct configurations of PR:KLF9 FFLs. Intriguingly, varied chemistry of PR ligands altered the transcriptional effects of PR:KLF9 cross-talk (Zhang et al., 2003), indicating that ligand chemistry may exert selective effects on NR-mediated gene regulation in part through perturbing FFL function, a concept we will explore in greater detail later in the review.
Beyond providing supporting evidence of direct regulation of KLF9 by PR, data from a genome-wide study of PR occupancy identified another transcription factor termed SRY (sex determining region Y)-box 17 (Sox17) as a direct target of PR signaling in the mouse uterus (Rubel et al., 2012). SOX17 binding site motifs were significantly enriched near PR-binding sites of known PR target genes, and further analysis revealed PR:SOX17 co-occupancy at a subset of these genes (Rubel et al., 2012). Another study in rabbit endometrium (Garcia et al., 2007) characterized the secretoglobin, family 1A, member 1 (uteroglobin; SCGB1A1) gene as a direct PR target with a co-localized SOX17 binding site in its promoter region. Although additional experiments would be required to confirm direct regulatory effects of PR and SOX17 on these putative target genes, the results support the notion that PR:SOX17 feed-forward architecture regulates a sector of endometrial transcriptional responses to progesterone.
3.2. ERα forms FFLs with FOXA1 and other targets
ERα signaling is a major driver of breast cancer proliferation, and pharmacologic inhibition of ERα activity is employed widely in the clinic to reduce the risk of breast cancer recurrence. Research on ERα signaling led to the discovery by several groups that the transcription factor named forkhead box A1 (FOXA1) is a key co-regulator of ERα activity (Robyr et al., 2000; Carroll et al., 2005; Laganiere et al., 2005). A number of studies provide evidence that FOXA1 expression is directly induced by ERα at the transcriptional level, including data showing that ERα knockdown reduces FOXA1 expression (Al Saleh et al., 2011), and that ERα occupies chromatin within/near the FOXA1 coding region (Laganiere et al., 2005; Fullwood et al., 2009) in MCF-7 breast cancer cells. Moreover, substantial genome-wide overlap between ERα and FOXA1 occupancy has been observed in clinical samples of breast cancer tissue and in breast cancer cell lines (Ross-Innes et al., 2012), where FOXA1 regulates a subset of ERα-dependent gene expression responses, including genes implicated in estrogen-mediated proliferation. For example, trefoil factor 1 (TFF1) was identified as a direct ERα and FOXA1 gene target in MCF-7 cells whose induction by estrogen was markedly reduced with FOXA1 knockdown (Carroll et al., 2005; Laganiere et al., 2005), consistent with coherent-type 1 feed-forward regulation of TFF1 by ERα and FOXA1 (see Fig. 1). ERα and FOXA1 may thus be poised to utilize feed-forward signaling to regulate the expression of multiple therapeutically relevant effectors of estrogen signaling.
While most work investigating ER-regulated transcription has focused on estrogen-induced genes, repression of gene expression constitutes a significant portion of all ER-dependent transcriptional changes in a cell (Carroll et al., 2006), although the underlying mechanisms are less well understood. In this regard, it has been found that ERα uses the corepressor termed nuclear receptor interacting protein 1 (NRIP1) to influence estrogen-mediated gene repression through a mechanism corresponding to coherent-type 3 feed-forward circuitry (see Fig. 1). Specifically, NRIP1 was directly bound and induced by ERα in MCF-7 cells (Carroll et al., 2005), where it was required for subsequent regulation of several late estrogen-repressed target genes. This subset included breast carcinoma amplified sequence 4 (BCAS4), iroquois homeobox 4 (IRX4), glucuronidase, beta (GUSB) and mucin 1, cell surface associated (MUC1), the regulatory regions of which were co-occupied by ERα and NRIP1 (Carroll et al., 2006). More recently, ERα-mediated induction of growth regulation by estrogen in breast cancer 1 (GREB1) was shown to result in ERα:GREB1 regulatory complexes that occupied genomic loci corresponding to subsets of both ER-induced and -repressed genes, such as chemokine (C-X-C motif) ligand 12 (CXCL12) and tripartite motif containing 5 (TRIM5), respectively (Mohammed et al., 2013). Coherent-type 3 FFLs may thus represent a novel mechanism through which ER and likely other nuclear receptors mediate target gene repression. However, verifying that a putative NR-FFL meets the criteria for a coherent-type 3 circuit may be complicated by accumulating genome-wide evidence that relatively few NR-repressed genes are located within realistic proximity to genomic NR binding sites to be direct targets (e.g. Santos et al., 2010). Nevertheless, these studies of ERα cross-talk with FOXA1, NRIP1, and GREB1 collectively suggest that FFLs feature prominently in the molecular control of ERα-mediated transcriptional regulation.
3.3. Induction of NKX3-1 by AR drives feed-forward gene regulation in prostate cancer cells
Similar to the important role of ERα in breast cancer, AR signaling drives prostate cancer cell proliferation. Two recent studies indicate that the transcription factor NK3 homeobox 1 (NKX3-1) is an important co-regulator of AR-mediated transcriptional responses in cell line models of prostate cancer. For example, Thomas et al. (2010) showed that AR directly binds to and induces expression of NKX3-1 in LnCAP prostate cancer cells. This was followed by a genome-wide characterization of AR:NKX3-1 cross-talk in LnCAP cells (Tan et al., 2012) that confirmed direct regulation of NKX3-1 by AR, which subsequently co-regulates the expression of a subset of AR target genes. In fact, overlap between AR and NKX3-1 genomic occupancy occurred at ~10% of all AR binding regions in this study. Notably, this subset of AR:NKX3-1 co-occupied genes contained many targets found in advanced prostate cancer, and was particularly enriched for genes involved in protein trafficking, including several members of the RAS oncogene family (RAB3B, RAB36, RAB20). Tan et al. (2012) reported reduced androgenic induction of RAB3B with NKX3-1 knockdown, suggestive of coherent-type 1 regulation of RAB3B by AR:NKX3-1, and further showed that RAB3B knockdown resulted in significant LnCAP cell death, indicating a critical role for RAB3B in prostate cancer cell survival. These findings suggest that AR forms feed-forward loops with NKX3-1 that modulate the transcriptional regulation of AR targets with important roles in prostate cancer survival and treatment responses.
3.4. PPARγ:TLE3 feed-forward regulation of adipogenesis
PPARγ is a central regulator of adipogenesis and also is targeted as an insulin sensitizer to treat type II diabetes. Villanueva and colleagues found that mRNA and protein expression of the transcription factor transducin-like enhancer of split 3 (TLE3) was dynamically increased during PPARγ-dependent adipogenesis of cultured murine preadipocytes (10T1/2 and 3T3-L1) and was further enhanced by treatment with a PPARγ (Villanueva et al., 2013). They mined existing genome-wide PPARγ binding data (Nielsen et al., 2008) to identify putative PPARγ binding sites in the mouse Tle3 locus, and then demonstrated through chromatin immunoprecipitation that Tle3 is a direct transcriptional target of PPARγ. They used cultured preadipocytes to show that Tle3 knockdown diminished adipogenic gene programming whereas stable overexpression of Tle3 promoted adipogenesis in a PPARγ-dependent process, and an in vivo model to illustrate that forced expression of TLE3 in murine adipose tissue altered fatty acid oxidation, improved systemic glucose homeostasis, and led to enhanced insulin sensitivity (Villanueva et al., 2013). Moreover, microarray analysis revealed that TLE3 regulated the expression of ~25% of PPARγ-regulated genes in 10T1/2 cells, including many established direct targets of PPARγ, such as fatty acid binding protein 4, adipocyte (Fabp4) and CD36 molecule (thrombospondin receptor; Cd36). These, and several other up-regulated adipocyte-selective genes (e.g. aquaporin 7 (Aqp7), 1-acylglycerol-3-phosphate O-acyltransferase 2 (Agpat2), lipoprotein lipase (Lpl)) were further shown to be co-occupied by PPARγ and TLE3 at genomic PPARγ response elements, indicating that multiple adipogenic genes are subject to PPARγ:TLE3 coherent-type 1 feed-forward regulation. Together, these findings by Villanueva et al. (2011), many of which were validated in a subsequent study (Villanueva et al., 2013), establish that PPARγ and TLE3 form FFLs that regulate adipogenic differentiation and may determine both therapeutic and pathologic consequences of PPARγ signaling.
3.5. Glucocorticoid receptor (GR):KLF15 FFLs influence transcriptional dynamics
Despite the growing number of reports indicating that NRs interact with secondary transcription factors in a manner consistent with FFL architecture, few studies have determined whether FFLs endow properties to NR-driven gene regulation that mirror the functions of these circuits in lower organisms. Our work (Sasse et al., 2013), and also work from Chen and colleagues (Chen et al., 2013), has begun to address this largely unexplored area. We showed that induction of the transcription factor Kruppel-like factor 15 (KLF15) by GR leads to both coherent (e.g. aminoadipate-semialdehyde synthase (AASS), glycine N-methyltransferase (GNMT), proline dehydrogenase (oxidase) 1 (PRODH)) and incoherent (e.g. metallothionein 2A (MT2A), TCDD-inducible poly(ADP-ribose) polymerase (TIPARP)) feed-forward gene regulation, and that ~7% of the GR-regulated transcriptome in murine lungs depended on Klf15 for correct expression responses to systemic injection of dexamethasone (dex), a synthetic GR agonist widely used in the clinic. The expression dynamics (i.e. expression changes as a function of time) of genes with reduced expression in Klf15−/− mice following dex treatment, a pattern consistent with GR:KLF15 coherent feed-forward circuitry, were distinct from those of GR:KLF15 target genes that followed incoherent logic (those with enhanced expression in Klf15−/− mice after dex treatment). Interestingly, the sets of genes under coherent versus incoherent feed-forward regulatory control had distinct gene ontologies, with coherent targets of the GR:KLF15 axis remarkably enriched for genes implicated in amino acid catabolism, and incoherent targets strongly enriched for genes implicated in metal binding and N-glycosylation, indicating that functional significance is associated with specific patterns of GR:KLF15 dependency. Moreover, we found that KLF15 expression was induced by acute serum deprivation in cultured human airway epithelial cells, corresponding with enhanced responses to GR induction for a subset of coherent GR:KLF15 targets. This suggests that regulation of KLF15 through GR-independent mechanisms may be integrated to modulate catabolic gene programming responses to GR activation. Taken together, these data indicate that many of the regulatory properties associated with FFLs in bacteria and yeast appear to be similarly enabled in mammalian cells by GR:KLF15 FFLs.
4. Feed-forward circuitry and NRs: implications for therapeutics
As we have reviewed above, a growing number of feed-forward circuits have been identified as contributing to NR-mediated gene regulation. Moreover, the prevalence of transcription factors among direct and/or strongly induced targets of various NRs, as revealed by genome-wide expression profiling and occupancy analyses (Carroll et al., 2006; Reddy et al., 2009; Masuno et al., 2011; Rubel et al., 2012), suggests a much broader potential footprint for this ancient regulatory motif within the architecture of NR signaling networks. That (secondary) transcription factors can be regulatory targets of multiple nuclear receptors adds an additional layer of complexity when viewing these circuits and their potential for cross-talk in the context of a global transcriptional regulatory network. For the GR:KLF15 network, FFL motifs appear to confer many of the specific timing properties to client gene regulation that are predicted by elegant work in lower organisms. Similarly, genes under control of ERα:NRIP1 feed-forward circuitry exhibited late repression, a predicted outcome of coherent-type 3 FFL gene regulation (Mangan & Alon, 2003). We therefore propose that feed-forward regulation is a major mechanism through which the temporal properties of NR-regulated gene programming are controlled. Further, FFLs provide an important system through which non-ligand signals can be integrated into NR-regulated gene expression responses. Specifically, signals that control the “factor Y” in any NR-driven FFL would be predicted to exert control over the subset of genes subordinate to regulation through FFLs comprised of the NR and factor Y.
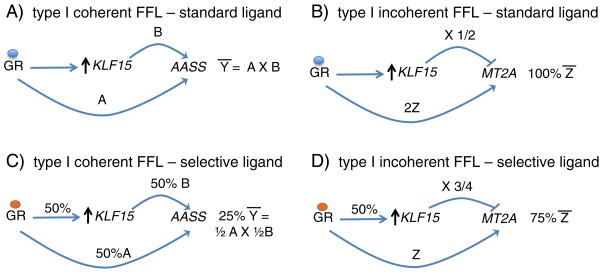
The potentially broad role of FFLs within NR networks has important implications for pharmacologic manipulation of individual NRs. For example, a range of so-called selective GR ligands that enable only a subset of the transcriptional effects of GR activation by standard ligands have been developed, with a goal of minimizing GR-associated side effects associated with the induction of specific GR targets (De Bosscher, Haegeman, and Elewaut, 2010). Many of these ligands act as partial agonists with respect to gene induction by GR (for example, see Bungard et al., 2011). In Fig. 2, in a highly simplified model, we consider the theoretical transcriptional responses to a hypothetical selective GR ligand in the context of GR:KLF15 FFLs. Although the structures of the FFLs we depict have been established experimentally, the combinatorial effects of GR and KLF15 on target gene transcription represented in this model should be construed as a heuristic tool rather than a direct reflection of GR:KLF15 composite interactions on transcriptional rates. As seen in Fig. 2A, in this model, a standard GR ligand enables the GR: KLF15 coherent-type 1 feed-forward target, AASS, to achieve a specific expression level, Ȳ, secondary to combined inductive activity of GR and KLF15 on AASS expression. In Fig. 2B, the incoherent-type 1 feed-forward target, MT2A, is expressed at level Z̄, which is a result of the inductive effects of GR combined with repressive effects of KLF15. In Fig. 2C–D, the hypothetical effects of a selective ligand, which enables only 50% of GR-mediated gene induction, are illustrated. In this scenario, regulation of AASS would be reduced to 25% of Y (Fig. 2C), secondary to combinatorial effects of reduced direct induction of AASS by GR, and reduced GR-mediated induction of KLF15. In contrast, MT2A expression would be at 75% of the level attained with a traditional ligand, as reduced induction of MT2A by GR would be offset by reduced KLF15-driven inhibition (Fig. 2D). Thus, a selective ligand that simply enables reduced GR inductive activity at primary targets could generate distinct transcriptional responses at GR targets regulated through coherent versus incoherent feed-forward logic.
Fig. 2.
A simplified, theoretical model comparing the effects of a hypothetical selective GR ligand on the expression of a GR:KLF15 coherent feed-forward target, AASS, and a GR:KLF15 incoherent feed-forward target, MT2A. A. With a standard GR ligand, shown as a blue circle, AASS expression is induced to level Ȳ, based on the combinatorial activity of both GR and KLF15. B. Similar to A, MT2A is induced to level Z̄, which represents combined inductive effects of GR and repressive effects of GR-induced KLF15. C. Here, a selective GR ligand, shown as an orange circle, causes GR to induce 50% of the expression level of KLF15 and AASS that was achieved with the standard ligand. However, the final level of AASS expression is 25% Ȳ, since lower GR-induced KLF15 levels lead to further decreases in AASS expression. D. Here the putative effects of the selective GR ligand are shown on the expression of the incoherent feed-forward target, MT2A. In this case, reduced GR-mediated induction of KLF15 in part balances the reduced inductive effect of GR on MT2A, leading to 75% expression of MT2A.
In addition to implications for selective ligands, feed-forward gene regulation could enable threshold responses to standard NR ligands that do not correlate with traditional dose response characteristics. Specifically, in line with the reasoning outlined above, dose response effects of traditional ligands on gene expression would be expected to vary between coherent and incoherent targets. It is also conceivable that clients of putative NR:factor Y coherent-type 1 FFLs may require prolonged exposure to an NR ligand, or a secondary signal that increases factor Y activity, for efficient induction to occur. An extension of this notion is that selected side effects of NRs that are regulated through the activity of FFLs may only occur if specific cross-regulatory pathways are also activated. For example, metabolic stress induces KLF15 in a GR-independent process (Gray et al., 2007), and KLF15 is implicated in mediating aspects of GR-induced muscle atrophy (Shimizu et al., 2011). Thus, patients with elevated KLF15 expression, perhaps secondary to critical illness-related alterations in metabolism, may be especially susceptible to GR-induced muscle atrophy. This, and many other potential implications of feed-forward circuitry in mediating side effects and modulating therapeutic effects of NR ligands, awaits further study.
In summary, FFLs represent an evolutionarily conserved system for controlling gene expression dynamics and facilitating the integration of multiple signals at the transcriptional level. Emerging evidence indicates that FFLs are prevalent in NR-mediated transcriptional regulation. A small number of studies have also linked NR-driven FFLs to established properties of FFLs in lower organisms. Future research is needed to fully establish the roles that FFLs play in regulating varied gene expression responses to selective NR ligands and in modulating treatment responses through non-ligand signal integration.
Acknowledgments
This work was supported in part by the National Institutes of Health R01HL109557 (A.N.G).
Abbreviations
- FFL
feed-forward loop
- NR
nuclear receptor
- AR
androgen receptor
- ER
estrogen receptor
- PR
progesterone receptor
- PPAR
peroxisome proliferator-activated receptor
- GR
glucocorticoid receptor
Footnotes
The authors have no conflicts of interest to disclose.
References
- Abdel-Hafiz HA, Horwitz KB. Post-translational modifications of the progesterone receptors. J Steroid Biochem Mol Biol. 2014;140:80–89. doi: 10.1016/j.jsbmb.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Saleh S, Al Mulla F, Luqmani YA. Estrogen receptor silencing induces epithelial to mesenchymal transition in human breast cancer cells. PLoS One. 2011;6:e20610. doi: 10.1371/journal.pone.0020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- Bagamasbad P, Denver RJ. Mechanisms and significance of nuclear receptor auto- and cross-regulation. Gen Comp Endocrinol. 2011;170:3–17. doi: 10.1016/j.ygcen.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chen SH, Masuno K, Cooper SB, Yamamoto KR. Incoherent feed-forward regulatory logic underpinning glucocorticoid receptor action. Proc Natl Acad Sci U S A. 2013;110:1964–1969. doi: 10.1073/pnas.1216108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- Deblois G, Giguere V. Nuclear receptor location analyses in mammalian genomes: from gene regulation to regulatory networks. Mol Endocrinol. 2008;22:1999–2011. doi: 10.1210/me.2007-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CL, Lightman SL, Biddie SC. Transcription factor interactions in genomic nuclear receptor function. Epigenomics. 2011;3:471–485. doi: 10.2217/epi.11.66. [DOI] [PubMed] [Google Scholar]
- Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 2007;5:305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SM, Mostaghel EA, Nelson PS. Androgen action and metabolism in prostate cancer. Mol Cell Endocrinol. 2012;360:3–13. doi: 10.1016/j.mce.2011.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross KL, Lu NZ, Cidlowski JA. Molecular mechanisms regulating glucocorticoid sensitivity and resistance. Mol Cell Endocrinol. 2009;300:7–16. doi: 10.1016/j.mce.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald M, Johnson S, Lu D, Wang Z, Lomberk G, Albert PR, et al. Mechanistic role for a novel glucocorticoid-KLF11 (TIEG2) protein pathway in stress-induced monoamine oxidase A expression. J Biol Chem. 2012;287:24195–24206. doi: 10.1074/jbc.M112.373936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins LS, Depaoli AM. Selective peroxisome proliferator-activated receptor gamma (PPARgamma) modulation as a strategy for safer therapeutic PPARgamma activation. Am J Clin Nutr. 2010;91:267S–272S. doi: 10.3945/ajcn.2009.28449E. [DOI] [PubMed] [Google Scholar]
- Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranfar N, Fuller D, Loomis WF. Transcriptional regulation of post-aggregation genes in Dictyostelium by a feed-forward loop involving GBF and LagC. Dev Biol. 2006;290:460–469. doi: 10.1016/j.ydbio.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Jagannathan V, Robinson-Rechavi M. The challenge of modeling nuclear receptor regulatory networks in mammalian cells. Mol Cell Endocrinol. 2011;334:91–97. doi: 10.1016/j.mce.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Johnson NP, Hummelshoj L. Consensus on current management of endometriosis. Hum Reprod. 2013;28:1552–1568. doi: 10.1093/humrep/det050. [DOI] [PubMed] [Google Scholar]
- King HA, Trotter KW, Archer TK. Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim Biophys Acta. 2012;1819:716–726. doi: 10.1016/j.bbagrm.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson TP, Lange CA. Dynamic regulation of steroid hormone receptor transcriptional activity by reversible SUMOylation. Vitam Horm. 2013;93:227–261. doi: 10.1016/B978-0-12-416673-8.00008-3. [DOI] [PubMed] [Google Scholar]
- Krejci A, Bernard F, Housden BE, Collins S, Bray SJ. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci Signal. 2009;2:1. doi: 10.1126/scisignal.2000140. [DOI] [PubMed] [Google Scholar]
- Kremoser C, Albers M, Burris TP, Deuschle U, Koegl M. Panning for SNuRMs: using cofactor profiling for the rational discovery of selective nuclear receptor modulators. Drug Discov Today. 2007;12:860–869. doi: 10.1016/j.drudis.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the cover: location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Magnani L, Brunelle M, Gevry N, Lupien M. Chromatin landscape and endocrine response in breast cancer. Epigenomics. 2012;4:675–683. doi: 10.2217/epi.12.64. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuno K, Haldar SM, Jeyaraj D, Mailloux CM, Huang X, Panettieri RA, Jr, et al. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2011;45:642–649. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster A, Ray DW. Modelling the glucocorticoid receptor and producing therapeutic agents with anti-inflammatory effects but reduced side-effects. Exp Physiol. 2007;92:299–309. doi: 10.1113/expphysiol.2006.036194. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Mohammed H, D’Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, et al. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat Immunol. 2002;3:652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- Oakley RH, Cidlowski JA. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol. 2013;132:1033–1044. doi: 10.1016/j.jaci.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabona JM, Simmen FA, Nikiforov MA, Zhuang D, Shankar K, Velarde MC, et al. Kruppel-like factor 9 and progesterone receptor coregulation of decidualizing endometrial stromal cells: implications for the pathogenesis of endometriosis. J Clin Endocrinol Metab. 2012;97:E376–E392. doi: 10.1210/jc.2011-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Gegonne A, Wolffe AP, Wahli W. Determinants of vitellogenin B1 promoter architecture. HNF3 and estrogen responsive transcription within chromatin. J Biol Chem. 2000;275:28291–28300. doi: 10.1074/jbc.M002726200. [DOI] [PubMed] [Google Scholar]
- Rogers PA, D’Hooghe TM, Fazleabas A, Giudice LC, Montgomery GW, Petraglia F, et al. Defining future directions for endometriosis research: workshop report from the 2011 World Congress of Endometriosis In Montpellier, France. Reprod Sci. 2013;20:483–499. doi: 10.1177/1933719113477495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenson RS, Wright RS, Farkouh M, Plutzky J. Modulating peroxisome proliferator-activated receptors for therapeutic benefit? Biology, clinical experience, and future prospects. Am Heart J. 2012;164:672–680. doi: 10.1016/j.ahj.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel CA, Lanz RB, Kommagani R, Franco HL, Lydon JP, DeMayo FJ. Research resource: genome-wide profiling of progesterone receptor binding in the mouse uterus. Mol Endocrinol. 2012;26:1428–1442. doi: 10.1210/me.2011-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Tindall DJ. Androgen receptor rediscovered: the new biology and targeting the androgen receptor therapeutically. J Clin Oncol. 2011;29:3651–3658. doi: 10.1200/JCO.2011.35.2005. [DOI] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, et al. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133:1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- Sasse SK, Mailloux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, et al. The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol. 2013;33:2104–2115. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever R, Glass CK. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol. 2013;5:a016709. doi: 10.1101/cshperspect.a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, Takeda S, et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011;13:170–182. doi: 10.1016/j.cmet.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Swiers G, Patient R, Loose M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol. 2006;294:525–540. doi: 10.1016/j.ydbio.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32:399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde MC, Iruthayanathan M, Eason RR, Zhang D, Simmen FA, Simmen RC. Progesterone receptor transactivation of the secretory leukocyte protease inhibitor gene in Ishikawa endometrial epithelial cells involves recruitment of Kruppel-like factor 9/basic transcription element binding protein-1. Endocrinology. 2006;147:1969–1978. doi: 10.1210/en.2005-1419. [DOI] [PubMed] [Google Scholar]
- Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011;13:413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Fujiki R, Ohtake F, Kato S. Regulated histone methyltransferase and demethylase complexes in the control of genes by nuclear receptors. Cold Spring Harb Symp Quant Biol. 2011;76:165–173. doi: 10.1101/sqb.2011.76.010736. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang XL, Michel FJ, Blum JL, Simmen FA, Simmen RC. Direct interaction of the Kruppel-like family (KLF) member, BTEB1, and PR mediates progesterone-responsive gene expression in endometrial epithelial cells. Endocrinology. 2002;143:62–73. doi: 10.1210/endo.143.1.8590. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Zhang D, Michel FJ, Blum JL, Simmen FA, Simmen RC. Selective interactions of Kruppel-like factor 9/basic transcription element-binding protein with progesterone receptor isoforms A and B determine transcriptional activity of progesterone-responsive genes in endometrial epithelial cells. J Biol Chem. 2003;278:21474–21482. doi: 10.1074/jbc.M212098200. [DOI] [PubMed] [Google Scholar]