Abstract
Polyelectrolyte complex micelles have great potential as gene delivery vehicles because of their ability to encapsulate charged nucleic acids forming a core by neutralizing their charge, while simultaneously protecting the nucleic acids from non-specific interactions and enzymatic degradation. Furthermore, to enhance specificity and transfection efficiency, polyelectrolyte complex micelles can be modified to include targeting capabilities. Here, we describe the design of targeted polyelectrolyte complex micelles containing inhibitors against dys-regulated microRNAs (miRNAs) that promote atherosclerosis, a leading cause of human mortality and morbidity. Inhibition of dys-regulated miRNAs in diseased cells associated with atherosclerosis has resulted in therapeutic efficacy in animal models and has been proposed to treat human diseases. However, the non-specific targeting of microRNA inhibitors via systemic delivery has remained an issue that may cause unwanted side effects. For this reason, we incorporated two different peptide sequences to our miRNA inhibitor containing polyelectrolyte complex micelles. One of the peptides (Arginine-Glutamic Acid-Lysine-Alanine or REKA) was used in another micellar system that demonstrated lesion-specific targeting in a mouse model of atherosclerosis. The other peptide (Valine-Histidine-Proline-Lysine-Glutamine-Histidine-Arginine or VHPKQHR) was identified via phage display and targets vascular endothelial cells through the vascular cell adhesion molecule-1 (VCAM-1). In this study we have tested the in vitro efficacy and efficiency of lesion- and cell-specific delivery of microRNA inhibitors to the cells associated with atherosclerotic lesions via peptide-targeted polyelectrolyte complex micelles. Our results show that REKA-containing micelles (fibrin-targeting) and VHPKQHR-containing micelles (VCAM-1 targeting) can be used to carry and deliver microRNA inhibitors into macrophages and human endothelial cells, respectively. Additionally, the functionality of miRNA inhibitors in cells was demonstrated by analyzing miRNA expression as well as the expression or the biological function of its downstream target protein. Our study provides the first demonstration of targeting dys-regulated miRNAs in atherosclerosis using targeted polyelectrolyte complex micelles and holds promising potential for translational applications.
Keywords: micelle, atherosclerosis, polyelectrolyte complex, self-assembly, microRNA, gene delivery, endothelium, macrophage, miR-33, miR-92a
Introduction
The in vivo delivery of therapeutic nucleotides, such as small interfering RNAs (siRNA) and inhibitors targeting microRNAs (miRNA), has often been problematic due to the small size, charge and instability of the molecules1. For this reason, polyelectrolyte complexes composed of nucleic acids and positively charged polymers have been explored as a possibility to neutralize the charge on the molecule and protect it from enzymatic degradation2. Furthermore, addition of a neutral hydrophilic polymer block to either the polyanion or polycation prevents macroscopic phase separation and allows for the stabilization of nanometer sized polyelectrolyte complex micelles, referred to as polyion complex micelles3,4, interpolyelectrolyte complex micelles5,6, or complex coacervate core micelles7,8. These micelles are ideal candidates for the delivery of nucleic acids because of their ability to encapsulate charged therapeutics in the core of the micelle, which is covered by a protective corona that allows for increased resistance against non-specific interactions with proteinacious components in serum9 and enzymatic degradation10.
Several studies have used polyelectrolyte complex micelles to encapsulate DNA, plasmid DNA9,11, antisense DNA12 and siRNA13,14. Some even incorporate cell penetrating peptides, fusogenic peptides, integrin binding domains and cleavable linkers to facilitate cellular uptake and endosomal escape15,16. However, few have focused on combining the advantages of a nanocarrier and targeting of specific cells. One example of a targeting and nucleic acid containing polyelectrolyte complex micelle involved a lactose group attached to the outside of the micelle corona14. The system was designed to target asialoglycoprotein receptors (which recognize terminal galactose moieties) located on the surface of human hepatoma cells for the treatment of liver cancer14. Oishi et al. demonstrated that through the delivery of siRNA, their targeted micelle exhibited enhanced gene silencing14 and further validated the benefits of this approach via the growth inhibition of a hepatic multicellular tumor spheroid13 . However, application of nano-carriers in treating cardiovascular diseases, particularly for delivering miRNA-modifying agents, has not been fully explored17.
Atherosclerosis, development of atheromatous plaques in the inner lining of the arteries, is the leading cause of human mortality and morbidity worldwide18. Current pharmacotherapy for atherosclerosis, such as statins, principally target systemic risk factors and remains suboptimal, leaving an unmet need for more effective therapies aimed at the pathophysiology of the vessel wall. Atherosclerotic plaques occur at specific arterial sites of curvature, branching and bifurcation where chronic inflammation leads to series of cellular events within the arterial wall18 . For instance, build-up of atherosclerotic lesions initiates at predictable arterial sites where endothelial cells are activated by local disturbed flow in combination with systemic risk factors (e.g., hypercholesterolemia, hypertension), leading to lipoprotein accumulation and monocyte recruitment to the arterial intima19. The recruited monocytes then differentiate into lesion macrophages which internalize native or modified lipoproteins. Lipid-laden macrophage foam cells form as a consequence of the disruption of lipid cholesterol homeostatic mechanisms that regulate the uptake, intracellular metabolism, and efflux of cholesterol20. Therefore, pharmacotherapy that suppresses endothelial activation and promotes macrophage cholesterol efflux is expected to reduce atherosclerotic burdens.
miRNA (miR), highly conserved noncoding small RNAs of 19 to 26 nucleotides that posttranscriptionally suppress their target genes, have recently emerged as important pathophysiological mediators of atherogenesis21. Studies by our group and others have demonstrated that miR-92a is a major contributor to the pro-inflammatory endothelial phenotype by inhibiting transcription factors Kruppel-like factor 2 (KLF2) and Kruppel-like factor 4 (KLF4) 22,23. Moreover, miR-33 has been implicated in promoting atherosclerosis by inhibiting ATP-binding cassette transporters, ABCA1 and ABCG1, that are key components of macrophage cholesterol efflux in the process of reverse cholesterol transport (the trafficking of cellular cholesterol from peripheral tissues back to the liver via the plasma) 24–26. Systemic inhibition of miR-92a and miR-33a was shown to alleviate atherosclerosis27,28; however, specific arterial wall targeting of macrophage miR-33 and endothelial miR-92 in atherosclerotic plaques that is predicted to improve the benefit to risk ratio compared to systemic therapy, has not been achieved.
Previously, delivery of a fluorophore and small molecule drug to plaque has been achieved using functionalized polymeric micelles conjugated with the fibrin-binding peptide CREKA (Cys-Arg-Glu-Lys-Ala)29,30 or its shorter form REKA (Arg-Glu-Lys-Ala)31. The aforementioned micelles are formed via the self-assembly of molecules containing a hydrophobic tail, a polyethylene glycol (PEG) linker, and the peptide binding sequence. However, two major challenges remain for using these micelles to accomplish intracellular delivery of miRNA-modifying agents. First, this micelle contains a hydrophobic core, which is inappropriate for ionic cargos such as therapeutic nucleotides. Second, uptake of such nanoparticles by vascular cells and cellular internalization of functional miRNA-modifying agents has not been demonstrated32,33.
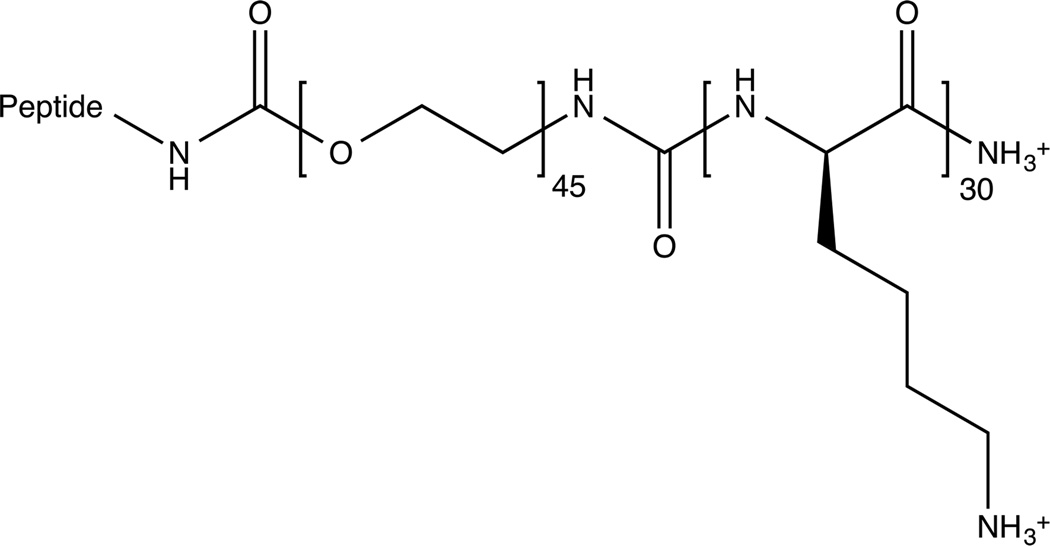
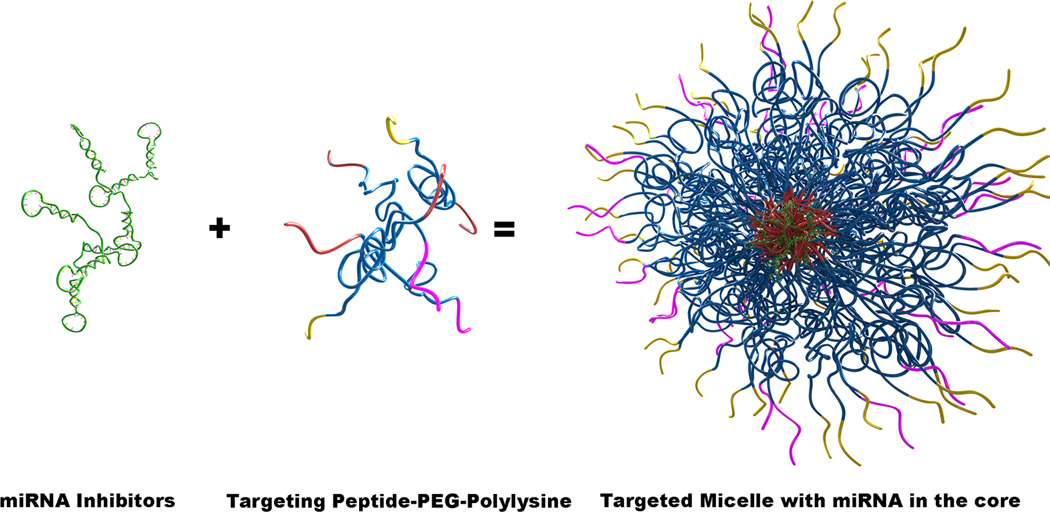
To circumvent these limitations, here we engineered polyelectrolyte complex micelles to inhibit dys-regulated vascular miRNAs that promote atherosclerosis. To the best of our knowledge, this has not been attempted before. Our approach is to encapsulate miRNA inhibitors in the core of the micelle and use two different peptides on the periphery of the nanoparticle corona to target markers known to be a part of atherosclerotic lesions. One piece of the strategy consists of the fibrin-binding peptide, arginine-glutamic acid-lysine-alanine (REKA)-conjugated polyelectrolyte micelles engineered to deliver miR-33a inhibitors to macrophages. The second strategy targets vascular cell adhesion molecule 1 (VCAM-1) on the surface of endothelial cells using micelles conjugated with VCAM1-binding peptides that have the sequence valine-histidine-proline-lysine-glycine-histidine-arginine, or VHPKQHR34. The formation of these miRNA inhibitor-containing, peptide-targeted micelles is achieved via the electrostatic complexation between the negatively charged miRNA inhibitors and polycations containing targeting peptides that we have designed and synthesized in our laboratory. Both polycation molecules (Figure 1A) contain three functional domains consisting of targeting peptides for cellular or plaque localization, a polyethylene glycol (PEG) domain to prevent macrophase separation, and a polylysine domain to complex with the negatively charged miRNA inhibitors. Combining the miRNA inhibitors with these targeting peptide-PEG2000-polylysine molecules should result in the formation of electrostatically driven, self-assembled micelles with a polylysine-miRNA inhibitor core, protected by a PEG corona that is decorated with the targeting peptide (Figure 1C). By taking advantage of the benefits of self-assembly, we can tailor the micelle corona to enable the targeting of diverse cell types and load the micelles with specific miRNA inhibitors, thereby providing the means to target various pathological mechanisms in a wide range of cells and contexts. Here we characterize these micelle constructs and show their ability to inhibit miRNA function in vitro using both macrophages and human aortic endothelial cells.
Figure 1. Construction of targeted polyelectrolyte complex micelles.
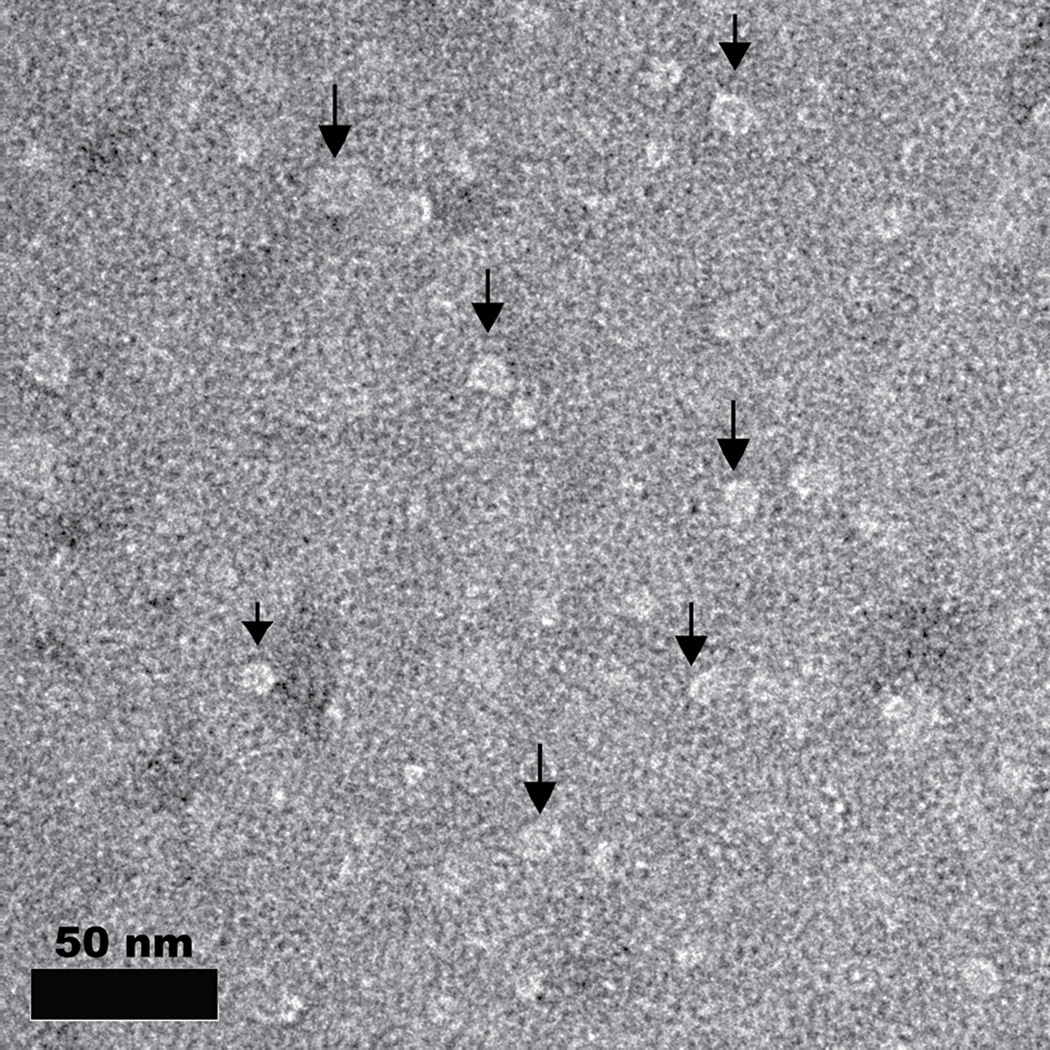
(A) Individual peptide-PEG(2000)-polylysine molecules are made up of a targeting peptide for cellular or plaque localization, a polyethylene glycol (PEG) domain to prevent macrophase separation, and a polylysine (K30) domain to complex with the negatively charged miRNA inhibitors. (B) Negatively stained TEM image of micelles formed via complexation of REKA-PEG-K30 and miR-92a inhibitor. (C) A schematic of the formation of targeted polyelectrolyte complexes with miRNA inhibitors in the core.
Materials and Methods
Material Synthesis and Purification
Targeting peptide-PEG(2000)-poly-L-lysine with a degree of polymerization of 30 (Peptide-PEG-K30) was synthesized using standard fluorenylmethyloxycarbonyl (FMOC) solid phase synthesis methods35 on an automated PS3 peptide synthesizer from Protein Technologies Inc (Tucson, AZ, USA). FMOC protected amino acids were also purchased from Protein Technologies Inc. The heterobifunctional molecule, FMOC-PEG2000-COOH was purchased from JenKem Technology USA (Allen, TX, USA). Two different targeting peptides were used in this study, REKA, which is named after the four amino acids in its sequence and a peptide targeting VCAM1 which has the sequence: VHPKQHR. The targeting peptide-PEG-K30 molecule was subsequently purified using reverse phase high performance liquid chromatography (HPLC, Shimadzu Corporation, Japan) and confirmed using a Bruker UltrafleXtreme (Fremont, CA, USA) matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF). The miRNA inhibitor molecules (miRIDIAN microRNA Hairpin Inhibitors, Dharmacon, USA) are single-stranded, chemically enhanced oligonucleotides diluted to a working stock solution of 100 µM.
Micelle Formation
Micelles of varying concentrations were formed by complexing the targeting-peptide-PEG-K30 and miRNA inhibitors at an equal charge molar ratio. First, the appropriate amount of deionized water was added, followed by the miRNA inhibitor, and finally the targeting-peptide-PEG-K30. The sample was vortexed after each polymer addition.
Dynamic Light Scattering
Micelle solutions containing 5 µM miRNA inhibitors were measured at 90° using a BI-200SM goniometer containing a red laser diode with a wavelength of 637nm and TurboCorr digital correlator, all from Brookhaven Instruments (Holtsville, NY, USA). Brookhaven Instruments Dynamic Light Scattering software was used to analyze the inverse Laplace transforms of the intensity autocorrelation functions using the non-negatively constrained least-squares (NNLS) algorithm to obtain multimodal size distribution data.
Transmission Electron Microscopy (TEM)
Negatively stained TEM samples were prepared by placing micelles on 400 mesh lacey carbon grids (Ted Pella, Redding, CA, USA) and then staining with a 1 wt% uranyl acetate solution. Images were obtained using a FEI Tecnai F30 electron microscope operated at 300 kV.
Zeta Potential
Zeta potential was determined by measuring the polyanions: miR92a inhibitor, miR33a inhibitor, and miR inhibitor control at a concentration 4 µM and the polycations: VHPKQHR-PEG-K30 and REKA-PEG-K30 at a concentration of 9 µM (equal in molar charge to the miRNA inhibitors). Micellar solutions of the combined polyanion and polycation were measured at the combined concentration of 13 µM (equal molar charge). All samples were dissolved in MQ water and measured at 25°C (Zetasizer Nano ZS, Malvern, Worcestershire, United Kingdom, N=3).
Cell culture
Mouse macrophages, J774 cells, were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS). Human aortic endothelial cells (HAECs) were purchased from Lonza (Allendale, NJ, USA) and cultured in EGM2 medium. Cells were cultured at 37°C in a humidified incubator under 5% CO2. Cells at passage X were used and media was changed every 2–3 days.
Micelle treatment
To separate unbound, free polymers from the micelle preparation, the micelle solution was separated into filtered and concentrated fractions by a 50-kDa cut-off membrane (Amicon® Ultra-0.5 centrifugal filter devices, Millipore). The concentrated fraction was further washed with nuclease-free water and the volume of concentrate was evaluated after centrifugation and concentration of concentrated fraction calculated. Assuming there is no free miR-33a inhibitor in the filtrate fraction, J774 cells (1E5 cells/well) were treated with same amount (200 nM) of miR-33a inhibitor or miR inhibitor control via REKA-conjugated micelles from 2 filtrate and micelle containing fractions. RNA was isolated 1 day after treatment and the expression of miR-33a was analyzed by real-time PCR. For analysis of the effect of miR-33a inhibition on ABC transporters expression, J774 cells were treated with 400 nM of miR-33a inhibitor or miR inhibitor control via REKA-conjugated micelle followed by 10 µM T0901314 (Cayman Chemical) treatment for another day. RNA was isolated after T0901317 treatment and protein samples were obtained 3 days after micelle treatment. For the treatment in HAECs (1E5 cells/well), 200 nM miR-92a inhibitor or miR inhibitor control carried by various micelles were applied to cells in the presence of 10 µM LPA (Santa Cruz).
Distribution of Dy547-labeled miR inhibitor control in cells
To monitor the uptake of micelles by cells in vitro, Dy547-labelled miR inhibitor control (Dharmacon) was encapsulated in REKA-, VHPKQHR-, or VHPKQHR/REKA-conjugated micelles. Cells were also transfected with Dy547-labelled miR inhibitors using Lipofectamine-RNAiMAX transfection reagent (Life Technologies). For fluorescence detection, cells grown on glass cover-slips were washed with DPBS and fixed with 4% paraformaldehyde 24 hr after micelle treatment. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). Images were randomly taken from the center (ten images) of each sample using a Nikon fluorescent microscope equipped with a CCD camera.
Quantitative real-time PCR
Total RNAs were isolated from cells utilizing the Direct-zol RNA isolation kit (Zymo Research). Expression of miR-92a was quantified by two-step quantitative real-time PCR using the TaqMan miRNA reverse transcription kit, TaqMan miRNA assay kits (Applied Biosystems), and LightCycler® 480 RNA Master Hydrolysis Probes (Roche). Expression of miR-33a was determined using miScript II RT kit and miScript SYBR Green PCR kit (Qiagen). miRNA expression was normalized in relation to expression of small nuclear U6 RNA or housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
For cDNA quantitative real-time PCR, total RNAs were reverse-transcribed using high capacity cDNA reverse transcription kit (Life Technologies) and amplified employing LightCycler® 480 SYBR Green I Master (Roche) the gene expression was normalized to the geometric mean of GAPDH, β-actin (ACTB), and large ribosomal protein P0 (RPLP0). PCR primers for genes of interest are listed in Table 1.
Table I.
Primer sequence for quantitative real-time PCR
| gene | Forward primer (5’ to 3’) | Reverse primer (5’ to 3’) |
|---|---|---|
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
| ACTB | TCCCTGGAGAAGAGCTACGA | AGGAAGGAAGGCTGGAAGAG |
| P0 | TCGACAATGGCAGCATCTAC | ATCCGTCTCCACAGACAAGG |
| KLF2 | GAACCCATCCTGCCGTCCTT | CACGCTGTTGAGGTCGTCG |
Western blot
Protein samples from cells were solubilized in sample buffer (NuPAGE® LDS Sample Buffer) and separated using Novex® NuPAGE® SDS-PAGE gel system followed by transferring to PVDF membrane using iBlot® Gel Transfer Device (Life Technologies). Antibodies against ABCA1 (NB400-105, Novus Biologics), ABCG1 (NB400-132, Novus Biologics), and β-actin (Abcam) were applied to PVDF membrane after being blocked with 5% non-fat milk. HRP-conjugated antibodies were applied and the chemiluminescent signal was detected using enhanced chemiluminescence substrate (Pierce).
Cholesterol measurement and efflux assay
For cholesterol mass measurements, total cholesterol was measured using 20 µL extracted sample and 20 µL cholesterol standard (0–1000 mg/mL) in 9:1 isopropanol:tergitol and 200 µL Chol reagent (Roche Diagnostics). Free cholesterol was measured using 200 µL Free Chol E reagent (Wako Diagnostics) in place of Chol reagent. Total protein was measured using the Pierce BCA protein assay (Pierce).
For cholesterol efflux measurement, J774 cells were induced with 10 µM T0901317 LXR agonist and at the same time labeled with 2 µCi/ml [3H]-Cholesterol (Perkin Elmer) for 16 hrs. Cells were washed 3 times with PBS and incubated for 24 hrs with 1 mL 2% mouse (FVB/N) plasma in DMEM +1% pen/strep antibiotic. Media was harvested from plates after 24 hrs and centrifuged for 5 min at 12,000 x g to remove any floating cells. 50% of the media volume was transferred to scintillation vial and counted in UltimaGold (Perkin Elmer) as described previously. Cells were extracted 2 times with 2 mL 3:2 hexane:isopropanol directly in the tissue culture dish with gentle swirling for 1 hr at room temperature. The solvent was transferred to a 2 mL borosilicate screw cap vial and dried under air, combining sequential extractions. Dried extracts were resuspended in 0.2 mL 9:1 isopropanol (molecular biology grade):tergitol and frozen for later analysis36. Extracted cell pellets were resuspended in 0.2 mL 2% SDS and incubated for 1 hr at 37°C to dissolve protein and frozen for later analysis. For cellular [3H]-cholesterol label measurements, 20 µL of the re-solubilized cellular extracts were counted in UltimaGold scintillation fluid (Perkin Elmer, Waltham, MA, USA). % efflux was calculated as 100% x media [3H]-cholesterol counts/total [3H]-cholesterol counts (media + cell).
Cell viability and proliferation
Cell viability was determined in J774 and HAECs treated with 200 µM polyelectrolyte complex micelles containing miR inhibitor control for 24 hrs employing a LIVE/DEAD cytotoxicity kit (Life Technologies). Briefly, confluent J774 and HAEC were cultured within 96-well plates and 24 hrs after the micelle treatment, dead cells were labeled with Ethidium homodimer-1 followed by fluorescence intensity measurements (excitation: 495 nm; emission: 520 nm). Cell proliferation was determined by a CyQANT Cell Proliferation Assay Kit (Life Technologies) according to the manufacturer’s protocol. Briefly, subconfluent J774 cells and HAECs were cultured within 96-well plates followed by the treatment of 200 µM polyelectrolyte complex micelles containing miR inhibitor control for 24–48 hrs. Cell density was measured 24 hrs or 48 hrs after the micelle treatment employing CyQUANT GR dye, which exhibits strong fluorescence (ex/em 480 nm/520 nm) when bound to cellular nucleic acids.
Statistical analysis
Data represent mean ± SEM. For the analysis of miR-33a and miR-92a expression, a paired-end, two-tailed student t test was applied. For the cholesterol analysis and western blot quantification, an unpaired, one-tailed student t tests was applied. For [3H]-cholesterol analysis, an unpaired, two-tailed student t test was applied. For the analysis of KLF2 expression, a paired-end, one-tailed student t test was applied. For the cell viability and proliferation, two-tailed student t test was applied. For analysis involving more than 2 groups, one-way ANOVA was applied. P value of less than 0.05 was considered statistical significant.
Results
Micelle characterization
The complexation of targeting-peptide-PEG-K30 with miR inhibitors was performed under charge neutral conditions in which the number of charged monomer units from the polylysine equaled the number of charged units in the miR inhibitors. Micelles were made by complexing REKA-PEG-K30 with miR-33a inhibitors, miR-92a inhibitors and miR inhibitor controls as well as by complexing VHPKQHR-PEG-K30 with miR-92a inhibitors and miR inhibitor controls. Additionally, mixed micelles were made containing equal parts VHPKQHR-PEG-K30 and REKA-PEG-K30 complexed with miR inhibitor 92a and miR inhibitor control. DLS was used to confirm the formation and measure the hydrodynamic diameter of all micelles (Table IIA). The size of polyelectrolyte complex micelles is determined by the length of the charged block covalently linked to the neutral non-charged polymer, as well as the ratio between them37. Therefore we expect only slight variations in the size of all the micelle constructs we have designed, which is what we observe in Table IIA. The average size of all the micelles is 21.1± 4 nm. In addition to DLS, negatively stained TEM confirmed our DLS observations. A characteristic TEM image is shown in Figure 1B. We observe micelles between 15–20 nm in diameter using TEM, which is in agreement with our DLS results. As another characterization method, we measured the zeta-potential of the individual polymers prior to complexation and the micelle solutions. The results, shown in Table IIB , indicate that the targeting-peptide-PEG-K30 molecules are highly positively charged and the miR inhibitors are highly negatively charged. These results are expected since targeting-peptide-PEG-K30 contains positively charged poly-L-lysine and miR inhibitors contain the negatively charged phosphate backbone. The micellar solutions all have values in between the miR inhibitors and the peptide-PEG-K30 molecules indicative of a mixture of the two components.
Table II.
Micelle Characterization Data. (A) DLS of micelles. (B) Zeta potential of miR inhibitors and PEG complexes.
| A | ||
|---|---|---|
| Micelle | Dh(nm) | Polydispersity |
| REKA-micelle miR-92a inhibitor | 23.0 | 0.280 |
| REKA-micelle miR-33a inhibitor | 23.9 | 0.140 |
| REKA-micelle miR inhibitor control | 28.1 | 0.132 |
| VHPKQHR-micelle miR-92a inhibitor | 18.2 | 0.103 |
| VHPKQHR-micelle miR inhibitor control | 15.5 | 0.257 |
| REKA/VHPKQHR-micelle miR-92a inhibitor | 17.8 | 0.175 |
| REKA/VHPKQHR-micelle miR inhibitor control | 20.9 | 0.299 |
| B | |
|---|---|
| Polymer or Micelle | Zeta potential |
| miR-92a inhibitor | −29.6 ± 1.9 |
| miR-33a inhibitor | −22.2 ± 2.9 |
| REKA-PEG-K30 | 44.0 ± 2.2 |
| VHPKQHR -PEG-K30 | 30.9 ± 3.1 |
| REKA-micelle miR-92a inhibitor | 19.9 ± 2.8 |
| REKA-micelle miR-33a inhibitor | −15.5 ± 1.6 |
| REKA-micelle miR inhibitor control | 0.5 ± 0.1 |
| VHPKQHR-micelle miR-92a inhibitor | 7.2 ± 1.0 |
| VHPKQHR-micelle miR inhibitor control | 18.3 ± 2.1 |
| REKA/VHPKQHR-micelle miR-92a inhibitor | −7.6 ± 2.1 |
| REKA/VHPKQHR-micelle miR inhibitor control | 10.2 ± 1.1 |
Delivery of miRNA inhibitors to macrophages employing REKA-conjugated polyelectrolyte micelles
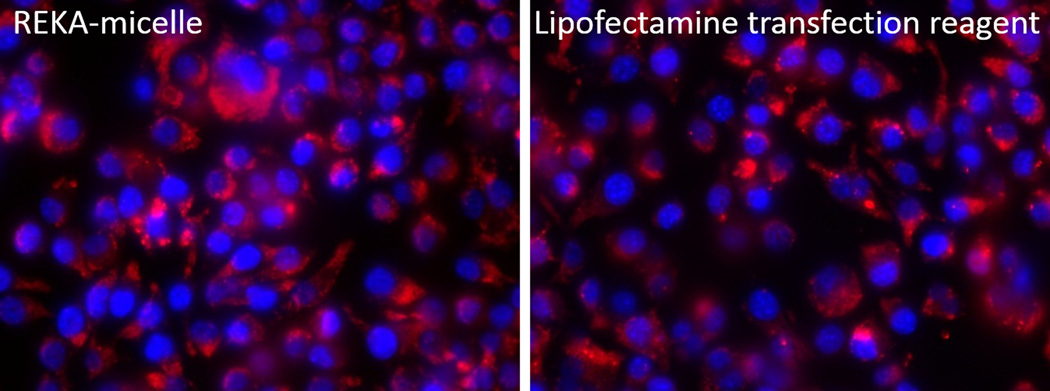
To test the effectiveness of lesion-targeting micelles in delivering inhibitors against atherogenic miRNAs in macrophages, murine macrophages (J774 cells) were treated with REKA-conjugated micelles that encapsulate negatively-charged miRNA inhibitors, as described in Figure 1. Dy547-labled miR inhibitor controls were first used to visualize the internalization of miRNA-modifying agents in macrophages. Fluorescence-labeled miRNA inhibitors were delivered in parallel to macrophages employing Lipofectamine RNAiMAX, a well-established transfection reagent that has efficiently delivered miRNA inhibitors to vascular cells in our previous studies22,38. As demonstrated in Figure 2A, significant fluorescence signal in the cytoplasm was detected in J774 cells incubated with miRNA inhibitor-encapsulated REKA micelles and in cells transfected with fluorescence-labeled miRNA inhibitors employing Lipofectamine RNAiMAX after 24 hrs. Similar fluorescence intensity observed in REKA-micelle and Lipofectamine treated cells indicate comparable delivery efficiency.
Figure 2. Delivery of miR inhibitor into mouse macrophages via REKA-micelle.
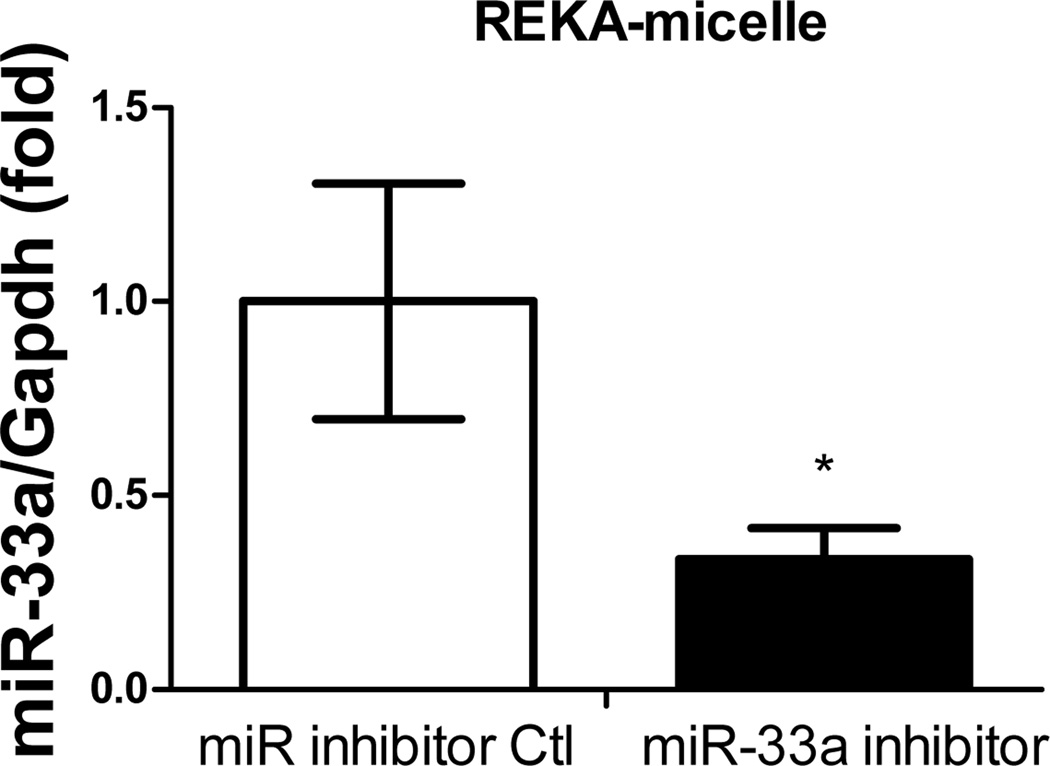
(A) Distribution of Dy547-miR inhibitor control in mouse macrophages (J774 cells). Left: Mouse macrophages incubated with 10 nM Dy547-miR inhibitor control using REKA/Dy-547 miR inhibitor micelles. Right: Mouse macrophages incubated with 10 nM Dy547-miR inhibitor control using Lipofectamine-RNAiMAX transfection reagent Red signal indicates Dy547-miR inhibitor control. Blue signal indicates DAPI staining for the nucleus. (B) Inhibition of miR-33a expression using REKA/miR-33a inhibitor micelle analyzed by real-time PCR and normalized to housekeeping gene GAPDH.
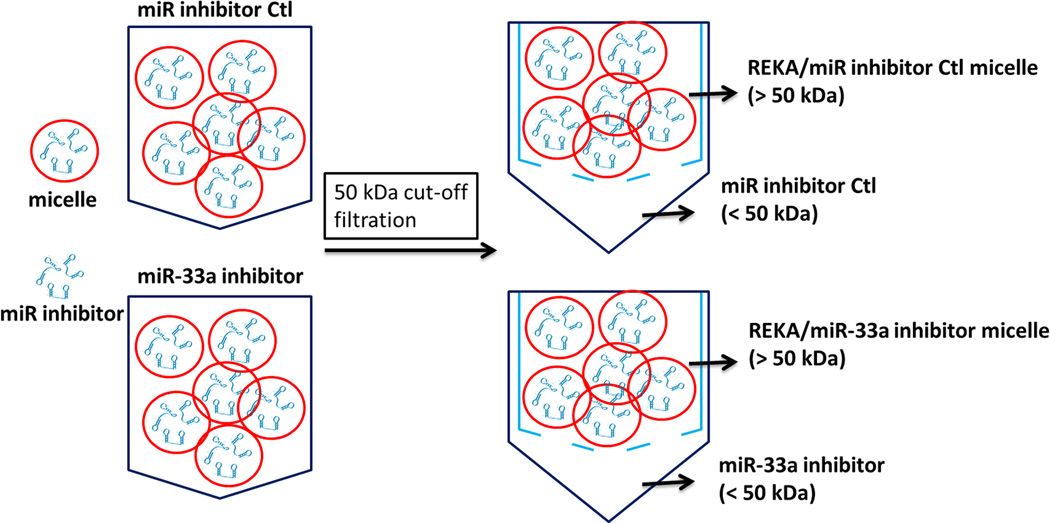
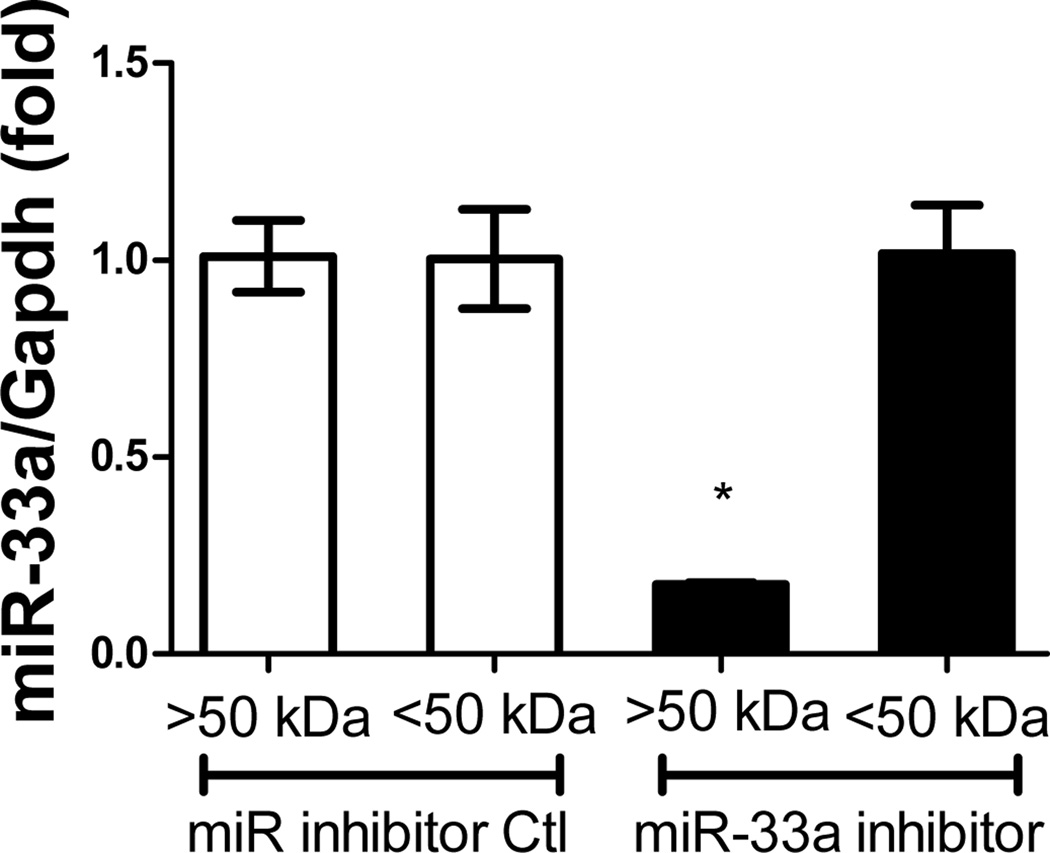
Given the atherogenic role of miR-33a, which suppresses ATP-binding cassette transporters in macrophages24–26, the effectiveness of REKA micelles in delivering miR-33a inhibitors to J774 cells was further investigated. Here we assembled both REKA/miR-33a inhibitor and REKA/inhibitor control micelles and incubated them with J774 cells followed by total RNA isolation and real-time PCR measurement of miR-33a expression. Our data suggests that miR-33a activity was significantly reduced (66 ± 8% lower) in murine macrophages treated with REKA-micelles carrying miR-33a inhibitors when compared to micelles containing miR inhibitor controls (Figure 2B). To eliminate any possibility that miR-33a inhibitors were delivered by un-encapsulated nucleotides, we performed a fractionation to separate micelles from the preparation solution. Given the molecular weight of miR inhibitors (~18,500 g/mole), a 50-kDa cut-off membrane was utilized to divide the solution into two fractions that were then incubated separately with J774 cells for 24 hr (Figure 3A). Both fractions from the REKA-micelles containing miR inhibitor controls did not affect miR-33a expression in J774 cells. However, significant inhibition of miR-33a activity (82 ± 0.3% lower) was detected in murine macrophages treated with the fraction containing >50KDa molecules purified from the REKA-micelles encapsulating miR-33a inhibitors (Figure 3B). The fraction containing <50KDa molecules isolated form miR-33a inhibitor-containing micelles had no effect on the miR-33a activity in J774 cells. This data collectively demonstrated an effective delivery of miR-33a inhibitors to the macrophage cytoplasm employing lesion-targeting, REKA-conjugated polyelectrolyte complex micelles.
Figure 3. Evaluation of miR inhibitor activity in micelle.
(A) A schematic of the molecular weight separation of micelles from excess peptide-PEG-polylysine or miR inhibitor. REKA-micelles containing either miR-33a inhibitor or miR inhibitor negative control were separated into 2 fractions by a 50-kDa cut-off membrane and then used to treat J774 cells with equal amounts from each fraction solution for 1 day. (B) Measurement of miR-33a expression in J774 cells using real-time PCR shows that miR-33a inhibition comes from the >50kDa fraction, implying that the micelle is effective in delivering miR-33a inhibitor. .
Inhibition of atherogenic miR-33a and activation of cholesterol efflux in macrophages employing REKA-conjugated polyelectrolyte micelles
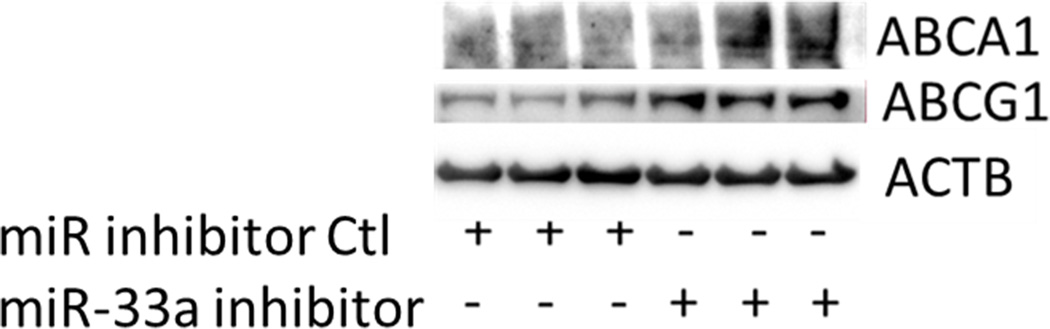
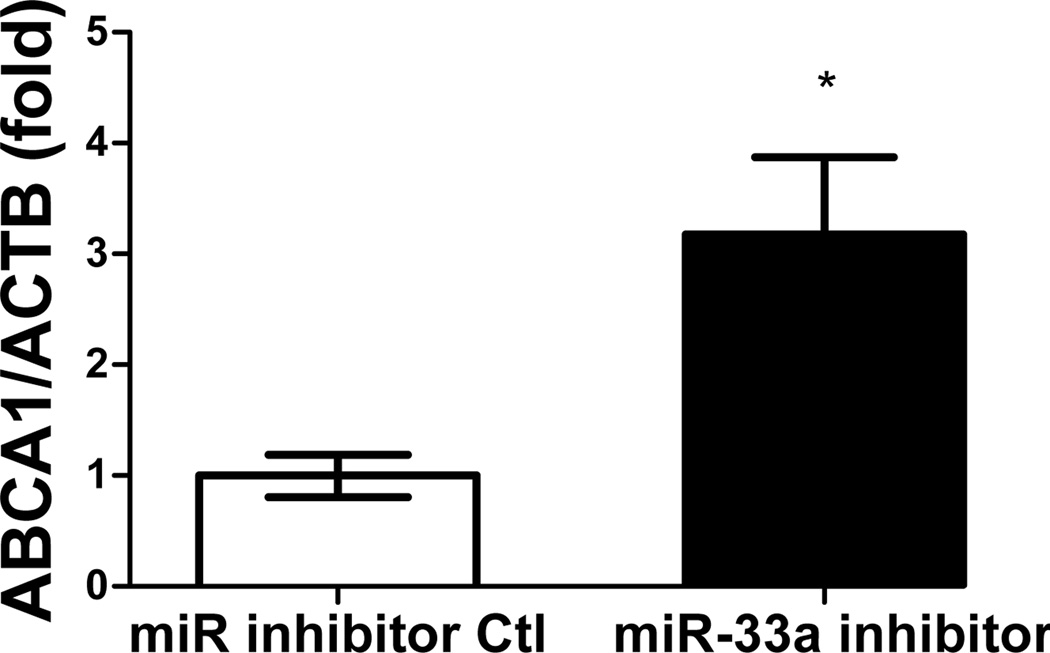
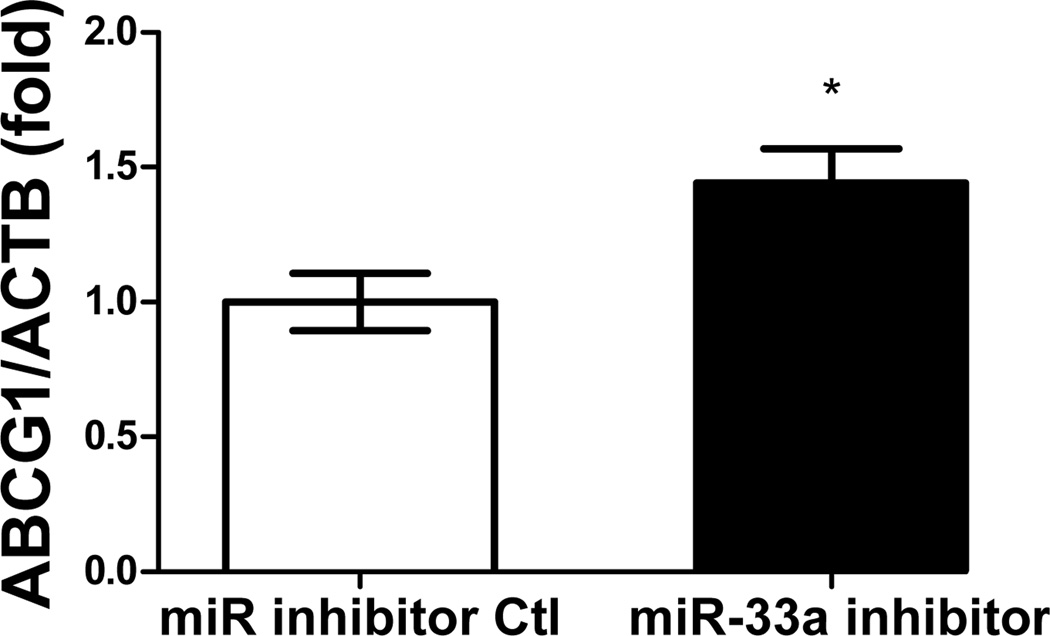
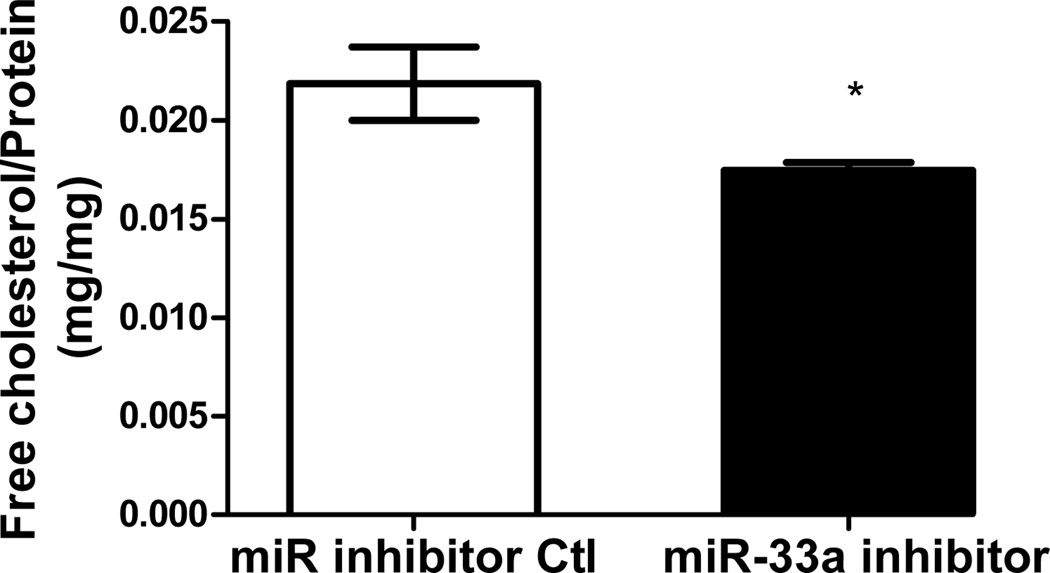
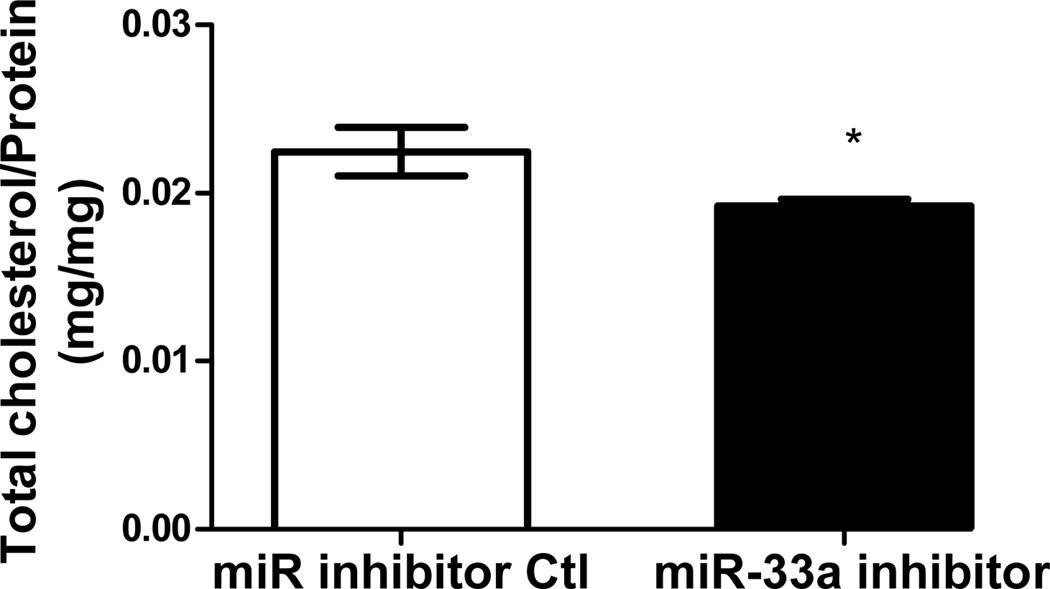
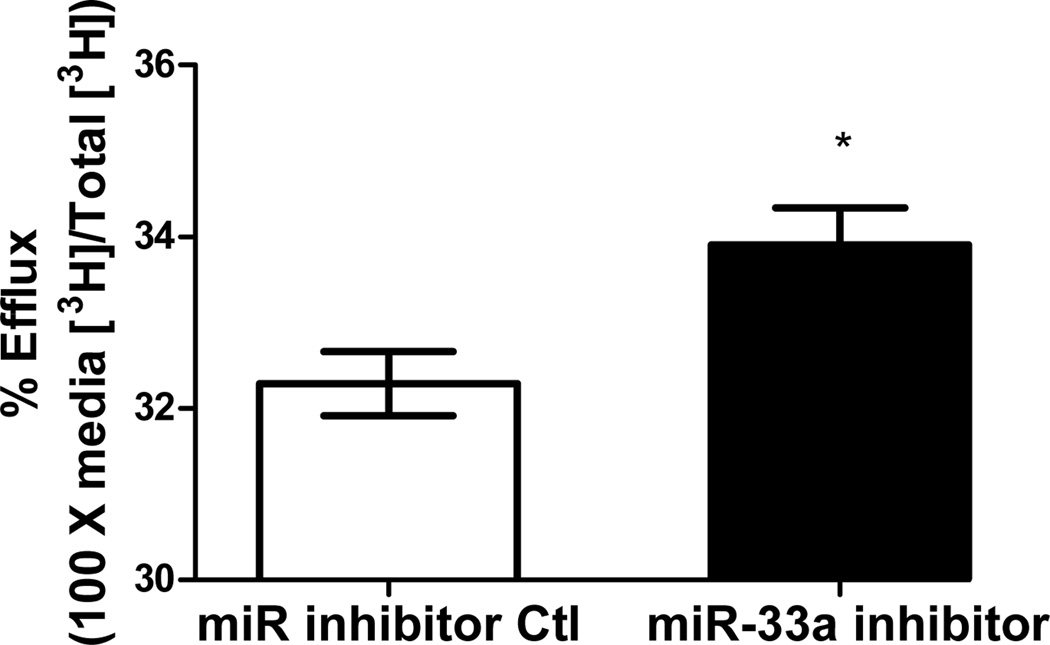
Functionality of miRNA inhibitors delivered by polyelectrolyte complex micelles is contingent upon the release of inhibitors from micelle cores and more importantly, the ability to interfere with miRNA-mediated functions in the targeted cells. To determine the feasibility of manipulating miR-33a-dependent cellular functions employing polyelectrolyte complex micelles, miR-33a-targeted genes and cellular cholesterol content that are associated with miR-33a activity were determined in J774 cells treated with REKA-micelles containing miR-33a inhibitors. miR-33a has been shown to directly suppress ATP-binding cassette transporters, ABCA1 and ABCG1, which contain miR-33a binding sites in the 3′ untranslated regions (UTR)24–26. ABCA1 and ABCG1 elevation, as the result of miR-33a inhibition, is predicted to stimulate cholesterol efflux and therefore decrease cholesterol content in macrophages39. Shown in Figure 4A-4C, western blots demonstrated increased protein expression of ABCA1 (3.2 fold) and ABCG1 (1.4 fold) in J774 cells treated with REKA-micelles containing miR-33a inhibitors when compared to inhibitor control-containing micelles. Moreover, miR-33a inhibitors delivered by REKA-conjugated micelles resulted in an increase of both free and total cholesterol in the macrophages (Figures 5D and 5E), indicating elevated cholesterol efflux. To measure cholesterol efflux, J774 cells treated with micelles containing miR inhibitor controls and miR-33a inhibitors were labeled with [3H] cholesterol in the presence of an LXR agonist to further upregulate ABCA1 and ABCG1 expression. [3H]-cholesterol was then measured in culture media and J774 cells and % efflux was calculated as 100% x media [3H]-cholesterol counts/total [3H]-cholesterol counts (media + cell). Cholesterol efflux was increased in the J774 cells treated with micelles containing miR-33a inhibitor (Figure 5F), consistent with the increase of ABCA1 and ABCG1 in these cells. Collectively, these data demonstrate the effectiveness of lesion-targeting micelles in inhibiting atherogenic miR-33a activating downstream ATP-binding cassette transporters, and stimulating cholesterol efflux in macrophages.
Figure 4. Functional analysis of miR-33a inhibition delivered by REKA-micelle into mouse macrophage, J774 cells.
After J774 cells were treated with REKA/miR-33a for 1 day and LXR agonist for 1 day, (A) western blot analysis of ABCA1 and ABCG1 show increased expression 3 days post-miR-33a inhibition. ACTB, actin beta, was used as an internal control. Quantification of (B) ABCA1 and (C) ABCG1 expression from western blot results, and (D) free and (E) total cholesterol measurement. (F) % efflux of cholesterol increases 3 days after miR-33a inhibition.
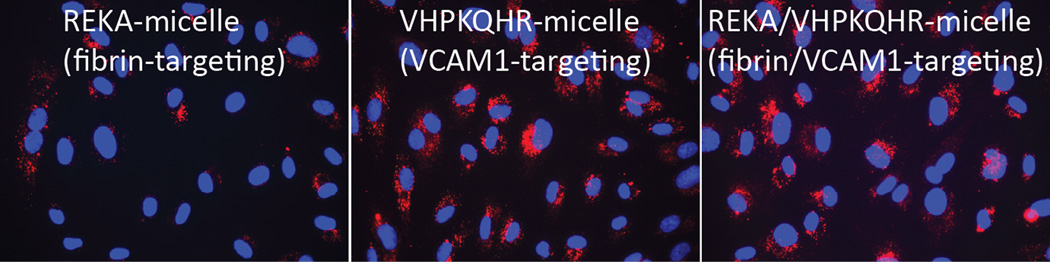
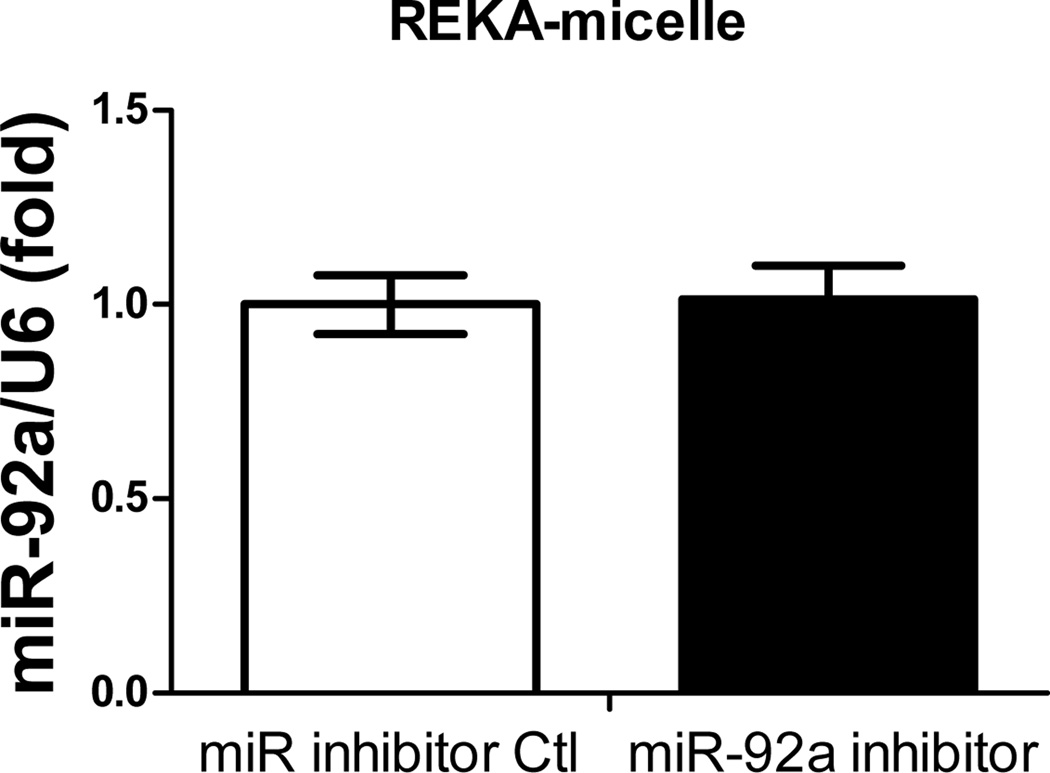
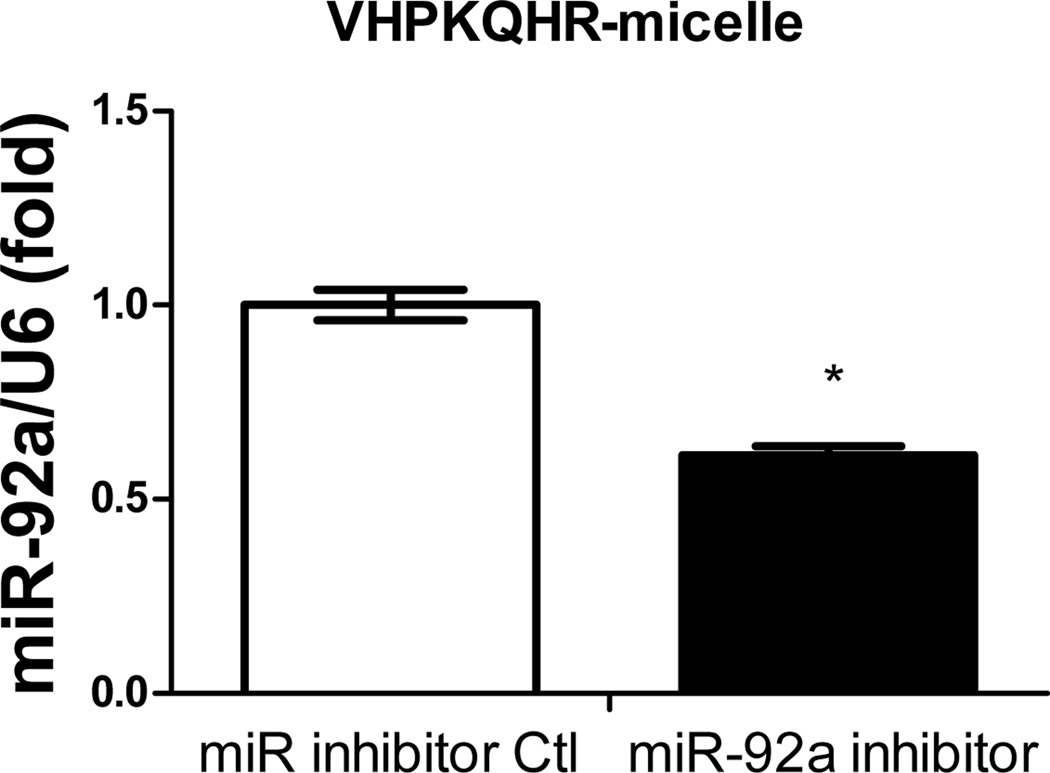
Figure 5. Cell-specific delivery of miR inhibitor into human aortic endothelial cells (HAECs) via VCAM1-targeting-micelles.
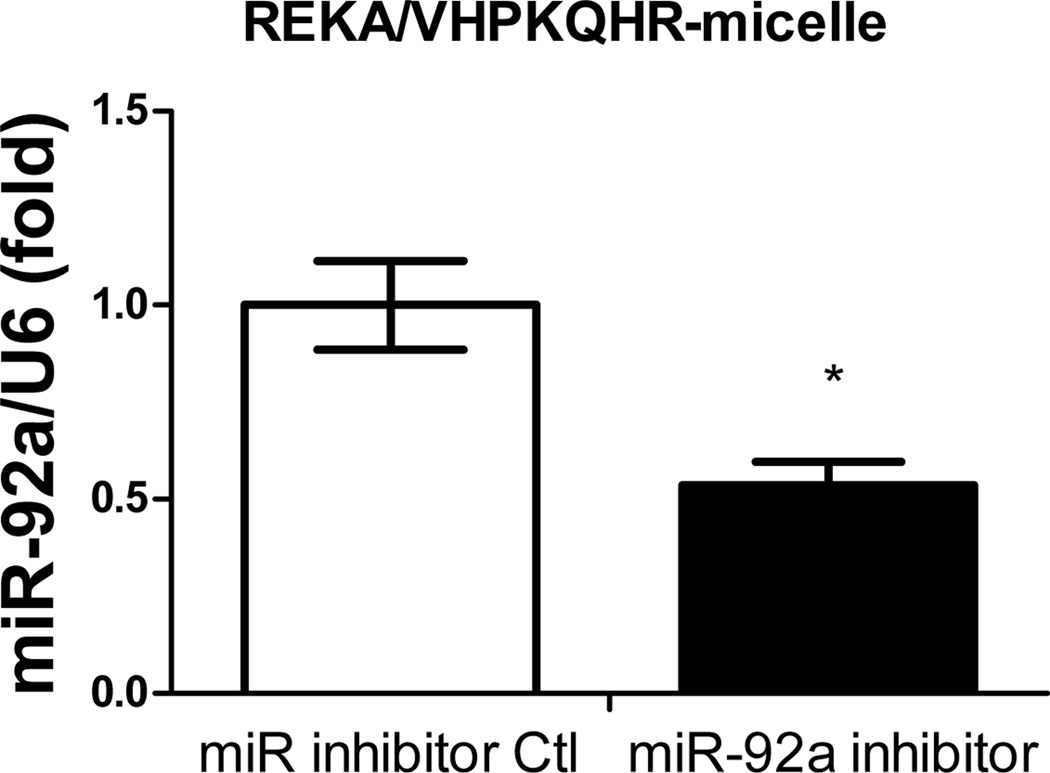
(A) Distribution of Dy547-miR inhibitor control in HAECs incubated with REKA-micelle, VHPKQHR-micelle, or REKA/VHPKQHR-micelle containing Dy547-miR inhibitor control shows increased uptake via micelles containing the VHPKQHR peptide. Red signal indicates Dy547-miR inhibitor control. Blue signal indicates DAPI staining for the nucleus. HAECs treated with miR-92a inhibitor inside (B) REKA-micelle, (C) VHPKQHR-micelle, or (D) REKA/VHPKQHR-micelle confirm that miR-92a expression is only inhibited using micelles containing the VCAM-1 targeting moiety.
Inhibition of atherogenic miR-92a in endothelial cells employing VHPKQHR-conjugated polyelectrolyte micelles
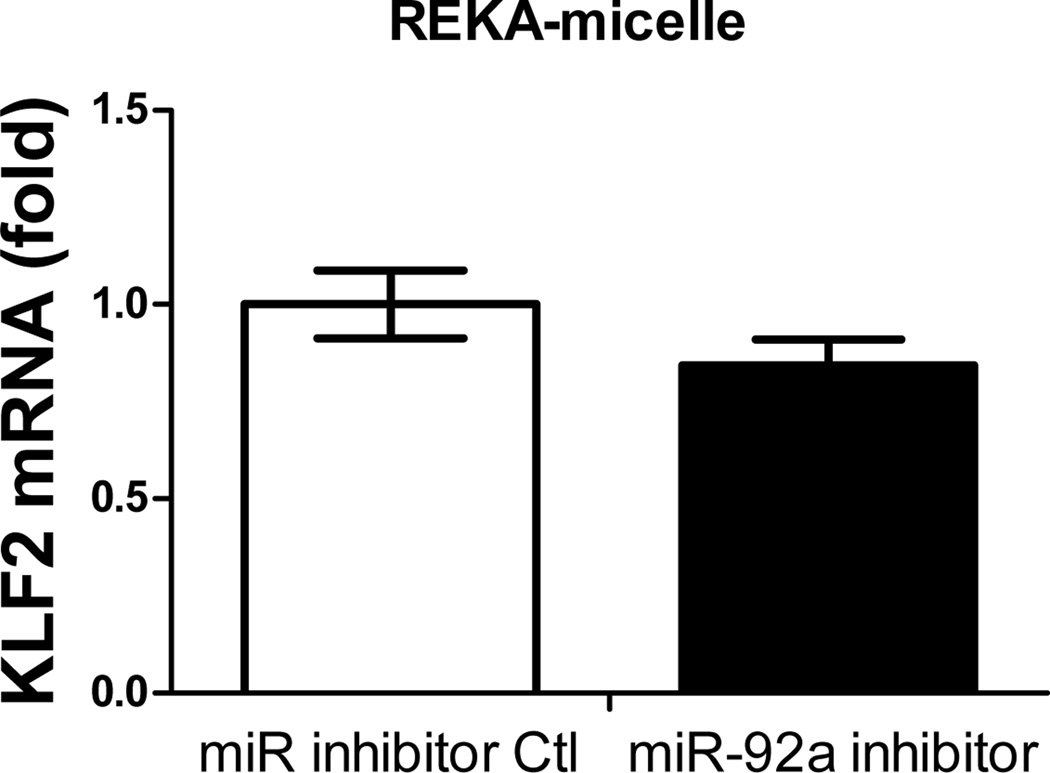
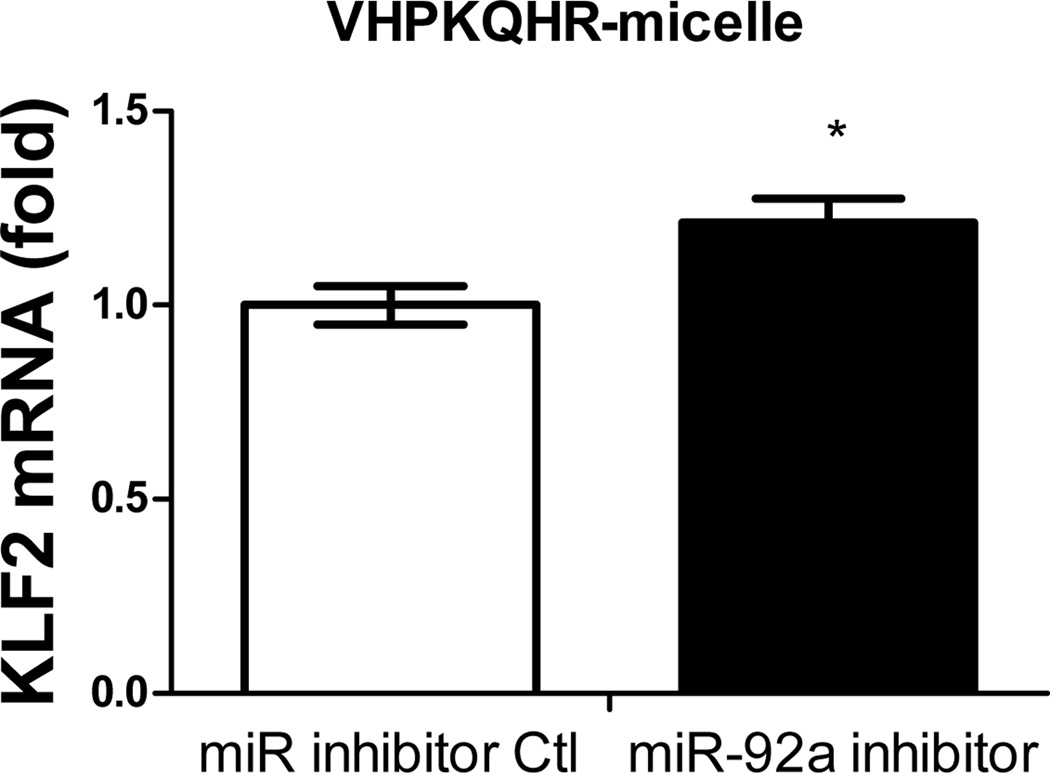
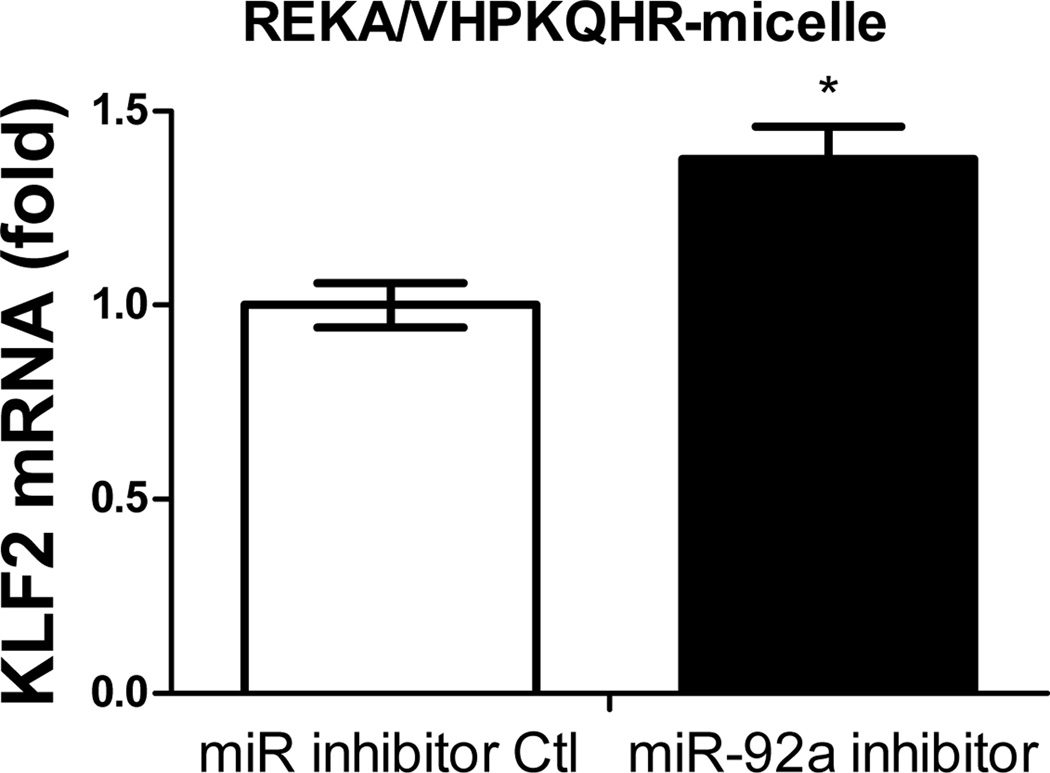
Previous studies have demonstrated the atherogenic role of endothelial miR-92a in activating vascular inflammation and promoting atherogenesis22,23,27. Due to the lack of ligands to trigger receptor-mediated endocytosis, it is foreseeable that intracellular internalization of REKA/miR-92a inhibitor micelles by endothelial cells, which requires the delivery miR-92a inhibitor to the cytoplasm, will be low. To overcome this obstacle, a phage display-identified peptide, VHPKQHR, that exhibits high affinity for vascular cell adhesion molecule-1 (VCAM-1), a cell adhesion molecule expressed in inflamed endothelium40, was utilized as a targeting molecule. The VHPKQHR peptide is predicted to increase endothelium-specific targeting as well as facilitate intracellular internalization34,41. Three types of polyelectrolyte micelles were engineered and tested for their effectiveness to deliver miRNA inhibitors to human aortic endothelial cells (HAECs): micelles conjugated with the fibrin-binding peptide, REKA, micelles conjugated with the VCAM1-binding peptide, VHPKQHR, and micelles conjugated with both VHPKQHR (50%) and REKA (50%). Lysophosphatidic acid (LPA), a well-known stimulator of vascular inflammation and atherosclerosis42,43, was used to activate VCAM-1 expression in HAECs. Dy547-labeled miR inhibitor controls were encapsulated in the aforementioned micelles to determine the cellular uptake and endothelial internalization. Significant levels of intracellular internalization were observed utilizing VHPKQHR- and VHPKQHR/REKA-conjugated micelles but not REKA-coated micelles in LPA-stimulated HAECs (Figure 5A). To further determine the feasibility to suppress atherogenic miR-92a in inflamed endothelial cells employing targeted polyelectrolyte micelles, HAECs were incubated with REKA-, VHPKQHR- or VHPKQHR/REKA-conjugated micelles that encapsulate miR inhibitor controls or miR-92a inhibitors. As shown in Figures 5B-5D, delivery of miR-92a inhibitors by VHPKQHR- and VHPKQHR/REKA-conjugated micelles significantly suppressed the endogenous expression of miR-92a by 39.8% and 46.5%, respectively. In contrast, the miR-92a inhibitor-containing REKA-micelle is unable to inhibit miR-92a activity in LPA-treated HAECs. Functionality of miR-92a inhibitors delivered by VHPKQHR- and VHPKQHR/REKA-conjugated micelles was further established by real-time PCR demonstrating the up-regulation of kruppel like factor 2 (KLF2), an anti-inflammatory transcription factor whose suppression by miR-92a has been previously reported22,23.
Biocompatibility of targeted polyelectrolyte complex micelles
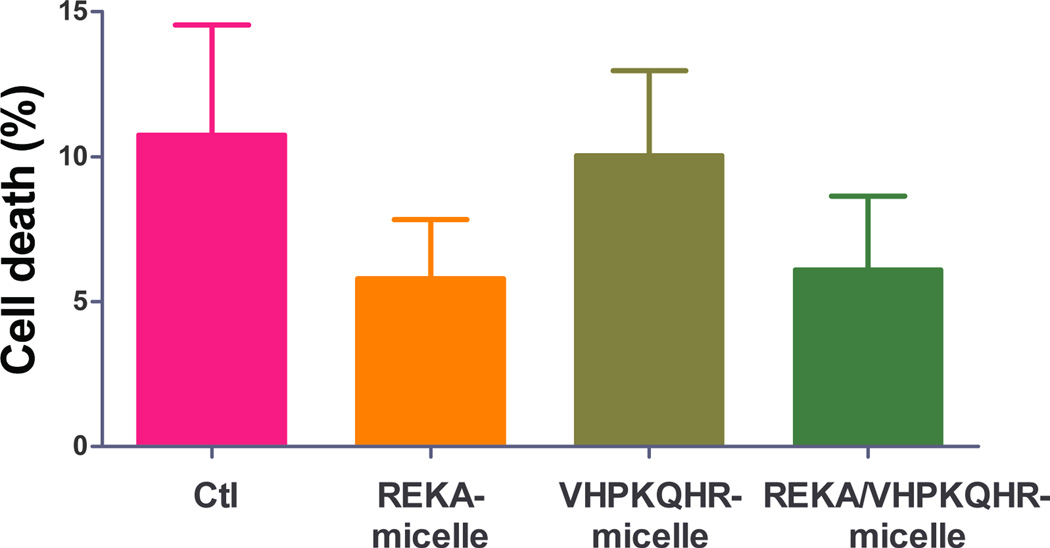
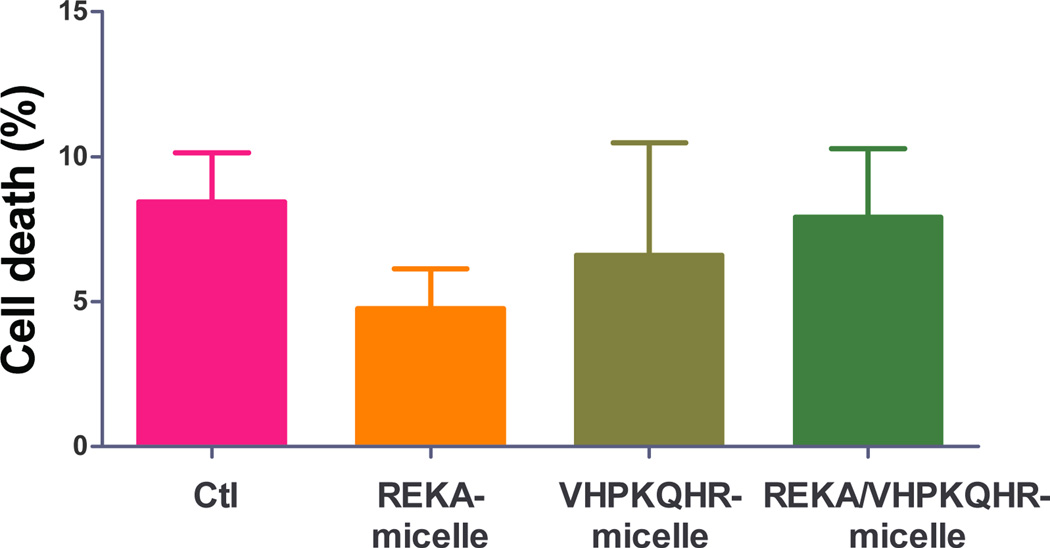
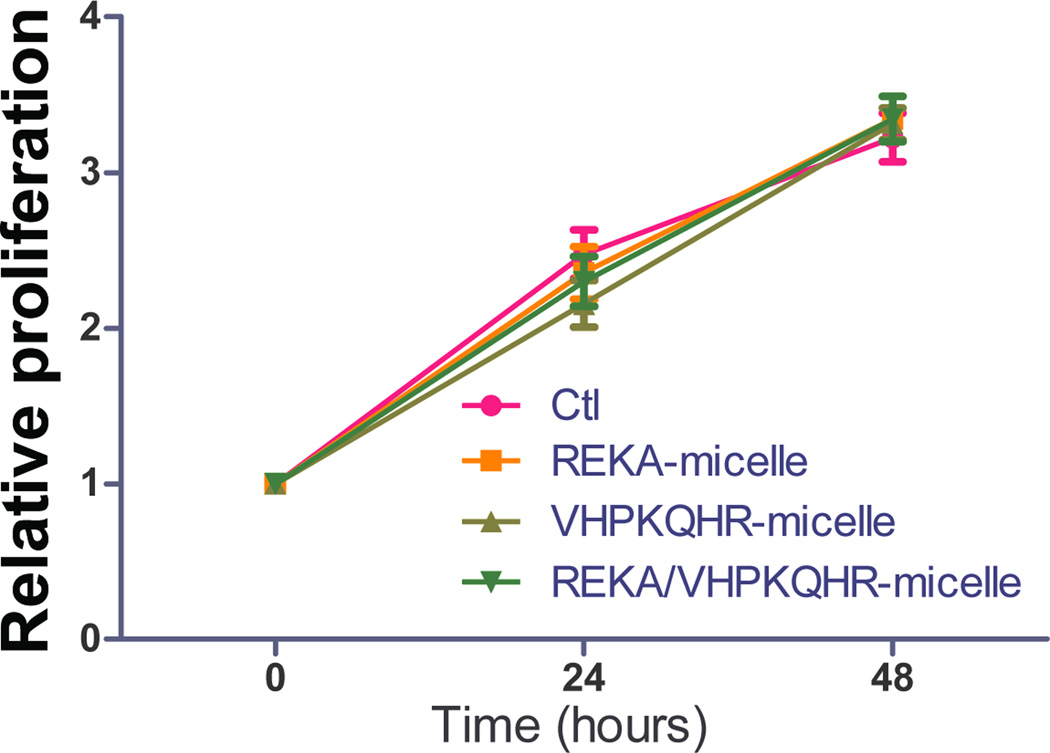
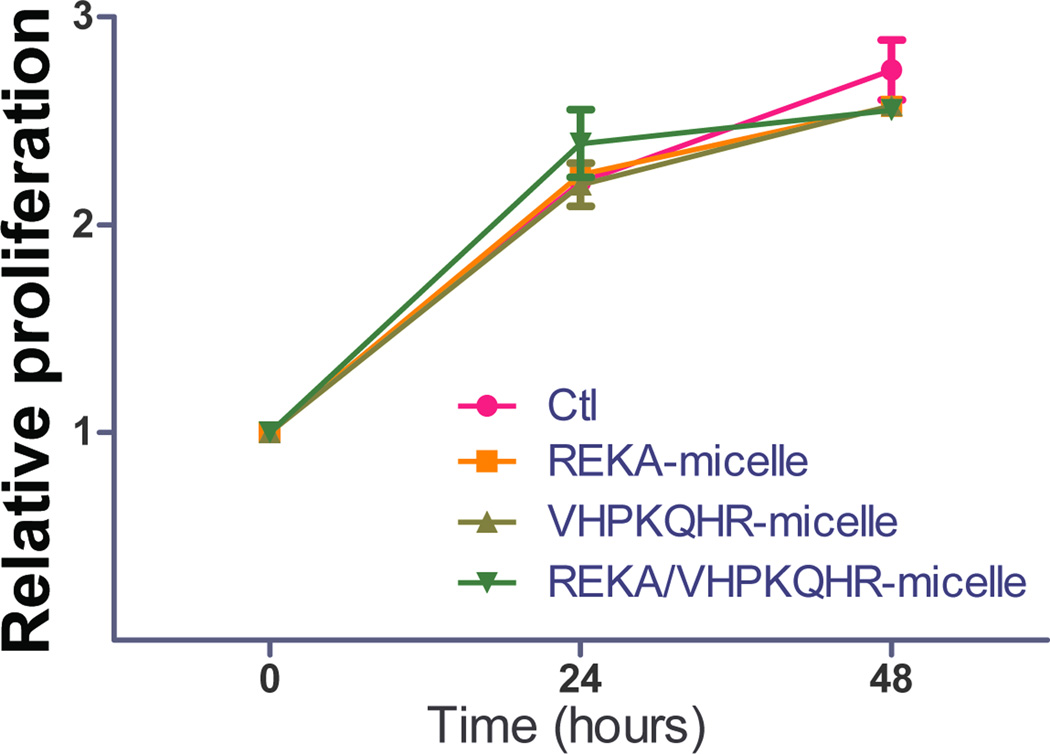
To evaluate the potential cytotoxic effect of polyelectrolyte complex micelles, cell viability and cell proliferation assays were performed in J774 cells and in HAECs treated with targeted polyelectrolyte complex micelles that contain miR inhibitor control. Cell viability was determined in confluent J774 cells and HAECs incubated with REKA-, VHPKQHR-, or VHPKQHR/REKA-conjugated micelles for 24 hrs followed by the quantification of live and dead cells. Comparable cell death rates were measured in J774 cells (Figure 7A) and in HAECs (Figure 7B) treated with a given polyelectrolyte complex micelle when competed to untreated cells, demonstrating low cytotoxicity of miRNA inhibitor-containing coacervates. Moreover, the pro-/anti- proliferative property of polyelectrolyte complex micelles was determined in subconfluent J774 cells and in HAECs incubated with a given targeted micelles for 24–48 hrs. As demonstrated in Figures 7C and 7D, treatment of the miRNA inhibitor-containing coacervates had no effect on proliferation of J744 cells and HAECs when compared to untreated cells. Collectively, these data demonstrate the biocompatibility and safety of targeted polyelectrolyte complex micelles.
Figure 7. Biocompatibility of targeted polyelectrolyte complex micelles.
Quantification of dead cells of confluent (A) J774 cells and (B) HAECs treated with 200 µM targeted polyelectrolyte complex micelles containing miR inhibitor control for 1 day. Relative cell proliferation of subconfluent (C) J774 cells and (D) HAECs treated with 200 µM targeted polyelectrolyte complex micelles containing miR inhibitor control for 1–2 days.
Discussion
Nanoparticle-based diagnostic and therapeutic applications have been available for clinical uses of a variety of human diseases such as cancer, infectious diseases, and muscular degeneration44. However, nanomedicine in cardiovascular diseases, particularly for delivering miRNA-modifying agents to atherosclerotic plaques in diseased arteries, has not been explored. Here, we engineered novel polyelectrolyte complex micelles that effectively deliver functional miR inhibitors to macrophages and endothelial cells in vitro and moreover, successfully modulate the expression of targeted miRNAs whose functions are critical to the pathology of atherosclerosis.
Characterization of our micellar constructs via DLS (Table 2a) and TEM (Figure 1b) demonstrated the formation of 21.1± 4 nm, spherical particles. The size of the micelles was approximately half the size of polyelectrolyte complex micelles formed by complexing antisense oligodeoxynucleotides containing between 15 and 20 base pairs and a PEG-polylysine molecule with a polylysine degree of polymerization of 18 and a PEG molecular weight of 5000 g/mol45. The size difference between these polyelectrolyte complex micelles and the micelles used in this study is attributed to the different molecular weight of the PEG segment. Subsequent experiments using micelles containing Dy547 labeled miR inhibitor molecules (Figure 2) showed significant cellular internalization of the labeled miR inhibitors in macrophages employing micelles coated with the fibrin-targeting peptide, REKA. Although the exact mechanisms contributing to uptake of micelles by macrophages is not the emphasis of this study, we speculate that the micelles may have entered the cells via phagocytosis and/or fluid-phase endocytosis since in macrophages there is no known expressed membrane protein that can be targeted by REKA peptides to trigger receptor-mediated endocytosis. In contrast, the internalization of polyelectrolyte complex micelles by endothelial cells seems to be mediated by receptor-mediated endocytosis since higher levels of internalized miR inhibitors were observed utilizing micelles conjugated with VCAM-1 targeting peptides when compared to those coated with fibrin-binding peptides. Consistent with these observations, VHPKQHR peptide has been shown to facilitate VCAM1-mediated intracellular internalization of nano-materials in the endothelium in vitro and in vivo34.
Beyond cellular uptake, the miR inhibitor polyelectrolyte complex micelles used in this study also successfully inhibited the function of their target miR as shown in Figures 2,3, and 5. Furthermore, similar to cellular uptake, Figure 5 illustrates that miR inhibition was also specific to the targeting moiety appropriate for HAECs. These results indicate that the miR inhibitor molecules were released from the micelle to perform their function. It has been shown using complexes of polyethyleneimine (PEI) and siRNA embedded with quantum dots, that the entire complex was internalized and the polylectrolyte complex began disassociating near the endosomal compartment close to the inner cell membrane46. However, internalization, endosomal escape, and release of the nucleic acid are thought to be polymer specific mechanisms. Although still open to question, PEI-based complexes may enable endosomal escape via the “proton sponge effect” in which protonation of the branched polyamine causes an influx of Cl- ions and water leading to osmotic and PEI swelling that could rupture the endosome47. As a way to mimic this effect, Oishi et al. used acid cleavable linkers between the PEG and siRNA to enable detachment of the PEG chain inside the acidic endosome and facilitate endosomal release14. Furthermore, certain studies involving DNA and polylysine complexes have shown that chloroquine, which swells and disrupts the endosome, is necessary to facilitate endosomal escape48. The side chain of polylysine used in our micellar complexes has a pKa ~10 and therefore does not contain this pH buffering capability49 or any acid cleavable linkers. However, we know that the miR inhibitor molecules are able to leave the endosome because of their functionality. Fortunately, the ability of polylysine based complexes to release nucleic acids scales with both the molecular weight of the polymer and the nucleic acid50. For this reason, we believe that our relatively short polylysine sequence and miR inhibitors facilitate miR release without the use of pH based release mechanisms or endosomal disrupting agents such as chloroquine. However, the exact mechanism of cellular uptake, endosomal escape, and separation of the complexed molecules remains unclear and is the subject of a future study.
Although the targeting of the fibrin binding micelles is not cell specific, it still offers a unique ability to deliver miR inhibitors preferentially to atherosclerotic plaques where fibrin deposits29. The success of managing pro-atherogenic miRNAs in human atherosclerosis has been hindered by the ineffective delivery of miRNA-modifying agents to diseased arteries of interest. In this study, we have demonstrated that lesion-targeting REKA-micelles effectively deliver inhibitors against miR-33a, which in turn elevate ATP-binding cassette transporters and stimulate cholesterol efflux in macrophages in vitro. Monocyte-derived macrophages/foam cells play a central role in the initiation and progression of atherosclerosis20. Lesion macrophages internalize native or modified lipoproteins via macropinocytosis or pathways mediated by scavenger receptors such as scavenger receptor A1 (SR-A1) and CD36. Meanwhile, ATP-binding cassette transporters ABCA1 and ABCG1 export excess cellular cholesterol into the high-density lipoprotein pathway and reduce cholesterol accumulation in macrophages39. When cellular cholesterol influx exceeds the capacity of efflux, lipid-laden macrophage foam cells can form as the result of compromised cholesterol homeostasis. Macrophage aberrations and foam cell accumulation contribute to the non-resolving inflammatory responses in the artery and the subsequent formation of necrotic cores in vulnerable lesions. Therapeutically activating macrophage cholesterol efflux, such as via inhibition of miR-33a, is expected to at least partially restore macrophage cholesterol physiology, decrease athero-promoting inflammation, reduce plaque lipid content and facilitate macrophage emigration. Moreover, herein we demonstrated successful inhibition of atherogenic miR-92a that promotes endothelial inflammation employing polyelectrolyte complex micelles conjugated with VCAM-1-targeting peptide: VHPKQHR. Endothelial miR-92a is synergistically up-regulated by oxidized LDL and disturbed flow27, two major risk factors of atherosclerosis. Furthermore, elevated endothelial miR-92a and accompanied suppression of Kruppel-like factors have been reported in human atherosclerotic plaques27 and in arterial regions susceptible to atherosclerosis in swine22. Therefore, inhibition of dys-regulated endothelial miR-92a regionally in atherosclerotic plaques is proposed to increase Kruppel-like factors, promote vascular health, and interfere with lesion development. The VCAM1-binding peptide, VHPKQHR, was chosen to facilitate endothelium-targeting and intracellular internalization of the lesion-targeting micelles. VCAM-1 expression increases in activated endothelial cells but remains low in healthy endothelium51,52, making it an ideal target for imaging and therapy of atherosclerosis that occurs and develops in predicted arterial sites associated with inflamed endothelia. Notably, the high surface area to volume ratio of nanoparticles allows us to engineer micelles with dual display of peptides targeting VCAM-1 and fibrin without compromising endothelial internalization, indicating the possibility to achieve multi-targeting of diverse molecules and proteins.
Conclusions
In summary, we have successfully engineered lesion-targeting polyelectrolyte complex micelles as an innovative delivery system for therapeutic nucleotides and tested their effectiveness in inhibiting critical athero-promoting miRNAs in macrophages and endothelial cells in vitro. Atherosclerotic vascular disease continues to be the leading cause of morbidity and mortality worldwide and vascular wall-based therapy, targeted specifically to dysfunctional macrophages and inflamed endothelial cells in the diseased arteries may provide a novel complementary strategy to fill the current treatment gap. Here we demonstrated that modular, multi-functional polyelectrolyte complex micelles can effectively deliver inhibitors against atherogenic miR-33a in macrophages and pro-inflammatory miR-92a in endothelial cells, leading to increased macrophage cholesterol efflux and elevated endothelial KLF2 expression, which are critical steps in restoring vascular health. The effectiveness of these innovative polyelectrolyte complex micelles in treating atherosclerosis in vivo is a subject of future studies. Moreover, the modularity of our platform allows the capability to present a variety of different cell targeting mechanisms on the micelle corona and to encapsulate various nucleotides in the micelle core. For instance, the micelles can be utilized to target various types of cells where distinct targetable membrane receptors are expressed. Additional therapeutic nucleotides, such as miRNA mimics, small interfering RNAs, and transcripts can be selectively encapsulated for accommodating a wide range of therapeutic strategies.
Supplementary Material
Figure 6. Upregulation of KLF2 expression in human endothelial cells via miR-92a inhibitor inside different micelles.
HAECs treated with miR inhibitor inside (A) REKA-micelle, (B) VHPKQHR-micelle, or (C) REKA/VHPKQHR-micelle show that KLF2 expression was upregulated only in micelles containing the VCAM-1 targeting moiety as quantified via real-time PCR.
Acknowledgements
The authors would like to acknowledge Ryan Klein for his assistance in purification of the peptide-based materials, Dr. Seungmin Hwang who kindly provided J774 cells, and Dr. Marion Hofmann-Bowman who generously shared antibodies. We thank Dr. Francis Alenghat for critical reading of the manuscript. This work was supported by the University of Chicago, the U.S. Department of Energy Office of Science program in Basic Energy Sciences and the Materials Sciences and Engineering Division (L.L. and M.T.), National Science Council of Taiwan NSC 102-2917-I-564-018 (C.H.K.), National Institutes of Health R00HL103789 (Y.F.), and American Heart Association 11BGIA7080012 (Y.F).
References
- 1.Höbel S, Aigner A. WIREs Nanomed. Nanobiotechnol. 2013;5:484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 2.Miyata K, Nishiyama N, Kataoka K. Chem. Soc. Rev. 2012;41:2562–2574. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 3.Harada A, Kataoka K. Science. 1999;283:65–67. doi: 10.1126/science.283.5398.65. [DOI] [PubMed] [Google Scholar]
- 4.Harada A, Kataoka K. Macromolecules. 1995;28:5294–5299. [Google Scholar]
- 5.Novoa-Carballal R, Pergushov DV, Müller AH. Soft Matter. 2013;9:4297–4303. [Google Scholar]
- 6.Baulin VA, Trizac E. Soft Matter. 2012;8:6755–6766. [Google Scholar]
- 7.Cohen Stuart MA, Besseling NAM, Fokkink RG. Langmuir. 1998;14:6846–6849. [Google Scholar]
- 8.Hofs B, Voets IK, de Keizer A, Stuart MAC. Phys. Chem. Chem. Phys. 2006;8:4242–4251. doi: 10.1039/b605695d. [DOI] [PubMed] [Google Scholar]
- 9.Itaka K, Yamauchi K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Biomaterials. 2003;24:4495–4506. doi: 10.1016/s0142-9612(03)00347-8. [DOI] [PubMed] [Google Scholar]
- 10.Katayose S, Kataoka K. Bioconjugate Chem. 1997;8:702–707. doi: 10.1021/bc9701306. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Qu W, Pan D, Ren Y, Williford J-M, Cui H, Luijten E, Mao HQ. Adv. Mater. 2013;25:227–232. doi: 10.1002/adma.201202932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kataoka K, Togawa H, Harada A, Yasugi K, Matsumoto T, Katayose S. Macromolecules. 1996;29:8556–8557. [Google Scholar]
- 13.Oishi M, Nagasaki Y, Nishiyama N, Itaka K, Takagi M, Shimamoto A, Furuichi Y, Kataoka K. ChemMedChem. 2007;2:1290–1297. doi: 10.1002/cmdc.200700076. [DOI] [PubMed] [Google Scholar]
- 14.Oishi M, Nagasaki Y, Itaka K, Nishiyama N, Kataoka K. J. Am. Chem. Soc. 2005;127:1624–1625. doi: 10.1021/ja044941d. [DOI] [PubMed] [Google Scholar]
- 15.Choi SW, Lee SH, Mok H, Park TG. Biotechnol. Progr. 2010;26:57–63. doi: 10.1002/btpr.310. [DOI] [PubMed] [Google Scholar]
- 16.Xiong X-B, Uludağ H, Lavasanifar A. Biomaterials. 2010;31:5886–5893. doi: 10.1016/j.biomaterials.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 17.Lobatto ME, Fuster V, Fayad ZA, Mulder WJM. Nat. Rev. Drug Discov. 2011;10:835–852. doi: 10.1038/nrd3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber C, Noels H. Nat Med. 2011;17:1410–1422. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- 19.Davies PF, Civelek M, Fang Y, Fleming I. Cardiovasc. Res. 2013;99:315–327. doi: 10.1093/cvr/cvt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore KJ, Tabas I. Cell. 2011;145:341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small EM, Olson EN. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Y, Davies PF. Arterioscl. Throm. Vasc. Biol. 2012;32:979–987. doi: 10.1161/ATVBAHA.111.244053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Wang K-C, Geary GG, Zhang Y, Wang W-C, Huang H-D, Zhou J. Circulation. 2011;124:633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernández-Hernando C. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquart TJ, Allen RM, Ory DS, Baldan A. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loyer X, Potteaux S, Vion A-C, Guérin CL, Boulkroun S, Rautou P-E, Ramkhelawon B, Esposito B, Dalloz M, Paul J-L, Julia P, Maccario J, Boulanger CM, Mallat Z, Tedgui A. Circ. Res. 2014;114:399–401. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 28.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suárez Y. J. Clin. Invest. 2011;121:2921. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters D, Kastantin M, Kotamraju VR, Karmali PP, Gujraty K, Tirrell M, Ruoslahti E. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9815–9819. doi: 10.1073/pnas.0903369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung EJ, Cheng Y, Morshed R, Nord K, Han Y, Wegscheid ML, Auffinger B, Wainwright DA, Lesniak MS, Tirrell MV. Biomaterials. 2014;35:1249–1256. doi: 10.1016/j.biomaterials.2013.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lempens EHM, Merkx M, Tirrell M, Meijer EW. Bioconjugate Chem. 2011;22:397–405. doi: 10.1021/bc100403e. [DOI] [PubMed] [Google Scholar]
- 32.Missirlis D, Teesalu T, Black M, Tirrell M. PLoS ONE. 2013;8:e54611. doi: 10.1371/journal.pone.0054611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Missirlis D, Krogstad DV, Tirrell M. Mol. Pharmaceutics. 2010;7:2173–2184. doi: 10.1021/mp100193h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nahrendorf M, Jaffer FA, Kelly KA, Sosnovik DE, Aikawa E, Libby P, Weissleder R. Circulation. 2006;114:1504–1511. doi: 10.1161/CIRCULATIONAHA.106.646380. [DOI] [PubMed] [Google Scholar]
- 35.Fields GB, Noble RL. Int. J. Pep. Prot. Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 36.Robinet P, Wang Z, Hazen SL, Smith JD. J. Lipid Res. 2010;51:3364–3369. doi: 10.1194/jlr.D007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada A, Kataoka K. Macromolecules. 2003;36:4995–5001. [Google Scholar]
- 38.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westerterp M, Bochem AE, Yvan-Charvet L, Murphy AJ, Wang N, Tall AR. Circ. Res. 2014;114:157–170. doi: 10.1161/CIRCRESAHA.114.300738. [DOI] [PubMed] [Google Scholar]
- 40.Cybulsky MI, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos J-C, Connelly PW, Milstone DS. J. Clin. Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly KA, Nahrendorf M, Yu AM, Reynolds F, Weissleder R. Mol Imaging Biol. 2006;8:201–207. doi: 10.1007/s11307-006-0043-6. [DOI] [PubMed] [Google Scholar]
- 42.Rizza C, Leitinger N, Yue J, Fischer DJ, Wang DA, Shih PT, Lee H, Tigyi G, Berliner JA. Lab. Invest. 1999;79:1227–1235. [PubMed] [Google Scholar]
- 43.Siess W, Zangl KJ, Essler M, Bauer M, Brandl R, Corrinth C, Bittman R, Tigyi G, Aepfelbacher M. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6931–6936. doi: 10.1073/pnas.96.12.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Etheridge ML, Campbell SA, Erdman AG, Haynes CL, Wolf SM, McCullough J. Nanomedicine: NBM. 2013;9:1–14. doi: 10.1016/j.nano.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harada A, Togawa H, Kataoka K. Eur. J. Pharm. Sci. 2001;13:35–42. doi: 10.1016/s0928-0987(00)00205-0. [DOI] [PubMed] [Google Scholar]
- 46.Lee H, Kim I-K, Park TG. Bioconjugate Chem. 2010;21:289–295. doi: 10.1021/bc900342p. [DOI] [PubMed] [Google Scholar]
- 47.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. Mol. Ther. 2012;21:149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupický D, Barrett LB, Spice R, Kendall M, Berry M. Nucleic Acids Res. 2005;33:e86–e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Li C, Wang H-Y, Zhang X-Z, Zhuo R-X. Chem. Eur. J. 2012;18:2297–2304. doi: 10.1002/chem.201102704. [DOI] [PubMed] [Google Scholar]
- 50.Gary DJ, Puri N, Won Y-Y. J. Controlled Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 51.Davies MJ, Gordon JL, Gearing AJH, Pigott R, Woolf N, Katz D, Kyriakopoulos A. J. Pathol. 1993;171:223–229. doi: 10.1002/path.1711710311. [DOI] [PubMed] [Google Scholar]
- 52.Nakashima Y, Raines EW, Plump AS, Breslow JL, Ross R. Arterioscl. Throm. Vasc. Biol. 1998;18:842–851. doi: 10.1161/01.atv.18.5.842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.