Abstract
8-Hydroxyguanine (8-OH-G) was discovered in 1983 in our laboratory at the National Cancer Center Research Institute, Tokyo. Since it could be formed in DNA not only in vitro but also in vivo by oxygen radical forming agents, we immediately hypothesized the importance of this discovery in connection with its biological consequence. Further intensive efforts by us from 1983 to 1990 confirmed that 8-OH-G is a highly significant oxidated DNA lesion involved in mutation and/or carcinogenesis in mammals, including humans. With the subsequent entry of many investigators to this research field the number of publications on 8-OH-G increased exponentially, reaching more than several thousands by the end of 2005. In this article, a summary is given of the important works carried out in the early days, and further notable contributions by many investigators are reviewed, focusing on 8-OH-G in the mammalian system. A special emphasis is given to research on knockout mice that are deficient in genes involved in the repair systems of the 8-OH-G lesion. Lastly, our own recent work is summarized involving a one-year carcinogenesis study of Ogg1 (the gene for 8-OH-G specific glycosylase/AP lyase*1) knockout mice that have been exposed to oxidative stress.
Keywords: 8-Hydroxyguanine; oxygen radical; DNA repair; mutagenesis, carcinogenesis; Mut M
Discovery of 8-hydroxyguanine
In the early 1980s, Takashi Sugimura (Director of the National Cancer Center Research Institute of Tokyo and Chief of the Biochemistry Division at the time) discovered that when fish and meat are broiled by cooking, large amounts of mutagens are produced. Sugimura and his colleagues subsequently defined the mutagenic properties as Glu-P-1, Glu-P-2, Trp-P-1 and Trp-P-2 which were isolated from pyrolysis products of pure glutamic acid and tryptophan, respectively. This finding heralded the beginning of a new research field, namely the exploration of carcinogenic heterocyclic amines produced by cooking.1)–3) In order to characterize mutagens present in broiled fish and meat, a collaborative study was initiated between the Biology Division, which I directed at the time, and the Biochemistry Division.
The scientists of the Biology Division had considerable experiences in chemistry from previous studies that involved the structural characterization of modified nucleosides present in tRNA. Minako Nagao of the Biochemistry Division prepared quite a large amount of extract from broiled fish as well as broiled beef (minced beef used for hamburgers). Hiroshi Kasai in our Division subsequently attempted to purify and identify active mutagenic components from these materials through various purification procedures. This work resulted in the successful isolation of IQ and MeIQ from fish and MeIQx from beef.4)–6) When assayed by the Ames’ Salmonela test using TA 98 strain, these isolates proved to be the most active mutagens discovered so far and were later demonstrated to be carcinogenic in rodents and monkey.1)–3)
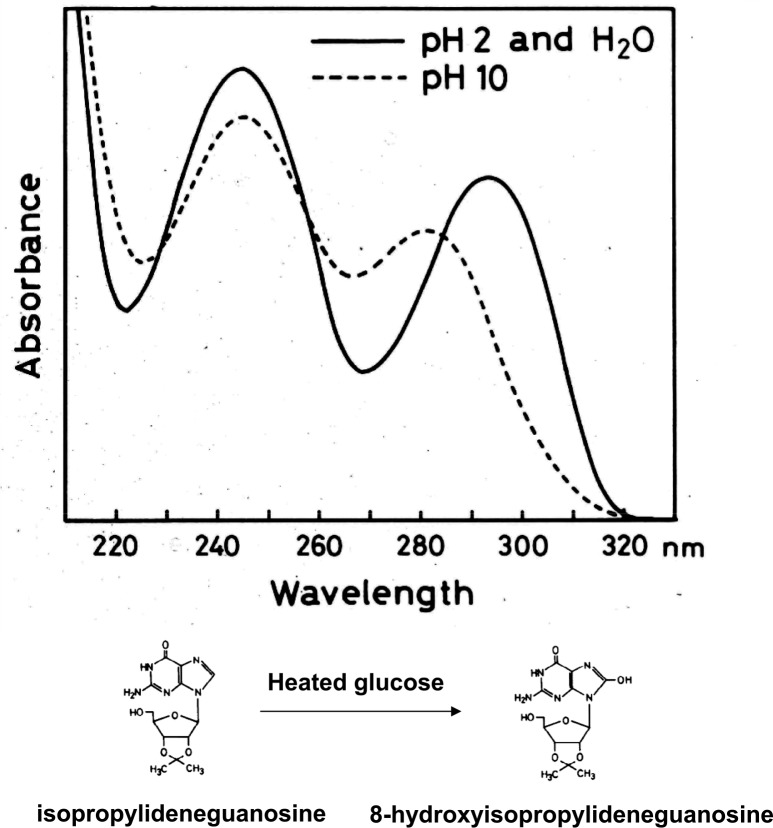
During the course of the characterization of mutagenic isolates, Kasai became aware of large amounts of mutagenic activity not requiring activation by microsomal enzymes that were lost during their enrichment, presumably due to their instability. Thus, Kasai adopted an alternative approach to identify such unstable mutagens, that is, isolation of guanine adducts by HPLC. Among the four DNA constituents, deoxyguanosine is the most important for adduct formation with mutagens/carcinogens. Isopropylideneguanosine instead of deoxyguanosine was used for identification of guanine adducts, because it is easily separated by organic solvent extraction. Serving as a model compound from broiled food, heated glucose was first tested. When isopropylideneguanosine was mixed with heated glucose and then fractionated by HPLC, two additional peaks could be distinguished. Surprisingly, one of them turned out to be 8-hydroxyisopropylideneguanosine, a simple guanine adduct having just one oxygen atom (Fig. 1).7) The chemical structure was established by NMR and massspectral analysis and the comparison with authentic 8-hydroxyisopropylideneguanosine which was prepared from 8-bromoguanosine.7) This finding suggested that oxygen radicals are involved in the reaction.*2 Reactive oxygen species are generated endogenously by cellular oxygen metabolism and exogenously by ionizing radiation, environmental mutagens and carcinogens. Thus, we quickly realized the importance of this discovery, and Kasai set about showing, by in vitro reaction, that various kinds of oxygen radical forming agents, including x-ray irradiation, had the ability to catalyze 8-hydroxylation of deoxyguanosine as well as the deoxyguanosine residue present in DNA (Fig. 2).8)–12) Interestingly, 8-hydroxydeoxyguanosine was a new compound not previously reported, although 8-hydroxyguanosine was previously synthesized by Morio Ikehara and his colleagues. It was rather surprising that 8-hydroxyguanine had not already been found in DNA, considering the numerous studies carried out in the radiation biology field. Apparently, such studies had been focusing on pyrimidine modifications following from the discovery of the biological significance of thymine dimers by Richard B. Setlow.13)
Fig. 1.
Structure and unique UV spectra of 8-hydroxyisopropylidenguanosine.
Fig. 2.
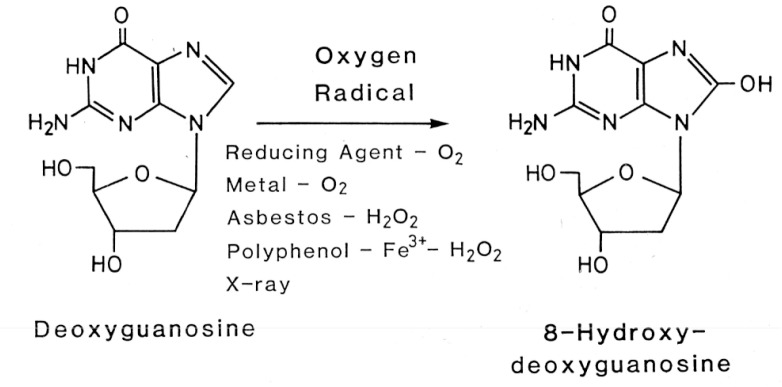
Formation of 8-OH-G in DNA by oxygen radical-forming agents.
Early accomplishments in our laboratory
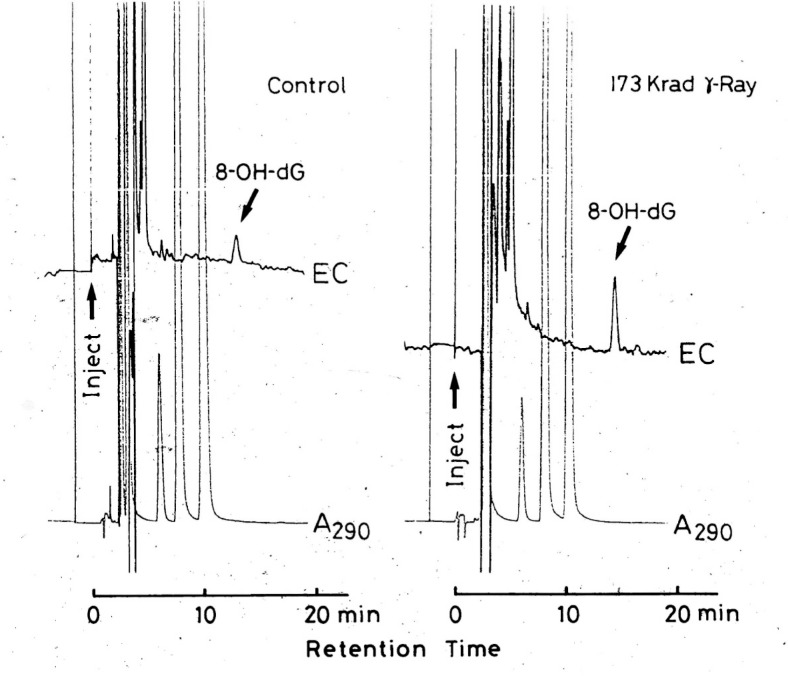
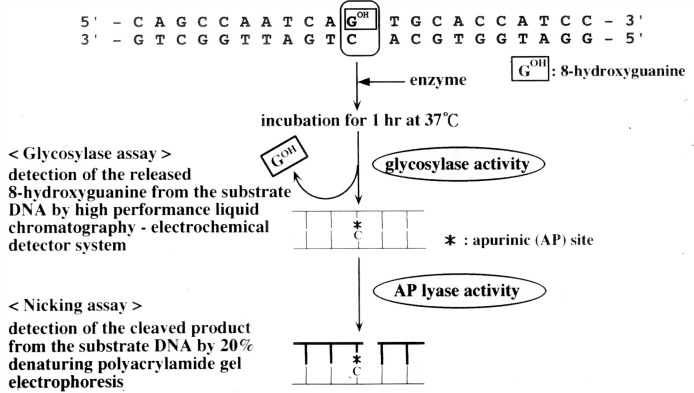
Whether 8-OH-G could be formed by oxidative stress in vivo was a key question at the time of discovery of 8-OH-G, because many modified bases that can be produced in a test tube, may not necessarily be found in nature. The challenge was quickly contended with by the breakthrough discovery of Robert A. Floyd of University of Oklahoma, who showed that the use of an electrochemical detection system instead of ultraviolet absorption gave an enhanced sensitivity of more than one thousand fold.14) We immediately adopted this technique and found that 8-OH-G was naturally present in DNA and could be increased in liver DNA when the whole body of mouse was exposed to γ -radiation (Fig. 3).15) It was also shown that 8-OH-G in liver DNA decreased during recovery phase from γ -radiation, indicating that a repair system(s) for the removal of 8-OH-G from DNA exists in mice.15) This was a fascinating finding suggesting that 8-OH-G has an important role in biological processes. Myong C. Chung, who came from Seoul National University as a visiting scientist supported by the First 10-Year Strategy of Cancer Control Program of the Japanese government, became involved in isolating a specific enzyme for the repair of 8-OH-G in DNA. To simplify the experiment we used Escherichia coli instead of mammalian cells. Detection of an 8-OH-G specific repair enzyme was achieved using a chemically synthesized deoxyribopolynucleotide with 8-OH-G located in a specific position (Fig. 4).16) This substrate was prepared by Eiko Ohtsuka and her colleagues at Hokkaido University. Such a deoxyribopolynucleotide, having a modified base in a specific position, was the first to be chemically synthesized. Ultimately, an 8-OH-G specific enzyme was successfully isolated from E. coli.16) This 8-OH-G specific glycosylase/AP lyase was later found to be identical to formamidepyrimidine DNA glycosylase (FPG protein) which was previously discovered as an enzyme with the ability to remove formamidopyrimidine.17)
Fig. 3.
Detection of 8-OH-G in mouse liver DNA by HPLC-coupled electrochemical detection system.
Fig. 4.
Glycosylase and nicking assay for MutM protein by using synthetic deoxyribopolynucleotide having 8-OH-G.
Further important discoveries on 8-OH-G from these early days, mostly accomplished within our laboratory, will be briefly summarized.
1). Misreading of 8-OH-G during DNA replication:
We firstly showed that 8-OH-G is misread by E. coli Klenow fragment when 8-OH-G containing deoxyribopolynucleotide is used as a template. In this reaction, 8-OH-G was read not only by cytosine (C), but also by adenine (A) and other bases. Significantly, neighboring bases in the vicinity of the 8-OH-G residue were also found to be misread.18) Later Arthur P. Grollman and his colleague extended this study using a highly elegant approach19) and showed that an 8-OH-G residue is principally misread as adenine, when mammalian polymerase α is used.
2). Conformation of 8-OH-G and its electrostatic potential:
By using an Ab initio molecular orbitol method, it was shown that the favored conformation of 8-OH-G is the 6,8-diketo form (8-oxo form) rather than the 8-hydroxy form (Fig. 5 and Fig. 6).20) It also showed that the addition of an oxygen atom to the 8 position of guanine base changed the electrostatic potential of the molecule entirely and gave it a negative character (Fig. 5).20) X-ray crystallographic analysis of 9-ethyl-8-hydroxyguanine also showed its 8-oxo form.21)
Fig. 5.
Conformation of 8-hydroxyguanine obtained by ab initio molecular orbital method. Map of electrostatic potential (ESP) at the molecular surface, 1.5 Å distant from van der Waals spheros. Dot: red, 9 ≦ ESP; pink, 3 ≦ ESP<9; yellow, −3<ESP<3; light blue, −9<ESP ≦ −3; blue, −15 < ESP ≦ −9; green, ESP ≦ −15(Kcal/mole).
Fig. 6.
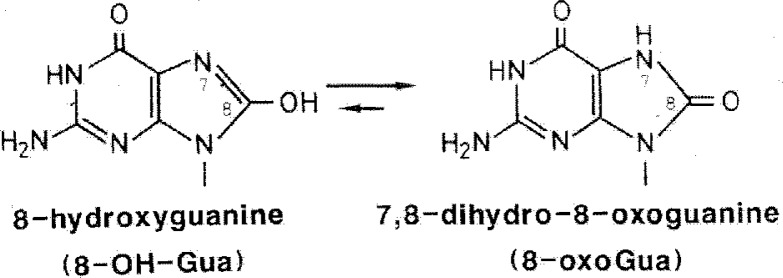
Tautomeric form of 8-OH-G.
3). NMR study of double-stranded deoxyribooligonucleotides having 8-OH-G:
Our NMR study indicated that an 8-OH-G residue located in a deoxyribooligonucleotide can be paired with a cytosine in the opposite strand.22) Later Michael Kouchakdfian et al. showed by NMR that if the opposite strand had adenine instead of cytosine, 8-OH-G can also make a base-pair with adenine, in a syn conformation with the 8-oxo form (Fig. 7).23) This was also confirmed by X-ray crystallography.24) These observations indicated that the conformation of 8-OH-G in DNA can be altered depending upon the identity of the base paired with 8-OH-G in the opposite strand.
Fig. 7.
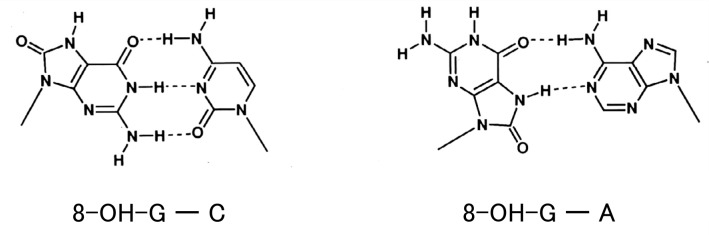
Formation of base pairs of 8-OH-G with C or A in the opposite strands.
4). Formation of 8-OH-G in kidney DNA by oral administration of potassium bromate (KBrO3) to rats:
In collaboration with Yuji Kurokawa and his coworkers, we showed that oral administration of the renal carcinogen KBrO3 to rats induced the formation of 8-OH-G in kidney DNA, but with far less being generated in liver DNA.25) These results suggested that formation of 8-OH-G in tissue DNA is target organ specific for carcinogenesis. Later, many investigators adopted this approach in the assessment of target organ specificity during carcinogenesis.26)
5). Formation of 8-OH-G by asbestos:
We showed that the carcinogenic potential of asbestos correlates with the extent of 8-OH-G formation by in vitro reaction, when it was incubated with DNA and hydrogen peroxide.27) The results strongly suggested that the carcinogenic activity of asbestos could be explained by its potential to produce 8-OH-G in DNA.
6). Detection of an 8-OH-G specific glycosylase/AP lyase in various rat and mouse tissues:
An 8-OH-G specific glycosylase/AP lyase, similar to MutM/FPG protein, was identified in various tissues from rats and confirmed the ubiquitous presence of the 8-OH-G specific repair enzyme(s) not only in bacteria, but also in mammals.28),29)
In March 1986, the US-Japan Cooperative Cancer Research Program on “Oxygen Radicals in Cancer” was held in Honolulu, Hawaii. This meeting permitted the first presentation of our work on 8-OH-G to an international audience.30) Bruce N. Ames from the USA side, among other participants from the USA, took considerable interest in our work and in fact, Ames also directed some of his efforts towards investigating this important subject.
The presence of the three genes involved in 8-OH-G repair
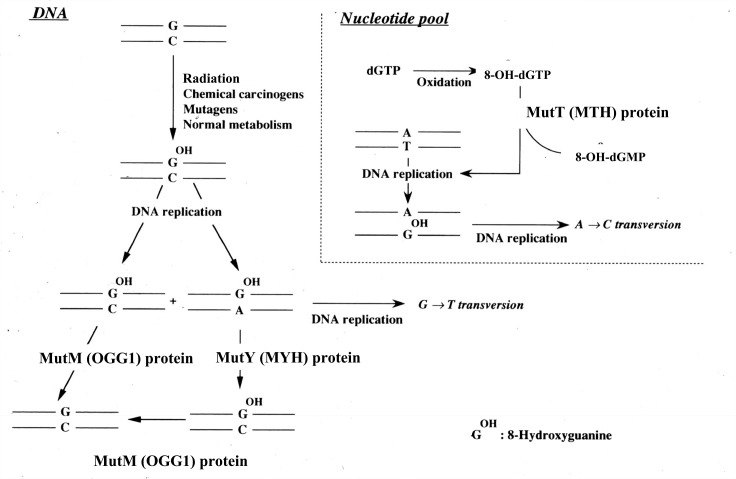
From around 1990 there was a large influx of investigators to the 8-OH-G research field. An outcome of particular notice from this period was the discovery of three genes that are engaged in 8-OH-G repair both in microorganisms and mammals (Fig. 8). Initially, much work had been done in E. coli to clarify the mechanism of 8-OH-G repair in DNA. As mentioned previously, FPG (the MutM gene product) is a specific glycosylase/AP lyase for 8-OH-G, and following its inactivation, an enhancement of G to T transversions is observed.31),32) MutY, identified by Jeffrey F. Miller and his colleagues, is a gene encoding a specific mismatch repair glycosylase enzyme which removes an adenine base when paired to 8-OH-G.33) MutT, discovered by Mutsuo Sekiguchi and his coworkers, codes for a specific pyrophosphatase with the ability to cleave 8-OH-dGTP to 8-OH-dGMP.34) Oxygen radicals produce not only 8-OH-G in DNA, but also 8-OH-dGTP from dGTP in the nucleotide pool. 8-OH-dGTP is incorporated into DNA opposite adenine (A) instead of cytosine (C), and thus, if not repaired prior to a subsequent round of replication, induces a mutation. Therefore, inactivation of MutT can result in A to C transversion. Lawrence A. Loeb and his colleagues in collaboration with us also showed that an A to C transversion takes place when using a plasmid vector system in E. coli.35) The existence of three gene products in E. coli responsible for the repair of 8-OH-G in DNA, sometimes called the GO system, demonstrates a functional cooperation to minimize mutagenesis36) and is a reflection of the importance of 8-OH-G repair to living organisms.
Fig. 8.
DNA repair systems for 8-OH-G in E. coli and mammalian cells. The names of the enzymes in parenthesis are that from mammalian cells.
An important question is whether a similar mechanism for the repair of 8-OH-G exists in mammalian cells. It has been shown that homologs of both MutY and MutT are present in mammalian cells.37),38) As described in the previous section,28),29) we found a similar enzymatic activity to the MutM product (FPG protein) in mouse and rat tissues. An activity with a similar function was also found in human leukocytes.39) Subsequent attempts to purify the enzyme from human cells showed the presence of at least two isoforms.40) The gene coding for the MutM homolog was not identified in mammalian cells until 1997. Meanwhile, a gene encoding a functional MutM homolog called OGG1 was isolated from the yeast Saccharomyces cerevisiae in 1996 by Serge Boiteux and his coworkers,41) and independently by Gregory L. Verdine and his colleagues.42) Suddenly in 1997, seven groups independently obtained a human or mouse homolog of OGG1 through similarity searches of the respective EST database using the yeast OGG1 sequence.43)–49) Among them, Hiroyuki Aburatani and his group of the University of Tokyo in collaboration with us, isolated four isoforms of cDNA produced by alternative splicing, as well as the related genomic DNA from human and mouse cells.43) This gene is referred to as hMMH/ hOGG1 or mMMH/ mOGG1 in human and mouse, respectively.*3 One of the human isoforms, type 1a, was expressed in E. coli and its gene product was isolated. It was shown to function as a glycosylase/Ap lyase specific for the 8-OH-G residue. Transfection of each of the four cDNA isoforms into MutM−and MutY− E. coli caused a reduced mutation frequency from G to T transversions. The type 1a isoform encodes a nuclear localization signal at its 3′ end whereas the other does not have such signal, suggesting that the type 1a variant is involved in the repair of 8-OH-G in nuclear DNA, and the other is directed towards the repair of mitochondrial DNA. Later, this was confirmed by Akira Yasui and his colleagues.50) Although expression of human and mouse Ogg1 was confirmed at mRNA level, it had not yet been shown at the protein level. In addition, it had not been clarified as to whether hOGG1 type 1a, type 1b, type 1c, type 2, or some other enzyme ranks as the most important member in the repair of 8-OH-G lesions. To answer this question, we prepared an OGG1 type 1a specific antibody. Using this antibody, we confirmed that the hOGG1 type 1a protein is indeed expressed in many human cell lines and that endogenous hOGG1 type 1a protein has repair activity for 8-OH-G lesions. In addition, we showed that upon depletion of the type 1a protein in a whole cell extract by its antibody, most of Ap lyase activity was lost, indicating that the type 1a protein is a major contributor to the repair of 8-OH-G lesions.51)
Studies using OGG1 knockout mice
An important outstanding question was whether OGG1 is in fact involved in repair of 8-OH-G lesions in vivo. To answer this question, we generated Ogg1 knockout mice (C57BL/6J) in collaboration with Tetsuo Noda of the Cancer Institute (Tokyo) and Aburatani.52) Genotype analysis of F2 offspring revealed typical Mendelian ratios of wild-type, heterozygous, and homozygous pups amounting to 25, 48, and 25% respectively, indicating that the Ogg1 gene product is not essential for embryonic development. Both Ogg1−/− and Ogg1+/− mice appeared healthy with respect to their survival and gross phenotypic appearance and male and female homozygous mice were found to be fertile. A significant increase of 8-OH-G was observed in the liver DNA of Ogg1−/− mice. At 9 weeks of age, the level of 8-OH-G in liver DNA of Ogg1 homozygous mice was 4.2-fold higher than that of wild-type mice. A further increase in 8-OH-G levels was observed at 14 weeks representing a 7-fold increase over wild-type mice of the same age. The mutation frequency of gpt liver tissue of Ogg1−/− mice was 2.3-fold higher than that of the wild type mice, indicating that 8-OH-G in DNA stimulates the induction of mutations. It is worth highlighting that Tomas Lindahl and his colleagues also reported similar results from an independently derived gene-targeted mouse line.53)
High accumulation of 8-OH-G in kidney DNA of Ogg1 knockout mice by chronic oxidative stress
Since the mutation frequency of Ogg1 knockout mice was marginal, it was difficult to clarify the type of mutation induced by 8-OH-G in vivo. To obtain more information regarding the involvement of 8-OH-G in mutation and/or carcinogenesis, it was necessary to promote oxidative stress in mice. We concluded that potassium bromate (KBrO3) was best suited for this purpose because of its specific oxygen radical forming property without formation of complexed DNA adduct and the fact it is a known renal carcinogen for rodents.54),55) Ogg1−/−, −/+, and +/+ mice carrying a gpt marker were treated with KBrO3 in their drinking water over a long period (12 weeks). The level of 8-OH-G in kidney DNA of Ogg1−/− mice was tremendously increased in a time-dependent manner (∼70 times above that of Ogg1+/+ mice treated with KBrO3). KBrO3 treatment also gave rise to an increased mutation frequency. Another interesting observation is that the accumulated 8-OH-G did not decrease following discontinuation of KBrO3 treatment, indicating the absence of significant repair in the total genome.56)
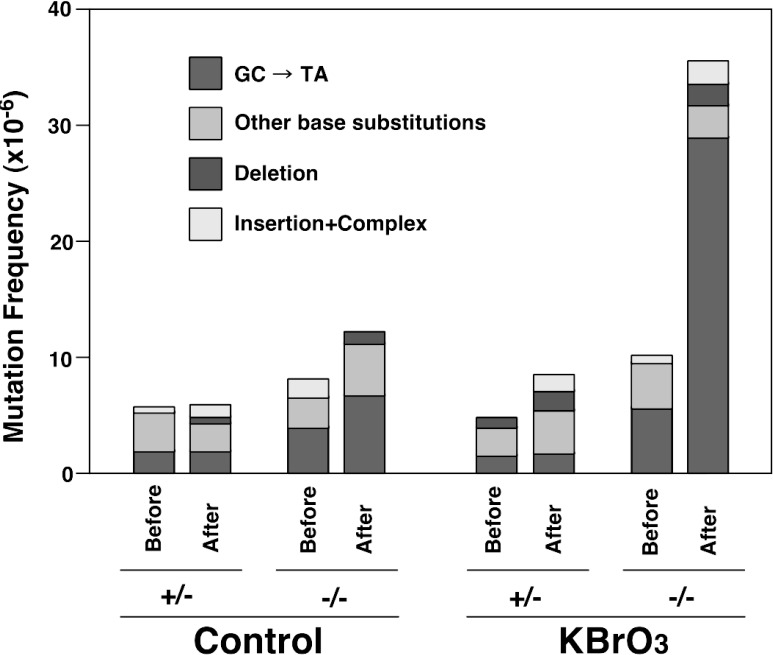
With the extensive exposure to KBrO3 as described, the amounts of 8-OH-G in the liver DNA of Ogg1−/− mice also showed significant increases; one fifth that of kidney DNA. Such accumulated DNA lesions did not decrease even 4 weeks after termination of KBrO3 treatment. Therefore, in order to assess whether cell proliferation influences the extent of mutation when 8-OH-G is present in DNA, we performed partial hepatectomy on Ogg1−/− and Ogg1+/− mice after being treated with KBrO3. The remnant liver from Ogg1−/− mice treated with KBrO3 regenerated to the same extent as nontreated Ogg1+/− mice. Interestingly, 8-OH-G was not repaired during cell proliferation following partial hepatectomy, indicating that there is no appreciable replication coupled repair of preexisting 8-OH-G, induced by DNA replication or cell proliferation. The mutation frequency after the regeneration of liver from treated Ogg1−/− mice showed a 3.5-fold increase as compared with that before regeneration. The proliferation of cells with accumulated amounts of 8-OH-G caused mainly GC→TA transversions, as would be predicted from previous in vitro experiments (Fig. 9).57)
Fig. 9.
Mutation spectra of gpt gene from liver of gpt/Ogg1 mutant mice which had been exposed to KBrO3, followed by partial hepatectomy.
Next, in order to assess the effect of KBrO3 on tumor induction, a one year carcinogenesis study was performed. Ogg1−/− and Ogg1+/+ mice were chronically exposed to KBrO3 for 29 weeks. After termination of the treatment, mice were kept for an additional 23 weeks. The amount of 8-OH-G in kidney DNA after 29 weeks of KBrO3 exposure reached 500 residues of 8-OH-dG/106dG, almost 250 fold that of untreated wild type mice. During the course of the study the mice appeared normal, although a decrease of body weight gain and some kidney malfunction was observed. When Ogg1−/− and Ogg1+/+ mice were sacrificed at 52 weeks, tumor formation could not be found in the kidney or other organs, such as the lung, liver, spleen, thymus, stomach and intestine. Microscopic examination also failed to discern precancerous foci in any tissues from Ogg1−/− and Ogg1+/+ mice.58)
Evidence showing the involvement of 8-OH-G in carcinogenesis in rodents and human
Kunihiko Sakumi et al.59) reported that Ogg1 deficient mice derived from C57BL/6J kept under normal conditions developed a greater number of lung adenoma and carcinoma compared to wild type mice at an age of one and half years. A plausible explanation for the discrepancy between the results of our groups may arise from the fact that Sakumi et al.59) showed that even wild type mice produced tumors in various organs, although the number of the tumors was less than that found in Ogg1−/− mice. On the contrary, in our experiment, no tumors were found in control wild type mice whether or not they had been exposed to KBrO3. One could argue that in the study of Sakumi et al., the mice were kept for a longer period of time (78 weeks compared to 60 weeks in our experiment). KBrO3 is known to be a specific oxygen radical forming agent. Thus, one can speculate that mutation of the genome alone is insufficient for tumor formation, and a promoting process or inflammation is necessary for tumor induction. In order to explore this possibility, we are currently performing a carcinogenesis study in which partially hepatectomized Ogg1−/− mice are kept for a long period of time after treatment with KBrO3, since partial hepatectomy is known to have tumor promoting activity.
Other knockout mice strains related to 8-OH-G repair, such as MYH and MTH also display a higher tumor incidence compared to wild type mice, even when not exposed to oxidative stress.60)–63) MYH or MTH deficiency may be of greater importance to tumor induction in comparison to Ogg1 deficiency. However, the actual increase of 8-OH-G and the mutation frequency in these mice without applied oxidative stress is comparable or less than that found in Ogg1−/− mice exposed to KBrO3. Incidentally, Yoshimichi Nakatsu et al. recently reported that Mutyh deficient mice exposed to KBrO3 showed increased G→T mutation frequency and subsequently produce a large number of tumors in the intestine.64)
Other 8-OH-G repair systems are known to exist in addition to Ogg1. Tapas K. Hazra et al. found an OGG-2 enzyme which cleaves 8-OH-G/G and 8-OH-G/A lesions more efficiently than 8-OH-G/C.65) The same group also identified another repair enzyme, NEH 1 which has a wide substrate specificity.66) The nucleotide excision pathway may also play a role in the repair of 8-OH-G, because an 8-OH-G base was shown to be removed from synthetic oligonucleotides by a reconstituted system consisting of XPA, RPA, TFIIH, XPC-HHR23B, XPG and XPF-ERCCI.67) Transcription-coupled repair system can remove 8-OH-G in the transcription strand of Ogg1−/− cell lines, though at a slower rate than Ogg1+/+ cell lines.68),69) In addition, reports have shown that other enzymes, such as N-methylpurine-DNA glycosylase70) and ribosomal protein S369) have the capacity to cleave 8-OH-G containing DNA. Therefore, the possibility exists that although the contribution of other enzymes to the removal of 8-OH-G within the overall genome is marginal, their activity could be of greater relevance to preventing carcinogenesis.
In the case of humans, it is now clearly established that the presence of 8-OH-G in DNA correlates with carcinogenesis. Allelic polymorphisms in the Ogg1 exonic region, which is translated to an altered enzymatic activity of the OGG1 protein, were found to correlate with the incidence of lung cancer in humans.71)–73) A strong association was also found between a reduced activity in OGG1 protein and lung cancer incidence.74) Some hereditary colorectal cancer patients were found to have a deficiency in the MYH gene.75)–77)
The induction of cancers in human such as that occurs in lung and colon, is the results of multiple processes over a long period of time. Under these circumstances the significance of 8-OH-G in carcinogenesis might be clearly seen. Therefore, a more sophisticated animal model to elucidate the molecular mechanism of 8-OH-G in human cancer may be needed.
Transcriptional misreading of a human c-Ha-ras fragment within an 8-OH-G hot spot by DNA polymerase η and κ
Recent studies have provided interesting data on the participation of non-replicative DNA polymerases in mutagenesis. A group of minor polymerases were found to have the unique ability of translesion DNA synthesis. Enzymes of particular interest are the Y-family DNA polymerases ι, η and κ, which were shown to bypass various forms of DNA damage.78) This prompted us to examine the mechanism of how 8-OH-G residues in DNA are recognized by such Y-family polymerases.
We previously reported that a chemically synthesized form of the human c-Ha-ras gene having 8-OH-G in codon 12 or 61 showed remarkable transforming activity when transfected into NIH3T3 cells.79)–81) An interesting observation was that the mutation found in transfected human c-Ha-ras consisted not only of G→T transversions on the position of 8-OH-G, but also G→C transversions. In addition, neighboring bases to the 8-OH-G residue were also found to be mutated (Table I). To examine this phenomenon further, fragments of the human c-Ha-ras gene that contained 8-OH-G in codon 12 were used as a template for DNA polymerase η using an in vitro reaction.82) It was shown that DNA polymerase η not only misincorporated dAMP, but also dGMP, while DNA polymerases α and β exclusively led to misincorporation of dAMP. In addition, we observed that when using polymerase η, an 8-OH-G residue was capable of propagating a mutation as a result of an “action-at-a-distance” effect. The replication catalyzed by this enzyme resulted in misincorporation of dAMP, dTMP and dGMP opposite a normal guanine that was flanked on the 3′-position by an 8-OH-G residue. Intriguingly, with two adjacent 8-OH-G residues the specificity of polymerase η was even more compromised, leading to the incorporation all four nucleotides. By contrast, two adjacent 8-OH-G residues almost completely blocked synthesis by polymerases α and β. These results suggest an important role for polymerase η in conferring hypermutability to codon 12. Similar results were also obtained in the case of polymerase κ.83) Louise Prakash and her colleagues previously reported that polymerase η is able to carry out an efficient and acculate translesion replication at the site of the 8-OH-G residue.84) Apparently, misreading of 8-OH-G by polymerase η depends on the sequence context of DNA.
Table I.
Mutations induced by 8-OH-G containing c-Ha-ras genes transfected into NIH3T3 cells
| Position of 8-OH-G | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 34 | 35 | 181 | 35 | ||||
| G*→Ta | 14 | G*→T | 22 | G*→T | 35 | G*→T | 8 |
| G*→A | 1 | G*→A | 8 | G*→A | 0b | G*→A | 3 |
| G*→C | 0 | G*→C | 1 | G*→C | 0b | G*→C | 1 |
| 3′-G→T | 0 | 5′-G→T | 4 | 5′-T→A | 1 | 5′-G→T | 1 |
| 3′-G→A | 0 | 5′-G→A | 5 | 5′-T→C | 4 | 5′-G→A | 1 |
| 3′-G→C | 0 | 5′-G→C | 2 | 5′-T→G | 0b | 5′-G→C | 0 |
| total mutants | 15 | total mutants | 42 | total mutants | 40 | total mutants | 14 |
G* represents 8-OH-G.
Mutations at this position do not activate the c-Ha-ras gene.
Motoya Katsuki and his colleagues in collaboration with us previously reported that transgenic mice having a normal human c-Ha-ras gene are quite sensitive to spontaneous tumor induction or by various chemical carcinogens and the resulting tumors always contained activated human c-Ha-ras that had been altered at either at codon 12 or 61.85),86) Thus, experiments are now underway to test whether Ogg1 knockout mice harboring a normal human c-Ha-ras gene develop tumors upon exposure to compounds that induce oxidative stress. Should this be the case, it would provide direct proof that mutagenesis by 8-OH-G in DNA is linked to carcinogenesis.
Future aspects of 8-OH-G involvement in carcinogenesis
From what has been described in the preceding sections, it is reasonable to conclude that 8-OH-G is involved in the induction of cancer in humans, although some discrepancy has been found with the rodent model. Obviously, a more suitable rodent experimental model is required to elucidate how 8-OH-G is involved in cancer induction at a molecular level. However, the negative results with respect to carcinogenesis in OGG1 knockout mice exposed to high oxidative stress might offer some clues; for example, the possibility that another process in addition to a point mutation in the genome is necessary for the induction of cancer.
Giving consideration to future approaches, several other factors need to be addressed. It is possible that 8-OH-G formation in DNA by oxygen radicals is not evenly distributed throughout the genome, and indeed, the distribution of 8-OH-G in DNA also may differ depending upon which reagents are used to generate oxygen radicals. It is also true that the pattern of 8-OH-G repair in DNA by OGG1 is not uniform through the genome, as it has been shown that 8-OH-G residues in the transcribed DNA strand are resistant to repair by OGG1.68),69) Therefore, studies based on other 8-OH-G repair enzymes and further genetically engineered mouse models are needed. It is also important to assess the effect of post-translational modification(s) on OGG1 activity and other 8-OH-G related repair enzymes.87)
Oxygen radicals are generated not only by exogeneous reagents such as radiation and environmental chemical carcinogens, but also by internal cellular metabolism. Oxygen radicals have the ability to react with various cellular components such as proteins and lipids; being in some cases beneficial to the cell. Therefore, the administration of antioxidants may not be a wholly appropriate solution for the prevention of carcinogenesis. The inhibition of 8-OH-G formation in DNA by more specific means may be a better alternative. In other words, it would be interesting to find specific reagents that could inhibit formation of 8-OH-G in DNA by oxygen radicals, or inhibit misreplication in translesion DNA synthesis at the site of the 8-OH-G residue. Another possible alternative would be to use an agonist(s) to enhance the enzymatic activity of OGG1. Although admittedly the successful attainment of such goals would be a challenge, it is certainly worthwhile to consider the pursuit of these long-term objectives.
Acknowledgments
I thank my colleagues who were involved in the studies on 8-OH-G described in this article, while I was in the National Cancer Center Research Institute, Tokyo from 1965 to 1992 and Tsukuba Research Institute, Banyu Pharmaceutical Co. Ltd., Tsukuba from 1992 to 2004. Particularly I thank Dr. H. Kasai who did his pioneering work of discovery of 8-OH-G. I also thank Drs. E. Ohtsuka, H. Aburatani, T. Noda, F. Hanaoka, H. Ohmori, S. Mitra, A. P. Grollman and L. A. Loeb for their important and crucial collaborations with us. I am also grateful to Drs. T. Lindahl, P. C. Hanawalt, R. A. Floyd, and M. Dizdarouglu for their interest and discussion on our work.
Abbreviations:
- 8-OH-G
8-hydroxyguanine;
- 8-OH-dG
8-hydroxydeoxyguanosine;
- HPLC
high performance liquid chromatography.
Profile
Susumu Nishimura was born in 1931 in Tokyo. He graduated from Department of Chemistry, Faculty of Science, University of Tokyo in 1955 and received PhD in Department of Biochemistry and Biophysics, Faculty of Science, University of Tokyo in 1960. After briefly joining in Cancer Institute, Tokyo as a research associate, he did his postdoctral training in Biology Division, Oak Ridge National Laboratory under the late G. David Novelli in 1961–1963, and then in Institute for Enzyme Research, University of Wisconsin in 1963–1965 under H. Gobind Khorana, where he was involved in elucidation of genetic code.

After his postdoctral training in US, he joined National Cancer Center Research Institute, Tokyo in 1965. He worked there for 26 years. In the earlier phase of his study in National Cancer Center Research Institute, he discovered and characterized many hyper-modified nucleosides present in tRNA and elucidated their biological roles in tRNA functions. The example is discovery of queosine with J. A. McClosky, and elucidation of its unique biosynthesis catalyzed by tRNA-guanine transglycosylase.
Later his studies were shifted to cancer. By collaboration with T. Sugimura, Nishimura’s group was involved in characterization of IQ, MeIQ and MeIQx which are potent mutagens/carcinogens produced by broiling fish and meat. In the field of oncogene research, Y. Taya and Nishimura first showed combination of K-ras and myc amplification accompanied by point mutational activation of K-ras in human lung cancer cells. Nishimura was also involved in elucidation of crystal structure of human c-Ha-ras protein, P21, carried out with S.-H. Kim of University of California, Berkeley, and E. Ohtsuka of Hokkaido University. Another notable finding by Nishimura was the discovery of 8-hydroxyguanine with H. Kasai in his laboratory as reviewed in this article.
S. Nishimura moved to Tsukuba Research Institute of Banyu Pharmaceutical Co., Ltd. in 1992 as Director, after mandatory retirement from National Cancer Center Research Institute at the age of 60. There he continued to work on the basic problem on cancer, but also participated in discovery of a new anticancer agent with indolocarbazole structure, targeting topoisomerase 1. From 2004, he moved to University of Tsukuba, and he is now a Visiting Professor at Center for TARA of University of Tsukuba.
S. Nishimura has published more than 450 papers and his work has been cited more than 15,000 times. He is one of the most cited 100 researchers in the field of Biochemistry. He is a foreign honorary member of the American Society for Biochemistry and Molecular Biology, and American Academy of Arts and Sciences. He is also a honorary member of the Japanese Cancer Association, the Japanese Biochemical Society and the Chemical Society of Japan. He was awarded Naito Science Award in 1980, Princess Takamatsu Cancer Research Fund Prize in 1988, and Fujihara Prize in 1990. He is also a recipient of the Imperial Prize and Japan Academy Prize in 1988.
Appendix
Analysis of 8-hydroxydeoxyguanosine:
It is worth emphasizing the need to properly analyze the amount of 8-hydroxydeoxyguanosine (8-OH-dG) present in DNA, isolated from various sources including mammalian tissues, with the minimal amount of artifacts. The lowest amount of background 8-OH-dG (for example, DNA from wild type mice not exposed to exogenous oxidative stress) is approximately 1∼2 8-OH-dG residue per 106 dG of DNA. With such a low background of 8-OH-dG, an increase of 8-OH-dG in tissue DNA can be observed in OGG1 knockout mice with or without exposure to oxidative stress.52) The method utilized in our studies is presently the best procedure to avoid the artificial generation of 8-OH-dG during isolation of deoxynucleosides from tissue DNA. Dai Nakae et al. were the first to show the usefulness of NaI in extracting DNA from tissue.89) Later, Ames and his colleagues endorsed this procedure in a subsequent detailed study.90) It does not appear to matter whether the final quantitative analysis is made by means of HPLC-coupled electrochemical detection or HPLC-MS since the crucial steps in avoiding artifacts are the isolation of DNA and the subsequent conversion of DNA to deoxynucleosides. Consequently, many previous reports have inadvertently expressed the background 8-OH-dG levels as 10 to 100 times above that which it should be. In these cases, the conclusion of the report must be cautiously considered.
Nomenclature:
8-Hydroxyguanine was also later referred to as 7,8-dihydro-8-oxoguanine, since its favored tautomeric confirmation is the 8-oxo-form. However, personally I prefer to use 8-hydroxyguanine rather than 7,8-dihydro-8-oxoguanine. As a comparison, malonaldehyde which is a known DNA modifier has been widely adopted as the name, even though its favoured tautomeric conformation is a hydroxyl form. Additionally, 7,8-dihydro-8-oxoguanine does not actually exist in nature, whereas 8-hydroxyguanine is derived from guanine. Perhaps, 7,8-dihydro-8-oxoguanine is a more biologically biased name. Finally, 8-oxoguanine is an abbreviated term, and therefore not formally correct.
Analysis of urinary 8-OH-dG:
The amount of 8-OH-dG present in the urine of animals is regarded as a reliable marker to assess the extent of exposure to oxidative stress. For example, it was shown that the urinary level of 8-OH-dG from smokers is statistically higher than that of non-smokers.91) However, there is no direct relationship between the amounts of 8-OH-dG in urine and the extent of 8-OH-dG formation in the DNA of tissues, since 8-OH-G is removed from DNA in the form of the base, 8-hydroxyguanine. It is very likely that the removal of 8-OH-G residues in DNA is principally carried out by base-excision repair enzymes such as OGG1. Nucleotide excision repair enzymes, involved in transcription-coupled repair processes may not be a major contributor to 8-OH-G repair in DNA. 8-Hydroxydeoxyguanosine found in urine is perhaps derived from the free nucleotide pool.
Footnotes
The enzyme OGG1, as well as MutM, has two enzymatic activities, glycosylase and AP lyase in a single molecule; see Fig. 4.
The actual mutagens present in heated glucose were later defined as methylreductic acid and hydroxymethylreductic acid.88)
Other groups refer to this as OGG1, because it was isolated by a similarity search using yeast OGG1. We named it MMH based on its similarity to the MutM activity of E. coli. Hereafter, to avoid confusion, we will use the term OGG1.
References
- 1.Sugimura, T. (2004) Establishment of the concept that cancer is a disease of DNA: Serendipitous discoveries in my research career concerning the science of carcinogenesis. Selected Topics in the History of Biochemistry: Personal Recollections VIII. In Comprehensive Biochemistry, Vol 43 (eds. by Semenza G., and Turner A. J.). Elsevier B. V., pp. 355–392. [Google Scholar]
- 2.Sugimura, T., Wakabayashi, K., Nakagama, H., and Nagao, M. (2004) Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 95, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugimura, T. (1997) Overview of carcinogenic heterocyclic amines. Mutation Res. 376, 211–219. [DOI] [PubMed] [Google Scholar]
- 4.Kasai, H., Yamaizumi, Z., Wakabayashi, K., Nagao, M., Sugimura, T., Yokoyama, S., Miyazawa, T., Springan, N. E., Weisburger, J. H., and Nishimura, S. (1980) Potent novel mutagens produced by broiling fish under normal conditions. Proc. Jpn. Acad., Ser. B 56, 278–283. [Google Scholar]
- 5.Kasai, H., Yamaizumi, Z., Wakabayashi, K., Nagao, M., Sugimura, T., Yokoyama, S., Miyazawa, T., and Nishimura, S. (1980) Structure and chemical synthesis of Me-IQ, a potent mutagen isolated from broiled fish. Chem. Lett., 1931–1934. [Google Scholar]
- 6.Kasai, H., and Yamaizumi, Z., Shiomi. T., Yokoyama, S., Miyazawa, T., Wakabayashi, K., Nagao, M., Sugimura, T., and Nishimura, S. (1981) Structure of a potent mutagen isolated from fried beef. Chem. Lett., 485–488. [Google Scholar]
- 7.Kasai, H., Hayami, H., Yamaizumi, Z., Saito, H., and Nishimura, S. (1984) Detection and identification of mutagens and carcinogens as their adducts with guanosine derivatives. Nucl. Acids Res. 12, 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasai, H., and Nishimura, S. (1983) Hydroxylation of the C-8 position of deoxyguanosine by reducing agents in the presence of oxygen. Nucl. Acids Res., Symp. Ser., No 12, s165–s167. [PubMed] [Google Scholar]
- 9.Kasai, H., and Nishimura, S. (1984) Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucl. Acids Res. 12, 2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasai, H., and Nishimura, S. (1984) Hydroxylation of deoxyguanosine at the C-8 position by polyphenols and aminophenols in the presence of hydrogen peroxide and ferric ion. Gann 75, 565–566. [PubMed] [Google Scholar]
- 11.Kohda, K., Tada, M., Kasai, H., Nishimura, S., and Kawazoe, Y. (1986) Formation of 8-hydroxyguanine residues in cellular DNA exposed to the carcinogen 4-nitroquinoline 1-oxide. Biochem. Biophys. Res. Commun. 139, 626–632. [DOI] [PubMed] [Google Scholar]
- 12.Kasai, H., Tanooka, H., and Nishimura, S. (1984) Formation of 8-hydroxyguanine residues in DNA by X-irradiation. Gann 75, 1037–1039. [PubMed] [Google Scholar]
- 13.Setlow, R. B., and Setlow, J. K. (1962) Evidence that ultraviolet-induced thymine dimers in DNA cause biological damage. Proc. Natl. Acad. Sci. USA, 48, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floyd, R. A., Watson, J. J., Wong, P. K., Altmiller, D. H., and Rickard, R. C. (1986) Hydroxyl free radical adduct of deoxyguanosine: sensitive detection and mechanism of formation. Free Radical Res. Commun. 1, 163–172. [DOI] [PubMed] [Google Scholar]
- 15.Kasai, H., Crain, P. F., Kuchino, Y., Nishimura, S., Ootsuyama, A., and Tanooka, H. (1986) Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 7, 1849–1851. [DOI] [PubMed] [Google Scholar]
- 16.Chung, M. H., Kasai, H., Jones, D. S., Inoue, H., Ishikawa, H., Ohtsuka, E., and Nishimura, S. (1991) An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutation Res. 254, 1–12. [DOI] [PubMed] [Google Scholar]
- 17.Tchou, J., Kasai, H., Shibutani, S., Chung, M.-H., Laval, J., Grollman, A. P., and Nishimura, S. (1991) 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. USA 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchino, Y., Mori, F., Kasai, H., Inoue, H., Iwai, S., Miura, K., Ohtsuka, E., and Nishimura, S. (1987) Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 327, 77–79. [DOI] [PubMed] [Google Scholar]
- 19.Shibutani, S., Takeshita, M., and Grollman, A. P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 20.Aida, M., and Nishimura, S. (1987) An ab initio molecular orbital study on the characteristics of 8-hydroxyguanine. Mutation Res. 192, 83–89. [DOI] [PubMed] [Google Scholar]
- 21.Kasai, H., Nishimura, S., Toriumi, Y., Itai, A., and Iitaka, Y. (1987) The crystal structure of 9-ethyl-8-hydroxyguanine. Bull. Chem. Soc. Jpn. 60, 3799–3800. [Google Scholar]
- 22.Oda, Y., Uesugi, S., Ikehara, M., Nishimura, S., Kawase, Y., Ishikawa, H., Inoue, H., and Ohtsuka, E. (1991) NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucl. Acids Res. 19, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouchakdjian, M., Bodepudi, V., Shibutani, S., Eisenberg, M., Johnson, F., Grollman, A. P., and Patel, D. J. (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-oxo-7H-dG (syn), dA (anti) alignment at lesion site. Biochemistry 30, 1403–1412. [DOI] [PubMed] [Google Scholar]
- 24.Lipscomb, L. A., Peek, M. E., Morningstar, M. L., Verghis, S. M., Miller, E. M., Rich, A., Essigmann, J. M., and Williams, L. D. (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. USA 92, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasai, H., Nishimura, S., Kurokawa, Y., and Hayashi, Y. (1987) Oral administration of the renal carcinogen, potassium bromate, specifically produces 8-hydroxydeoxyguanosine in rat target organ DNA. Carcinogenesis 8, 1959–1961. [DOI] [PubMed] [Google Scholar]
- 26.Kasai, H., Okada, Y., Nishimura, S., Rao, M. S., and Reddy, J. K. (1989) Formation of 8-hydroxydeoxyguanosine in liver DNA of rats following long-term exposure to a peroxisome proliferator. Cancer Res. 49, 2603–2605. [PubMed] [Google Scholar]
- 27.Kasai, H., and Nishimura, S. (1984) DNA damage induced by asbestos in the presence of hydrogen peroxide. Gann 75, 841–844. [PubMed] [Google Scholar]
- 28.Yamamoto, F., Kasai, H., Bessho, T., Chung, M. H., Inoue, H., Ohtsuka, E., Hori, T., and Nishimura, S. (1992) Ubiquitous presence in mammalian cells of enzymatic activity specifically cleaving 8-hydroxyguanine-containing DNA. Jpn. J. Cancer Res. 83, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto, F., Kasai, H., Togashi, Y., Takeichi, N., Hori, T., and Nishimura, S. (1993) Elevated level of 8-hydroxydeoxyguanosine in DNA of liver, kidney and brain of Long-Evans cinnamon rats. Jpn. J. Cancer Res. 84, 508–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura, S., and Ames, B. N. (1986) US-Japan meeting on “Oxygen radicals in cancer”. Jpn. J. Cancer Res. (Gann) 77, 843–848. [Google Scholar]
- 31.Michaels, M. L., Pham, L., Cruz, C., and Miller, J. H. (1991) MutM, a protein that prevents GC → TA transversions, is formamidopyrimidine-DNA glycosylase. Nucl. Acids Res. 19, 3629–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bessho, T., Tano, K., Kasai, H., and Nishimura, S. (1992) Deficiency of 8-hydroxyguanine DNA endonuclease activity and accumulation of the 8-hydroxyguanine in mutator mutant (MUTM) of Escherichia coli. Biochem. Biophys. Res. Commun. 188, 372–378. [DOI] [PubMed] [Google Scholar]
- 33.Michaels, M. L., Cruz, C., Grollman, A. P., and Miller, J. H. (1992) Evidence that MutY and MutM combine to prevent mutation by oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89, 7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maki, H., and Sekiguchi, M. (1992) Mut T protein specifically hydrolyzes a potent mutagenic substrate for DNA synthesis. Nature 355, 273–275. [DOI] [PubMed] [Google Scholar]
- 35.Cheng, K. C., Cahill, D. S., Kasai, H., Nishimura, S., and Loeb, L. A. (1992) 8-Hydroxyguanine, an abundant product of oxidative DNA damage, causes G → T and A → C substitutions. J. Biol. Chem. 267, 166–172. [PubMed] [Google Scholar]
- 36.Michaels, M. L., and Miller, J. H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydrooxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174, 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slupska, M. M., Baikalov, C., Luther, W. M., Chiang, J.-H., Wei, Y. F., and Miller, J. H. (1996) Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol. 178, 3885–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakumi, K., Furuichi, M., Tsuzuki, T., Kakuma, T., Kawabata, S., Maki, H., and Sekiguchi, M. (1993) Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxodGTP, mutagenic substrate for DNA synthesis. J. Biol. Chem. 268, 23524–23530. [PubMed] [Google Scholar]
- 39.Chung, M.-H., Kim, H.-S., Ohtsuka, E., Kasai, H., Yamamoto, F., and Nishimura, S. (1991) An endonuclease activity in human polymorphonuclear neutrophils that removes 8-hydroxyguanine residues from DNA. Biochem. Biophys. Res. Commun. 178, 1472–1478. [DOI] [PubMed] [Google Scholar]
- 40.Bessho, T., Tano, K., Kasai, H., Ohtsuka, E., and Nishimura, S. (1993) Evidence for two DNA repair enzymes for 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in human cells. J. Biol. Chem. 268, 19416–19421. [PubMed] [Google Scholar]
- 41.van der Kemp, P. A., Thomas, D., Barbey, R., de Oliveira, R., and Boiteux, S. (1996) Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93, 5197–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nash, H. M., Bruner, S. D., Schärer, O. D., Kawate, T., Addona, T. A., Spooner, E., Lane, W. S., and Verdine, G. L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA repair protein superfamily. Current Biology 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 43.Aburatani, H., Hippo, Y., Ishida, T., Takashima, R., Matsuba, C., Kodama, T., Takao, M., Yasui, A., Yamamoto, K., Asano, M.et al. (1997) Cloning and characterization of mammalian 8-hydroxyguanine-specific glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 57, 2151–2156. [PubMed] [Google Scholar]
- 44.Rosenquist, T. A., Zharkov, D. I., and Grollman, A. P. (1997) Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA 94, 7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodlan-Arjona, T., Wei, Y.-F., Carter, K. C., Klungland, A., Anselmino, C., Wang, R.-P., Augstutand, M., and Lindahl, T. (1997) Molecular cloning and functional expression of a human cDNA encoding the antitumor enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA 94, 7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radicella, J. P., Dherin, C., Desmaze, C., Fox, M. S., and Boiteux, S. (1997) Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94, 8010–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arai, K., Morishita, K., Shinmura, K., Kohno, T., Kim, S.-R., Nohmi, T., Taniwaki, M., Ohwada, S., and Yokota, J. (1997) Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene 14, 2857–2861. [DOI] [PubMed] [Google Scholar]
- 48.Lu, R., Nash, H. M., and Verdine, G. L. (1997) A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Current Biology 7, 397–407. [DOI] [PubMed] [Google Scholar]
- 49.Bjørås, M., Luna, L., Johnsen, B., Hoff, E., Haug, T., Rognes, T., and Seeberg, E. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic site. EMBO J. 16, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takao, M., Aburatani, H., Kobayashi, K., and Yasui, A. (1998) Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucl. Acids Res. 26, 2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monden, Y., Arai, T., Asano, M., Ohtsuka, E., Aburatani, H., and Nishimura, S. (1999) Human MMH (OGG1) type 1 a protein is a major enzyme for repair of 8-hydroxyguanine lesions in human cells. Biochem. Biophys. Res. Commun. 258, 605–610. [DOI] [PubMed] [Google Scholar]
- 52.Minowa, O., Arai, T., Hirano, M., Monden, Y., Nakai, S., Fukuda, M., Itoh, M., Takano, H., Hippou, Y., Aburatani, H.et al. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. USA 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klungland, A., Rosewell, I., Hollenbach, S., Larsen, E., Daly, G., Epe, B., Seeberg, E., Lindahl, T., and Barnes, D. E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurokawa, Y., Hayashi, Y., Maekawa, A., Takahashi, M., Kokubo, T., and Odashima, S. (1983) Carcinogenicity of potassium bromate administered orally to F344 rats. J. Natl. Cancer Inst. 71, 965–972. [PubMed] [Google Scholar]
- 55.Kurokawa, Y., Aoki, S., Matsushima, Y., Takamura, N., Imazawa, T., and Hayashi, Y. (1986) Dose-response studies on the carcinogenicity of potassium bromate in F344 rats after long-term oral administration. J. Natl. Cancer Inst. 77, 977–982. [PubMed] [Google Scholar]
- 56.Arai, T., Kelly, V. P., Minowa, O., Noda, T., and Nishimura, S. (2002) High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis 23, 2005–2010. [DOI] [PubMed] [Google Scholar]
- 57.Arai, T., Kelly, V. P., Komoro, K., Minowa, O., Noda, T., and Nishimura, S. (2003) Cell proliferation in liver of Mmh/Ogg1-deficient mice enhances mutation frequency because of the presence of 8-hydroxyguanine in DNA. Cancer Res. 63, 4287–4292. [PubMed] [Google Scholar]
- 58.Arai, T., Kelly, V. P., Minowa, O., Noda, T., and Nishimura, S. (2006) The study using wild-type and Ogg1 knockout mice exposed to potassium bromate shows no tumor induction despite an extensive accumulation of 8-hydroxyguanine in kidney DNA. Toxicology 221, 179–186. [DOI] [PubMed] [Google Scholar]
- 59.Sakumi, K., Tominaga, Y., Furuichi, M., Xu, P., Tsuzuki, T., Sekiguchi, M., and Nakabeppu, Y. (2003) Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 63, 902–905. [PubMed] [Google Scholar]
- 60.Russo, M. T., De Luca, G., Degan, P., Parlanti, E., Dogliotti, E., Barnes, D. E., Lindahl, T., Yang, H., Miller, J. H., and Bignami, M. (2004) Accumulation of the oxidative base lesion of 8-hydroxyguanine in DNA of tumor-prone mice defective in both Myh and Ogg1 DNA glycosylases. Cancer Res. 64, 4411–4414. [DOI] [PubMed] [Google Scholar]
- 61.Xie, Y., Yang, H., Cunanan, C., Okamoto, K., Shibata, D., Pan, J., Bernes, D. E., Lindahl, T., McIlhatton, M., Fishel, R., and Miller, J. H. (2004) Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-Ras oncogene in lung tumors. Cancer Res. 64, 3096–3102. [DOI] [PubMed] [Google Scholar]
- 62.Cheadle, J. P., Dolwani, S., and Sampson, J. R. (2003) Inherited defects in the DNA glycosylase MYH cause multiple colorectal adenoma and carcinoma. Carcinogenesis 24, 1281–1282. [DOI] [PubMed] [Google Scholar]
- 63.Tsuzuki, T., Egashira, A., Igarashi, H., Iwakuma, T., Nakatsuru, Y., Tominaga, Y., Kawate, H., Nakao, K., Nakamura, K., Ide, Fet al. (2001) Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxodGTPase. Proc. Natl. Acad. Sci. USA 9811456–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakatsu, Y., Yamauchi, K., Isoda, T., Nakabeppu, Y., and Tsuzuki, T. (2005) Oxidative stress-induced intestinal tumors in Mutyh-deficient mice. The Proceedings of Sixty-Fourth Annual Meeting of the Japanese Cancer Association, pp. 1–337. [Google Scholar]
- 65.Hazra, T. K., Izumi, T., Maidt, L., Floyd, R. A., and Mitra, S. (1998) The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucl. Acids Res. 26, 5116–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hazra, T. K., Izumi, T., Boldogh, I., Imhoff, B., Kow, Y. W., Jaruga, P., Dizdaroglu, M., and Mitra, S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. USA 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reardon, J. T., Bessho, T., Kung, H. C., Bolton, P. H., and Sancar, A. (1977) In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in Xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA 94, 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Page, F., Randrianarison, V., Marot, D., Cabannes, J., Perricaudet, M., Feunteun, J., and Sarasin, A. (2000) BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. 60, 5548–5552. [PubMed] [Google Scholar]
- 69.Le Page, F., Klungland, A., Barnes, D. E., Sarasin, A., and Boiteux, S. (2000) Transcription coupled repair of 8-oxoguanine in murine cells: the Ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl. Acad. Sci. USA 97, 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bessho, T., Roy, R., Yamamoto, K., Kasai, H., Nishimura, S., Tano, K., and Mitra, S. (1993) Repair of 8-hydroxyguanine in DNA by mammalian N-methyl-DNA glycosylase. Proc. Natl. Acad. Sci. USA 90, 8901–8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yacoub, A., Augeri, L., Kelley, M. R., Doetsch, P. W., and Deutsch, W. A. (1997) A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J. 15, 2306–2312. [PMC free article] [PubMed] [Google Scholar]
- 72.Ishida, T., Takashima, R., Fukayama, M., Hamada, C., Hippo, Y., Fujii, T., Moriyama, S., Matsuba, C., Nakahori, Y., Morita, H.et al. (1999) New DNA polymorphism of human MMH/OGG1 gene: prevalence of one polymorphism among lung-adenocarcinoma patients in Japanese. Int. J. Cancer 8, 18–21. [DOI] [PubMed] [Google Scholar]
- 73.Sugimura, H., Kohno, T., Wakai, K., Nagura, K., Genka, K., Igarashi, H., Morris, B. J., Baba, S., Ohno, Y., Gao, C. M.et al. (1999) hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol. Biomakers Prev. 8, 669–674. [PubMed] [Google Scholar]
- 74.Paz-Elizur, T., Krupsky, M., Blumenstein, S., Elinger, D., Schechtman, E., and Livneh, Z. (2003) DNA repair activity for oxidative damage and risk of lung cancer. J. Natl. Cancer Inst. 95, 1312–1319. [DOI] [PubMed] [Google Scholar]
- 75.Sieber, O. M., Lipton, L., Crebtree, M., Heiniman, K., Fidalgo, P., Phillips, R. K. S., Bisgaard, M.-L., Orntoft, T. F., Aaltonen, L. A., Hodgson, S. V.et al. (2003) Multiple colorectal adenoma, classic adenomatous polyposis, and germ-line mutations in MYH. New Engl. J. Med. 348, 791–799. [DOI] [PubMed] [Google Scholar]
- 76.Al-Tassan, N., Chemiel, N. H., Maynard, J., Fleming, N., Livingston, A. L., Williams, G. T., Hodges, A. K., Davies, D. R., David, S. S., Sampson, J. R., and Cheadle, J. P. (2002) Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nature Genet. 30, 227–232. [DOI] [PubMed] [Google Scholar]
- 77.Cheadle, J. P., Dolwani, S., and Sampson, J. R. (2003) Inherited defects in the DNA glycosylase MYH cause multiple colorectal adenoma and carcinoma. Carcinogenesis 24, 1281–1282. [DOI] [PubMed] [Google Scholar]
- 78.Prakash, S., and Prakash, L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16, 1872–1883. [DOI] [PubMed] [Google Scholar]
- 79.Kamiya, H., Miura, K., Ohtomo, N., Koda, T., Kakinuma, M., Nishimura, S., and Ohtsuka, E. (1989) Transformation of NIH3T3 cells with synthetic c-Ha-ras genes (1989) Jpn. J. Cancer Res. 80, 200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamiya, H., Murata-Kamiya, N., Koizumi, S., Inoue, H., and Nishimura, S. (1995) 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene: effect of sequence contexts on mutation spectra. Carcinogenesis 16, 883–889. [DOI] [PubMed] [Google Scholar]
- 81.Kamiya, H., Miura, K., Ishikawa, H., Inoue, H., Nishimura, S., and Ohtsuka, E. (1992) c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res. 52, 3483–3485. [PubMed] [Google Scholar]
- 82.Jałloszyński, P., Masutani, C., Hanaoka, F., Perez, A. B., and Nishimura, S. (2003) 8-Hydroxyguanine in a mutational hotspot of the c-Ha-ras gene causes misreplication, ‘action-at-a-distance’ mutagenesis and inhibition of replication. Nucl. Acids Res. 31, 6085–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jałoszyński, P., Ohashi, E.Ohmori, H., and Nishimura, S. (2005) Error-prone and inefficient replication across 8-hydroxyguanine (8-oxoguanine) in human and mouse ras gene fragments by DNA polymerase κ. Genes to Cells 10, 543–550. [DOI] [PubMed] [Google Scholar]
- 84.Haracska, L., Yu, S.-L., Johnson, R. E., Prakash, L., and Prakash, S. (2000) Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nature Genet. 25, 458–461. [DOI] [PubMed] [Google Scholar]
- 85.Saitoh, A., Kimura, M., Takahashi, R., Yokoyama, M., Nomura, T., Izawa, M., Sekiya, T., Nishimura, S., and Katsuki, M. (1990) Most tumors in transgenic mice with human c-Ha-ras gene contained somatically activated transgenes. Oncogene 5, 1195–1200. [PubMed] [Google Scholar]
- 86.Ando, K., Saitoh, A., Hino, O., Takahashi, R., Kimura, M., and Katsuki, M. (1992) Chemically induced forestomach papillomas in transgenic mice carry mutant human c-Ha-ras transgenes. Cancer Res. 52, 978–982. [PubMed] [Google Scholar]
- 87.Bhakat, K. K., Mokkapati, S. K., Boldogh, I., Hazra, T. K., and Mitra, S. (2006) Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 26, 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kasai, H., Nakayama, M., Toda, N., Yamaizumi, Z., Oikawa, J., and Nishimura, S. (1989) Methylreductic acid and hydroxymethylreductic acid: oxygen radical-forming agents in heated starch. Mutation Res. 214, 159–164. [DOI] [PubMed] [Google Scholar]
- 89.Nakae, D., Mizumoto, Y., Kobayashi, E., Noguchi, O., and Konishi, Y. (1995) Improved genomic/nuclear DNA extraction for 8-hydrodeoxyguanosine analysis of small amounts of rat liver tissue. Cancer Lett. 97, 233–239. [DOI] [PubMed] [Google Scholar]
- 90.Helbock, H. J., Beckman, K. B., Shigenaga, M. K., Walter, P. B., Woodall, A. A., Yeo, H. C., and Ames, B. N. (1998) DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-guanosine and 8-oxo-guanine. Proc. Natl. Acad. Sci. USA 95, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loft, S., Vistisen, K., Ewertz, M., Tjønneland, A., Overvad, K., and Poulsen, H. E. (1992) Oxidative DNA-damage estimated by 8-hydroxydeoxyguanosine excretion in humans: influence of smoking, gender and body mass index. Carcinogenesis 13, 2241–2247. [DOI] [PubMed] [Google Scholar]