Abstract
Mexico has one of the highest incidences of childhood leukemia worldwide and significantly higher mortality rates for this disease compared with other countries. One possible cause is the high prevalence of gene rearrangements associated with the etiology or with a poor prognosis of childhood acute lymphoblastic leukemia (ALL). The aims of this multicenter study were to determine the prevalence of the four most common gene rearrangements [ETV6-RUNX1, TCF3-PBX1, BCR-ABL1, and MLL rearrangements] and to explore their relationship with mortality rates during the first year of treatment in ALL children from Mexico City. Patients were recruited from eight public hospitals during 2010–2012. A total of 282 bone marrow samples were obtained at each child's diagnosis for screening by conventional and multiplex reverse transcription polymerase chain reaction to determine the gene rearrangements. Gene rearrangements were detected in 50 (17.7%) patients. ETV6-RUNX1 was detected in 21 (7.4%) patients, TCF3-PBX1 in 20 (7.1%) patients, BCR-ABL1 in 5 (1.8%) patients, and MLL rearrangements in 4 (1.4%) patients. The earliest deaths occurred at months 1, 2, and 3 after diagnosis in patients with MLL, ETV6-RUNX1, and BCR-ABL1 gene rearrangements, respectively. Gene rearrangements could be related to the aggressiveness of leukemia observed in Mexican children.
1. Introduction
Leukemia is the most common cancer in children worldwide, and acute lymphoblastic leukemia (ALL) is the most common subtype, accounting for 80% of all cases [1]. Mexico has two major problems in relation to childhood leukemia: it has one of the highest incidences of childhood leukemia in the world [2], and it has significantly higher mortality rates for this disease compared with other countries [3].
The related factors for these two problems in Mexico are not completely understood. However, it has been suggested that one factor could be the high prevalence of gene rearrangements associated with the etiology or with a poor prognosis of children with ALL [4, 5].
In Mexico City, three studies have reported the frequencies of gene rearrangements in children with ALL. They were single hospital studies based on a small number of cases [4–6]. Pérez-Vera et al. [4] reported a low frequency of the ETV6-RUNX1 gene rearrangement in 57 Mexican patients with leukemia from the Instituto Nacional de Pediatría. In another study by Jiménez-Morales et al. [5], a high proportion of TCF3-PBX1 cases was reported in 53 ALL patients. Finally, Daniel-Cravioto et al. [6], in one of the hospitals at the Instituto Mexicano del Seguro Social (IMSS), reported a high frequency of the MLL-AF4 gene rearrangement, which has been associated with leukemia of a poor prognosis.
When a child is diagnosed with leukemia in Mexico City, the detection of gene rearrangements is not routinely performed. Therefore, the prognostic stratification and the choice of chemotherapy treatment are based on clinical characteristics, laboratory tests, and the immunophenotype [7].
There is no available population level information on the prevalence of gene rearrangements in Mexican patients with childhood ALL.
The aims of this multicenter study were to determine the prevalence of the four most common gene rearrangements [ETV6-RUNX1, TCF3-PBX1, BCR-ABL1, and MLL rearrangements] and to explore their relationship with mortality rates during the first year of treatment in ALL children from Mexico City.
2. Materials and Methods
2.1. Patients
The Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia (MIGICCL) conducted a prospective study of newly diagnosed ALL patients below the age of 19 years between January 1, 2010, and December 31, 2012, in eight public hospitals in Mexico City. The diagnosis of ALL was based on bone marrow morphology and immunophenotyping. Patients were treated with existing local treatment protocols, which differ from one hospital to another. The present study was approved by the National Ethics and Scientific Committees with the following number: 2009-785-001. Informed consent was obtained from the children's parents in accordance with the Declaration of Helsinki.
2.2. Hospitals
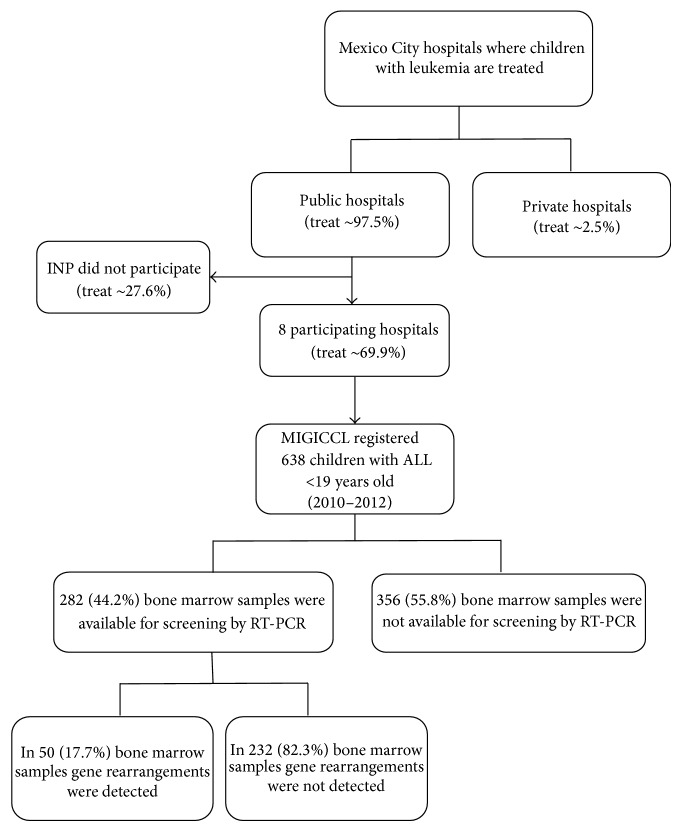
It has been estimated that the majority (97.5%) of children with leukemia are treated in nine public hospitals of Mexico City (Figure 1). The remaining cases are treated at private institutions [2]. The Instituto Nacional de Pediatría (INP) did not participate in this study because approval from the INP Institutional Review Board was not granted.
Figure 1.
Flow chart of the selection process. Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia (MIGICCL) study of newly diagnosed ALL patients below the age of 19 years between January 1, 2010, and December 31, 2012, in eight public hospitals in Mexico City.
Participating hospitals represent four different Mexican Health Institutions: the Hospital de Pediatría, Centro Médico Nacional (CMN) “Siglo XXI,” the Hospital General “Gaudencio González Garza,” CMN “La Raza” and the Hospital General Regional “Carlos McGregor Sánchez Navarro” from the Instituto Mexicano del Seguro Social (IMSS), the Hospital Infantil de México Federico Gómez, the Hospital General de México and the Hospital Juárez de México from the Secretaria de Salud (SSa), the Hospital Pediátrico de Moctezuma from the Secretaría de Salud del Distrito Federal (SSDF), and the Hospital CMN “20 de Noviembre” from the Instituto de Seguridad Social al Servicio de los Trabajadores del Estado (ISSSTE).
2.3. Clinical Data and Definitions
The following clinical data were collected from patient records: information regarding sex, age at diagnosis, immunophenotype classification, white blood cell (WBC) count at diagnosis, and date of the patient's last visit to the hospital or the date of the patient's death as of one year after ALL diagnosis.
According to the National Cancer Institute (NCI) risk classification, patients were classified as having a standard risk [with ages ranging from 1 to 9.99 years and initial white blood cell count (WBC) < 50 × 109/L)] or a high risk [age <1 or ≥10 years and/or initial WBC ≥ 50 × 109/L] [8].
Early mortality was defined as a patient's death within the first year from the time of ALL diagnosis.
2.4. Detection of Gene Rearrangements
The identification of gene rearrangements was carried out at the Unidad de Investigación Médica en Inmunología e Infectología, Hospital de Infectología “Dr. Daniel Méndez Hernández,” “La Raza” from the IMSS.
2.4.1. Cell Lines
In this study, cell lines were obtained from the American Type Culture Collection (ATCC) and used as positive controls for RT-PCR: the t(12; 21) positive cell line Reh (ATCC CRL-8286) (having the ETV6/RUNX1 fusion gene); SUP-B15 (ATCC CRL-1929) (having t(9;22)(q34;q11), t(4;14) (p11;q24), der(4)t(1;4) (p11;q33), t(9;22) (q34;q11), der(10)t(3;10) (q25;q26), and (16); Philadelphia chromosome is present); RS4;11 (ATCC CRL-1873) (with t(4;11)(q21;q23); K-562 (ATCC CCL-243); p210Bcr-Abl p185Bcr-Abl positive cell line for BCR-ABL major and minor, respectively. B1H1 (ATCC 59171) HL-60 promyelocytic leukemia was used as a negative control. The t(1;19) (q23;p13) positive cell line 697 was obtained from DSMZ-German collection of microorganisms and cell cultures (Braunschweig, Germany). All cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics (Invitrogen, Carlsbad, CA) and maintained according to the manufacturer's instructions. The total RNA was extracted from cell lines and used as internal controls (positive and negative).
2.4.2. RNA Isolation and Synthesis of Complementary DNA (cDNA)
Total RNA was extracted from leukemic and normal cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The quality of RNA was confirmed by the presence of intact ribosomal RNA (28s and 18s bands) by denaturing agarose gel electrophoresis and visualized by UV illumination. A total of 3 μg of total RNA was reverse-transcribed using the Superscript one-step RT-PCR with Platinum Taq Kit (Invitrogen Life Technologies, Carlsbad, CA, USA). The cDNA was incubated at 94°C for 10 min to inactivate the reverse transcriptase. Finally, the synthesized cDNA was stored at −20°C until use.
2.4.3. Detection of Gene Rearrangements by Polymerase Chain Reaction (PCR)
Gene rearrangements were detected with a conventional RT-PCR assay as described in Daniel-Cravioto et al. [6] in a previous study. In addition, a commercial Multiplex RT-PCR kit was used according to the manufacturer's instructions (Hemavision, DNA-Technology A/S, Aarhus, Denmark). This screening assay covering 28 different fusion transcripts was used to test for the presence of more than 80 fusion transcript variants. After cDNA synthesis, the PCR amplification was performed in two steps: first, a master PCR amplification followed by nested PCR to screen for the presence of fusion transcripts and second a split-out PCR amplification followed by nested PCR to identify specific fusion transcripts [9, 10]. We obtained one hundred percent concordance between our in-house procedure and the commercially available kit. Because we were only interested in the four rearrangements, we decided, once validated, to use the conventional RT-PCR assay as described in Daniel-Cravioto et al. [6].
3. Statistical Analysis
Statistical analyses were performed using SPSS IBM (Statistical Package for the Social Sciences, Inc., Version 21, Chicago, IL, USA) and relative frequencies regarding the four most frequent gene rearrangements were obtained.
4. Results
From 2010 to 2012, the MIGICCL registered 638 pediatric patients newly diagnosed with ALL from 8 public hospitals of Mexico City of which 334 (52.4%) were male. The entire population median age was 6.2 years (75 months; range from 2 to 222 months). According to the NCI risk classification, 356 patients (55.8%) were classified as standard risk and 282 (44.2%) as high risk. The median WBC at diagnosis was 10.07 × 109/L (range from 0.46 to 970 × 109/L). According to their immunophenotype, 543 (85.1%) patients were classified as having B-cell precursor ALL, 69 (10.8%) patients as having the T-cell immunophenotype, and 26 (4.1%) patients as having biphenotypic leukemia. Early mortality occurred in 86 (13.5%) patients in our cohort study.
The analysis to detect the most frequent gene rearrangements was carried out on 282 (44.2%) available samples (Figure 1), and gene rearrangements were detected in 50 (17.7%) from these patients. Patient characteristics according to molecular subgroups are displayed in Table 1.
Table 1.
Patient characteristics in molecular subgroups of childhood acute lymphoblastic leukemia (ALL). Results from the Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia (MIGICCL).
| Patient characteristics | Gene rearrangement |
Samples not available for screening (n = 356) |
||||
|---|---|---|---|---|---|---|
|
ETV6-RUNX1
(n = 21) |
TCF3-PBX1
(n = 20) |
BCR-ABL1
(n = 5) |
MLL
rearrangements (n = 4) |
Undetected (n = 232) |
||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Gender | ||||||
| Male | 7 (23.3) | 10 (50) | 4 (80) | 1 (25) | 115 (49.5) | 197 (55.3) |
| Female | 14 (66.7) | 10 (50) | 1 (20) | 3 (75) | 117 (50.4) | 159 (44.7) |
| Age (in months) | ||||||
| Range | 31–144 | 9–187 | 15–185 | 3–137 | 5–222 | 2–203 |
| Median age | 75 | 95.5 | 102 | 43 | 84.5 | 68.5 |
| Age groups | ||||||
| <1 year | — | 1 (5) | — | 2 (50) | 4 (1.7) | 14 (3.9) |
| 1–9 years | 18 (85.7) | 12 (60) | 3 (60) | 1 (25) | 158 (68.1) | 237 (66.6) |
| 10–14 years | 3 (14.3) | 5 (25) | 1 (20) | 1 (25) | 51 (22) | 78 (21.9) |
| ≥15 years | — | 2 (10) | 1 (20) | — | 19 (8.2) | 27 (7.6) |
| WBC count ×109/L | ||||||
| Range | 2.2–306 | 0.9–137.6 | 4.6–149.5 | 17.4–970 | 0.4–910 | 0.6–697.8 |
| Median | 15.8 | 21.2 | 31.2 | 210.7 | 10.8 | 9.0 |
| Immunophenotype | ||||||
| B-cell precursor | 20 (95.2) | 19 (95) | 4 (80) | 3 (75) | 205 (88.4) | 292 (82) |
| T-cell | — | — | — | 1 (25) | 24 (10.3) | 44 (12.4) |
| Biphenotypic | 1 (4.8) | 1 (5) | 1 (20) | — | 3 (1.3) | 20 (5.6) |
| NCI risk classification | ||||||
| Standard | 14 (66.7) | 10 (50) | 3 (60) | 1 (25) | 121 (52.2) | 207 (58.1) |
| High | 7 (33.3) | 10 (50) | 2 (40) | 3 (75) | 111 (47.8) | 149 (41.9) |
| Early death (1st year) | ||||||
| Yes | 1 (4.8) | 1 (5) | 2 (40) | 1 (25) | 31 (13.4) | 50 (14) |
| No | 20 (95.2) | 19 (95) | 3 (60) | 3 (75) | 201 (86.6) | 306 (86) |
ETV6-RUNX1 was detected in 21 patients (7.4%), of whom 14 (66.7%) patients were female. The ETV6-RUNX1 gene rearrangement was observed in 18 (85.7%) children under the age of 10 and B-cell precursor was the predominant immunophenotype (Table 1). Moreover, the majority of patients (66.7%) had a standard risk and early mortality occurred in 1 (4.8%) patient with this fusion gene.
The TCF3-PBX1 translocation was present in 20 (7.1%) patients. The B-cell precursor immunophenotype was observed in 19 (95%) patients and 1 patient (5%) had biphenotypic leukemia. In addition, ten patients (50%) with this rearrangement were classified as high risk and 1 (5%) patient died.
The frequency of patients with the MLL rearrangements was 1.4% (n = 4) of which 3 (75%) were female. Three patients presented the MLL-AF4 rearrangement, and one case presented the MLL-AF17 rearrangement. The patients' age ranged between 3 and 137 months with a median of 43 months (3.5 years). These patients had a median WBC of 210.7 × 109/L (range from 17.4 to 970 × 109/L) and 75% (n = 3) of MLL cases were classified as high risk patients. One of the four patients (25%) presented early mortality (Table 1).
The BCR-ABL1 gene rearrangement was detected in five (1.8%) patients, four (80%) were male. The median age at diagnosis was 102 months (range from 15 to 185). Four (80%) patients had a B-cell precursor immunophenotype, and in this molecular subgroup early death occurred in two patients (40%).
Interestingly, the earliest deaths occurred at months 1, 2, and 3 after diagnosis in patients with the MLL, ETV6-RUNX1, and BCR-ABL1 gene rearrangements, respectively.
5. Discussion
This is the first report regarding the prevalence of the four most frequent gene rearrangements in pediatric patients with ALL from eight public hospitals where ~69.9% of the all Mexico City's children with leukemia are treated [2].
5.1. ETV6-RUNX1 Gene Rearrangement
The frequency of ETV6-RUNX1 in the present study (7.4%) is consistent with reports from developing countries. In a previous study from 26 pediatric cases from the Hospital La Raza in Mexico City, the frequency was 3.8% [6]. In India [14] and Brazil [15], reported frequencies were 7% and 11.3%, respectively (Table 2). However, we observed a lower frequency of the ETV6-RUNX1 rearrangement in comparison to reports from developed countries [11–13, 20]. Interestingly, the disparities regarding gene rearrangement prevalence among countries could be explained by environmental factors as playing an important role in the development of childhood leukemia, as would be the case of Mexico [1, 6].
Table 2.
Proportions of the four molecular subgroups reported in the present study, other Mexican studies, and studies from other countries.
| Author, year of publication [Reference] | Country | Number of patients |
ETV6-RUNX1
n(%) |
TCF3-PBX1
n (%) |
MLL rearrangements n (%) |
BCR-ABL1
n (%) |
|---|---|---|---|---|---|---|
| MIGICCL study, 2014 [present study] | Mexico City | 287 | 24 (7.4) | 20 (7.1) | 4 (1.4) | 5 (1.8) |
| Other Mexican studies | ||||||
| Pérez-Vera et al., 2008 [4] | Mexico City | 59 | 5 (8.7) | — | 5 (8.7) | 1 (1.7) |
| Jiménez-Morales et al., 2008 [5] | Mexico City | 53 | 7 (13.5) | 6 (11.5) | — | 2 (3.8) |
| Daniel-Cravioto et al., 2009 [6] | Mexico City | 26 | 1 (3.8) | — | 17 (65.4) | 1 (3.8) |
| Studies from other countries | ||||||
| Amor et al., 1998 [11] | Australia | 66 | 22 (33.0) | — | — | — |
| Zuna et al., 1999 [12] | Czech Republic | 190 | 41 (21.6) | — | — | — |
| Codrington et al., 2000 [13] | England | 56 | 22 (39.0) | — | — | — |
| Siraj et al., 2003 [14] | India | 259 | 18 (7.0) | 18 (7.0) | 2 (4)∗ | 14 (5.0) |
| Mesquita et al., 2003 [15] | Brazil | 88 | 10 (11.4) | 5 (5.7) | 1 (1.1) | 1 (1.1) |
| Pui et al., 2003 [16] | ||||||
| White children | USA | 338 | 64 (18.9) | 10 (3.0) | 10 (3.0) | 8 (2.4) |
| Black children | USA | 68 | 9 (13.2) | 8 (11.8) | 1 (1.5) | 4 (5.9) |
| Aldrich et al., 2006 [17] | ||||||
| Non-Hispanic White | USA | 140 | 34 (24.3) | 6 (4.3) | 4 (2.9) | 1 (0.7) |
| Hispanic | USA | 151 | 19 (12.6) | 7 (4.6) | 3 (2.0) | 2 (1.3) |
| Lazic et al., 2010 [18] | Serbia | 70 | 12 (17.1) | 6 (8.6) | 0 | 7 (10.0) |
| Alonso et al., 2012 [19] | Argentina | 380 | 49 (12.9) | 19 (5.0) | 40 (10.5) | 6 (1.6) |
MIGICCL: Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia.
— Not screened.
∗Other MLL rearrangements were detected in two (4%) of 50 samples analyzed by Southern blot.
Interestingly, ETV6-RUNX1 patients with a B-cell precursor immunophenotype were predominantly female. This finding is consistent with previous reports [21, 22]. Notably, in our study, the majority of ETV6-RUNX1 cases were under the age of 10. It has been reported that ETV6-RUNX1 translocation is most commonly observed in children with leukemia under this age [23].
5.2. TCF3-PBX1 Gene Rearrangement
The proportion of positive patients for the TCF3-PBX1 gene rearrangement represents one of the highest reported worldwide [14–19, 24] (Table 2).Our findings agree with results reported by Jiménez-Morales et al. [5] who found that the frequency of TCF3-PBX1 was 11.5% in children treated at a single institution in Mexico City. In our cohort, the proportion of TCF3-PBX1 is intermediate with respect to those reported for white children (3.0%) and black children (11.8%) [16]. In addition, it has been reported that the TCF3-PBX1 fusion gene is associated with a poor outcome in ALL patients [25, 26]. Therefore, the high frequency of TCF3-PBX1 could possibly explain the high mortality rates observed in Mexican children with ALL.
5.3. MLL Rearrangements
With regards to the translocations involving the chromosome 11q23 gene MLL, a lower frequency was observed (1.4%) compared to that reported by Daniel-Cravioto et al. [6] at the Hospital La Raza of Mexico City (65.4%). However, our findings are consistent with the reported in developed countries (Table 2). The results reported in the work by Daniel-Cravioto et al. [6] regarding the frequency of this gene rearrangement could be explained by an event called random variability, which is very probable when frequencies of extremely rare diseases are reported. For example, in 1996, the incidence of childhood leukemia corresponded to 80 cases per million in Mexico City, and this value was an exceptional and astonishing observation that has not registered again [27]. On the other hand, the median age of the MLL rearrangements subgroup was 42 months, and two patients were less than 1 year of age. These findings are consistent with the literature surmising that this type of gene rearrangement is associated with leukemia in early life [28].
5.4. BCR-ABL1 Gene Rearrangement
We observed a relatively low frequency (1.8%) of the BCR-ABL1 gene rearrangement in comparison to previously reported studies carried out in Mexico City (Table 2). In addition, our findings are similar to those reported in Brazil (1.1%) [15] and Argentina (1.6%) [19]. Interestingly, two BCR-ABL1 cases died during the first year of treatment. This gene rearrangement confers a poor prognosis in ALL patients [24] unless treated with tyrosine kinase inhibitors [TKIs] or hematopoietic stem cell transplant [HSCT]) [26].
5.5. Early Mortality during the First Year of Treatment
In our cohort (n = 638), the mortality rate during the first year of treatment was high (13.5%) considering that 5-year survival rates in developed countries, currently, are higher than 90% [29]. Early mortality was observed in patients with positive gene rearrangements too. In a study conducted by Curado et al. [3] they reported that Mexico is among the few countries worldwide that has failed to reduce the mortality caused by childhood leukemia. The authors also mentioned that in recent years there has been a significant increase in mortality rates for childhood leukemia in this country [3]. Children who are classified as having a lower risk are treated with less intensive chemotherapy, while those classified as having a higher risk might be given a higher dose of chemotherapy than required leading to a higher rate of toxicity and higher mortality rates [26]. The inclusion of gene rearrangement detection in risk-adapted therapy has significantly contributed to improve the survival rate of pediatric patients with ALL in developed countries [25, 30]. However, in the majority of Mexico City public hospitals, current risk allocation schemes do not include the detection of gene rearrangements [7]. This might explain why Mexican children with ALL have higher mortality rates compared to developed countries where molecular techniques are routinely carried out in all patients with leukemia [24]. However, to draw solid conclusions about the impact of gene rearrangements on the survival of Mexican children with ALL, a long-term followup is necessary.
6. Conclusions
We present results from a multicenter study carried out by the MIGICCL on the frequency of the four most common gene rearrangements in Mexican pediatric ALL patients. We found a particularly high prevalence of the TCF3-PBX1 gene rearrangement (associated with a poor prognosis) and a low prevalence of the ETV6-RUNX1 fusion gene (associated with a better prognosis) in comparison to developed countries [25, 31]. Studies on the frequency of gene rearrangements in children with ALL are very heterogeneous; this is most likely due to ethnic and/or environmental factors. Moreover, this is a significant finding, as it may help to explain the high mortality rates observed in Mexican children with ALL; however, further research is needed to elucidate this hypothesis. We recommend the routine detection of gene rearrangements at ALL diagnosis in Mexican children to provide a better prognostic stratification and help to decide the most appropriate treatment, as already implemented in developed countries.
Acknowledgments
This work was supported by the Consejo Nacional de la Ciencia y la Tecnología (CONACYT) through its program, Fondo Sectorial de Investigación en Salud y Seguridad Social (SALUD 2007-1-71223/FIS/IMSS/PROT/592), by the Fondo Sectorial de Investigación para la Educación (CB-2007-1-83949/FIS/IMSS/PROT/616) and by the Instituto Mexicano del Seguro Social (FIS/IMSS/PROT/G10/846, FIS/IMSS/PROT/G12/1134, FIS/IMSS/PROT/PRIO/11/017, and FIS/IMSS/PROT/G11/951). The authors thank the Coordinación de Investigación en Salud of the IMSS for covering the cost of the translation and publishing.
Conflict of Interests
The authors declare that they have no conflict of interests.
References
- 1.Mejía-Aranguré J. M. Novel Aspects in Acute Lymphoblastic Leukemia. 1st. InTech. Rijeka, Croatia: 2011. Childhood acute leukemias in Hispanic population: differences by age peak and immunophenotype; pp. 3–32. [Google Scholar]
- 2.Pérez-Saldivar M. L., Fajardo-Gutiérrez A., Bernáldez-Ríos R., et al. Childhood acute leukemias are frequent in Mexico City: descriptive epidemiology. BMC Cancer. 2011;11:p. 355. doi: 10.1186/1471-2407-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curado M. P., Pontes T., Guerra-Yi M. E., de Camargo Cancela M. Leukemia mortality trends among children, adolescents, and young adults in Latin America. Revista Panamericana de Salud Pública. 2011;29(2):96–102. doi: 10.1590/S1020-49892011000200004. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Vera P., Salas C., Montero-Ruiz O., et al. Analysis of gene rearrangements using a fluorescence in situ hybridization method in Mexican patients with acute lymphoblastic leukemia: experience at a single institution. Cancer Genetics and Cytogenetics. 2008;184(2):94–98. doi: 10.1016/j.cancergencyto.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Jiménez-Morales S., Miranda-Peralta E., Saldaña-Alvarez Y., et al. BCR-ABL, ETV6-RUNX1 and E2A-PBX1: Prevalence of the most common acute lymphoblastic leukemia fusion genes in Mexican patients. Leukemia Research. 2008;32(10):1518–1522. doi: 10.1016/j.leukres.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Daniel-Cravioto A., Gonzalez-Bonilla C. R., Mejia-Arangure J. M., et al. Genetic rearrangement MLL/AF4 is most frequent in children with acute lymphoblastic leukemias in Mexico City. Leukemia and Lymphoma. 2009;50(8):1352–1360. doi: 10.1080/10428190903015636. [DOI] [PubMed] [Google Scholar]
- 7.Dorantes-Acosta E., Zapata-Tarrés M., Miranda-Lora A., et al. Comparación de las características clínicas al diagnóstico de niños con leucemia linfoblástica aguda afiliados al Seguro Popular, con respecto al desenlace. Boletín Médico del Hospital Infantil de México. 2012;69(3):190–196. [Google Scholar]
- 8.Smith M., Arthur D., Camitta B., et al. Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. Journal of Clinical Oncology. 1996;14(1):18–24. doi: 10.1200/JCO.1996.14.1.18. [DOI] [PubMed] [Google Scholar]
- 9.User Manual HemaVision HV01-28N Multiplex RT-PCR test Screens for 28 leukemia causing translocations. 2014, http://www.dna-diagnostic.com/media/10841/manual_hv01_screen_rev2013.12.02.pdf.
- 10.Johnsen H. E., Hoffmann M. H., Klausen T. W., et al. Health technology assessment OF multiplex reverse transcriptase polymerase chain reaction screening FOR translocations AT diagnosis IN acute myeloid leukaemia. European Haematology. 2010;4:39–42. [Google Scholar]
- 11.Amor D. J., Algar E. M., Slater H. R., Smith P. J. High frequency of t(12;21) in childhood acute lymphoblastic leukemia detected by RT-PCR. Pathology. 1998;30(4):381–385. doi: 10.1080/00313029800169666. [DOI] [PubMed] [Google Scholar]
- 12.Zuna J., Hrusák O., Kalinová M., Muzíková K., Starý J., Trika J. TEL/AML1 positivity in childhood ALL: average or better prognosis? Czech paediatric haematology working group. Leukemia. 1999;13(1):22–24. doi: 10.1038/sj.leu.2401256. [DOI] [PubMed] [Google Scholar]
- 13.Codrington R., O'Connor H. E., Reza Jalali G., et al. Analysis of ETV6/AML1 abnormalities in acute lymphoblastic leukaemia: Incidence, alternative spliced forms and minimal residual disease value. British Journal of Haematology. 2000;111(4):1071–1079. doi: 10.1046/j.1365-2141.2000.02464.x. [DOI] [PubMed] [Google Scholar]
- 14.Siraj A. K., Kamat S., Gutierrez M. I., et al. Frequencies of the major subgroups of precursor B-cell acute lymphoblastic leukemia in Indian children differ from the west. Leukemia. 2003;17(6):1192–1193. doi: 10.1038/sj.leu.2402931. [DOI] [PubMed] [Google Scholar]
- 15.Mesquita D. R., Córdoba J. C., Magalhães I. Q., et al. Molecular and chromosomal mutations among children with B-lineage lymphoblastic leukemia in Brazil's Federal District. Genetics and Molecular Research. 2009;8(1):345–353. doi: 10.4238/vol8-1gmr582. [DOI] [PubMed] [Google Scholar]
- 16.Pui C. H., Sandlund J. T., Pei D., et al. Results of therapy for acute lymphoblastic leukemia in black and white children. The Journal of the American Medical Association. 2003;290(15):2001–2007. doi: 10.1001/jama.290.15.2001. [DOI] [PubMed] [Google Scholar]
- 17.Aldrich M. C., Zhang L., Wiemels J. L., et al. Cytogenetics of Hispanic and White children with acute lymphoblastic leukemia in California. Cancer Epidemiology Biomarkers & Prevention. 2006;15(3):578–581. doi: 10.1158/1055-9965.EPI-05-0833. [DOI] [PubMed] [Google Scholar]
- 18.Lazic J., Tosic N., Dokmanovic L., et al. Clinical features of the most common fusion genes in childhood acute lymphoblastic leukemia. Medical Oncology. 2010;27(2):449–453. doi: 10.1007/s12032-009-9232-x. [DOI] [PubMed] [Google Scholar]
- 19.Alonso C. N., Gallego M. S., Rossi J. G., et al. RT-PCR diagnosis of recurrent rearrangements in pediatric acute lymphoblastic leukemia in Argentina. Leukemia Research. 2012;36(6):704–708. doi: 10.1016/j.leukres.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Lightfoot T. J., Roman E. Causes of childhood leukaemia and lymphoma. Toxicology and Applied Pharmacology. 2004;199(2):104–117. doi: 10.1016/j.taap.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 21.Pui C., Carroll W. L., Meshinchi S., Arceci R. J. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. Journal of Clinical Oncology. 2011;29(5):551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enshaei A., Schwab C. J., Konn Z. J., et al. Long-term follow-up of ETV6-RUNX1 ALL reveals that NCI risk, rather than secondary genetic abnormalities, is the key risk factor. Leukemia. 2013;27(11):2256–2259. doi: 10.1038/leu.2013.136. [DOI] [PubMed] [Google Scholar]
- 23.Walsh K. M., de Smith A. J., Welch T. C., et al. Genomic ancestry and somatic alterations correlate with age at diagnosis. American Journal of Hematology. 2014;89(7):721–725. doi: 10.1002/ajh.23727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pui C. H., Mullighan C. G., Evans W. E., Relling M. V. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120(6):1165–1174. doi: 10.1182/blood-2012-05-378943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorman A. V. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Reviews. 2012;26(3):123–135. doi: 10.1016/j.blre.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Teachey D. T., Hunger S. P. Predicting relapse risk in childhood acute lymphoblastic leukaemia. British Journal of Haematology. 2013;162(5):606–620. doi: 10.1111/bjh.12442. [DOI] [PubMed] [Google Scholar]
- 27.Fajardo-Gutiérrez A., Juárez-Ocaña S., González-Miranda G., et al. Incidence of cancer in children residing in ten jurisdictions of the Mexican Republic: importance of the Cancer registry (a population-based study) BMC Cancer. 2007;7, article 68 doi: 10.1186/1471-2407-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greaves M. F., Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nature Reviews Cancer. 2003;3(9):639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 29.Hunger S. P., Lu X., Devidas M., et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. Journal of Clinical Oncology. 2012;30(14):1663–1669. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel R., Desantis C., Virgo K., et al. Cancer treatment and survivorship statistics, 2012. CA: A Cancer Journal for Clinicians. 2012;62(4):220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 31.Maloney K. W., McGavran L., Murphy J. R., et al. TEL-AML1 fusion identifies a subset of children with standard risk acute lymphoblastic leukemia who have an excellent prognosis when treated with therapy that includes a single delayed intensification. Leukemia. 1999;13(11):1708–1712. doi: 10.1038/sj.leu.2401548. [DOI] [PubMed] [Google Scholar]