Abstract
Variation in endocrine signaling is proposed to underlie the evolution and regulation of social life histories, but the genetic architecture of endocrine signaling is still poorly understood. An excellent example of a hormonally influenced set of social traits is found in the honey bee (Apis mellifera): a dynamic and mutually suppressive relationship between juvenile hormone (JH) and the yolk precursor protein vitellogenin (Vg) regulates behavioral maturation and foraging of workers. Several other traits cosegregate with these behavioral phenotypes, comprising the pollen hoarding syndrome (PHS) one of the best-described animal behavioral syndromes. Genotype differences in responsiveness of JH to Vg are a potential mechanistic basis for the PHS. Here, we reduced Vg expression via RNA interference in progeny from a backcross between 2 selected lines of honey bees that differ in JH responsiveness to Vg reduction and measured JH response and ovary size, which represents another key aspect of the PHS. Genetic mapping based on restriction site-associated DNA tag sequencing identified suggestive quantitative trait loci (QTL) for ovary size and JH responsiveness. We confirmed genetic effects on both traits near many QTL that had been identified previously for their effect on various PHS traits. Thus, our results support a role for endocrine control of complex traits at a genetic level. Furthermore, this first example of a genetic map of a hormonal response to gene knockdown in a social insect helps to refine the genetic understanding of complex behaviors and the physiology that may underlie behavioral control in general.
Keywords: Apis mellifera, complex trait genetics, genetic architecture juvenile hormone, social evolution, vitellogenin
The evolution and mechanistic control of complex behaviors is an important question in biology. An emerging model of behavioral control suggests that crosstalk between peripheral tissues and the central nervous system can be important for the regulation of complex behaviors (Badisco et al. 2008; Wang et al. 2009; Page et al. 2012). How such signaling impacts the genetic architecture of the relevant behaviors is not yet well understood, but pleiotropy via endocrine regulation likely plays a key role (Flatt et al. 2005; Amdam et al. 2007). The foraging behavior of worker honey bees (Apis mellifera) is a well-studied example of a complex behavioral trait influenced by hormone cascades regulated by signaling between diverse tissues including brain, fat body, and ovary (Robinson 1987; Amdam et al. 2007; Ament et al. 2008; Wang et al. 2009; Nilsen et al. 2011). The age-associated progression of behavioral roles in honey bee workers is also influenced by the interplay of JH and Vg (Amdam and Omholt 2003; Ihle et al. 2010) The age at which a worker transitions from inside-the-nest tasks to outside foraging is flexible and known to be affected by a variety of factors including genotype (Calderone and Page 1988), the presence of older foragers (Huang and Robinson 1992), reproductive physiology (Nelson et al. 2007), and hormonal dynamics (Robinson 1987). The influence of juvenile hormone and vitellogenin, 2 central physiological regulators of the behavioral transition to foraging, and their mutually suppressive relationship has been particularly well-studied (Robinson 1987; Huang et al. 1994; Amdam et al. 2007; Nelson et al. 2007).
JH is produced in the corpora allata, paired glands at the base of the honey bee brain. It has broad effects on insect development, physiology, and behavior (Robinson 1987; Schmidt-Capella and Hartfelder 1998; Flatt et al. 2005). Vg is a yolk precursor protein synthesized in the honey bee fat body, a loose tissue analogous to vertebrate liver and adipose tissue. This reproduction-associated protein is synthesized at high levels in the facultatively sterile honey bee workers despite their lack of reproduction and undeveloped ovaries (Engels 1974; Rutz and Lüscher 1974). In many insects, JH acts as a gonadotropin and is positively correlated with Vg expression (Flatt et al. 2005). However, in honey bees this dynamic is reversed, and JH and Vg have a mutually suppressive relationship (Page et al. 2012). Young nest bees have low JH titers and high Vg expression. This relationship shifts in foragers as JH titers rise and Vg expression declines (Engels 1974).
This dynamic has been confirmed experimentally: suppression of Vg expression releases high JH titers, although treatment with a JH analogue reduces Vg expression (Pinto et al. 2000; Guidugli et al. 2005). Further, early foraging behavior can be elicited by treatment with JH and its analogs (Robinson 1987), as well as by experimental reduction of Vg via RNA interference (Nelson et al. 2007). However, the mechanisms by which Vg suppresses JH synthesis in the corpora allata and JH inhibits Vg production in the fat body are not yet known.
Experiments with 2 selected lines of honey bees have highlighted the importance of the Vg/JH interaction in the regulation of foraging onset (Amdam et al. 2007). The high and low pollen hoarding strains of Page and Fondrk (1995; reviewed in Page et al. 2012; Page 2013) were selected for the amount of stored pollen in the colony. The disruptive selection for low and high amounts of stored pollen in the colony resulted in strains with divergent behavior and life history, physiology, and gene expression collectively known as the pollen hoarding syndrome (PHS) (Page et al. 2012; Page 2013). Among the traits that vary between the strains are differences in ovary size and the strength of the Vg/JH relationship. Both of these traits have been implicated in the coordination of the PHS (Amdam et al. 2004; Amdam et al. 2007).
The feedback relationship between Vg and JH appears to be strong and intact in the high pollen hoarding strain, whereas it has been disrupted or weakened in the low pollen hoarding strain (Amdam et al. 2007). RNAi-mediated Vg knockdown induces increased JH titers in the high pollen hoarding strain but not in the low pollen hoarding strain (Amdam et al. 2007). Additionally, the strength of the JH response to Vg knockdown is closely correlated with ovary size in the high pollen hoarding strain while in the low pollen hoarding strain this relationship is absent (Amdam et al. 2007). The strain-specific physiological response to Vg knockdown may explain their differences in behavioral response to Vg knockdown. Like wild–type (unselected) workers, high pollen hoarding strain workers respond to Vg knockdown with early foraging onset and increased nectar collection (Ihle et al. 2010). In contrast, in the low pollen hoarding strain Vg knockdowns do not differ behaviorally from controls (Ihle et al. 2010). The differential JH responsiveness to Vg expression in the pollen hoarding strains could thus link ovary size with behavioral maturation and foraging initiation, but the molecular architecture of this relationship is not known (Amdam et al. 2007).
The genetic basis of the PHS has been well-studied (Page et al. 2012). Quantitative trait loci (QTL) have been identified for several traits in the PHS: pln1-4 for pollen hoarding and foraging loading decisions (Hunt et al. 1995, Page et al. 2000, Rüppell et al. 2004); aff3, aff4, and affnew for age of first foraging (Rueppell et al. 2004, Rueppell 2009); wos1-5 for worker ovary size (Graham et al. 2011, Rueppell et al. 2011), and per1 for sucrose responsiveness (Rueppell et al. 2006). In general, these studies have found pleiotropic effects of previously identified QTL and also identified new QTL that are potentially trait-specific (Rueppell 2014). The genetic architecture of the PHS is complex because QTL interact epistatically (Rüppell et al. 2004), pleiotropic relations may be complicated (Page et al. 2012), and transgressive phenotypes have been discovered (Linksvayer et al. 2009).
The PHS is one of the best described animal behavioral syndromes, but its genetic characterization is hampered by genetic heterogeneity because honey bees cannot be inbred. However, the pollen hoarding strains, with their well-studied genetic background and differential JH responsiveness to Vg, provide a unique opportunity to identify the molecular mechanisms that regulate the mutually suppressive relationship between Vg and JH in honey bees. Here, we used a backcross design between the strains to produce a mapping population of workers with broad distributions of JH responsiveness to Vg knockdown. We also studied ovary size, a previously mapped trait with potential endocrine function that may be central to the PHS (Amdam et al. 2007; Wang et al. 2009; Graham et al. 2011). This mapping population was analyzed for QTL with regard to both traits using restriction site-associated DNA (RAD) sequencing (Baird et al. 2008). Our results confirm a number of previously identified QTL for ovary size and reveal pleiotropic effects of some previously mapped QTL on the responsiveness of JH to Vg knockdown. This overlap at the QTL level suggests that worker ovary size and JH responsiveness are at least partly influenced by the same segregating loci. However, the overall phenotypic correlation between ovary size and JH responsiveness was nonsignificant, demonstrating that other genetic and environmental factors may influence either or both traits. Nevertheless, the results reinforce the importance of pleiotropy in the regulation of complex traits, which may in turn lead to trade-offs and constrain trait evolution.
Methods
High Strain Backcross
We derived the mapping population from a backcross of the high and low pollen hoarding strains of the Western honey bee (Page et al. 2012) kept at the Arizona State University Apiaries in Mesa, AZ. A hybrid queen was bred through an experimental cross between a high pollen hoarding strain queen and a drone from the low pollen hoarding strain. Our mapping population was produced by mating this hybrid queen to a male from her maternal high strain colony resulting in a high-strain back cross (HBC) colony. We chose to use a HBC as genetic maps produced for various traits in the PHS have demonstrated that the back-cross to the high strain has been most informative due to dominance interactions and genetic background effects (Hunt et al. 1995; Page et al. 2000; Rueppell et al. 2006; Page et al. 2012). The backcross queen was caged overnight on each of 3 successive nights to allow easy collection of same aged workers. Twenty days post-caging, we collected brood frames and allowed the workers to emerge in an incubator kept at 34 °C. Approximately 1000 newly emerged bees were injected with dsRNA against Vg and 50 injected with control dsRNA against green fluorescent protein (GFP). Bees were injected between the fourth and fifth abdominal tergites and received 10 μg of dsRNA in a 2-μl injection volume as described before (Guidugli et al. 2005; Nelson et al. 2007).
We placed treated bees in 1 of 6 small Plexiglas® cages and returned them to the incubator. Cages were divided in half by a wire-mesh screen. Treated workers were introduced to one-half of the cage and approximately 200 wild–type bees collected from comb containing open brood were put into the other. Bees near open brood cells are likely to be performing brood care or nursing tasks. This arrangement was designed to approximate a social environment and allow the newly emerged, experimental bees to receive nourishment from nurse bees (Nelson et al. 2007). All bees we fed an ad libitum diet of 30% sucrose solution, water, and pollen dough. We allowed bees to mature for 7 days before sample collection and phenotyping.
Preparation of dsRNA
We prepared dsRNA against Vg and GFP as described previously (Guidugli et al. 2005). Briefly, we derived the dsRNA constructs for Vg from cDNA clone AP4a5, and for GFP from the pGFP vector (Clontech, Palo Alto, CA). Primers were fused with T7 promoter sequence (underlined): for Vg:
Fwd: 5′-TAATACGACTCACTATAGGGCGAAC GACTCGACCAACGACTT-3′
Rev: 5′-TAATACGACTCACTATAGGGCGAAA CGAAAGGAACGGTCAATTCC-3′;
and for pGFP:
Fwd: 5′-TAATACGACTCACTATAGGGCGATT CCATGGCCAACACTTGTCC-3′
Rev: 5′-TAATACGACTCACTATAGGGCGATC AAGAAGGACCATGTGGTC-3′.
PCR reactions were performed according to standard procedures. Products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA), and RNA was prepared using the Promega RiboMax T7 system (Promega, Madison, WI), and purified using TRIzol LS reagent (Invitrogen, San Diego, CA) before renaturation and resuspension.
Sample Collection and Phenotyping
We collected 7-day old bees for phenotyping and sample collection. From each bee, we collected fat body tissue for knockdown verification, hemolymph for JH measurement, determined the number of ovarioles per ovary, and collected the thorax for DNA extraction.
Knockdown Verification
We confirmed the efficiency of the Vg knockdown as described before (Amdam et al. 2010; Ihle et al. 2010). Briefly, we isolated RNA from fat body tissue using a protocol combining Trizol (Invitogen) and the RNeasy kit (Qiagen). We measured RNA quality and concentration using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). Relative gene expression levels were determined by one-step reverse transcription polymerase chain reaction (RT-qPCR) using QuantiTect SYBR Green RT-PCR Master Mix kit (Qiagen) and ABI Prism 7500 (Applied Biosystems, Foster City, CA). Relative expression was calculated relative to β-actin. Primers for Vg are: 5′-GTTGGAGAGCAACATGCAGA-3′ and 5′-TCGATCCATTCCTTGATGGT-3′. Primers for β-actin are: 5′-TGCCAACACTGTCCTTTCTG-3′ and 5′-AGAATTGACCCACCAATCCA-3′.
Ovary Size
Ovary dissections were performed as described before (Rueppell et al. 2011). In brief, we removed the abdomen from the rest of the bee carcass and pinned it securely at the anterior and posterior ends to a dissecting tray. We then made incisions along the sides of the abdomen to remove the dorsal tergites. We exposed the ovaries by removing the gut tract. The left and right ovaries were removed, and transferred to a microscope slide for counting. Total ovary size was measured as the sum of ovarioles in the left and right ovaries.
Quantification of JH
JH was extracted and measured using an established radioimmunoassay protocol for honey bee hemolymph (Huang et al. 1994). Briefly, we collected hemolymph samples from individual worker bees using 5μl capillary tubes. Samples were stored in acetonitrile (Sigma-Aldrich, St Louis, MO) at −80 °C until analysis. To extract JH, we added 0.9% NaCl and hexane to the sample which was then separated by centrifugation. The hexane phase containing JH was removed, and the extraction was repeated. We pooled and dried both hexane phases for each sample. The hormone residue was resuspended in 100 μl methanol. Radioimmunoassay was performed according to Jassim et al (2000) and JH titers were calculated according to Huang et al. (1996).
DNA Isolation
The thorax from each bee were flash frozen in liquid nitrogen and stored at −80 °C until DNA extraction. We extracted DNA using DNeasy Blood and Tissue kits (Qiagen). DNA integrity and quantity were measured using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE).
Sequencing and SNP Identification
SNP Discovery and Genotyping
SNP discovery and genotyping were performed by Floragenex/Biota Sciences (Eugene, OR) using RAD tag technology (Baird et al. 2008). Genomic DNA from the hybrid queen mother and 200 worker offspring was digested with the 5-methylcytosine sensitive restriction endonuclease PstI and RAD libraries were constructed according to Floragenex/Biota Sciences standard procedures as previously reported (Baird et al. 2008). The RAD library derived from the queen was then run on a Genome Analyzer II (Illumina, San Diego, CA) using a paired end 2×54bp protocol resulting in 1 528 475 reads. The RAD long-read (LR) protocol (Florogenex/Biota Sciences) was then used to build short DNA contigs from which a large number of SNPs could be identified.
In samples from the worker mapping populations, an individual barcode was added to the resulting small fragments of DNA from each worker. Multiplexed samples were sequenced with a Genome Analyzer II using a single-end 60bp read protocol. More than 100 000 reads were recovered from each individual amounting to a total of 98 447 195bp of raw sequence data. An average of 20 209 RAD-tag markers were initially identified per individual that could be used to call SNP genotypes.
Assembly and QTL Analysis
Assembly
SNP data were quality filtered based on Phred scores, resulting in approximately 7000 identified SNPs in the diploid mother. After further marker exclusion based on biased allele distribution and missing data in the worker offspring, 1415 markers remained that were used to construct a genomic linkage map. This map was assembled using Kosambi’s map function with Mapmaker 3.0b (Lander and Botstein 1989), according to previously described procedures (Rueppell et al. 2004; Rueppell 2009). We combined de novo linkage analysis with information from the marker order on the physical honey bee genome (NCBI taxid: 7460), as determined by BLASTn with standard parameters. Inconsistencies between the 2 methods were resolved on a case by case basis to minimize local map size and markers that expanded the genetic map by more than 5 cM or 5% of the respective interval were excluded (Rueppell et al. 2011).
QTL Mapping
We used MapQTL 4.0 (van Ooijen et al. 2002) for QTL mapping based on the constructed linkage map. In a 2-step evaluation procedure, we first tested the SNPs most closely linked to previously identified QTL for social behavior (Hunt et al. 1995) and ovary size (Graham et al. 2011, Rueppell et al. 2011) for effects on the traits “JH titer” and “Total ovary size” with simple Kruskal–Wallis tests to investigate the a priori hypothesis of pleiotropy. Second, the presence of new QTL was evaluated by subsequent interval mapping, using genome-wide thresholds to determine significant (logarithm of the odds [LOD] > 3) and suggestive (LOD > 2) QTL (Rueppell et al. 2004; Graham et al. 2011). In concordance with the policies of the journal the primary data have been deposited on Dryad (Baker 2013).
Results
Knockdown Verification
We first confirmed that the Vg knockdown was effective. Vg expression was significantly lower in the Vg dsRNA injected bees (n = 12) relative to GFP dsRNA injected controls (n = 12, Mann–Whitney U: U = 26, Z = 2.66, P < 0.008).
Ovary Size Distribution
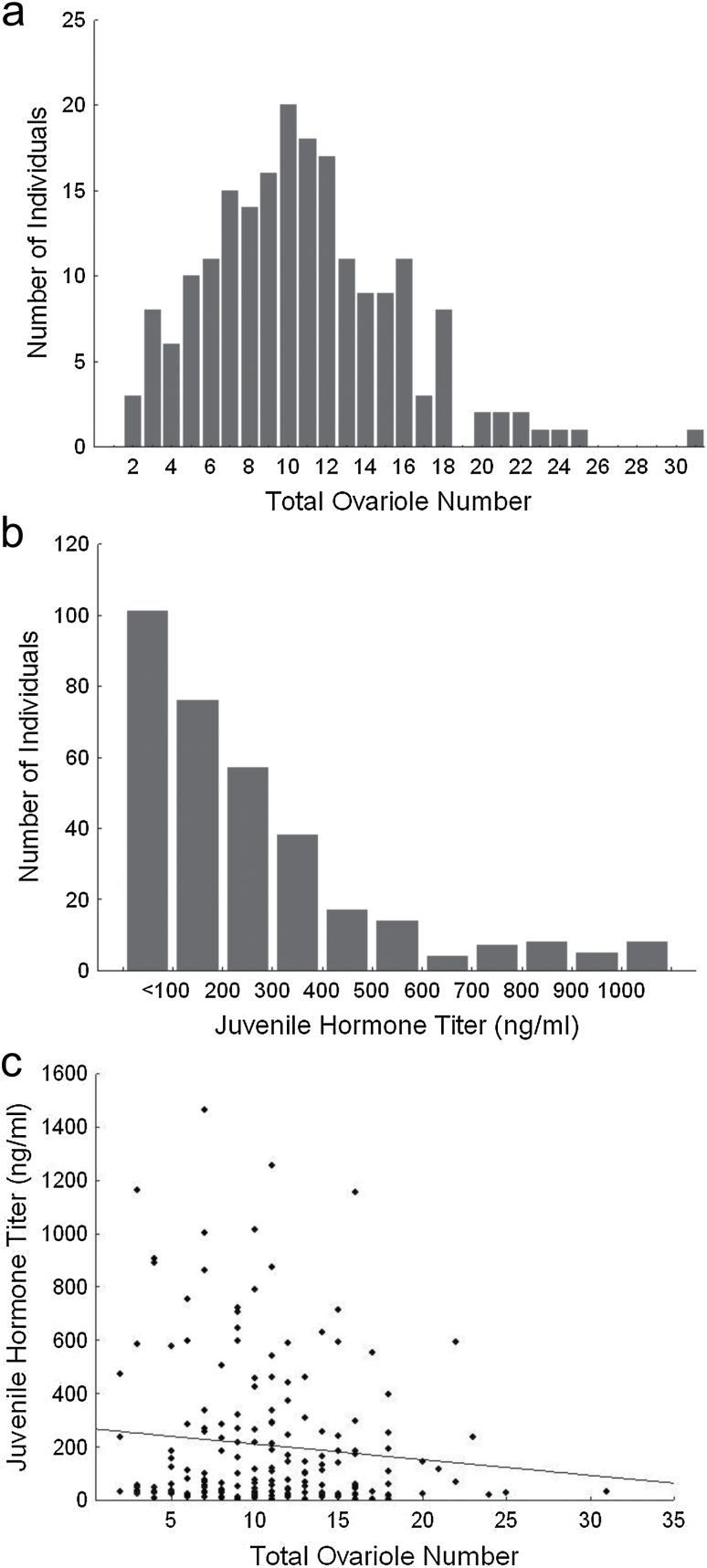
There was broad variation in the distribution of ovary sizes in the mapping population of 200 workers, measured as total number of ovarioles in the left and right ovary. Minimum ovary size was 2 ovarioles and maximum size was 31 ovarioles. The data fit a normal distribution (Kolmogorov–Smirnov: d = 0.9217, P < 0.10). Mean ovary size was 10.9 (SD = 5.1) ovarioles (Figure 1a).
Figure 1.
Phenotype distributions and correlation. (a) Ovary size distribution. Ovary size for each individual is measured as the sum of total ovarioles from the left and right ovary for each individual worker from the approximately 700 workers assayed. Mean ovary size was 10.89 ± SD. 5.078. (b) Juvenile hormone titer distribution. Juvenile hormone titer as measured for individual workers from the overall mapping population. Bars represent bins of individuals with titers greater or equal to the lower bound. Mean juvenile hormone titer was 257.9±253.3ng/ml. (c) Total ovary size and juvenile hormone titer after Vg knockdown were not correlated in our sample (r = −0.103, P = 0.161).
JH Titers
Juvenile hormone titer after Vg knockdown varied widely across the mapping population. Mean JH titer was 257.9ng/mL (SD = 253.3, Figure 1b). While the mean JH titer in our sample is relatively high for young bees (Huang et al. 1994; Guidugli et al. 2005; Amdam et al. 2007), it is similar to the high JH titers previously found in 7-day-old high strain bees after Vg knockdown (Amdam et al. 2007). The individuals with the 100 highest and 100 lowest titers were chosen for SNP genotyping. A final population of 189 individuals was included in the final analyses. Despite the 2 traits’ association in the parental high pollen hoarding line, JH titer was not significantly correlated with ovary size (total ovary size: n = 189, r = −0.103, P = 0.161; Figure 1c).
Genomic Map and QTL Analysis
Linkage Map
After iterative evaluation of the linkage data, 1125 markers and 189 individuals were included in the linkage map, which remained significantly larger than previously published records (Table 1).
Table 1.
Marker distribution and calculated size of each chromosome based on linkage data derived from 1125 markers and 189 individuals
| Chromosome | cM | Markers |
|---|---|---|
| C1 | 802.7 | 129 |
| C2 | 382.4 | 71 |
| C3 | 358.6 | 64 |
| C4 | 390.4 | 71 |
| C5 | 376.8 | 83 |
| C6 | 396.7 | 89 |
| C7 | 328.8 | 62 |
| C8 | 340.8 | 77 |
| C9 | 302.2 | 55 |
| C10 | 313.6 | 79 |
| C11 | 379 | 68 |
| C12 | 296.6 | 63 |
| C13 | 304.6 | 68 |
| C14 | 299.4 | 57 |
| C15 | 251.2 | 44 |
| C16 | 172.2 | 45 |
| All: | 5696 | 1125 |
Effects of Previously Mapped QTL
Single markers linked to 8 of the 12 previously identified QTL of the PHS exhibited significant effects on total ovary size and JH responsiveness to Vg knockdown. All previously identified QTL for honey bee ovary size were associated with an effect in this study. Consistent with ovary size QTL previously identified from the high and low pollen hoarding strains (Rueppell et al. 2011), we confirmed ovary size effects in our samples on chromosomes 2, 3, and 4 (for all P < 0.05; Table 2). We also found ovary size effects in QTL identified in crosses between European and African honey bee stocks (Linksvayer et al. 2009; Graham et al. 2011) on chromosomes 4 (P < 0.05), 6 (P < 0.01), 11 (P < 0.0005 peak 1, P < 0.005 peak 2; Table 2), and 13 (P < 0.05). The ovary size effects on chromosome 11 also overlap the QTL affnew, a region associated with age of first foraging (Rueppell 2009). The ovary QTL on chromosome 13 coincides with pln1 (Hunt et al. 1995; Page et al. 2000). Ovary size was also correlated with the genotype of a marker/SNP near pln4 (Hunt et al. 2007: chromosome 13; P < 0.01; Table 2).
Table 2.
Genetic effects of previously identified QTL on worker ovary size and juvenile hormone response to vitellogenin knockdown
| Trait | QTL | Chromosome | Marker | Mann–Whitney |
|---|---|---|---|---|
| Ovary | wos2 | 2 | B3019 | P < 0.05 |
| wos1 | 3 | C8782 | P < 0.05 | |
| wos3 | 4 | D5963 | P < 0.05 | |
| wos5 | 6 | F1637 | P < 0.01 | |
| wos4 | 11 | K20330 | P < 0.0005 | |
| AFFnew | 11 | K7714 | P < 0.05 | |
| pln1 | 13 | M8560 | P < 0.05 | |
| pln4 | 13 | M7102 | P <0.01 | |
| JH responsiveness | wos2 | 2 | B14160 | P < 0.005 |
| wos1 | 3 | P < 0.0005 | ||
| pln3 | 1 | A222 | P < 0.01 |
We found significant effects on JH responsiveness in QTL regions mapped for ovary size in high and low strain populations on chromosomes 2 (P < 0.005) and 3 (P < 0.0005) (Rueppell et al. 2011). We also found a significant JH responsiveness effect in pln3 (chromosome 1; P < 0.01; Table 2 (Page et al. 2000; Rüppell et al. 2004)).
Whole Genome Scan for QTL by Interval Mapping
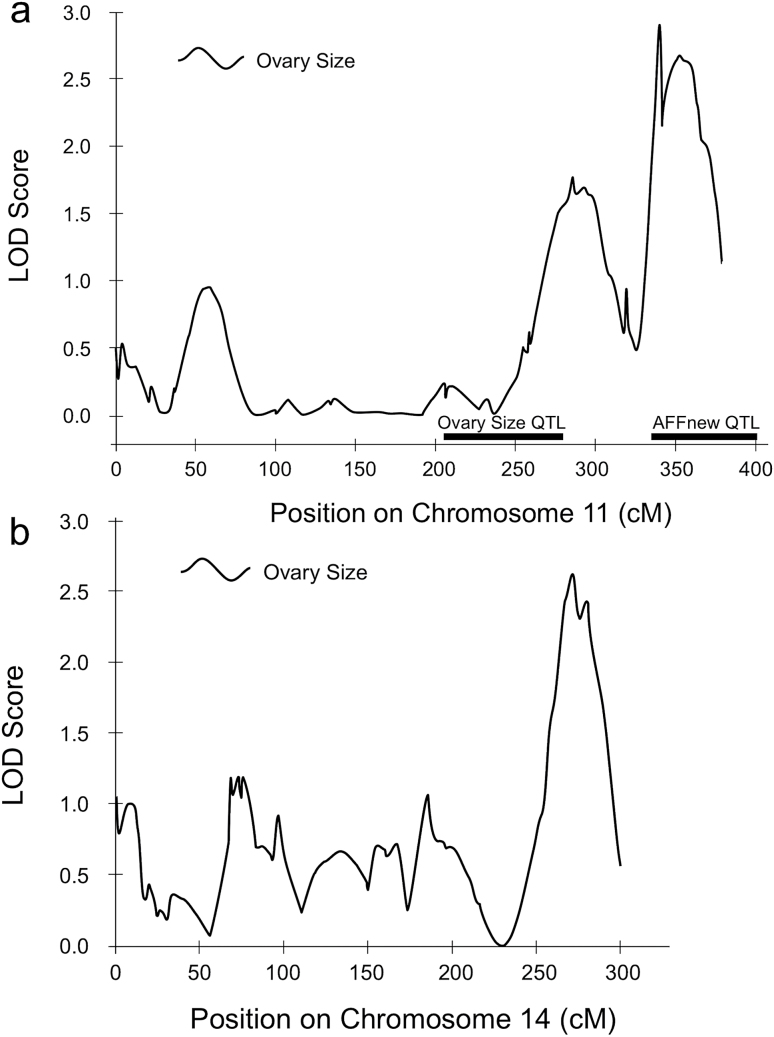
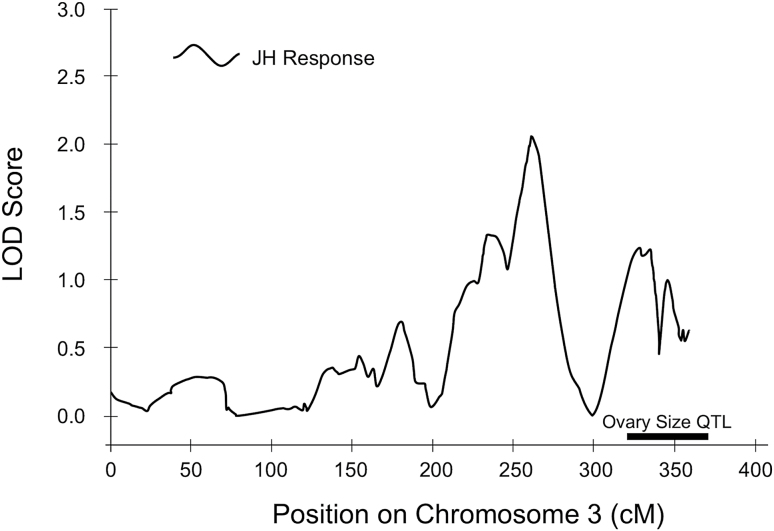
No novel, significant QTL were detected. Two suggestive QTL for total ovary size were identified on chromosome 11 (contigs 11.18; LOD = 2.9 and 11.20; LOD = 2.7; Figure 2a) in a region overlapping QTL previously linked to both ovary size (Graham et al. 2011) and age of first foraging (Rueppell 2009). A third suggestive QTL for total ovary size was located on contig 14.15 (LOD = 2.6; Figure 2b). One suggestive QTL for JH responsiveness to Vg knockdown was located on contig 3.14 (LOD = 2.1; Figure 3), coinciding with a previously mapped QTL for ovary size. Only the suggestive QTL on contig 14.15 was unmapped before this study. Its one-LOD support interval contained only 19 annotated genes. We identified promising genes for future study from the suggestive QTL and especially near significant effects in previously identified QTL for components of the PHS. Especially of interest were genes associated with pathways known to affect the PHS traits (Table 3).
Figure 2.
Suggestive QTL for ovary size on chromosomes 11 (a) and 14 (b). On chromosome 11, there is evidence for overlap with previously identified QTL for both ovary size and age of first foraging. The 1.0 LOD support intervals for the previously identified QTL are denoted by black-labeled bars.
Figure 3.
Suggestive QTL for juvenile hormone responsiveness to vitellogenin knockdown located near a previously identified QTL for worker ovary size on chromosomes 3. The 1.0 LOD support intervals for the previously identified QTL is denoted by a black-labeled bar.
Table 3.
Genes of interest for future study from suggestive QTL or from near markers showing significant effects of JH responsiveness or ovary size in previously identified QTL
| Trait | Chromosome | Gene | Function | Reference(s) |
|---|---|---|---|---|
| Ovary size | 11 (Contig 11:18) | Ceramide kinase-like (GB408315) | Antiapoptotic effects | Tuson et al. (2009) |
| 11 (Contig 11:20) | Nuclear factor related to kappaB- binding protein (GB43163) | Behavioral maturation honey bees | Kucharski and Maleszka (2002) | |
| Inositol 1,4,5-triphosphate kinase (GB43168) | Behavioral maturation honey bees | Kucharski and Maleszka (2002) | ||
| RAC serine/threonine-protein kinase (Akt1: GB43135) | IIS/TOR signaling | Jacinto et al. (2006) | ||
| Target of rapamycin (TOR, GB44905) | Nutrient sensing and growth | Oldham and Hafen (2003); Mutti et al. (2011) | ||
| 14 | Daughterless (GB41727) | ovary follicle development | Cummings and Cronmiller (1994) | |
| JH response | 3 | PH domain leucine-rich repeat protein phosphatase (Phlpp, GB49184) | dephosphorylation of Akt/PKB and PKC | O'Neill et al. (2012) |
| Buffy (GB49154) | Apoptotic/antiapoptotic effects | Quinn et al. (2003); Dallacqua and Bitondi (2014) | ||
| 2 | Big bang (GB55426) | Border follicle cell migration | Aranjuez et al. (2012) | |
| Odorant-binding protein 1 (GB55593) | Pheromone binding | Pesenti et al. (2008) |
Discussion
Worker ovary size is correlated with variation in foraging behavior and has been demonstrated to be a central trait in the PHS (Graham et al. 2011; Page et al. 2012). Ovary size has also been shown to correlate with JH responsiveness to Vg knockdown in a genotype-specific manner (Amdam et al. 2007). In this study we confirmed numerous, small effects of previously identified QTL on ovary size and JH responsiveness to Vg knockdown. Some of these QTL show simultaneous effects on ovary size and JH responsiveness, suggesting a pleiotropic link between the 2 traits, even though we did not find a significant correlation between these traits at the phenotypic level. This discrepancy may be due to other genetic or environmental effects on either trait that are not shared between traits. On the genetic level, we may have identified such effects indicated by the single markers or suggestive QTL that only affect one of the traits. Previously, high strain workers have revealed a phenotypic association between JH responsiveness and ovary size, in contrast to their low strain counterparts (Amdam et al. 2007). We selected our high backcross design based on the previous results but the introgression of low strain genetic material may have disrupted the overall phenotypic correlation despite pleiotropic effects of single loci. Furthermore, environmental effects that uncouple the link between ovary size and JH responsiveness may include nutrition during development or pheromone signals from nestmates.
The effects of previously identified QTL accounted for the majority of the genetic variation in the studied backcross population. Interval mapping, which combines a more sophisticated statistical test with higher significance thresholds than our specific markers tests, identified 3 suggestive QTLs for total ovary size and one suggestive QTL for JH responsiveness to Vg knockdown. One of the 4 was located in a novel genome region, while the other 3 coincided with previously identified QTL. In sum, our results confirm all previously identified QTL for worker ovary size and are consistent with prior findings that the different aspects of the PHS are connected by partial overlap of their genetic architecture (Rüppell et al. 2004; Page et al. 2012).
Ovary size in honey bee workers is highly variable relative to other bee species (Michener 2000), and is influenced by nutritional and physiological, and genetic factors (Beetsma 1985; Linksvayer et al. 2009; Graham et al. 2011; Rueppell et al. 2011). JH itself rescues ovarian progenitor cells from apoptosis late in larval development to influence adult ovary size (Schmidt-Capella and Hartfelder 1998). Because all bees in the mapping population were reared at the same time, in the same colony, we were able to minimize variation due to environmental and indirect genetic effects as much as possible. Nevertheless, our sample included bees with both very small ovaries (2 ovarioles) and large ovaries (31 ovarioles), exceeding typical phenotypes of the parental sources (Rueppell et al. 2011). Highly variable worker ovary sizes, including transgressive phenotypes, have also been identified in backcrosses between European and Africanized honey bees and indicate the disruption of coadapted gene combinations (Linksvayer et al. 2009).
This is the first ovary size mapping study in honey bees that did not identify new major effect QTL (Linksvayer et al. 2009; Graham et al. 2011; Rueppell et al. 2011). Interval mapping identified suggestive QTL for ovary size on chromosome 14 and on chromosome 11 overlapping a region previously liked to both ovary size (Graham et al. 2011) and age of first foraging (Rueppell 2009). Targeted analyses confirmed ovary size effects in all of the 6 QTL for worker ovary size previously identified from crosses between the pollen hoarding strains as well as from commercial stocks of European and Africanized honey bees (Wang et al. 2009; Rueppell et al. 2011; Graham et al. 2011). Earlier studies have reported little or no overlap between ovary size QTL from different crosses (Linksvayer et al. 2009; Graham et al. 2011; Rueppell et al. 2011). Therefore, the results are quite remarkable and may suggest that we have identified most or all regions containing genes with major effects on ovary size. More targeted studies of positional candidate genes are now needed to build a comprehensive understanding of the genetics of ovary size in honey bee workers.
From our data, we identified several promising genes for worker ovary size from the suggestive QTL regions located on chromosomes 11 and 14. Within these regions, there are 36 genes with expression differences between high and low strain worker ovaries (Wang et al. 2012). Daughterless, which is involved in ovary follicle formation and development (Cummings and Cronmiller 1994), is more highly expressed in the ovaries of high strain workers (Wang et al. 2012). Expression of Akt1 (or PKB) is also higher in the high strain (Wang et al. 2012), and it is upregulated during the period of ovary size determination in larvae that will develop into queens relative to those that will become workers (Chen et al. 2012). Akt1 is downstream of both the insulin/insulin-like signaling (IIS) and target of rapamycin (TOR) pathways which are known to affect ovary size in honey bee workers (Mutti et al. 2011). Akt1 integrates signals from both pathways to affect cell growth and other downstream processes through suppression of the forkhead transcription factor Foxo (Jacinto et al. 2006). TOR itself is also present in this region and influences ovary size during development (Mutti et al. 2011).
As predicted from the consistent mutual segregation of traits in the PHS, overlap of QTL and interaction effects between several of these regions suggest pleiotropic connections between phenotypic aspects of the PHS. Consistent with this prediction, we found a high degree of genetic overlap between regions associated with ovary size and foraging behaviors. We identified ovary size effects in pln1, pln4, and affnew linking genetic regulation of ovary size to the previously mapped traits: foraging loading, pollen hoarding and age of first foraging (Hunt et al. 1995; Page et al. 2000; Rueppell et al. 2004; Rueppell 2009). Including our results, direct effects on ovary size have now been identified in all pln (pollen collection and hoarding) QTL as well as in 2 of 3 aff (age of first foraging) QTL (Wang et al. 2009; Graham et al. 2011; Rueppell et al. 2011), confirming that ovary size, a reproductive character, is central to the PHS.
Contrary to our expectations, we did not find any sharply defined QTL for JH responsiveness to Vg knockdown, suggesting that this trait is regulated not by simple genetic control via one or a few genes of major effect, but rather by many genes with small effects. The strongest effects for JH responsiveness were found in the previously identified QTL for ovary size wos1 and wos2 on chromosomes 3 and 2 respectively. Genes from these regions could potentially be involved in the regulation of ovary size and JH responsiveness. On chromosome 3, we identified PHLPP and buffy as promising genes for future study. PHLPP is a phosphatase that can deactivate Akt /PKB and PKC (O’Neill et al. 2012). Both Akt and PKC are components of pathways associated with the PHS. PKC is differentially expressed in the brains of high and low pollen hoarding bees (Humphries et al. 2003), and is a candidate gene for age of first foraging regulation (Rueppell 2009). Akt1, as detailed above integrates signals from the IIS and TOR pathways, which seem increasingly to be central to the regulation of the PHS (Wang et al. 2009; Page et al. 2012). Buffy, a member of the Bcl-2 family (Quinn et al. 2003) is expressed in nearly all developing ovarioles in queen-destined honey bee larvae but in only a few ovarioles in worker destined larvae (Dallacqua and Bitondi 2014). This suggests that Buffy may function antiapoptotically in honey bee larvae. On chromosome 2, big bang and odorant binding protein 1 (Obp1) are potentially interesting for future study. big bang is involved in the migration of follicular border cells during late oogenesis in the fly ovary (upregulated in the ovaries of low strain workers (Aranjuez et al. 2012), and is upregulated in the ovaries of low strain workers (Wang et al. 2012). Obp1 interacts with the main component of queen mandibular pheromone (QMP; Pesenti et al. 2008). QMP influences the behavior and physiology of workers in several ways including delaying behavioral maturation and reducing JH biosynthesis (reviewed in Jarriault and Mercer 2012).
In addition to pleiotropy between ovary size and behavioral components of the PHS, we found JH responsiveness effects in previously known QTL for ovary size (wos1, 2) and foraging behavior (pln3). This along with the presence of candidate genes in pathways known to influence ovary size and social behaviors, in particular the IIS and TOR pathways, suggests that despite a low correlation between ovary size and JH responsiveness in our mapping population, these traits may be under partially overlapping genetic control. Presumably, there are components of JH responsiveness both linked to and independent from ovary size. Phenotypic associations of complex traits that are predicted based on parental phenotype correlations may be difficult to measure (Solovieff et al. 2013) because coadapted gene combinations may be broken up, or the amount of segregating genetic variation in the particular cross is insufficient. Further studies are needed to clarify the relationship between JH responsiveness and ovary size.
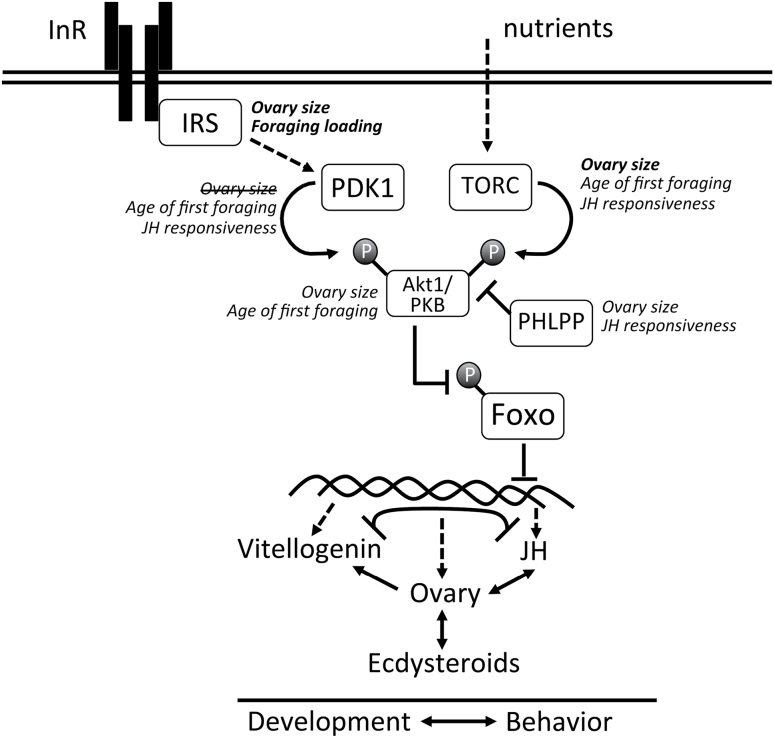
Components of 2 conserved and interacting pathways, IIS and TOR, have been identified as candidate genes for ovary size (Wang et al. 2009; Graham et al. 2011; Rueppell et al. 2011), foraging behavior (Rueppell 2009), and JH responsiveness (Figure 4 in this study). Experimental manipulations have further demonstrated a direct effect of TOR and the insulin receptor substrate (IRS) ovary size (Mutti et al. 2011) and of IRS on foraging behavior (Wang et al. 2010). Experimental work also indicates that these pathways are upstream of Vg and JH in honey bees (Mutti et al. 2011; Nilsen et al. 2011). The abundance of positional candidate genes at the intersection of these 2 pathways suggests that the integration of IIS and TOR signaling is a key mechanism regulating the collection of traits known as the PHS and may mediate the between-tissue signaling which has emerged as an important aspect of behavioral control (Figure 4).
Figure 4.
Simplified illustration of the effects of IIS and TOR pathway components on physiological, developmental and behavioral traits in the pollen hoarding syndrome. Components of both pathways have been identified as positional candidate genes influencing different aspects of worker phenotype (traits in italics). Experimental tests have confirmed (traits in bold) or excluded (traits struck through) a direct relationship between a few of the candidate genes and the traits.
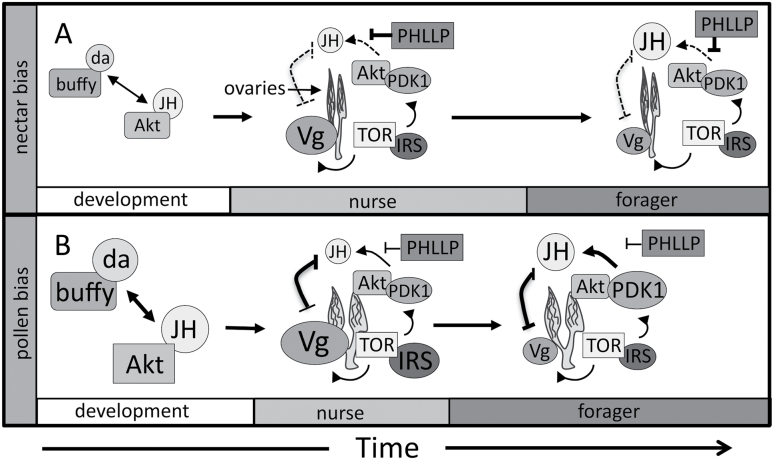
Here, we briefly outline a hypothesis for the mutual regulation of many of the traits that define the behavioral syndrome typified by the high and low pollen hoarding strains. We hypothesize that the complex foraging behavior of adult workers is regulated by cross-talk between ovary, fat body and brain, likely mediated by IIS/TOR signaling and JH action (Figure 5). We argue that the highly variable adult ovary sizes resulting from developmental differences in JH titer, apoptosis-associated genes and potentially hormone sensitivity in turn affect adult behavior, maturation, and hormonal dynamics (Schmidt-Capella and Hartfelder 1998; Amdam et al. 2007; Wang et al. 2009). Workers with larger ovaries, exemplified by the high strain, have higher peak titers of Vg. These initially high titers decrease sharply at the onset of foraging, resulting in increased JH synthesis. In the low strain, with smaller ovaries and decreased JH responsiveness, Vg titers decline slower (Amdam et al. 2007), corresponding with a later transition to foraging behavior. We argue that theses differences in JH responsiveness are mediated by ovary, IIS and TOR signaling (Wang et al. 2009; Ihle et al. 2010; Page et al. 2012; Figure 5). The IIS and TOR pathways converge on Akt1/PKB which requires phosphorylation by the TOR complex and PDK1, a candidate gene in pln3 located near JH responsiveness effects (Hunt et al. 1995; Wang et al. 2009), for activation (Jacinto et al. 2006). Larger ovaries signal increased expression of PDK1 in foragers (Wang et al 2009), potentially increasing JH synthesis downstream. Genes at the intersection of the IIS and TOR pathways TOR, Akt1/PKB, PHLPP, and PDK1, are all present in genetic regions associated with pollen hoarding traits (Rueppell 2009; Wang et al. 2009; Graham et al. 2011; Rueppell et al. 2011), and we hypothesize that functional differences at the intersection of the IIS and TOR pathways mediate the mutual regulation of many of the traits that comprise the PHS. This set of hypotheses must be tested by future studies.
Figure 5.
Hypothesis for the mutual regulation of the pollen hoarding syndrome traits by IIS/TOR signaling and JH action in: (A) late foraging workers who bias foraging loads toward nectar (low strain) and (B) early foraging workers who bias the foraging loads toward pollen (high strain). Differences in JH titer, apoptosis-associated genes, and potentially hormone sensitivity during larval development interact resulting in highly variable adult ovary sizes. Ovary size then influences adult behavior, maturation, and hormonal dynamics. Several genes at the intersection of the IIS and TOR pathways are present in genetic regions associated with pollen hoarding traits. We argue that functional differences at the intersection of the IIS and TOR pathways could result in large differences in how information from these central pathways are integrated and result in the differences in JH responsiveness associated with behavioral maturation and foraging collection observed in the pollen hoarding strains.
Previous work has confirmed the genetic links between ovary size and social behaviors (Wang et al. 2009; Graham et al. 2011), but this study is the first to examine the genetic architecture of the hormonal response to knockdown of Vg, a gene hypothesized to influence complex behaviors through it’s role in intertissue signaling between ovary, fat body and brain. By combining next-generation sequencing with RNAi, we have extended the genetic characterization one of the best described behavioral syndromes in animals, confirming that a complete framework to understand complex behaviors must recognize the critical role of peripheral tissues in behavioral control.
Funding
National Science Foundation Doctoral Dissertation Improvement (0910330 to K.E.I.); Arizona State University. Smithsonian Tropical Research Institute, Research Council of Norway (216776/F11 to L.E.); United Stated Department of Agriculture (NIFA Award #2010-65104-20533 to O.R.); National Institutes of Health (NIGMS: R15GM102753 and NIA: R21AG046837).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Acknowledgments
We thank Colin Brent, Timothy Linksvayer, Julianna Bolzer, Osman Kaftanoglu, Adam Dolezal, and Teague O’Mara for practical help and advice.
References
- Amdam GV, Omholt SW. 2003. The hive bee to forager transition in honeybee colonies: the double repressor hypothesis. J Theor Biol. 223:451–464. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Page RE, Jr, Fondrk MK, Brent CS. 2010. Hormone response to bidirectional selection on social behavior. Evol Dev. 12:428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam GV, Nilsen KA, Norberg K, Fondrk MK, Hartfelder K. 2007. Variation in endocrine signaling underlies variation in social life history. Am Nat. 170:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam GV, Norberg K, Fondrk MK, Page RE., Jr 2004. Reproductive ground plan may mediate colony-level selection effects on individual foraging behavior in honey bees. Proc Natl Acad Sci USA. 101:11350–11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament SA, Corona M, Pollock HS, Robinson GE. 2008. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci USA. 105:4226–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranjuez G, Kudlaty E, Longworth MS, Mcdonald JA. 2012. On the role of PDZ domain-encoding genes in Drosophila border cell migration. G3: Genes Genomes Genetics. 2:1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Badisco L, Claeys I, Van Hiel M, Clynen E, Huybrechts J, Vandersmissen T, Van Soest S, Vanden Bosch L, Simonet G, Vanden Broeck J. 2008. Purification and characterization of an insulin-related peptide in the desert locust, Schistocerca gregaria: immunolocalization, cDNA cloning, transcript profiling and interaction with neuroparsin. J Mol Endocrinol. 40:137–150. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 3:e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetsma J. 1985. Feeding behaviour of nurse bees, larval food composition and caste differentiation in the honey bee (Apis mellifera L.). Fortschr Zool. 31: 407–410. [Google Scholar]
- Calderone N, Page R. 1988. Genotypic variability in age polyethism and task specialization in the honey bee Apis-mellifera (Hymenoptera, apidae). Behav Ecol Sociobiol. 22: 17–25. [Google Scholar]
- Chen X, Hu Y, Zheng H, Cao L, Niu D, Yu D, Sun Y, Hu S, Hu F. 2012. Transcriptome comparison between honey bee queen- and worker-destined larvae. Insect Biochem Mol Biol. 42:665–673. [DOI] [PubMed] [Google Scholar]
- Cummings CA, Cronmiller C. 1994. The daughterless gene functions together with Notch and Delta in the control of ovarian follicle development in Drosophila. Development. 120:381–394. [DOI] [PubMed] [Google Scholar]
- Dallacqua RP, Bitondi MM. 2014. Dimorphic ovary differentiation in honeybee (Apis mellifera) larvae involves caste-specific expression of homologs of ark and buffy cell death genes. PLoS One. 9:e98088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. 1974. Occurrence and significance of Vitellogenins in female castes of social hymenoptera. Integr Comp Biol. 14: 1229–1237. [Google Scholar]
- Flatt T, Tu MP, Tatar M. 2005. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 27:999–1010. [DOI] [PubMed] [Google Scholar]
- Graham AM, Munday MD, Kaftanoglu O, Page RE, Jr, Amdam GV, Rueppell O. 2011. Support for the reproductive ground plan hypothesis of social evolution and major QTL for ovary traits of Africanized worker honey bees (Apis mellifera L.). BMC Evol Biol. 11:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidugli K, Nascimento A, Amdam G, Barchuk A, Omholt S, Simoes Z, Hartfelder K. 2005. Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 579:4961–4965. [DOI] [PubMed] [Google Scholar]
- Huang ZY, Robinson GE. 1992. Honeybee colony integration: worker-worker interactions mediate hormonally regulated plasticity in division of labor. Proc Natl Acad Sci USA. 89:11726–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZY, Robinson GE, Borst DW. 1994. Physiological correlates of division of labor among similarly aged honey bees. J Comp Physiol A. 174:731–739. [DOI] [PubMed] [Google Scholar]
- Huang Z-Y, Robinson GE. 1996. Regulation of honey bee division of labor by colony age demography. Behav Ecol Sociobiol. 39:147–158. [Google Scholar]
- Humphries MA, Muller U, Fondrk MK, Page RE. 2003. PKA and PKC content in the honey bee central brain differs in genotypic strains with distinct foraging behavior. J Comp Physiol A. 189:555–562. [DOI] [PubMed] [Google Scholar]
- Hunt G, Amdam G, Schlipalius D, Emore C, Sardesai N, Williams C, Rueppell O, Guzman-Novoa E, Arechavaleta-Velasco M, Chandra S, et al. 2007. Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften. 94:247-267. [DOI] [PMC free article] [PubMed]
- Hunt GJ, Page RE, Jr, Fondrk MK, Dullum CJ. 1995. Major quantitative trait loci affecting honey bee foraging behavior. Genetics. 141:1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle KE, Page RE, Frederick K, Fondrk MK, Amdam GV. 2010. Genotype effect on regulation of behaviour by vitellogenin supports reproductive origin of honeybee foraging bias. Anim Behav. 79:1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault D, Mercer A. 2012. Queen mandibular pheromone: questions that remain to be resolved. Apidologie. 43:292–307. [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. 2006. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 127:125–137. [DOI] [PubMed] [Google Scholar]
- Jassim O, Huang ZY, Robinson GE. 2000. Juvenile hormone profiles of worker honey bees, Apis mellifera, during normal and accelerated behavioural development. J Insect Physiol. 46:243–249. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. 2002. Molecular profiling of behavioural development: differential expression of mRNAs for inositol 1,4,5-trisphosphate 3-kinase isoforms in naive and experienced honeybees (Apis mellifera). Brain Res Mol Brain Res. 99:92–101. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 121:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer TA, Rueppell O, Siegel A, Kaftanoglu O, Page RE, Jr, Amdam GV. 2009. The genetic basis of transgressive ovary size in honeybee workers. Genetics. 183:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener C. 2000. The bees of the world. Baltimore (MA) The John Hopkins University Press. [Google Scholar]
- Munoz-Torres MC, Reese J, Childers C, Bennett A, Sundaram J, Childs K, Anzola J, Milshina N, Elsik C. 2011. Hymenoptera Genome Database: integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res. 39:D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti N, Dolezal A, Wolschin F. 2011. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J Exp Biol. 214:3977–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. 2007. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 5:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen KA, Ihle KE, Frederick K, Fondrk MK, Smedal B, Hartfelder K, Amdam GV. 2011. Insulin-like peptide genes in honey bee fat body respond differently to manipulation of social behavioral physiology. J Exp Biol. 214:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill AK, Niederst M, Newton A. 2012. Suppression of survival signalling pathways by the phosphatase PHLPP. FEBS J. 280:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Hafen E. 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13:79–85. [DOI] [PubMed] [Google Scholar]
- Page RE. 2013. The Spirit of the hive. Cambridge (MA) Harvard University Press. [Google Scholar]
- Page R, Fondrk M. 1995. The effects of colony level selection of the social-organization of honey-bee (Apis melifera L) colonies - colony level components of pollen hoarding. Behav Ecol Sociobiol. 36:135–144. [Google Scholar]
- Page R, Fondrk M, Hunt G, Guzman-Novoa E, Humphries M, Nguyen K, Greene A. 2000. Genetic dissection of honeybee (Apis mellifera L.) foraging behavior. J Hered. 91: 474–479. [DOI] [PubMed] [Google Scholar]
- Page RE, Jr, Rueppell O, Amdam GV. 2012. Genetics of reproduction and regulation of honeybee (Apis mellifera L.) social behavior. Annu Rev Genet. 46:97–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet J-C, Tegoni M, Cambillau C. 2008. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 380:158–169. [DOI] [PubMed]
- Pinto LZ, Bitondi MM, Simões ZL. 2000. Inhibition of vitellogenin synthesis in Apis mellifera workers by a juvenile hormone analogue, pyriproxyfen. J Insect Physiol. 46:153–160. [DOI] [PubMed] [Google Scholar]
- Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S, Richardson H. 2003. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 22:3568–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. 1987. Regulation of honey-bee age polyethism by juvenile-hormone. Behav Ecol Sociobiol. 20: 329–338. [Google Scholar]
- Rueppell O. 2014. The architecture of the pollen hoarding syndrome in honey bees: implications for understanding social evolution, behavioral syndromes, and selective breeding. Apidologie. 45: 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O, Metheny JD, Linksvayer T, Fondrk MK, Page RE, Jr, Amdam GV. 2011. Genetic architecture of ovary size and asymmetry in European honeybee workers. Heredity (Edinb). 106:894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O, Pankiw T, Nielsen DI, Fondrk MK, Beye M, Page RE., Jr 2004. The genetic architecture of the behavioral ontogeny of foraging in honeybee workers. Genetics. 167: 1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O, Chandra SB, Pankiw T, Fondrk MK, Beye M, Hunt G, Page RE. 2006. The genetic architecture of sucrose responsiveness in the honeybee (Apis mellifera L.). Genetics. 172:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O. 2009. Characterization of quantitative trait loci for the age of first foraging in honey bee workers. Behav Genet. 39:541–553. [DOI] [PubMed] [Google Scholar]
- Rüppell O, Pankiw T, Page RE., Jr 2004. Pleiotropy, epistasis and new QTL: the genetic architecture of honey bee foraging behavior. J Hered. 95:481–491. [DOI] [PubMed] [Google Scholar]
- Rutz W, Lüscher M. 1974. The occurrence of vitellogenin in workers and queens of Apis mellifica and the possibility of its transmission to the queen. J Insect Physiol. 20:897–909. [DOI] [PubMed] [Google Scholar]
- Schmidt-Capella IC, Hartfelder K. 1998. Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J Insect Physiol. 44:385–391. [DOI] [PubMed] [Google Scholar]
- Seeley T. 1995. The wisdom of the hive. Cambridge (MA): Harvard University Press. [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. 2013. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 14:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner EA, Blute TA, Brachmann CB, McCall K. 2011. Bcl-2 proteins and autophagy regulate mitochondrial dynamics during programmed cell death in the Drosophila ovary. Development. 138:327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M, Garanto A, Gonzàlez-Duarte R, Marfany G. 2009. Overexpression of CERKL, a gene responsible for retinitis pigmentosa in humans, protects cells from apoptosis induced by oxidative stress. Mol Vis. 15:168. [PMC free article] [PubMed]
- Wang Y, Amdam GV, Rueppell O, Wallrichs MA, Fondrk MK, Kaftanoglu O, Page RE., Jr 2009. PDK1 and HR46 gene homologs tie social behavior to ovary signals. PLoS One. 4:e4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kocher S, Linksvayer T, Grozinger C, Page RJ, Amdam G. 2012. Regulation of behaviorally associated gene networks in worker honey bee ovaries. J Exp Biol. 215:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mutti NS, Ihle KE, Siegel A, Dolezal AG, Kaftanoglu O, Amdam GV. 2010. Down-regulation of honey bee IRS gene biases behavior toward food rich in protein. PLoS Genet. 6:e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ.1987. Flexible strategy and social evolution. In: Itô Y, Brown JL, Kikkawa J, editors. Animal societies: theories and fact. Tokyo, Japan: Japan Scientific Societies Press. p. 35–51. [Google Scholar]
- West-Eberhard MJ. 2003. Developmental plasticity and evolution. New York: Oxford University Press. [Google Scholar]