Abstract
Functional magnetic resonance imaging (fMRI) in the resting state, particularly fMRI based on the blood-oxygenation level-dependent (BOLD) signal, has been extensively used to measure functional connectivity in the brain. However, the mechanisms of vascular regulation that underlie the BOLD fluctuations during rest are still poorly understood. In this work, using dual-echo pseudo-continuous arterial spin labeling and MR angiography (MRA), we assess the spatio-temporal contribution of cerebral blood flow (CBF) to the resting-state BOLD signals and explore how the coupling of these signals is associated with regional vasculature. Using a general linear model analysis, we found that statistically significant coupling between resting-state BOLD and CBF fluctuations is highly variable across the brain, but the coupling is strongest within the major nodes of established resting-state networks, including the default-mode, visual, and task-positive networks. Moreover, by exploiting MRA-derived large vessel (macrovascular) volume fraction, we found that the degree of BOLD–CBF coupling significantly decreased as the ratio of large vessels to tissue volume increased. These findings suggest that the portion of resting-state BOLD fluctuations at the sites of medium-to-small vessels (more proximal to local neuronal activity) is more closely regulated by dynamic regulations in CBF, and that this CBF regulation decreases closer to large veins, which are more distal to neuronal activity.
Keywords: Cerebral blood flow (CBF), Resting-state BOLD, Arterial-spin labeling (ASL), MR angiography, Blood volume fraction
Introduction
Functional magnetic resonance imaging (fMRI) is a widely used neuroimaging technique for monitoring brain function. The fMRI method noninvasively measures the blood-oxygenation level-dependent (BOLD) effect as an indicator for underlying neuronal activity, reflecting changes in the concentration of paramagnetic deoxy-hemoglobin resulting from neurovascular coupling (Ogawa et al., 1990). More specifically, changes in neuronal activity lead to changes in cerebral metabolic rate of oxygen (CMRO2), with corresponding alterations in cerebral blood flow (CBF) (Fox and Raichle, 1986). fMRI studies of the brain have traditionally focused on task-related BOLD signal changes. However, by assessing the coherence in the spontaneous slow fluctuation of the resting-state BOLD signal, numerous studies have found particular brain regions to be inter-connected while at rest (Biswal et al., 1995; Fox et al., 2005, 2006; Hunter et al., 2006; Nir et al., 2006). Since the first discovery of synchronous low-frequency (<0.1 Hz) spontaneous oscillations of BOLD response in the motor cortex (Biswal et al., 1995), similar types of correlated BOLD signal fluctuations in separate brain regions have been observed, including those within the default-mode network (DMN) (Fox et al., 2005), as well as visual (Nir et al., 2006), auditory (Hunter et al., 2006), and attention system networks (Fox et al., 2006). Correspondingly, the brain in the resting state is known to account for 20% of the body's oxygen consumption (Shulman et al., 2004) and concurrently requires the blood supply for supporting the intrinsic neuronal signaling (Raichle et al., 2001).
Given the consistency of resting-state networks identified by spontaneous BOLD fluctuations, some important questions include whether these resting-state BOLD fluctuations are purely hemodynamic and to what extent they reflect neuronal activity. Although it is well known that task-evoked BOLD responses reflect changes in CBF, CMRO2 and venous blood volume (Davis et al., 1998), the neurovascular origins of resting-state BOLD connectivity are still poorly understood. While several studies have assessed functional connectivity based on resting-state fluctuations in CBF using the arterial-spin labeling (ASL) technique (Biswal et al., 1997; Chuang et al., 2008; Fukunaga et al., 2008; Liang et al., 2013; Viviani et al., 2011; Zou et al., 2009), there has been a limited amount of work focusing on the dynamic relationship between BOLD and CBF fluctuations underlying resting-state networks on either a global or local basis. Fukunaga et al. (2008) were among the first to demonstrate a significant correlation between resting-state BOLD and CBF measurements, but this study was restricted to the primary visual cortex, and the correlation patterns were not spatially specific. In a more recent study by Viviani et al. (2011), group-level CBF–BOLD correlations were estimated as the correlations between BOLD and CBF connectivity maps, obtained independently by seed-based analyses. However, these results did not directly address correlations between temporal dynamics of CBF and BOLD signals, therefore did not elucidate the interpretation of dynamic CBF contribution to the low-frequency resting-state BOLD fluctuations used in computing functional connectivity.
In this work, we aim to (i) quantify the spatio-temporal contribution of cerebrovascular fluctuations to the resting-state BOLD signal on a voxel-wise basis, and (ii) to understand the contribution of static macrovascular content to this resting-state CBF–BOLD relationship. Our hypothesis is that within a resting-state network, dynamic characteristics of spontaneous BOLD oscillations significantly correspond to fluctuations in CBF. This work is novel in several aspects. First, to simultaneously measure CBF and BOLD responses, we used a dual-echo pseudo-continuous arterial spin labeling (pCASL) technique, which also offers higher signal-to-noise ratio (SNR) than conventional pulsed ASL (Dai et al., 2008). Second, to quantitatively assess the dynamic contribution of CBF to the BOLD response, we approximated the BOLD–CBF relationship using a general linear model (GLM), encouraged by prior literature. Within this framework, global vascular fluctuations were included as a covariate to enhance the sensitivity for detecting local vascular contributions to BOLD signal fluctuations that are independent of global vascular pulsatility. Third, in light of the potential association of large draining veins with the BOLD signal, we used MR angiography to assess the influence of resting blood volume fraction in the macrovasculature on the resting-state CBF–BOLD relationship.
Materials and methods
Participants and experimental protocol
This study involved 9 healthy participants (3 men, 6 women), aged from 18 to 32years (mean=26.7, SD=4.3). Participants were recruited through the Baycrest Participants Database, consisting of individuals from the Baycrest and local communities. During the resting-state scan, all participants were instructed to keep their eyes closed and remain awake. The total scan time for the resting-state runs was 350 s. The study was approved by the research ethics board (REB) of Baycrest, and the experiments were performed with the understanding and written consent of each participant, according to REB guidelines.
MRI acquisition
All images were acquired using a Siemens TIM Trio 3 T System (Siemens, Erlangen, Germany). The scans employed 32-channel phased-array head coil reception and body-coil transmission. A 3D T1-weighted anatomical scan was acquired using MPRAGE, with resolution 1 × 1 × 1 mm, repetition time (TR) = 2400 ms, inversion time (TI) =1000 ms, echo time (TE) = 2.43 ms, flip angle = 8°, field of view = 256 × 256 mm (sagittal), matrix size = 256 × 256, 192 slices (ascending order), bandwidth = 180 Hz/pixel, and GRAPPA acceleration factor = 2.
Resting-state CBF and BOLD data were simultaneously acquired with a dual-echo pCASL sequence (Dai et al., 2008). Detailed scanning protocols are as follows: TR = 3500 ms, TECBF/TEBOLD = 10/25 ms, field of view=220×220mm, matrix size=64×64, 18 slices (ascending interleaved order), voxel size = 3.4 × 3.4 × 5.0 mm3, the number of time frames = 100, bandwidth = 2520 Hz/pixel, and GRAPPA acceleration factor = 2. The labeling duration was 1500 ms, and the post-labeling delay was 1000 ms (Alsop and Detre, 1996; Wu et al., 2007) with a mean Gz of 1 mT/m was selected to achieve transit time insensitivity.
To image the blood vessels in the brain, a 3D multi-slab whole-brain time-of-flight (TOF) MR angiography (MRA) images was used with TR = 20 ms, TE = 3.59 ms, field of view = 200 × 181 mm, matrix size = 768 × 696 × 200, spanning six slabs with a distance factor of 20%, TONE ramp=70%, voxel size=0.26×0.26×0.5mm3, bandwidth=165 Hz/pixel, and GRAPPA acceleration factor = 2. No superior saturation band was used in order to image venous contributions as well as arterial.
Physiological recording
Global cardiac pulsation was measured as the pulse oximetry waveform using the Siemens built-in pulse oximeter fixed onto the left index finger. The sampling rate was 50 Hz. Participants were instructed to keep the left hand still during the scans to minimize the movement artifact. The recording was synchronized with the beginning of the acquisition of the first fMRI volume via a scanner-generated trigger signal.
Data analysis
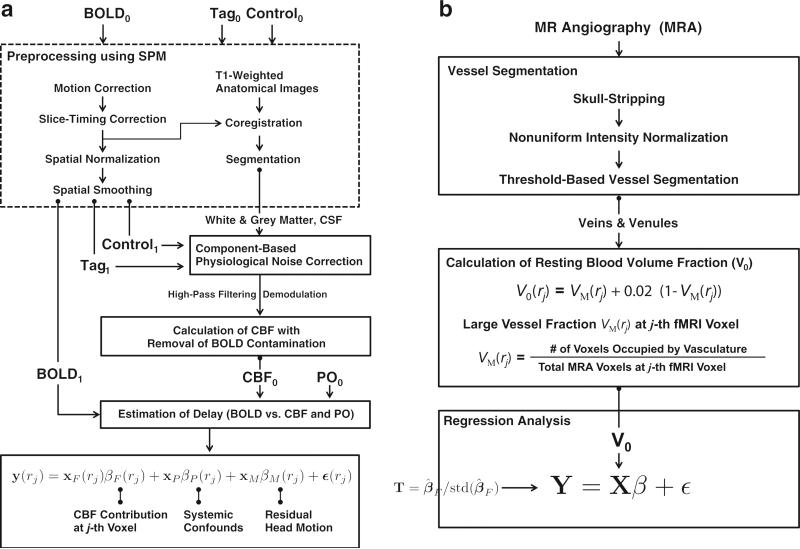
A schematic describing the proposed analysis method is shown in Fig. 1.
Fig. 1.
Schematics of proposed methods. The main modules are: (a) estimation of the CBF contribution to resting-state BOLD signal during resting-state, and (b) linear regression analysis with resting blood volume fraction.
Image preprocessing
The tag, control, and BOLD images in the dual-echo pCASL data were separately preprocessed using SPM8 (Wellcome Trust Centre for Neuro-imaging, London, UK, Friston et al., 2011). The first four time frames were discarded to ensure the MR steady state. The effects of head motion were reduced by spatially realigning all time frames to the first time frame using a least squares approach and a six parameter spatial transformation. A set of realignment parameters was saved for modeling the residual head motion effects on BOLD time frames in the proposed GLM framework. To compensate for slice-acquisition delays, the signal in each slice was realigned temporally to a reference slice (e.g., the middle slice) using sinc interpolation. All time frames were spatially normalized into the Montreal Neurological Institute (MNI) space, and resampled to 2-mm isotropic voxels. Spatial smoothing with a 6-mm full-width at half-maximum (FWHM) Gaussian kernel was applied.
As the contributions of physiological noise to the interleaved tag and control images are different (Restom et al., 2006), the tag and control images were subjected to physiological noise correction separately. Assuming that the physiological noise contribution arising from cardiac pulsation and respiration is globally distributed, and neuronal activity-related signals are low in the white matter and cerebrospinal fluid (CSF), the white matter and CSF were used as a noise regions-of-interest (ROIs) and the signals from these ROIs were used as sources that primarily reflect physiological noise. After segmenting the T1-weighted anatomical image into the different tissue classes, including gray matter, white matter and CSF, we derived four significant principal components from the mean signal in the noise ROI, transferred into the EPI volumes using singular-value decomposition. Physiological noise within the tag and control images was then removed by projecting onto the orthogonal complement of the range space of the noise regressors (Behzadi et al., 2007).
Dynamic CBF change estimation from pCASL data
One of the challenges of CBF estimation in ASL techniques is potential contamination of CBF by BOLD effects, because the CBF-weighted images contain weighting as well. In this work, to reduce BOLD contamination, the modulated CBF component, which is less affected by the BOLD-weighted tissue component, was extracted by high-pass filtering the ASL signal, followed by demodulation. This technique was introduced by Chuang et al. (2008), and was applied successfully in subsequent studies (Nasrallah et al. 2012; Wu et al., 2009; Zou et al., 2009). This approach is a more generalized version of direct subtraction of time-matched upsampled followed by sinc-interpolation of tag and control signals (Aguirre et al., 2002; Liu and Wong, 2005) — sinc subtraction is equivalent to filtering the demodulated ASL data with an ideal low-pass filter.
Specifically, the ASL time series with interleaved tag and control images, S[n], can be approximately described as the sum of an unmodulated component SN[n] and a modulated component SF[n] where the rapid modulation reflects the alternating tag and control images (Chuang et al., 2008; Liu and Wong, 2005),
| (1) |
where
| (2) |
| (3) |
where F[n] denotes CBF, B[n] denotes the BOLD contamination, n is the frame number (odd: tag, even: control); the subscript ‘0’ denotes baseline; M0 is the equilibrium magnetization; β is the signal decay during TR; and k is a constant, k = 2 α exp−δ(R1a −R1)} exp−R1w} (1−exp−R1 τ)}/λ R1, where α is the labeling efficiency, δ is the arterial blood transit time, R1 and R1a represent the longitudinal relaxation rate of tissue and blood, respectively, w is the post labeling delay, τ is the labeling duration, and λ is the brain/blood partition coefficient. Note that i) the BOLD contamination term, ΔB[n], is included only in the unmodulated component SN[n] because the BOLD component is equally present in the tag and control images, and ii) given that the resting-state BOLD and CBF signals are dominated by low-frequency content, the cosine-modulated CBF component SF[n] remains only within the frequency spectrum of S[n] over half of the Nyquist frequency (i.e., 1/4TR). Therefore, to estimate the dynamic CBF changes with removal of BOLD contamination, the unmodulated component SN[n] should be attenuated while the modulated component SF[n] will be preserved. By high-pass filtering (with a cutoff frequency of 0.071Hz) the ASL signal S[n] in Eq. (1), thus isolating it from the low-frequency BOLD contribution, we can extract the modulated component SF[n]. Then, by demodulating SF[n] to a low frequency range through multiplication by cos[πn], the changes in CBF with reduced BOLD contamination are given by
| (4) |
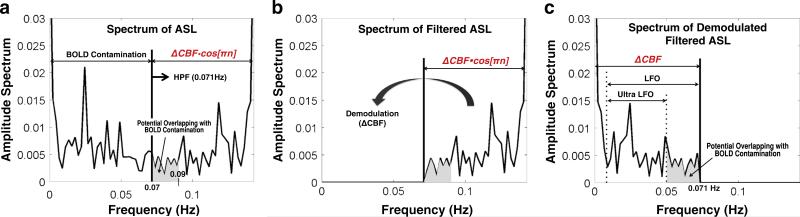
where model parameter c = 2/(kM0B0F0), S’F[n] denotes demodulated SF[n], and the frequency of ΔF[n] is within the range of 0–0.071 Hz. We illustrate this procedure in Fig. 2. Note that within ultra-low frequencies (0.009–0.05 Hz), the potential overlap between the CBF signal and BOLD contamination is minimized (Fig. 2c).
Fig. 2.
Estimation of CBF signals from pCASL data. Single-sided amplitude spectrum of (a) the initial ASL signal, (b) the high-pass filtered ASL signal, and (c) demodulated and high-pass filtered ASL signal. BOLD contamination of the CBF signal was substantially reduced using the procedure introduced in Chuang et al. (2008). However, within the frequency spectrum of the estimated CBF signal (i.e., 0–0.071 Hz), residual BOLD contamination might still be found in the range of 0.05–0.07 Hz (c). Therefore, in order to investigate the BOLD–CBF coupling during resting-state, we also tested the BOLD–CBF coupling using the CBF signal in the low-frequency oscillation range (LFO, 0.009–0.071 Hz) as well as the ultra-low frequency oscillation range (Ultra UFO, 0.009–0.05 Hz).
Dynamic cerebrovascular contributions: general linear model analysis
To assess the local vascular effects (in terms of CBF) on the resting-state BOLD fluctuations, we performed GLM analysis of the BOLD data using a voxel-wise CBF regressor. Our model is based on the assumption of approximate linearity between BOLD and CBF signals during resting state (Fukunaga et al., 2008). Let y(rj) and xF(rj) denote the vectors of BOLD and CBF time series at the j-th voxel, respectively:
| (5) |
where rj is the j-th voxel position, j = 1,..., J, tk is the k-th time frame, k = 1,..., K,ΔB(rj, tk) denotes the changes in BOLD signal, and ΔF(rj, tk) denotes the changes in CBF signal. In our study, BOLD signal fluctuations, ΔB(r, t), were estimated by surround addition of the tag and control signals acquired at the second TE such that at every time point the sum of each signal and the average of its two nearest neighbors is formed (Liu and Wong, 2005; Liu et al., 2013), and the changes in CBF signal, ΔF(r, t), were estimated from the tag and control signals acquired at the first TE using Eq. (4).
Let y(rj), the resting-state BOLD signal at the j-th voxel, be modeled by the following GLM:
| (6) |
where
| (7) |
Here, X(rj) denotes the K × 9 design matrix which contains a covariate of interest (i.e., the CBF signal at voxel j, xF(rj)) and nuisance variables (i.e., the pulse oximetry signal xP, six vectors of spatial transformation associated with residual head motion effects xM1,..., xM6 (Friston et al., 1996; Hutton et al., 2011), and a vector of ones, 1), β(rj) is a 9×1 vector containing unknown model parameters to be estimated, and ε(rj) is a K×1 vector of residuals independent and normally distributed with zero mean and a variance of σ2(rj). The pulse oximetry signal was down-sampled to match the imaging TR. The pulse oximetry signal was found to reflect systemic physiological confounds originating from both cardiac and respiration components of the BOLD signal (Verstynen and Deshpande, 2011). The inclusion of these signals in the GLM enhances the sensitivity for detecting the local contribution of CBF to BOLD. In a comparison to a similar method (Behzadi et al., 2007) which typically uses six regressors estimated from noise ROIs (e.g., white matter and CSF), a downsampled pulse oximetry regressor can provide a more direct assessment of changes in global vascular fluctuation (Mannheimer, 2007) and may reduce the risk of over-parameterization of the GLM estimation (McCullagh and Nelder, 1989; Verstynen and Deshpande, 2011). Therefore, in order to investigate the separate effects of local and global cerebrovascular fluctuation on resting-state BOLD more reliably, we used the pulse oximetry regressor in our linear model of the BOLD response while the noise regressor taken from the white matter and CSF was used for removing the physiological noise from CBF response. Here, all regressors were orthogonalized with respect to each other, and regressor intensities were normalized, setting the sum of squares to unity with zero-mean. In addition, potential time shifts between BOLD and the vascular regressors, including CBF and pulse oximetry, were estimated for each voxel, based on cross-correlation (Chang et al., 2008; Frederick et al., 2012; Fukunaga et al., 2008; Tong and Frederick, 2010, 2012). We selected the shift value that maximizes the statistical significance (in terms of t-statistic) within the physiological range. The ranges of −3.5 s to +3.5 s (Fukunaga et al., 2008) and −7.0 s to +7.0 s (Tong and Frederick, 2010) were considered as valid time shift ranges for CBF and pulse oximetry regressors, respectively. Both vascular regressors were then shifted in time to match the BOLD time course prior to the estimation of GLM parameters.
The magnitude of individual level regression coefficients at the j-th voxel position ( in Eq. (6)) and its variance were estimated with ordinary least squares (OLS) (Friston et al., 1994). Here, the OLS coefficient estimate is proportional to the covariance between BOLD and CBF, which is a measure of how much the two time series change together. The statistical significance was then quantified using t-statistics. In order to calculate the significance of CBF–BOLD associations across individuals, we performed group analysis based on a mixed-effect model (Mumford and Nichols, 2009; Penny and Holmes, 2006). Specifically, combining the individual-level OLS estimates, the group-level t-statistic was calculated using the OLS approach (assuming homogeneity of individual-level variance).
Static cerebrovascular contributions
To assess the contribution of baseline CBF to the resting-state BOLD–CBF relationship, average CBF values were calculated using the ASL Data Processing Toolbox (ASLtbx) (Wang et al., 2008). The association between these baseline CBF maps and the BOLD–CBF was assessed using a voxel-wise GLM. Moreover, we obtained angiographic measurements of macrovascular volume, as described below.
The gradient-echo BOLD signal is widely attributed to an intravascular and an extravascular component, both of which can arise from either the micro- or macrovasculature. The latter, particularly pial vessels, may have a substantial but undesirable weighting on the BOLD signal (Boxerman et al., 1995). While this is widely known, current BOLD acquisitions do not distinguish between macro- and microvascular contributions. Thus, to characterize the effects of vascular content on the resting-state dynamic BOLD–CBF relationship (quantified through t-statistic), we derived resting-state macrovascular fraction, V0.
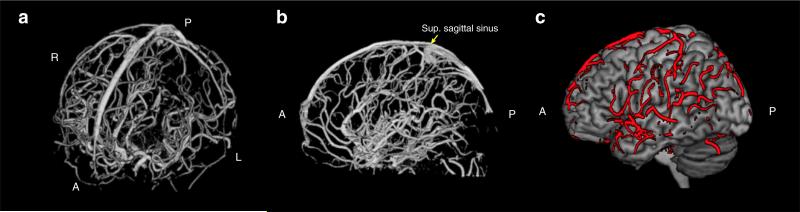
Because in TOF MRA data signal intensities within the blood vessels are higher than those of surrounding tissues, vessel structures were segmented using a threshold-based approach. More specifically, to improve our ability to accurately extract vasculature from the MRA image volume, we removed non-brain tissue such as scalp, skull, and dura (known as “skull-stripping”) using brain-extraction tool (BET2) of FSL software (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/, Jenkinson et al., 2005), and corrected intensity non-uniformity in MRA using nonpara-metric nonuniform intensity normalization (N3) (Sled et al., 1998). We then extracted the vessel structure from the remaining MRA image volume using thresholding. An example of the resulting vascular map is shown in Fig. 3, demonstrating that the MRA data was able to capture both pial vessels and intracranial vessels.
Fig. 3.
3D visualization of the vessel segmentation obtained from the MR angiography scan. (a) Anterior–dorsal view of the vascular tree; (b) sagittal view of the medial and intracranial vessels; and (c) sagittal view of the pial vessels located at the surface of cortex, outlined in red. Volume rendering of the blood vessels was performed using MRIcron (http://www.mccauslandcenter.sc.edu/mricro/mricron/). Abbreviations: A: anterior, P: posterior, L: left, R: right, Sup: superior.
Then, we performed a linear regression analysis. Resting blood volume fraction V0 is typically defined as the ratio of blood vessels to tissue volume in the fMRI measurements (Buxton et al., 1998), and V0 can be calculated as the combination of small-vessel fraction VS and macrovascular fractions VM (Hu et al., 2012a, b):
| (9) |
where rj is the j-th voxel position in the fMRI image volume, j = 1,..., J, the small-vessel fraction VS is assumed as 0.02 (Friston et al., 2000; Hu et al., 2012a, b), and macrovascular (large-vessel) fraction at the j-th voxel, VM(rj), can be assessed by using the vasculature segmentations based on the MRA image volumes:
| (10) |
where NV(rj) is the number of voxels occupied by the segmented vasculature at the j-th fMRI voxel and NA is the number of MRA voxels at each voxel of fMRI volume. Note that as the MRA images were spatially normalized into the MNI space, and resampled to a 0.5-mm isotropic grid, the resulting voxel size of the MRA data (0.5 × 0.5 × 0.5 mm3) is much smaller than the voxel size of our fMRI dataset (2 × 2 × 2 mm3 after re-sampling). Therefore, in our dataset, NA was 64 for all voxels, NV(rj) was within a range of 0 to1, and the corresponding value of VM(rj) was determined within a range of [0, 1/64, ...,63/64, 64/64], which leads to 64 bins of V0(rj). Using resting blood volume fraction V0 and group-average of corresponding t-statistics (that fall into each bin of V0), we performed linear regression analysis to investigate the static blood volume effects on the degree of coupling between CBF and BOLD fluctuations during resting-state.
Results
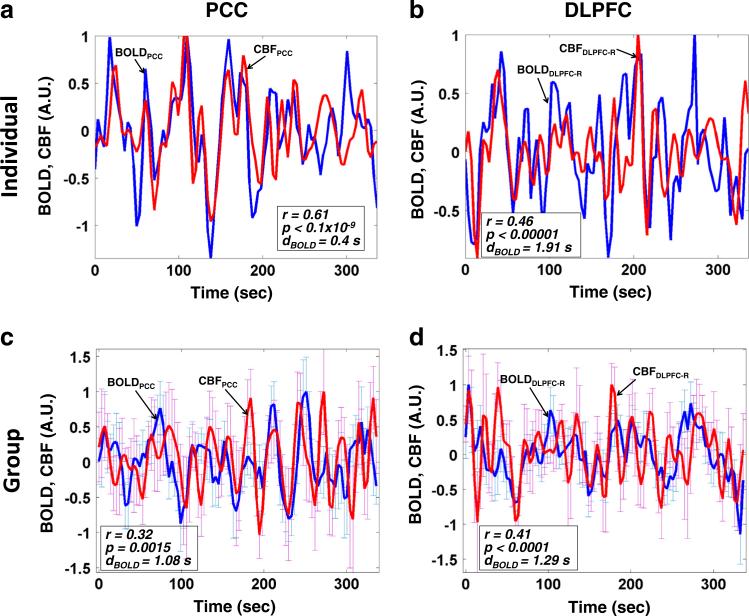
The average BOLD and CBF time-series in the posterior cingulate cortex (PCC) and the right dorsolateral prefrontal cortex (DLPFC) are shown in Fig. 4. PCC, a prominent and robustly observed node of the DMN, was selected as an 8 mm-radius sphere centered at coordinates x = −6, y = −58, z = 28 in MNI305 space (Toro et al., 2008). We also selected part of Brodmann area 46 as the right DLPFC (DLPFC-R), a main node of the so-called “task-positive network” — a set of regions that are positively correlated with cognitive and attention tasks but consistently and negatively correlated with the DMN (Fox et al., 2005). The task-positive network typically consist of the DLPFC, intraparietal sulcus (IPS), inferior precentral sulcus (IPCS), dorsal anterior cingulate cortex (dACC), middle temporal region (MT), premotor cortex, and supplementary motor area (Chang and Glover, 2009; Fox et al., 2005, 2009). In order to better visualize the comparison between the BOLD and CBF time courses, we normalized both signals to unity maximum in Fig. 4, which shows that the spontaneous BOLD fluctuations in PCC are consistent with the CBF fluctuation changes, in agreement with our hypothesis. Of note, in addition to the DMN, the positively correlated patterns between CBF and BOLD fluctuations during resting-state were also observed in the right DLPFC, part of the task-positive network. As a further validation of our findings, we also noted that the relationship between our pulse-oximetry regressor and the resting BOLD signal is similar to previous findings (Tong and Frederick, 2010, 2012; Verstynen and Deshpande, 2011).
Fig. 4.
Regional-mean time series of BOLD (blue) and CBF (red) in select brain regions. We examined the posterior cingulate cortex (PCC, (a, c)) and the right dorsolateral prefrontal cortex (DLPFC-R, (b, d)). The time series for one illustrative subjective are shown in (a) and (b), whereas the corresponding group-average time courses are shown in (c) and (d). The error bars represent the standard error of the mean (SEM) across individuals at each time point.
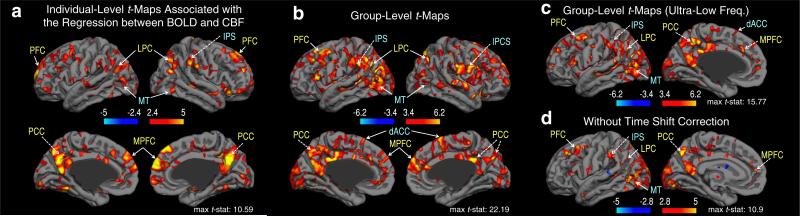
The t-statistic maps demonstrating the spatial variability in the strength of BOLD–CBF associations are shown in Fig. 5; a sample individual t-map (uncorrected p < 0.01) and the corresponding group t-maps (uncorrected p < 0.005) are shown in Figs. 5a and b, respectively. Volumetric t-statistics are overlaid on a cortical surface atlas (supplied by FreeSurfer (http://surfer.nmr.mgh.harvard.edu), Dale et al., 1999; Fischl et al., 1999). While the coupling between BOLD and CBF during resting-state was highly variable across the brain, the most significantly positive coupling was found in the regions of DMN, including PCC, me-dial prefrontal cortex (MPFC), and lateral parietal cortex (LPC). Moreover, we observed a strong positive coupling between BOLD and CBF fluctuations within the visual network (the primary visual area), and the task-positive network, including the IPS, IPCS, dACC and MT regions. We further tested the BOLD–CBF coupling within ultra-low frequencies (0.009–0.05 Hz, where there is minimal potential overlap between CBF signal and BOLD contamination), as shown in Fig. 5c. Moreover, a map of BOLD–CBF coupling without compensating for time shifts between BOLD and the vascular regressors (i.e. CBF and pulse oximetry waveforms) is shown in Fig. 5d. In all cases, while statistical significance varied, the spatial distribution of significantly positive BOLD–CBF coupling remained consistent with the major nodes of the DMN and the task-positive network (Figs. 5c and d). These results suggest that our estimated BOLD–CBF coupling during resting state is not likely to be dominated by BOLD–CBF cross-contamination and time-shift correction, and represents synchronized oscillations of BOLD and CBF.
Fig. 5.
Association between resting-state BOLD and CBF time course: t-statistics. (a) A sample individual t-map (uncorrected p < 0.01), and the corresponding group t-maps for testing the BOLD–CBF coupling of (b) low-frequency oscillations (0.009–0.071 Hz, uncorrected p < 0.005), (c) ultra-low frequency oscillations (0.009–0.05 Hz, uncorrected p < 0.005), and (d) low-frequency oscillations without shifting the BOLD signal with respect to the vascular regressors, including CBF and pulse oximetry (uncorrected p < 0.01). Note that CBF and BOLD are significantly consistent in prominent resting-state networks (including the DMN and visual network, labeled in yellow) as well as in the task-positive network (marked in cyan). Moreover, within ultra-low frequencies minimally overlapped with the frequency band of BOLD contamination, the spatial distribution of BOLD–CBF coupling remained consistent with the major nodes of DMN and task-positive network. In addition, the low-frequency BOLD–CBF coupling was preserved when the potential time delays between BOLD and vascular regressors were not compensated for. These suggest that our estimated BOLD–CBF coupling is not likely to be dominated by BOLD–CBF cross-contamination or by the time-delay estimations, but represents synchronized oscillations of BOLD and CBF. Abbreviations: PFC: prefrontal cortex, PCC: posterior cingulate cortex, MPFC: medial prefrontal cortex, LPC: lateral parietal cortex, IPS: intraparietal sulcus, IPCS: inferior precentral sulcus, dACC: dorsal anterior cingulate cortex, MT: medial temporal region.
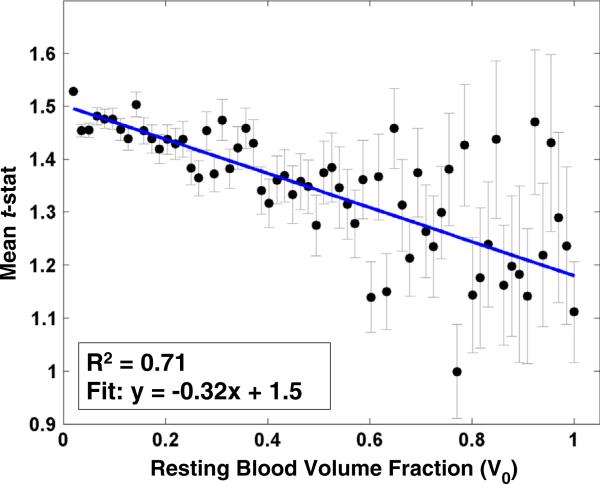
Linear regression of the group-average t-statistics (CBF vs. BOLD) against MRA-derived resting-state macrovascular volume fraction (V0) is shown in Fig. 6. Regression analysis results indicate that the degree of positive coupling between BOLD and CBF significantly increased as the macrovascular blood volume fraction decreased (R2 = 0.71). Incidentally, our voxel-wise paired t-test did not reveal a significant relationship between BOLD–CBF coupling and ASL-derived baseline perfusion values.
Fig. 6.
Linear regression of the regional mean t-statistics of the BOLD–CBF association against resting blood volume fraction (V0) associated with regional vasculature. The coefficient of determination (R2) was 0.71. The error bar indicates the standard error of the mean (SEM). The degree of the positive coupling between BOLD and CBF significantly decreased as the macrovascular volume fraction (V0) increased.
Discussion
Dynamic cerebrovascular contributions to resting-state BOLD fluctuations
Since the BOLD effect, based on both CBF and oxygen extraction, was initially introduced by Ogawa et al. (1992, 1993), several biophysical models of the cerebrovascular contribution to the BOLD signal have been proposed (Buxton et al., 1998; Davis et al., 1998; Hoge et al., 1999; Kim et al., 1999). According to the Balloon Model (Buxton et al., 1998), stimulus-evoked BOLD response is determined by two state variables (i.e. cerebral blood volume (CBV) and deoxy-hemoglobin content) and one input variable (CBF), with CBF being a major and undisputed contributor to BOLD signal changes. In addition, in calibrated BOLD (Davis et al., 1998; Hoge et al., 1999; Kim et al., 1999), the task-induced BOLD response was modeled as a function of CBF and CMRO2 changes. Although these BOLD models are based on stimulus-evoked neuronal activity (i.e., neurovascular coupling mechanism), we hypothesize that CBF changes are a primary contributor to the BOLD signal in intrinsic BOLD fluctuations as well.
While the dynamic relationship between CBF and BOLD signal fluctuations is not clear, several lines of evidence support our adoption of a linear relationship as a good approximation. Recent animal experiments (Hyder et al., 2010) showed a linear relationship between hemo-dynamic responses (such as BOLD, CBF, CBV) and neuronal activity in an event-related paradigm, suggesting that the linear relationship between low-level vascular and neuronal variations underlying steady-state BOLD may continue into the dynamic state. Furthermore, during the post-stimulus BOLD undershoot, a significant CBF participation was observed, and in re-analysis of the data from Chen and Pike (2009), the amplitudes of BOLD and CBF undershoots were successfully fit to a linear model (data not shown). The idea of approximate linearity between low-amplitude BOLD and CBF fluctuations was tested and adopted in a recent report (Fukunaga et al., 2008) for estimation of dynamic CMRO2 in resting-state data.
Our analysis approach exploits this approximate linearity between low-amplitude BOLD and CBF fluctuations. The first key finding of this work is that the dynamic characteristics of resting-state BOLD fluctuation significantly correspond to spontaneous fluctuations in CBF, as shown in Fig. 5. Our approach is novel in that we isolated the local cerebrovascular contribution (i.e. CBF) from the global vascular modulation of the BOLD signal (i.e., the pulse-oximetry recording). We found the coupling between BOLD and CBF during resting-state to be highly variable across the brain, but was most significant within major regions of resting-state networks (including the DMN, the visual network and the task-positive network). Of note, the physiological significance of the task-positive network is still controversial, being a potential artifact induced by “global signal regression” (i.e. removal of whole-brain average time-series from the data) (Chang and Glover, 2009; Fox et al., 2009; Murphy et al., 2009). However, in this work, we did not perform the debatable global signal regression, and limited our noise regressors to specific noise ROIs, thus precluding such artifacts. Furthermore, we observed consistent correlations between BOLD and CBF within both the task-positive network and the DMN, which are unlikely to result solely from artifacts, and attest to the neuronal significance of both networks (Fox et al., 2005, 2009; Chang and Glover, 2009).
Previous studies have argued that spontaneous CBF fluctuations in resting-state functional networks reflect neuronal activity (Biswal et al., 1997; Chuang et al., 2008; Fukunaga et al., 2008; Liang et al., 2013; Viviani et al., 2011; Zou et al., 2009). Specifically, using positron emission tomography (PET), Raichle et al, (2001) observed that the CBF values in a set of regions (including the PCC, MPFC, thalamus, and insula) are higher than the whole-brain average resting CBF. Similar regions were reported to exhibit higher resting CBF fluctuations as measured using ASL (Zou et al. (2009). Moreover, a recent study (Liang et al., 2013) showed a stronger correlation between mean CBF and functional connectivity strength in the DMN and executive-control networks. Together with these findings, our results suggest that within the major nodes of established resting-state brain networks, the mechanism of vascular regulation underlying resting-state BOLD fluctuations are highly reflective of CBF fluctuations, which have in turn been closely tied to neuronal activity (Boxerman et al., 1995). While we did not find a significant relationship between static quantitative CBF and the BOLD–CBF coupling strength in our particular dataset, it is conceivable that a larger dataset, spanning a wider range of CBF levels, may reveal such an association.
Relationship between resting-state CBF–BOLD coupling and regional static vascular content
A second key finding of this work is that the statistical strength and degree of coupling between resting-state BOLD and CBF signals is significantly reduced as macrovascular volume fraction increased — that is, BOLD and CBF appear to be less coupled in voxels near large vessels. While MRA captures both arterial and venous vessels, this finding is limited to regions exhibiting significant BOLD–CBF coupling, and is therefore relevant mainly for non-arterial vessels. In voxels containing the highest V0 values, the BOLD–CBF coupling is weakest, potentially due to a weak macrovascular contribution to ASL-derived CBF. More specifically, as the intravascular contribution to the BOLD response is often strongest in small-to-medium-sized vessels (Boxerman et al., 1995), we speculate that the resting-state CBF–BOLD link that we observe is not dominated by extravascular BOLD effects, which are maximal near the largest venous vessels. In other words, our results imply that the more the intravascular effect dominates, the stronger the CBF–BOLD coupling. In addition, given that smaller veins are more proximal to the sites of activation, our results suggest that the resting-state BOLD signal components that are more tightly regulated by CBF fluctuations may be more specific to neuronal activity.
These results are in keeping with observations during functional activation (Cho et al., 2012; Polimeni et al., 2010) and vascular challenges (Dagli et al., 1999). More specifically, also using MRA data, Cho et al. (2012) observed that the smaller the vessel size, the greater the increase in MRA signal intensity during neuronal stimulation, supporting an increasing neuronal specificity with decreasing vessel size. Conversely, several studies (Birn, 2012; Dagli et al., 1999; Polimeni et al., 2010) showed that the neuronal specificity of the BOLD response decreased near large blood vessels due to non-neuronal physiological noise. This is supported by a recent high spatial resolution study (Polimeni et al., 2010) demonstrating that the accuracy of the functional activation pattern degraded near the pial surface. In support of these past reports, our observation that BOLD and CBF dynamics are most discordant near large vessels is evidence for the microvascular dominance of CBF, and further demonstrates that CBF fluctuations are less likely to be influenced by non-neuronally-specific signals that are typical near large macrovessels. Furthermore, our results suggest that regional CBF fluctuations, presumably reflective of neuronal activity, may surpass the effect of global vascular fluctuations in strength, and may dominate the resting-state BOLD signal at the local scale in major functional networks.
Potential caveats
One important concern in a study of this nature is minimizing potential BOLD contamination of the CBF signal, and this was indeed the primary focus in developing our methodology. To minimize the BOLD contamination within a low-frequency range, we high-pass filtered the ASL signal at a cut-off frequency of 1/(4TR) (i.e., 0.071 Hz), and demodulated it to a low-frequency range. Arguably, these processing steps may not remove multiplicative BOLD contamination in the entire frequency band (Chuang et al., 2008), which may affect the correlation between regional BOLD and CBF fluctuations. However, many studies (Wu et al., 2008; Zou et al., 2009) showed that BOLD fluctuations during resting-state were dominant within low-frequency ranges (0.01–0.06 Hz), and that high frequency BOLD signals were relatively low in gray matter. Therefore, the effect of high-frequency BOLD contamination of CBF fluctuations is expected to be negligible and would not significantly bias the spatial distribution of coupling between BOLD and CBF during resting-state. This is further supported by the spatial heterogeneity of the CBF–BOLD associations as well as the global cardiac correlations with BOLD and CBF data.
Another potential limitation is the influence of physiological noise on the estimated coupling between BOLD and CBF fluctuations. To isolate physiological noise (mostly arising from cardiac and respiratory sources) from the BOLD signal, we included the pulse oximetry waveform as a nuisance regressor in our GLM (Verstynen and Deshpande, 2011). In addition, we regressed significant physiological noise out of the CBF fluctuations in the GLM. Nonetheless, other physiological factors may still contribute to the observed cerebrovascular fluctuations. For example, variations in breath-to-breath respiration depth cause changes in the arterial concentration of carbon dioxide (CO2), a potent vasodilator, which induces increases in CBF and BOLD (Birn, 2012). Although a recent study (Van Dijk et al., 2010) showed that regressing out the white matter and CSF signals could be sufficient for minimizing such effects, more elaborate methods involving direct physiological measures may enhance the sensitivity for detecting intrinsic neuronal activity-related signals. Regardless of the potential physiological biases, our objective in this work is to quantify the relationship between vascular and BOLD fluctuations. The precise origins of vascular fluctuations and their influence on neuronal activity are beyond the scope of this work, but will be the focus of our future work.
A third potential limitation is the accuracy of the vessel segmentations obtained from the MRA data, which might affect our static blood volume fraction estimation. Due to limitations in the spatial resolution and sensitivity of the 3D TOF MRA technique, certain vessels may not be detectable by MRA, and certainly not the microvasculature. In addition, suboptimal thresholding in the segmentation process may lead to a false-negative or false-positive detection of blood vessels. Nevertheless, our choice of the TOF MRA technique was based on its superiority over other available techniques. Moreover, as we restricted the use of MRA-segmented vessels to calculating large-vessel fraction, low sensitivity of MRA to the microvasculature is not likely to alter the main findings of this work. Furthermore, the strong correlation observed between vascular volume fraction and CBF–BOLD coupling precludes the possibility of dominance by random biases.
Lastly, the group size in this study is relatively small (N=9 subjects, 8 degrees of freedom), resulting in comparatively low statistical power and a long-tailed t-distribution (Desmond and Glover, 2002; Murphy and Garavan, 2004). This may account for the noisy appearance of the group-level t-statistic maps seen in Fig. 5. However, the regions exhibiting significant BOLD–CBF coupling (detected by p<0.00005) encompass the main nodes of the DMN and the task-positive network, suggesting true neuronal/physiological underpinnings of our findings despite the small sample size. Future work may utilize meta-analyses to boost sample size and ensure the generalizability of our findings.
Conclusion
Exploiting regionally specific CBF fluctuation and MRA-derived blood volume fraction, we observed a significant synchronization between spontaneous BOLD and CBF fluctuations, particularly within major resting-state networks. Moreover, we showed that this resting-state CBF–BOLD coupling was strongest in regions containing smaller blood vessels. Our results suggest that the component of the resting-state BOLD fluctuation that is more closely regulated by dynamic changes in CBF is likely to be more specific to neuronal activity.
Acknowledgments
This research was supported by the Natural Sciences and Engineering Council of Canada (NSERC) grant, the National Institutes of Health (NIH) grants NIBIB K01-EB011498, P50-HD-055784, R01-MH080892, R01-NS081077, and R01-EB014922 as well as by fellowship funding from the Rotman Research Institute of Baycrest and Heart and Stroke Foundation Centre for Stroke Recovery (S. Tak).
Footnotes
Disclosure/conflict of interest
The authors declare no conflict of interest.
References
- Aguirre GK, Detre JA, Zarahn E, Alsop DC. Experimental design and the relative sensitivity of BOLD and perfusion fMRI. Neuroimage. 2002;15(3):488–500. doi: 10.1006/nimg.2001.0990. [DOI] [PubMed] [Google Scholar]
- Alsop D, Detre J. Reduced transit-time sensitivity in noninvasive magnetic resonance imaging of human cerebral blood flow. J. Cereb. Blood Flow Metab. 1996;16(6):1236–1249. doi: 10.1097/00004647-199611000-00019. [DOI] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37(1):90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM. The role of physiological noise in resting-state functional connectivity. Neuroimage. 2012;62(2):1–7. doi: 10.1016/j.neuroimage.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Van Kylen J, Hyde JS. Simultaneous assessment of flow and BOLD signals in resting-state functional connectivity maps. NMR Biomed. 1997;10(4–5):165–170. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<165::aid-nbm454>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Boxerman JL, Hamberg LM, Rosen BR, Weisskoff RM. MR contrast due to intravascular magnetic susceptibility perturbations. Magn. Reson. Med. 1995;34(4):555–566. doi: 10.1002/mrm.1910340412. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: the balloon model. Magn. Reson. Med. 1998;39(6):855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- Chang C, Glover GH. Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. Neuroimage. 2009;47(4):1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Thomason ME, Glover GH. Mapping and correction of vascular hemo-dynamic latency in the BOLD signal. Neuroimage. 2008;43(1):90–102. doi: 10.1016/j.neuroimage.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Pike GB. Origins of the BOLD post-stimulus undershoot. Neuroimage. 2009;46(3):559–568. doi: 10.1016/j.neuroimage.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Cho ZH, Kang CK, Park CA, Hong SM, Kim SH, Oh ST, Kim YB. Microvascular functional MR angiography with ultra-high-field 7 T MRI: comparison with BOLD fMRI. Int. J. Imaging Syst. Technol. 2012;22(1):18–22. [Google Scholar]
- Chuang KH, van Gelderen P, Merkle H, Bodurka J, Ikonomidou VN, Koretsky AP, Duyn JH, Talagala SL. Mapping resting-state functional connectivity using perfusion MRI. Neuroimage. 2008;40(4):1595–1605. doi: 10.1016/j.neuroimage.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli MS, Ingeholm JE, Haxby JV. Localization of cardiac-induced signal change in fMRI. Neuroimage. 1999;9(4):407–415. doi: 10.1006/nimg.1998.0424. [DOI] [PubMed] [Google Scholar]
- Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn. Reson. Med. 2008;60(6) doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Davis TL, Kwong KK, Weisskoff RM, Rosen BR. Calibrated functional MRI: mapping the dynamics of oxidative metabolism. Proc. Natl. Acad. Sci. U. S. A. 1998;95(4):1834–1839. doi: 10.1073/pnas.95.4.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond JE, Glover GH. Estimating sample size in functional MRI (fMRI) neuro-imaging studies: statistical power analyses. J. Neurosci. Methods. 2002;118(2):115–128. doi: 10.1016/s0165-0270(02)00121-8. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fox PT, Raichle ME. Focal physiological uncoupling of cerebral blood flow and oxidative metabolism during somatosensory stimulation in human subjects. Proc. Natl. Acad. Sci. U. S. A. 1986;83(4):1140–1144. doi: 10.1073/pnas.83.4.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. U. S. A. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc. Natl. Acad. Sci. U. S. A. 2006;103(26):10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J. Neurophysiol. 2009;101(6):3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick B. d, Nickerson LD, Tong Y. Physiological denoising of BOLD fMRI data using Regressor Interpolation at Progressive Time Delays (RIPTiDe) processing of concurrent fMRI and near-infrared spectroscopy (NIRS). NeuroImage. 2012;60(3):1913–1923. doi: 10.1016/j.neuroimage.2012.01.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RS. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 1994;2(4):189–210. [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 1996;35(3):346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Mechelli A, Turner R, Price CJ. Nonlinear responses in fMRI: the balloon model, Volterra kernels, and other hemodynamics. Neuroimage. 2000;12(4):466–477. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Penny WD. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Academic Press; 2011. [Google Scholar]
- Fukunaga M, Horovitz SG, de Zwart JA, van Gelderen P, Balkin TJ, Braun AR, Duyn JH. Metabolic origin of BOLD signal fluctuations in the absence of stimuli. J. Cereb. Blood Flow Metab. 2008;28(7):1377–1387. doi: 10.1038/jcbfm.2008.25. [DOI] [PubMed] [Google Scholar]
- Hoge RD, Atkinson J, Gill B, Crelier GR, Marrett S, Pike GB. Investigation of BOLD signal dependence on cerebral blood flow and oxygen consumption: the deoxyhemoglobin dilution model. Magn. Reson. Med. 1999;42(5):849–863. doi: 10.1002/(sici)1522-2594(199911)42:5<849::aid-mrm4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Hu Z, Liu C, Shi P, Liu H. Exploiting magnetic resonance angiography imaging improves model estimation of BOLD signal. PLoS ONE. 2012a;7(2):e31612. doi: 10.1371/journal.pone.0031612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ni P, Liu C, Zhao X, Liu H, Shi P. Quantitative evaluation of activation state in functional brain imaging. Brain Topogr. 2012b;25(4):362–373. doi: 10.1007/s10548-012-0230-5. [DOI] [PubMed] [Google Scholar]
- Hunter MD, Eickhoff SB, Miller TWR, Farrow TFD, Wilkinson ID, Woodruff PWR. Neural activity in speech-sensitive auditory cortex during silence. Proc. Natl. Acad. Sci. U. S. A. 2006;103(1):189–194. doi: 10.1073/pnas.0506268103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton C, Josephs O, Stadler J, Featherstone E, Reid A, Speck O, Bernarding J, Weiskopf N. The impact of physiological noise correction on fMRI at 7 T. Neuroimage. 2011;57(1):101–112. doi: 10.1016/j.neuroimage.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyder F, Sanganahalli BG, Herman P, Coman D, Maandag NJG, Behar KL, Blumenfeld H, Rothman DL. Neurovascular and neurometabolic couplings in dynamic calibrated fMRI: transient oxidative neuroenergetics for block-design and event-related paradigms. Front. Neuroenerg. 2010;2 doi: 10.3389/fnene.2010.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Pechaud M, Smith S. Bet2: MR-based estimation of brain, skull and scalp surfaces. Eleventh Annual Meeting of the Organization for Human Brain Mapping. 2005;17 [Google Scholar]
- Kim SG, Rostrup E, Larsson HB, Ogawa S, Paulson OB. Determination of relative CMRO2 from CBF and BOLD changes: significant increase of oxygen consumption rate during visual stimulation. Magn. Reson. Med. 1999;41(6):1152–1161. doi: 10.1002/(sici)1522-2594(199906)41:6<1152::aid-mrm11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Liang X, Zou Q, He Y, Yang Y. Coupling of functional connectivity and regional cerebral blood flow reveals a physiological basis for network hubs of the human brain. Proc. Natl. Acad. Sci. U. S. A. 2013;110(5):1929–1934. doi: 10.1073/pnas.1214900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Wong EC. A signal processing model for arterial spin labeling functional MRI. Neuroimage. 2005;24:207–215. doi: 10.1016/j.neuroimage.2004.09.047. [DOI] [PubMed] [Google Scholar]
- Liu TX, Tak S, Wang JJ, Yan L, Chen JJ. Evaluation of dual-echo pseudo-continuous ASL for resting-state BOLD functional connectivity measurement.. Proceedings of the ISMRM 21st Scientific Meeting; Salt Lake City. 2013. p. 2227. [Google Scholar]
- Mannheimer PD. The light–tissue interaction of pulse oximetry. Anesth. Analg. 2007;105:S10–S17. doi: 10.1213/01.ane.0000269522.84942.54. [DOI] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. 2nd Ed. Chapman and Hall; New York: 1989. [Google Scholar]
- Mumford JA, Nichols T. Simple group fMRI modeling and inference. Neuroimage. 2009;47(4):1469–1475. doi: 10.1016/j.neuroimage.2009.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Garavan H. An empirical investigation into the number of subjects required for an event-related fMRI study. Neuroimage. 2004;22(2):879–885. doi: 10.1016/j.neuroimage.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44(3):893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah FA, Lee EL, Chuang K-H. Optimization of flow-sensitive alternating inversion recovery (FAIR) for perfusion functional MRI of rodent brain. NMR Biomed. 2012;25(11):1209–1216. doi: 10.1002/nbm.2790. [DOI] [PubMed] [Google Scholar]
- Nir Y, Hasson U, Levy I, Yeshurun Y, Malach R. Widespread functional connectivity and fMRI fluctuations in human visual cortex in the absence of visual stimulation. Neuroimage. 2006;30(4):1313–1324. doi: 10.1016/j.neuroimage.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. U. S. A. 1990;87(24):9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc. Natl. Acad. Sci. U. S. A. 1992;89(13):5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Menon R, Tank D, Kim S, Merkle H, Ellermann J, Ugurbil K. Functional brain mapping by blood oxygenation level-dependent contrast magnetic resonance imaging. A comparison of signal characteristics with a biophysical model. Biophys. J. 1993;64(3):803. doi: 10.1016/S0006-3495(93)81441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny W, Holmes A. Random effects analysis. In: Friston K, Ashburner J, Kiebel S, Nichols T, Penny W, editors. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Elsevier; London: 2006. [Google Scholar]
- Polimeni JR, Fischl B, Greve DN, Wald LL. Laminar analysis of 7 T BOLD using an imposed spatial activation pattern in human V1. Neuroimage. 2010;52(4):1334–1346. doi: 10.1016/j.neuroimage.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc. Natl. Acad. Sci. U. S. A. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restom K, Behzadi Y, Liu TT. Physiological noise reduction for arterial spin labeling functional MRI. Neuroimage. 2006;31(3):1104–1115. doi: 10.1016/j.neuroimage.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Shulman RG, Rothman DL, Behar KL, Hyder F. Energetic basis of brain activity: implications for neuroimaging. Trends Neurosci. 2004;27(8):489–495. doi: 10.1016/j.tins.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Tong Y, Frederick B. d. Time lag dependent multimodal processing of concurrent fMRI and near-infrared spectroscopy (NIRS) data suggests a global circulatory origin for low-frequency oscillation signals in human brain. NeuroImage. 2010;53(2):553–564. doi: 10.1016/j.neuroimage.2010.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Frederick B. d. Concurrent fNIRS and fMRI processing allows independent visualization of the propagation of pressure waves and bulk blood flow in the cerebral vasculature. NeuroImage. 2012;61(4):1419–1427. doi: 10.1016/j.neuroimage.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Fox PT, Paus T. Functional coactivation map of the human brain. Cereb. Cortex. 2008;18(11):2553–2559. doi: 10.1093/cercor/bhn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J. Neurophysiol. 2010;103(1):297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstynen TD, Deshpande V. Using pulse oximetry to account for high and low frequency physiological artifacts in the BOLD signal. Neuroimage. 2011;55(4):1633–1644. doi: 10.1016/j.neuroimage.2010.11.090. [DOI] [PubMed] [Google Scholar]
- Viviani R, Messina I, Walter M. Resting state functional connectivity in perfusion imaging: correlation maps with BOLD connectivity and resting state perfusion. PLoS ONE. 2011;6(11):e27050. doi: 10.1371/journal.pone.0027050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Aguirre GK, Rao H, Wang J, Fernandez-Seara MA, Childress AR, Detre JA. Empirical optimization of ASL data analysis using an ASL data processing tool-box: ASLtbx. Magn. Reson. Imaging. 2008;26(2):261–269. doi: 10.1016/j.mri.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W-C, Fernandez-Seara M, Detre JA, Wehrli FW, Wang J. A theoretical and experimental investigation of the tagging efficiency of pseudocontinuous arterial spin labeling. Magn. Reson. Med. 2007;58(5):1020–1027. doi: 10.1002/mrm.21403. [DOI] [PubMed] [Google Scholar]
- Wu CW, Gu H, Lu H, Stein EA, Chen J-H, Yang Y. Frequency specificity of functional connectivity in brain networks. Neuroimage. 2008;42(3):1047–1055. doi: 10.1016/j.neuroimage.2008.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CW, Gu H, Lu H, Stein EA, Chen J-H, Yang Y. Mapping functional connectivity based on synchronized CMRO2 fluctuations during the resting state. Neuroimage. 2009;45(3):694–701. doi: 10.1016/j.neuroimage.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Wu CW, Stein EA, Zang Y, Yang Y. Static and dynamic characteristics of cerebral blood flow during the resting state. Neuroimage. 2009;48(3):515–524. doi: 10.1016/j.neuroimage.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]