Abstract
Heterotrimeric serine/threonine protein phosphatase 2A (PP2A) consists of scaffolding (A), catalytic (C), and variable (B, B’, and B”) subunits. Variable subunits dictate subcellular localization and substrate specificity of the PP2A holoenzyme. The Bβ regulatory subunit gene is mutated in spinocerebellar ataxia type 12, and one of its splice variants, Bβ2, targets PP2A to mitochondria to promote apoptosis in PC12 cells. Here, we report that Bβ2 is localized to the outer mitochondrial membrane by a novel mechanism, combining a cryptic mitochondrial import signal with a structural arrest domain. Scanning mutagenesis demonstrates that basic and hydrophobic residues mediate mitochondrial association and the proapoptotic activity of Bβ2. When fused to green fluorescent protein, the N terminus of Bβ2 acts as a cleavable mitochondrial import signal. Surprisingly, full-length Bβ2 is not detectably cleaved and is retained at the outer mitochondrial membrane, even though it interacts with the TOM22 import receptor, as shown by luciferase complementation in intact cells. Mutations that open the C-terminal β-propeller of Bβ2 facilitate mitochondrial import, indicating that this rigid fold acts as a stop-transfer domain by resisting the partial unfolding step prerequisite for matrix translocation. Because hybrids of prototypical import and β-propeller domains recapitulate this behavior, we predict the existence of other similarly localized proteins and a selection against highly stable protein folds in the mitochondrial matrix. This unfolding-resistant targeting to the mitochondrial translocase is necessary but not sufficient for the proapoptotic activity of Bβ2, which also requires association with the rest of the PP2A holoenzyme.
Reversible phosphorylation by protein kinases and phosphatases is a key regulatory mechanism in eukaryotes. Protein phosphatase 2A (PP2A),1 together with protein phosphatase 1, accounts for the bulk of serine/threonine phosphatase activity in most cell types (1, 2). The predominant form of PP2A is a heterotrimer that consists of a core dimer of scaffolding (A) and catalytic (C) subunits and a variable third subunit. In vertebrates, at least 12 regulatory subunit genes impart substrate specificity and subcellular localization to PP2A holoenzymes. PP2A regulatory subunits fall into three to four gene families with little sequence conservation, except for a loose consensus motif involved in binding to the scaffolding subunit (3). The major catalytic (Cα) and scaffolding (Aα) subunits, the α4 regulatory subunit, as well as members of the B, B’, and B” subunit families are indispensable for mammalian cell viability (4–6).
B-family PP2A regulatory subunits contain seven imperfect WD repeats and are therefore predicted to adopt a β-propeller fold similar to β subunits of heterotrimeric G proteins (7). The region encompassing WD repeats 3 and 4 forms an extended interface with the A subunit, whereas a short divergent N terminus mediates isoform-specific functions (7–9). Roles ascribed to B-family regulatory subunits include regulation of cytoskeletal assembly, neuronal differentiation, mitogen-activated protein kinase signaling, and apoptosis (8–12).
Among the four genes coding for B-family regulatory subunits in vertebrates, the neuron-specific Bβ gene has received special attention because of its involvement in a neurodegenerative disorder. Spinocerebellar ataxia type 12 is caused by a trinucleotide repeat expansion in a non-coding, presumed promoter region of the human Bβ gene, PPP2R2B (13), indicating that proper regulation of Bβ expression is critical for neuronal survival. The Bβ gene gives rise to several N-terminal splice variants, including Bβ1 and Bβ2 (9, 14). We previously reported that Bβ2 is induced during postnatal brain development. The alternative N terminus of Bβ2 was found to target the PP2A heterotrimer to mitochondria in neuronal cells. Furthermore, transient or stable expression of Bβ2, but not Bβ1, potentiated apoptosis in growth factor-deprived PC6-3 cells (9), a subline of PC12 pheochromocytoma cells (15).
Here, we report that Bβ2 is localized to the outer mitochondrial membrane (OMM) by a previously undescribed mechanism. The N-terminal 26 residues of Bβ2 are sufficient to import green fluorescent protein (GFP) into the mitochondrial matrix, with conserved basic and hydrophobic amino acids playing critical roles. Full-length Bβ2 interacts with the mitochondrial import complex but is arrested at the OMM, unless its β-propeller domain is unraveled by small deletions that disrupt the “Velcro closure” between the first and last propeller blade. A chimeric protein consisting of the matrix import sequence of cytochrome oxidase subunit VIII (COX8) and the prototypical β-propeller of Gβ5 behaves identically, demonstrating that unfolding-resistant translocase targeting may be a general mechanism by which proteins with thermodynamically stable tertiary structures can be directed to the surface of mitochondria. Mitochondrial import of essential proteins appears not be compromised by overexpression of proteins targeted in this manner, judging by a lack of effect on mitochondrial membrane potential and cell viability. Our results also constrain the types of tertiary folds that can be adopted by nucleus-encoded proteins destined for the mitochondrial matrix.
EXPERIMENTAL PROCEDURES
Mutagenesis and Fusion Protein Construction
All deletion and amino acid substitution mutations in Bβ2 were generated by PCR using mutagenic primers harboring restriction sites. PCR products were li-gated into unique sites flanking the Bβ2 N terminus (residues 1–35) in the pEGFP-N1 vector (BD Biosciences). Most of the mutations incorporated to the Bβ21–35-GFP fusion protein were transferred into full-length Bβ2-GFP fusion proteins by restriction digest and ligation. N-terminal fusions of GFP with the import sequence of human COX8 (residues 1–31) (16) and the OMM anchor sequence of yeast MAS70 (residues 1–29) (17) were similarly generated by PCR using nested primers encoding the targeting sequences.
The cDNA of the Gβ5 short isoform (18) was kindly provided by Rory Fisher (University of Iowa). The full-length coding sequence of Gβ5, the Gβ5 coding sequence lacking the last 8 amino acids (Gβ5ΔC), and the β-propeller domain of Bβ2 (residues 28–447) were inserted between COX81–31 and GFP following PCR with primers carrying unique restriction sites to generate COX8-Gβ5-GFP, COX8-Gβ5ΔC-GFP, and COX8-Bβ-GFP, respectively. MAS70-Bβ was created by insertion of Bβ228 – 447 between MAS701–29 and GFP. Internal (Δ27–34) and C-terminal deletions (residues 1–311 and 1–437) of Bβ2 with and without N-terminal alanine substitutions (K2A and IL18AA) were created similarly. For luciferase complementation, fusions of an N-terminal fragment (residues 2–416) of Photinus luciferase to the C terminus of Bβ2 (wild-type and K2A mutant) and fusions of the C-terminal luciferase fragment (residues 398–550) to the N terminus of PP2A/Aα and TOM22 were constructed and ligated into pcDNA3.1, featuring an intervening poly(Gly-Ser) linker sequence as described by Luker et al. (19). All PCR-amplified sequences were verified by automated DNA sequencing.
Antibodies
The Bβ2 N terminus-directed polyclonal antibody was previously described (9). Rabbit anti-FLAG tag, mouse anti-FLAG tag (M2) conjugated to agarose, and rabbit anti-GFP antibodies were purchased from Affinity Bioreagents (Golden, CO), Sigma, and BD Biosciences, respectively.
Confocal Microscopy
PC6-3 cells were cultured in 2-well chambered slides (Nunc) and transfected with deletion and alanine substitution mutants in the context of Bβ21–35-GFP using Lipofectamine 2000 (BD Biosciences) as described previously (9). After 2 days, cells were stained with 100 nm MitoTracker dye (Molecular Probes) and imaged “live” with a Zeiss LSM 510 laser scanning confocal microscope at the University of Iowa Central Microscopy Facility.
To quantify colocalization of N-terminal Bβ2 mutants with mitochondria, 7–10 randomly selected images (each containing 4–8 transfected cells) were analyzed blind to the identity of the mutant. Each transfected cell was assigned a GFP/mitochondria colocalization score from 0 to 4 (0 = mutual exclusion; 4 = perfect overlap) (20).
Luciferase Complementation
COS-M6 cells cultured in 24-well plates were transfected with varying ratios of plasmids expressing N-and C-terminal luciferase fragment fusion proteins using Lipofectamine 2000. After 3 days, cells were gently scraped into 200 μl of Hanks’ balanced salt solution containing 1 mm d-luciferin. Light emission from the cell suspensions was measured with a tube luminometer (Berthold Sirius). Cells were then lysed and immunoblotted to compare expression levels of the transfected fusion proteins. Cells expressing single luciferase fragments displayed background luminescence.
Mitochondrial Membrane Potential Assay
PC6-3 cells cultured in 2-well chambered slides were loaded 3 days post-transfection with 50 nm of the potential sensitive dye tetramethylrhodamine methyl ester (TMRM; Molecular Probes) for 30 min at 37 °C. Red and green channel confocal images were acquired, and TMRM fluorescence in transfected cells was quantified using National Institutes of Health Image software, dividing the average pixel intensity of the mitochondria-containing cytosol by the background intensity measured within the nucleus (21).
Apoptosis Assay
Apoptosis of serum-starved PC6-3 cells was assessed by nuclear morphology as previously described (9), with minor modifications. Two days post-transfection, cells were washed three times with RPMI 1640 and cultured for an additional 24 h in the absence of serum. To include floating cells in the analyses, the chambered slides were centrifuged for 2 min at 12,000 × g prior to fixation of cells with 3.6% paraformaldehyde and staining with the DNA dye Hoechst 33342 (1 μg/ml; Sigma).
Proteolytic Processing Assay
PC6-3 cells transiently transfected with FLAG-GFP fusion proteins using Lipofectamine 2000 were lysed 3 days later in immunoprecipitation buffer containing 1% (v/v) Triton X-100, 0.5% (w/v) deoxycholate, 150 mm NaCl, 20 mm Tris (pH 7.5), 1 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 mm benzamidine. Lysates were briefly sonicated with a probe tip sonicator and cleared by centrifugation (20,000 × g, 15 min), and expressed proteins were immunoprecipitated with agarose-conjugated anti-FLAG tag antibody followed by immunoblotting for GFP. Digital densitometry was performed using National Institutes of Health Image software, dividing the band intensity of the major proteolytic fragment by total GFP immunoreactivity.
Mitochondrial Import Assay
Native PC6-3 cells or a PC6-3 cell line that inducibly overexpresses Bβ2 with a C-terminal tandem affinity purification tag (22) were transfected with FLAG-GFP fusion proteins and, in the case of the inducible cell line, treated with 1 μg/ml doxycycline. Three days after transfection and/or induction, cells were disrupted by N2 cavitation (1200 p.s.i., 15 min) in disruption buffer consisting of 250 mm sucrose, 20 mm KCl, 10 mm HEPES (pH 7.4), 2 mm EDTA, and 2 mm EGTA. Nuclei and unbroken cells were removed by two consecutive centrifugations (800 × g, 5 min). The resulting super-natant was centrifuged at high speed (20,000 × g, 15 min) to obtain a crude mitochondrial pellet, which was washed once in disruption buffer. Resupended mitochondrial fractions were incubated with increasing concentrations of trypsin in the presence or absence of 1% (v/v) Triton X-100 in disruption buffer for 25 min at 22 °C with intermittent shaking. The reactions were stopped by addition of SDS sample buffer. In some experiments, cell homogenates were directly digested with trypsin ± Triton X-100 without prior isolation of mitochondria. In this case, reactions were stopped by addition of immunoprecipitation buffer supplemented with 0.25 mg/ml soybean trypsin inhibitor, and fusion proteins were isolated with FLAG tag-directed antibodies conjugated to agarose. Mitochondrial fractions or immunoprecipitates were immuno-blotted for GFP.
RESULTS
Determinants of Mitochondrial Localization
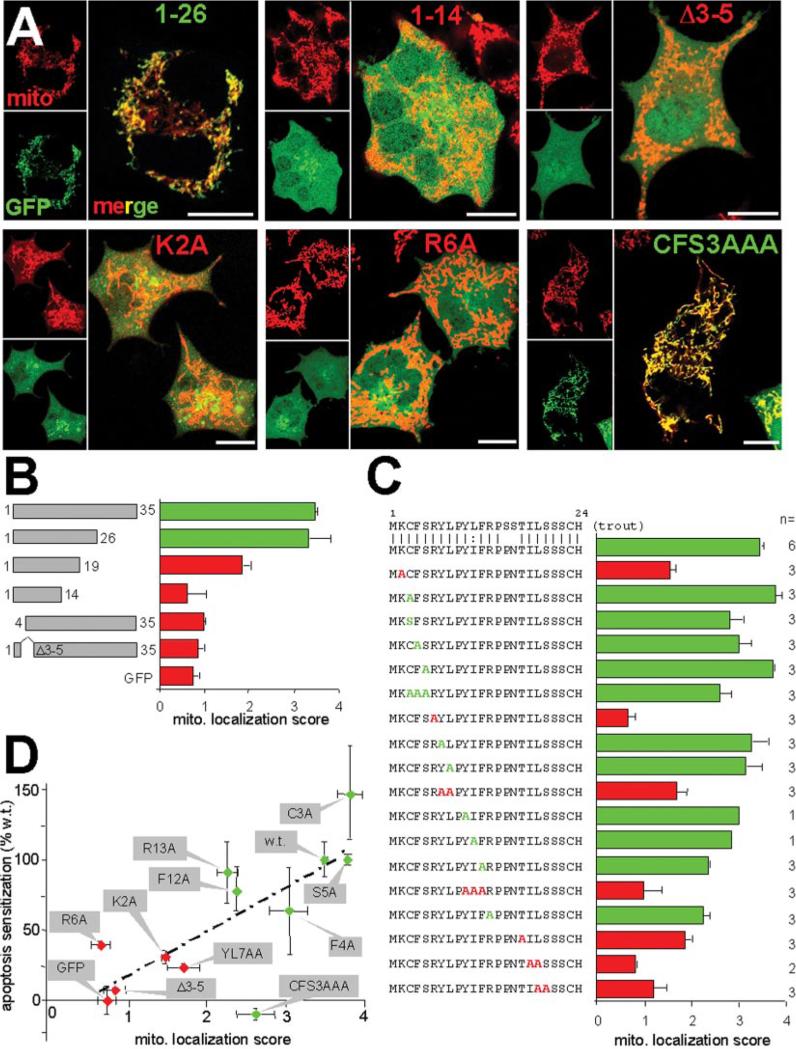
A fusion protein of the first 35 residues of Bβ2 to the N terminus of GFP (Bβ21–35-GFP) localizes to mitochondria (9). To further delineate critical regions, we carried out a systematic deletion and site-directed mutagenesis screen of the Bβ2 N terminus. The first 26 amino acids of Bβ2, which include the alternatively spliced region plus 2 amino acids shared with Bβ1, were sufficient to accurately localize GFP to MitoTracker-labeled mitochondria of transiently transfected PC6-3 cells. Representative confocal images are shown in Fig. 1A, and quantification of GFP and MitoTracker colocalization from three experiments is graphed in Fig. 1B. Bβ21–19-GFP displayed partial mitochondrial targeting, whereas an additional truncation by 5 amino acids resulted in localization indistinguishable from GFP alone. Deleting residues 1–4 or 3–5 also abrogated colocalization with mitochondria (Fig. 1, A and B). These results suggest that targeting information is distributed over the length of the divergent sequence of Bβ2.
FIG. 1. Determinants of mitochondrial association and proapoptotic activity of Bβ2.
A, representative confocal images of selected Bβ2 N terminus GFP fusion proteins (green) expressed in PC6-3 cells after labeling with the mitochondrial dye MitoTracker (mito). Point mutants and the Δ3–5 mutant were created in the context of Bβ21–35-GFP. Yellow in the larger, merged panels indicates colocalization. Scale bars = 10 μm. Throughout the figure, green fonts and graph coloring represent strong mitochondrial localization (score < 2), whereas red colors are reserved for critical mutations (score > 2). B, summary quantification of mitochondrial (mito.) localization of the indicated Bβ2 sequences fused to GFP (means ± S.E. of three or more experiments, <50 cells/experiment). C, summary of mitochondrial colocalization scores of the listed mutations in the context of Bβ21–35- or Bβ21–26-GFP (means ± S.E. of one to six experiments as indicated, <50 cells/experiment). D, positive correlation (r = 0.74) between mitochondrial localization (data from C) and proapoptotic activity. The mutations shown were introduced into full-length Bβ2 fused to GFP and analyzed for sensitization of apoptosis induced by 24 h of serum withdrawal. The percentage of sensitization is plotted relative to GFP alone (typically 5% apoptotic nuclei) and wild-type (w.t.) Bβ2 (~30% apoptosis) (means ± S.E. from three to six apoptosis assays, <50 cells/assay).
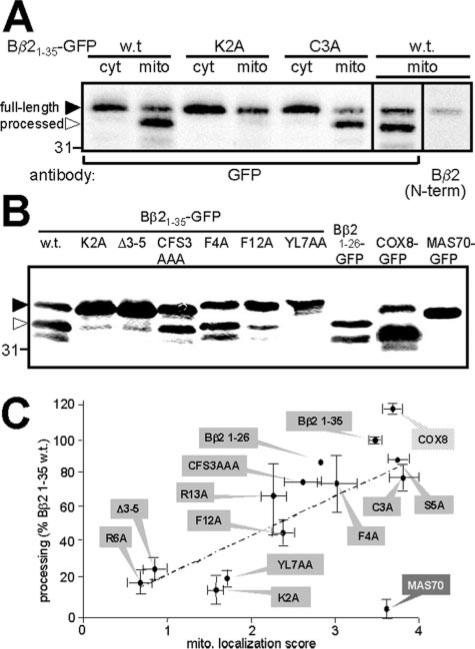
An alignment of Bβ2 orthologs from rainbow trout (Gen-Bank™ accession number CA376753) and mammals (e.g. Gen-Bank™ AY251277) was used as a guide for an alanine scan of the N terminus (Fig. 1C). We found that individual mutations of two conserved basic residues (K2A and R6A) greatly reduced the mitochondrial localization score of the GFP fusion proteins (Fig. 1, A and C). Neutralizing a more C-terminal arginine (R13A), on the other hand, had relatively modest consequences (Fig. 1C). Significantly, alanine or serine substitutions of the conserved Cys3 showed wild-type colocalization scores, ruling out a possible palmitoylation of this residue as contributing to membrane anchoring. The N terminus of Bβ2 is relatively hydrophobic (10 of 24 residues). Replacing single hydrophobic residues with alanine (F4A, Y7A, L8A, Y10A, I11A, and F12A) had little if any effect on mitochondrial localization (Fig. 1C). However, combining mutations of two or three hydrophobic residues (YL7AA, YIF10AAA, IL18AA) strongly impaired mitochondrial association (Fig. 1C). The T17A and LS19AA sub stitutions also confounded targeting significantly. A triple ala-nine substitution of residues 3–5 (CFS3AAA) showed only marginally worse targeting than the individual alanine substitutions, but deleting this region altogether (Δ3–5) reduced localization to levels of GFP alone (Fig. 1, A and C). We confirmed the colocalization results by subcellular fractionation, showing that mutating Lys2, but not Cys3, to Ala significantly reduced copurification of Bβ21–35-GFP with mitochondria (Fig. 2A). In aggregate, these experiments reveal the importance of properly spaced basic residues (Lys2 and Arg6), as well as polar and multiple hydrophobic residues.
FIG 2. Proteolytic processing of Bβ2 N terminus fusion proteins.
A, cytosolic (cyt) and mitochondrial fractions (mito) of PC6-3 cells transiently expressing Bβ21–35-GFP wild-type (w.t.) and substitution mutants were probed with the indicated antibodies. The position of the 31-kDa molecular mass marker is shown. B, the indicated fusion proteins were immunoprecipitated from total cell lysates and probed for GFP. In A and B, solid and open arrowheads point to the full-length and major processed fusion protein band, respectively. C, positive correlation (r = 0.93) between mitochondrial localization (data from Fig. 1C) and proteolysis of Bβ2 fusion proteins. Cleavage was quantified by densitometry of GFP blots (see B) and expressed relative to Bβ21–35-GFP wild-type (means ± S.E. from three to four independent experiments).
Mitochochondrial Association Determines Proapoptotic Activity
We have previously shown that transient or inducible expression of Bβ2 potentiates apoptosis when PC6-3 cells are deprived of serum (9). Bβ1 and a Bβ2 mutant unable to recruit the rest of the PP2A holoenzyme were inactive in this assay. To assess whether the apoptotic activity of Bβ2 depends on the ability of Bβ2 to localize to mitochondria, we introduced several of the alanine substitutions described above into the full-length Bβ2 coding sequence fused to the N terminus of GFP. All Bβ2 mutants could be transiently expressed to levels similar to the endogenous Bα subunit in PC6-3 cells (data not shown). 24 h following serum withdrawal, nuclei of transfected cells were scored for apoptotic morphology (Fig. 1D). Similar results were obtained when apoptotic cells were detected by annexin-V staining (data not shown). When apoptotic sensitization of each mutant is plotted against its mitochondrial localization score, a positive correlation is apparent (r = 0.74, Fig. 1D). Among the 10 mutants analyzed, the only exception to this rule was Bβ2 CFS3AAA, which localized fairly well to mitochondria but had no detectable proapoptotic activity. Therefore, mitochondrial association is prerequisite for the ability of Bβ2-containing PP2A holoenzymes to promote apoptosis.
The Bβ2 N Terminus Is a Cryptic Import Signal
The net positive charge (+4) of the Bβ2 N terminus, as well as the observation that basic and hydrophobic residues are important for mitochondrial targeting, led us to speculate that the N-terminal tail of Bβ2 is a matrix import signal. Most nucleus-encoded proteins destined for the mitochondrial matrix contain cleavable N-terminal presequences characterized by basic and hydrophobic residues (23, 24). These presequences are initially recognized by two receptor components of the TOM complex, TOM22 and TOM20, resulting in transfer of the preprotein through the pore-forming subunit TOM40 and the translocase of inner membrane (TIM) complex into the matrix, where pre-sequences are cleaved by the matrix processing peptidase (MPP) (24). Import is driven by the strongly negative membrane potential of mitochondria acting on the positively charged presequence and by ATP hydrolysis catalyzed by matrix heat shock protein 70 (mtHSP70) to partially unfold the imported protein.
Indeed, the MITOPROT signal prediction algorithm (25) assigns a high score to the Bβ2 N terminus and predicts cleavage by MPP after Pro15. Consistent with this prediction, we noted that transiently expressed Bβ21–35-GFP migrates as a doublet of 36 and 34 kDa and that the faster migrating band is enriched in a mitochondrial fraction. Only the upper band that migrates close to the predicted size of the fusion protein is recognized by an antibody raised against the extreme N terminus of Bβ2 (Fig. 2A). Because the Bβ2 N terminus is devoid of internal translation start sites, these data strongly suggest that the lower band is the product of N-terminal cleavage by MPP. Arguing against nonspecific degradation by cytosolic proteases, we observed that only mutants of Bβ21–35-GFP that scored high for mitochondrial colocalization underwent significant proteolytic processing (Fig. 2, A and B). Quantifying N-terminal proteolysis, we found a strong correlation to mitochondrial targeting of the Bβ2 mutants (r = 0.91, Fig. 2C). Positive and negative controls for signal peptide processing by MPP included the import signal of COX8 and the OMM anchor of MAS70, respectively (Fig. 2, B and C).
Full-length Bβ2 Is Arrested at the OMM
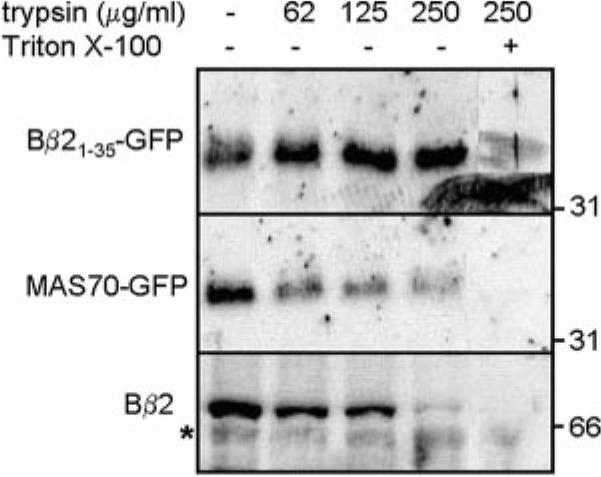
In contrast to Bβ21–35-GFP, full-length Bβ2 consistently migrates as a single protein species that is immunoreactive with both N and C terminus-directed antibodies (9). To resolve this conundrum, we carried out trypsin protection assays to determine the submitochondrial localization of ectopically expressed fusion proteins. Mitochondrial fractions were treated with increasing concentrations of trypsin in the presence or absence of Triton X-100. The processed (34-kDa) form of Bβ21–35-GFP is completely resistant to trypsinolysis, unless mitochondria are lysed by the detergent. This result confirms that the Bβ2 N terminus is a mitochondrial import signal. However, even without detergent permeabilization, trypsin degrades the full-length Bβ2 protein as readily as OMM-anchored MAS70-GFP (Fig. 3). Thus, Bβ2 is targeted to mitochondria via an N-terminal import signal but is somehow prevented from entering the matrix.
FIG 3. Submitochondrial localization of full-length Bβ2 and Bβ21–35-GFP.
Mitochondrial fractions of PC6-3 cells expressing the indicated proteins were incubated ± 1% Triton X-100 and the listed concentrations of trypsin, followed by probing with antibodies directed to GFP (top two panels) and the FLAG tag (Bβ2, bottom panel). The processed form of Bβ21–35-GFP is protected from trypsinolysis of intact mitochondria, whereas full-length Bβ2 and the OMM protein MAS70-GFP are degraded. The asterisk indicates a nonspecific band also detected in untransfected cells; the position of molecular mass markers is shown on the right.
Bβ2 Binds to TOM22 in Intact Cells
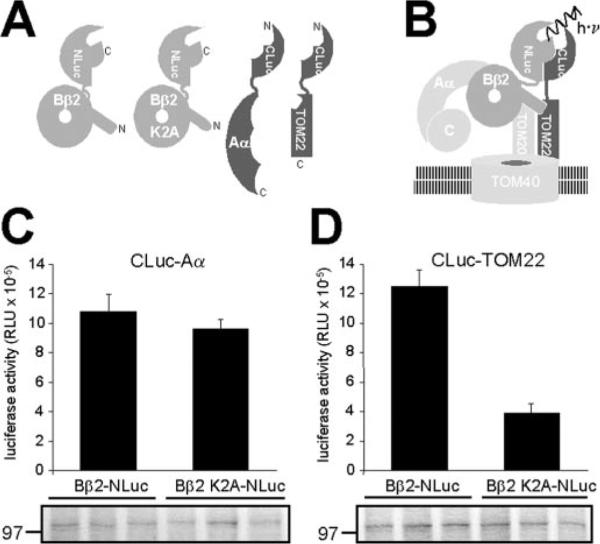
The unanticipated OMM localization of Bβ2 mandated that we confirm its interaction with the TOM translocase complex. The interaction of the translocase with its cargo is necessarily weak and transient and depends on the precise geometry of binding sites distributed over multiple integral membrane proteins. Therefore, rather than coimmunoprecipitation, we employed luciferase complementation as a sensitive in vivo protein-protein interaction assay (19). Complementary C- and N-terminal fragments of firefly luciferase (CLuc and NLuc) were fused to the N or C termini of Bβ2, the scaffolding Aα subunit, and the import receptor TOM22 as diagrammed in Fig. 4A. TOM22 binds positively charged residues in presequences and is inserted into the OMM via its C terminus, leaving the N terminus available for fusion to CLuc and complementation with Bβ2 with C-terminal NLuc (Fig. 4B). Coexpression of luciferase-tagged PP2A scaffolding subunits (CLuc-Aα) with either wild-type Bβ2-NLuc or a targeting defective point mutant (Bβ2 K2A-NLuc) resulted in robust light emission from intact COS-M6 cells (Fig. 4C), a cell line chosen for high expression levels. Luminescence was about 3-fold lower when CLuc-Aα was cotransfected with a previously described (9) monomeric mutant of Bβ2-NLuc (RR168EE) (data not shown). The reliable complementation between TOM22 and Bβ2 fusion proteins was significantly decreased by the Lys2 to Ala substitution in the Bβ2 N terminus (Fig. 4D), comparable to the effect of this mutation on mitochondrial localization (Fig. 1, A and C). Thus, OMM targeting of Bβ2 involves an interaction with import receptors of the TOM complex.
FIG 4. Bβ2 interacts with TOM22 in intact cells.
A, schematic of fusion proteins used for luciferase complementation. B, model of PP2A/ Bβ2 interaction with the TOM translocase complex. C- and N-terminal fragments of luciferase (NLuc and CLuc) reconstitute an active enzyme when their fusion partners Bβ2 and TOM22 interact. C, complementation between CLuc-Aα and Bβ2-NLuc in intact, transiently transfected COS-M6 cells is unaffected by the K2A mutation. D, binding to TOM22 requires Lys2 of Bβ2 (significant difference, p > 0.001 by Student's t test). The bar graphs show average luciferase activities ± S.E. from triplicate cotransfections of the indicated fusion proteins. C and D, the immunoblots with a pan-B subunit antibody were performed on cells lysed after the luciferase assays and show equal expression levels of wild-type and mutant Bβ2-NLuc. The position of the 97-kDa marker is shown.
The β-Propeller Resists the Unfolding Step of Import
OMM-anchored proteins commonly contain hydrophobic stop-transfer sequences C-terminal to an import signal, resulting in lateral release from the TOM complex into the outer membrane (24). A second, unusual form of anchoring was recently reported for a population of amyloid precursor protein (APP), a protein mutated in Alzheimer disease. APP arrests in the TIM/TOM complex by virtue of the mitochondrial membrane potential repelling an acidic stop-transfer sequence (26).
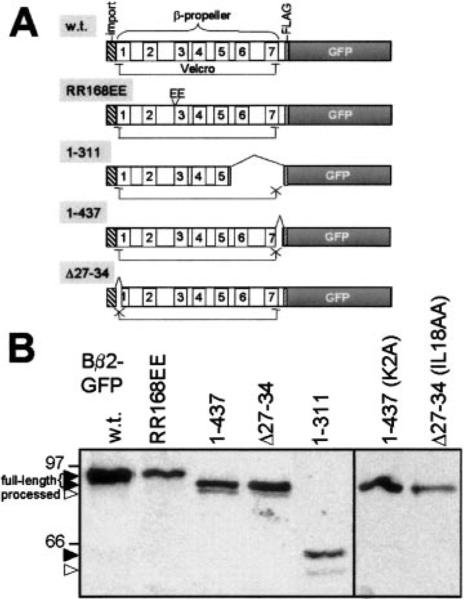
To identify the stop-transfer signal in Bβ2, we created a series of C-terminal truncations fused to the N terminus of GFP (Fig. 5A). Neither wild-type nor the monomeric Bβ2 mutant RR168EE was detectably processed in transiently transfected PC6-3 cells, indicating that PP2A holoenzyme association is not necessary for OMM arrest. In contrast, deleting the two C-terminal β-propeller blades (Bβ21–311) or just the last 10 amino acids (Bβ21–437) resulted in formation of a cleavage product with a size consistent with N-terminal processing by MPP (Fig. 5B). The yield of these presumed matrix-associated Bβ2 species was relatively low, which is consistent with large cytosolic pools of the near full-length proteins (9) (data not shown).
FIG 5. The β-propeller of Bβ2 is a stop-transfer fold.
A, schematic of β-propeller-mutant Bβ2 fusion proteins. Numbered boxes (1–7) represent WD repeats, and the brackets indicate intact or disrupted closures of the toroidal structure. B, the listed Bβ2-GFP fusion proteins were immunoprecipitated from total PC6-3 cell lysates via the FLAG tag and immunoblotted for GFP. All Velcro closure mutants are cleaved, unless mitochondrial targeting is also abolished (K2A, IL18AA).
The last 10 residues of Bβ2 are neither particularly hydro-phobic nor acidic. However, according to a tertiary structure model based on Gβ (7), their deletion removes the last β-strand of WD repeat 7, which interacts with the first β-strand of WD repeat 1 to close the β-propeller by a so-called Velcro mechanism (27). The import channel formed by TOM40 can only accommodate the width of a single α-helix (24). Combining these two observations, we surmised that the structurally rigid β-propeller fold of Bβ2 may represent a stop-transfer domain, which resists the partial unfolding required for translocation through the narrow import pore. To test this hypothesis, we removed 8 amino acids including the Velcro patch of WD repeat 1 (Bβ2Δ27–34, Fig. 5A). Similar N- and C-terminal Velcro patch deletions cause dissociation of the related Bγ subunit from the PP2A/A and C subunit, consistent with unraveling of the β-propeller torus (7). Bβ2Δ27–34 was processed to the same extent as Bβ21–437 (Fig. 5B). The proteolysis of both Velcro patch mutants is specific and depends on a functional import signal because no cleavage products were observed for Bβ21–437 K2A and Bβ2Δ27–34 IL18AA (Fig. 5B). These experiments show that an intact β-propeller structure is necessary for OMM retention of Bβ2.
A Prototypical Import Signal-β-Propeller Fusion Protein Re capitulates OMM Targeting of Bβ2
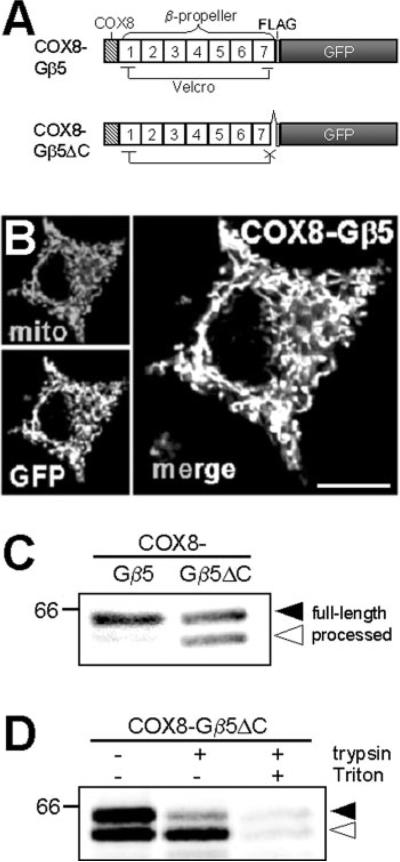
To further support this model and to generalize it to proteins other than Bβ2, we attached the well-characterized mitochondrial import sequence of COX8 to a prototypical β-propeller protein, the Gβ5 heterotrimeric G protein subunit. Equivalent to Bβ21–437, we also engineered a protein in which the Gβ5 β-propeller is destabilized by truncation of 8 residues (COX8-Gβ5ΔC, Fig. 6A). Both GFP-tagged proteins faithfully localized to mitochondria when expressed in PC6-3 cells (shown for COX8-Gβ5-GFP in Fig. 6B). However, only the Velcro-disrupted β-propeller mutant showed cleavage of the COX8 import sequence, indicative of quantitative OMM retention of the intact β-propeller, and ~50% mitochondrial matrix localization of the destabilized mutant (Fig. 6C). Only the processed form of COX8-Gβ5ΔC was resistant to trypsin digestion of intact mitochondria, providing further evidence for import of the unraveled β-propeller (Fig. 6D). These results demonstrate that β-propeller folds (and likely other domains with similarly rigid tertiary structures) are refractory to translocation by the TIM/TOM complex.
FIG 6. Unfolding-resistant translocase targeting of a model protein.
A, schematic of a mitochondria-targeted G protein β subunit (COX8-Gβ5) and its C-terminal truncation of 8 residues that unravels the β-propeller (COX8-Gβ5ΔC). WD repeats are numbered 1–7. B, confocal image of PC6-3 cell showing colocalization of COX8-Gβ5 (GFP) and MitoTracker-labeled mitochondria (mito). Scale bar = 10 μm. C, transfected fusion proteins were immunoisolated via their FLAG epitope and immunoblotted for GFP. The position of the 66-kDa marker is shown. D, PC6-3 cells expressing COX8-Gβ5ΔC were disrupted by N2 cavitation, and homogenates were incubated ± 100 μg/ml trypsin, ± 1% Triton X-100 as indicated. The fusion protein was immunoisolated from solubilized extracts via its FLAG tag and immunoblotted for GFP. In C and D, solid and open arrowheads point to full-length and processed COX8-Gβ5ΔC, respectively.
Translocase Targeting Is Necessary, but Not Sufficient for Apoptosis Induction by Bβ2—
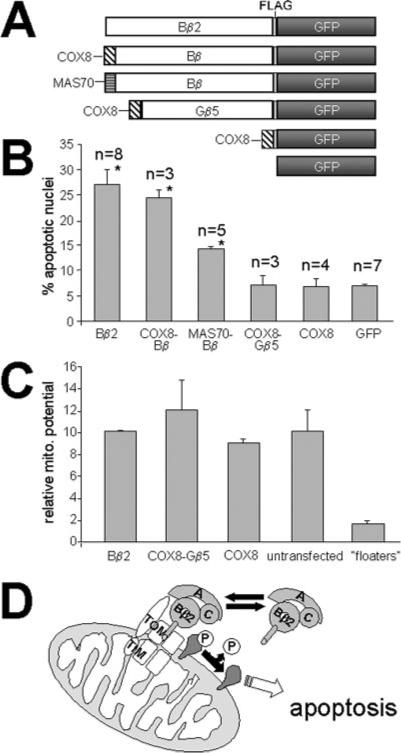
We previously showed that a Bβ2 mutant unable to recruit the PP2A holoenzyme (RR168EE) is inactive in apoptosis assays (9). An additional set of fusion proteins was constructed to examine the relationship between OMM localization and apoptosis induction (Fig. 7A). All Bβ2-based constructs were expressed to similar levels in PC6-3 cells; expression of the remaining, smaller fusion proteins was 3- to 5-fold higher (data not shown). The cryptic import sequence of Bβ2 was replaced with either the unrelated COX8 import sequence or the MAS70 OMM anchor sequence. Like Bβ2, both proteins coimmunoprecipitated with endogenous PP2A/A and C subunits, localized to mitochondria, and migrated as single protein species, characteristic of OMM localization (data not shown). Bβ2 and COX8-Bβ sensitized cells to serum starvation-induced apoptosis with equal potency, suggesting that the differentially spliced N terminus has no other function than to target Bβ2 to the import complex (Fig. 7B). In contrast, PP2A heterotrimers containing the transmembrane-anchored regulatory subunit (MAS70-Bβ) displayed significantly reduced proapoptotic activity (Fig. 7B), supporting a role for reversible targeting of PP2A to OMM microdomains containing TIM/TOM complexes.
FIG 7. Translocase targeting is necessary but not sufficient for toxicity of PP2A/Bβ2.
A, schematic of fusion proteins tested for effects on cell viability and mitochondrial function. B, PC6-3 cells transfected with the indicated fusion proteins were tested for sensitization to serum deprivation-induced apoptosis (see Fig. 1D) (means ± S.E. of three to eight assays as labeled, <50 cells/assay). Significant differences by Student's t test: *, p > 0.001 between Bβ2/COX8-Bβ and MAS70-Bβ, p > 0.0001 compared with GFP. C, PC6-3 cells expressing the listed GFP constructs, untransfected cells, and detached cells (“floaters”) were loaded with the mitochondrial (mito.) membrane potential-reporting dye tetramethylrhodamine methylester and analyzed by confocal microscopy (cytosolic to nuclear fluorescence ratio, means ± S.D. of two experiments with 20–30 cells each). D, model for OMM targeting and apoptosis sensitization by PP2A/Bβ2 (see the text).
Physical obstruction of the TIM/TOM complex by a subpopulation of Alzheimer disease APP has been proposed to compromise neuronal viability by interfering with import of essential proteins (26). To examine whether simply targeting a β-propeller to the TIM/TOM complex could kill cells, serum starvation assays were carried out with PC6-3 cells transfected with COX8-Gβ5 and control proteins. Despite higher expression levels than Bβ2, the OMM-targeted β-propeller was completely inert in these assays (Fig. 7B).
We lastly examined whether expression of OMM-targeted fusion proteins might reduce mitochondrial respiration, as has been shown for APP (26). Neither Bβ2 nor any of the artificial proteins affected mitochondrial membrane potential as determined by TMRM fluorescence (Fig. 7C). Together, these results suggest a model in which the cryptic import sequence and unfolding-resistant β-propeller domain of Bβ2 recruits PP2A to the OMM to dephosphorylate critical apoptosis regulators (Fig. 7D). Unlike APP, the interaction of β-propellers with receptor components of the TOM complex appears to be transient and readily reversible because it is necessary but not sufficient for the proapoptotic activity of Bβ2.
DISCUSSION
We describe here a detailed analysis of targeting determinants in Bβ2, a neuron-specific regulatory subunit of PP2A mutated in the neurodegenerative disorder spinocerebellar ataxia type 12, and we provide evidence for a novel mechanism of OMM localization. The differentially spliced N-terminal tail of Bβ2 constitutes a cryptic matrix import sequence with critical basic and hydrophobic residues. Luciferase complementation analyses in intact cells confirm an association of Bβ2 with the translocase complex that depends on Lys2. Matrix translocation and signal sequence cleavage of Bβ2 are prevented by a C-terminal β-propeller domain, which is resistant to the unfolding step required for import. We were able to recreate this behavior by fusing heterologous import and β-propeller domains. Thus, unfolding-resistant translocase targeting may be a general mechanism for bringing proteins to the OMM. Finally, we show that this targeting mechanism appears to be reversible because it does not impair mitochondrial function.
Mitochondrial PP2A in Apoptosis
Residues required for mitochondrial localization (e.g. Lys2 and Arg6) are also critical for the proapoptotic activity of PP2A/Bβ2. This is a significant finding because a sizable fraction of the GFP-tagged protein is diffusely localized and could therefore mediate dephosphorylation of cytosolic substrates (9). Moreover, only the translocase targeting function of the Bβ2 N terminus seems to be important because it is functionally interchangeable with the nonhomologous import sequence of COX8. Somewhat unexpectedly, the membrane-spanning sequence of MAS70 confers only limited proapoptotic activity. A possible explanation for this finding is that Bβ2 requires some degree of mobility around the TOM complex to dephosphorylate apoptosis-related substrates, whereas MAS70-Bβ is fixed near the plane of the OMM.
Recent evidence suggests that a fraction of Alzheimer disease-related APP obstructs the TIM/TOM complex to compromise mitochondrial respiration (26). A pore-blocking, stable translocation intermediate is formed, presumably because the cleavable import and acidic stop-transfer sequences of APP are separated by more than 200 residues of largely hydrophilic character. In contrast to APP, the signal sequences of Bβ2 and model proteins with intact β-propellers show no evidence of cleavage, and even high level expression of COX8-Gβ5 is not detrimental to cells. It is probable that a larger separation between import sequence and β-propeller would expose the N terminus to the matrix, resulting in MPP processing, irreversible blockade of the translocase, and impaired mitochondrial function similar to APP.
In a model based on previous (9) and present results, the neuronal PP2A/Bβ2 holoenzyme exists in equilibrium between cytosol and mitochondrial surface. Its N terminus transiently interacts with receptor components of the TOM complex, increasing the local PP2A concentration to promote dephosphorylation of unidentified OMM-associated proteins, which in turn sensitizes neurons to proapoptotic insults (Fig. 7D). Other proapoptotic PP2A regulatory subunits appear to substitute for Bβ2 in non-neuronal cells. Ruvolo et al. (28, 29) reported that ceramide-induced desphosphorylation of Bcl-2 in a lymphocyte cell line involves mitochondrial redistribution of PP2A containing the B’α regulatory subunit. B’α is structurally unrelated to Bβ2, has no recognizable import signal, and is therefore likely localized by a different mechanism.
A Novel OMM Targeting Mechanism and Its Implications
Partial unfolding of proteins destined for the mitochondrial matrix is catalyzed by mtHSP70, a component of the presequence-associated motor (PAM) complex at the matrix side of the TIM complex. mtHSP70 is thought to unfold preproteins by a ratchet mechanism (30, 31). Facilitated by thermal “breathing” of the tertiary structure of the preprotein, the positively charged signal sequence is electrostatically pulled into the matrix, where the partially linearized polypeptide is trapped by mtHsp70 and prevented from sliding back into the translocase pore. Assisted by chaperones, the imported protein then refolds into its original conformation.
Conformational stability is a well-established factor governing the efficiency of mitochondrial import. Fusion of various import signals to dihydrofolate reductase promotes efficient import of this cytosolic enzyme into the mitochondrial matrix. The dihydrofolate reductase inhibitor methotrexate stabilizes the conformation of the enzyme so that the fusion protein arrests in the TIM/TOM complex, an experimental trick commonly exploited in translocase structure/function studies (24, 32). To our knowledge, Bβ2 is the first non-artificial protein identified that employs a structure-based mechanism for OMM localization, and the β-propeller constitutes the first fold shown to resist import in a ligand-independent manner. We predict that additional examples of proteins targeted in this fashion will be discovered, utilizing β-propellers or other rigid tertiary structures as stop-transfer domains.
These results also imply that the diversity of folds that can be adopted by nucleus-encoded matrix proteins is constrained by the translocase. That is, β-strand-rich proteins with unfolding energies in the range of β-propellers (<10 kcal•mol−1) should be underrepresented in the mitochondrial matrix because they are poor translocase substrates. Indeed, only 2 of the 751 proteins (0.25%) constituting the mitochondrial proteome of Saccharomyces cerevisiae (33) are predicted β-propellers, including one membrane protein (YHR186C/KOG1). This contrasts with a ~1% representation of β-propellers in the entire yeast proteome (bmerc-www.bu.edu/wdrepeat/yeast.html).
Acknowledgments
We thank Michael Van Kanegan and Ron Merrill for critical reading of the manuscript and Audrey Dickey for help preparing two of the mutants.
Footnotes
This work was supported by National Institutes of Health Grant NS43254, American Heart Association Grant AHA0455653Z, a research grant from the United Mitochondrial Disease Foundation (to S. S.), and National Research Service Award Predoctoral Fellowship NS049659 (to R. K. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: PP2A, protein phosphatase 2A; APP, amyloid precursor protein; COX8, cytochrome oxidase subunit VIII; GFP, green fluorescent protein; MPP, matrix processing peptidase; mtHSP70, matrix heat shock protein 70; OMM, outer mitochondrial membrane; TIM, translocase of inner membrane; TOM, translocase of outer membrane.
REFERENCES
- 1.Janssens V, Goris J. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honkanen RE, Golden T. Curr. Med. Chem. 2002;9:2055–2075. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Virshup DM. Eur. J. Biochem. 2002;269:546–552. doi: 10.1046/j.0014-2956.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- 4.Strack S, Cribbs JT, Gomez L. J. Biol. Chem. 2004;279:47732–47739. doi: 10.1074/jbc.M408015200. [DOI] [PubMed] [Google Scholar]
- 5.Gotz J, Probst A, Ehler E, Hemmings B, Kues W. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12370–12375. doi: 10.1073/pnas.95.21.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong M, Fox CJ, Mu J, Solt L, Xu A, Cinalli RM, Birnbaum MJ, Lindsten T, Thompson CB. Science. 2004;22:695–698. doi: 10.1126/science.1100537. [DOI] [PubMed] [Google Scholar]
- 7.Strack S, Ruediger R, Walter G, Dagda RK, Barwacz CA, Cribbs JT. J. Biol. Chem. 2002;277:20750–20755. doi: 10.1074/jbc.M202992200. [DOI] [PubMed] [Google Scholar]
- 8.Strack S. J. Biol. Chem. 2002;277:41525–41532. doi: 10.1074/jbc.M203767200. [DOI] [PubMed] [Google Scholar]
- 9.Dagda RK, Zaucha JA, Wadzinski BE, Strack S. J. Biol. Chem. 2003;278:24976–24985. doi: 10.1074/jbc.M302832200. [DOI] [PubMed] [Google Scholar]
- 10.Sontag E, Nunbhakdi-Craig V, Bloom GS, Mumby MC. J. Cell Biol. 1995;128:1131–1144. doi: 10.1083/jcb.128.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sontag E, Nunbhakdi-Craig V, Lee G, Bloom GS, Mumby MC. Neuron. 1996;17:1201–1207. doi: 10.1016/s0896-6273(00)80250-0. [DOI] [PubMed] [Google Scholar]
- 12.Turowski P, Myles T, Hemmings BA, Fernandez A, Lamb NJ. Mol. Biol. Cell. 1999;10:1997–2015. doi: 10.1091/mbc.10.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes SE, O'Hearn EE, McInnis MG, Gorelick-Feldman DA, Kleiderlein JJ, Callahan C, Kwak NG, Ingersoll-Ashworth RG, Sherr M, Sumner AJ, Sharp AH, Ananth U, Seltzer WK, Boss MA, Vieria-Saecker AM, Epplen JT, Riess O, Ross CA, Margolis RL. Nat. Genet. 1999;23:391–392. doi: 10.1038/70493. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt K, Kins S, Schild A, Nitsch R, Hemmings B, Gotz J. Eur. J. Neurosci. 2002;16:2039–2048. doi: 10.1046/j.1460-9568.2002.02274.x. [DOI] [PubMed] [Google Scholar]
- 15.Pittman RN, Wang S, DiBenedetto AJ, Mills JC. J. Neurosci. 1993;13:3669–3680. doi: 10.1523/JNEUROSCI.13-09-03669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Kuilenburg AB, Muijsers AO, Demol H, Dekker HL, Van Beeumen JJ. FEBS Lett. 1988;240:127–132. doi: 10.1016/0014-5793(88)80353-3. [DOI] [PubMed] [Google Scholar]
- 17.McBride HM, Millar DG, Li JM, Shore GC. J. Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson AJ, Katz A, Simon MI. J. Biol. Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- 19.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strack S, McNeill RB, Colbran RJ. J. Biol. Chem. 2000;275:23798–23806. doi: 10.1074/jbc.M001471200. [DOI] [PubMed] [Google Scholar]
- 21.Frieden M, James D, Castelbou C, Danckaert A, Martinou JC, Demaurex N. J. Biol. Chem. 2004;279:22704–22714. doi: 10.1074/jbc.M312366200. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Omura T. J. Biochem. (Tokyo) 1998;123:1010–1016. doi: 10.1093/oxfordjournals.jbchem.a022036. [DOI] [PubMed] [Google Scholar]
- 24.Wiedemann N, Frazier AE, Pfanner N. J. Biol. Chem. 2004;279:14473–14476. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 25.Claros MG, Vincens P. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 26.Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. J. Cell Biol. 2003;161:41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith TF, Gaitatzes C, Saxena K, Neer EJ. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 28.Ruvolo PP, Deng X, Ito T, Carr BK, May WS. J. Biol. Chem. 1999;274:20296–20300. doi: 10.1074/jbc.274.29.20296. [DOI] [PubMed] [Google Scholar]
- 29.Ruvolo PP, Clark W, Mumby M, Gao F, May WS. J. Biol. Chem. 2002;277:22847–22852. doi: 10.1074/jbc.M201830200. [DOI] [PubMed] [Google Scholar]
- 30.Ungermann C, Guiard B, Neupert W, Cyr DM. EMBO J. 1996;15:735–744. [PMC free article] [PubMed] [Google Scholar]
- 31.Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 32.Eilers M, Schatz G. Nature. 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- 33.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]