Abstract
The role of peripheral blood mononuclear cells (PBMCs) in HBV intrauterine infection is not fully defined. Particularly the origin of PBMCs in HBV-infected neonates remains to be addressed. We carried out a population-based nested case-control study by enrolling 312 HBsAg-positive mothers and their babies. PBMC HBV DNA as well as serum HBsAg and HBV DNA was tested in cohort entry samples. Totally, 45.5% (142/312) of the newborns were found to be infected with HBV in perinatal transmission. 119 mother-infant pairs were identified to be different in the genetic profile of maternal and fetal PBMCs by AS-PCR and hemi-nested PCR. Among them, 57.1% (68/119) of the maternal PBMCs in index cases were positive for HBV DNA while 83.8% (57/68) of the HBV DNA positive maternal PBMCs passed the placental barrier and entered the fetus. Furthermore, maternal PBMC HBV infection was significantly associated with newborn infants HBV infection. PBMC traffic from mother to fetus resulted in a 9.5-fold increased risk of HBV infection in PBMC HBV DNA positive newborn infants. These data indicate that maternal PBMCs infected with HBV contribute to HBV intrauterine infection of newborn infants via PBMC traffic from mother to fetus.
Keywords: Hepatitis B virus, mother-to-infant transmission, peripheral blood mononuclear cell, fetomaternal cellular traffic.
Background
Hepatitis B virus (HBV) infection represents a major health problem worldwide. It is estimated that 350-400 million people are chronically infected with HBV in the world 1. People with chronic HBV infection are at high risk of developing serious sequelae, such as cirrhosis and hepatocellular carcinoma. Mother-to-child transfer (MTCT) or perinatal transmission of HBV is one of the major causes for HBV infection in endemic countries such as in China, accounting for 40-50% of the new cases of infection 2.
Administration of HBV immunoglobulin and HBV vaccine to the children of women positive for HBV surface antigen (HBsAg) to prevent viral transmission can effectively block MTCT of HBV during delivery and post-partum. However, passive-active immunoprophylaxis failure and breakthrough HBV infections are two unsolved issues for newborns with HBV intrauterine infection 2-4. Effective control of HBV infections should focus on preventing perinatal transmission of the virus 2.
In general, HBV intrauterine infection is determined by the presence of HBsAg and HBV DNA in the serum of infants. Recently, certain pregnant women were found positive for HBV DNA and/or HBsAg in their peripheral blood mononuclear cells (PBMCs), but negative for HBsAg and HBV DNA in serum 5,6. Their infants were infected with HBV, and even developed acute hepatitis 6,7. Some HBV-infected newborn infants were only positive for HBV DNA in neonatal PBMCs 6. These results suggest that maternal PBMC infected with HBV may play an important role during the transfer of HBV from mother to infants to cause intrauterine infection of HBV 6. PBMC HBV DNA and/or HBsAg may therefore serve as valuable biomarkers of neonatal HBV infection.
The overall prevalence of HBV intrauterine infection varies from 5% to 40% due to the differences of study population, testing methods and diagnostic criteria 8,9. The mechanism(s) of HBV intrauterine infection is still unclear. Maternal blood could enter the fetal circulation to cause HBV intrauterine infection through placental leakage due to uterine contraction during pregnancy 10-12. In addition, HBV could enter the infant blood circulation through the transfer of HBV-infected placental cells 13. A recent study suggested that maternal PBMCs infected with HBV could enter the fetal blood circulation and cause HBV intrauterine infection 6. This study raised great interest in the role of maternal PBMCs in HBV intrauterine infection. Indeed, two-way cell traffic between mother and fetus has been well documented, and may result in the vertical transfer of infectious agents 5,14. It has been reported that HBV can infect PBMCs and replicate in PBMCs 15. HBV-infected PBMCs can also release infectious viral particles 16. These results indicate that the traffic of maternal PBMCs infected with HBV may represent an additional route for mother-to-infant HBV infection. The aim of this study was to investigate the potential risk factors for mother-to-infant HBV transmission and to clarify the role of maternal PBMCs in MTCT. We recruited 312 HBsAg positive mothers and 312 newborns. Both serum and PBMC samples were collected from HBV-infected mothers and their infants, and tested for HBV biomarkers. The traffic of maternal PBMC to infant was determined by allele-specific polymerase chain reactions (AS-PCR) and hemi-nested polymerase chain reactions (hemi-nPCR) according to documents [5,7 28]. We found that the risk of infant HBV infection is significantly higher for mothers with HBV-infected PBMCs. These results highlighted the role of maternal PBMCs in the intrauterine infection of HBV.
Methods
Study design and Patients
This was a population-based nested case-control study. HBsAg-positive consecutive pregnant women aged 20-40 years old and their newborn infants were recruited in this study from the Hospitals in Beijing, China, between August 2008 and September 2012. Only HBsAg-positive pregnant women were selected; co-infection with HCV and/or HIV were excluded. Finally, 46 co-infections with HCV and 28 co-infections with HIV pregnant women were excluded. A total of 312 HBsAg-positive pregnant women and 312 newborn infants were enrolled to establish our subject cohorts. A detailed medical record was collected for each patient using a structured standard questionnaire. This questionnaire included demographic data, clinical characteristics, family and personal history of HBV infection, and details of reproductive history. The study protocol was approved by the Ethics Committee for Medical Research of the Academy of Military Medical Sciences. Written informed consent was given by all participants prior to the study.
Specimen Collection
Eight milliliters of venous blood was taken from pregnant women approximately1 hr before delivery, all infants received HBIG and hepatitis B vaccination within 24 hrs of birth. Six milliliters of blood was drawn from neonatal femoral vein within 24 hr after birth. Each blood specimen was divided into two aliquots. One tube was treated with anti-coagulant heparin sodium. PBMCs were prepared from heparinized blood using Ficoll-Hypaque (Amersham-Pharmacia, Buckinghamshire, UK) gradient centrifugation according to the manufacturer's instructions. Serum was collected from another tube without anti-coagulant.
Passive and active immunoprophylaxis were given to infants according to the standard Chinese HBV vaccination protocol. Within 12 hr of delivery, a 200-IU-dose of hepatitis B immunoglobulin (HBIG, Shanghai Institute of Biological Products, Shanghai, China) and a 10-mg dose of recombinant yeast-derived hepatitis B virus vaccine (manufactured by Tiantan Biological Products Co. Ltd, Beijing, China) were administrated intramuscularly after blood sample collection.
Detection of HBsAg, Serum HBV DNA, and PBMC HBV DNA
HBV serological markers e.g. HBsAg, hepatitis B e antigen (HBeAg), anti-HBs, and anti-HBe were detected using commercially available enzyme linked immunosorbent assay (ELISA) kits (Abbott Laboratories, Irving, TX).
For detection of HBV DNA, DNA was extracted using a “G'NOME DNA isolation kit” (BIO 101, Joshua Way, CA), according to the manufacturer's instructions, and amplified by nested PCR (nPCR) and selected PCR (sPCR). The PCR assay conditions have been previously described 17. Primer pairs were designed to bind the S region of HBV and included “outer primers” [P1,5'-CATCTTCTTGTTGGTTCTTCTG-3' (nucleotides 300-321), P2, 5'-AGGGTTTAAATGTACCC-3' (nucleotides 715-695)] and “inner primers” [P3, 5'-TCTATGTTTCCCTCTTGTTGC-3' (nucleotides 421-441), P4, 5'-TACCACATCATCCATATAACTG-3' (nucleotides 626-605)]. Amplified PCR products were analyzed by electrophoresis on 2% agarose gels. The 1st round of PCR produced a fragment of 416 base pairs (bps) while the 2nd round of PCR resulted in a 206-bp band. PBMCs were extensively washed with 1x phosphate-buffered saline (PBS) to avoid possible contamination of serum HBV DNA. Other preventative measures described by Cabrerizo 17 and Lo 14 were taken to avoid possible contamination of samples in PCR. Appropriate negative controls were included in each PCR. All of the samples were tested twice by different operators in two independent experiments, and 100% concordance was obtained in all cases.
Quantitation of Serum HBV DNA by Real-time PCR
Viral DNA was extracted from serum using QIAamp mini columns (Qiagen Ltd, Sussex, UK) according to the manufacturer's instructions. PCR amplification was performed with two sets of PCR primers and probes corresponding to the HBV core gene. Both primer sets were highly conserved among 44 known HBV sequences obtained from GenBank corresponding to the six viral genotypes (A-F) of HBV. The PCR primers and probes were purchased from TIB Molbiol (Berlin, Germany). The oligonucleotide sequences of primers were as follows. HBVF: 5'-GACCACCAAATGCCCCTAT-3' (2299-2317) and HBVR: 5'-CGAGATTGAGATCTTCTGCGAC-3' (2442-2421). Hybridization was performed with two different short oligonucleotide probes that hybridize to two adjacent internal sequences of the amplified PCR fragment. One probe, HBVLC, [5'-TCCCTCGCCTGCCAGACG(A/C)AG(A/G)TCTC (2386-2411)], was labeled at the 5' end with a Light Cycler red fluorophore (LC 640) and phosphorylated at the 3' end. The other probe, HBVFL, [5'-GA(G/C)GCAGGTCCCCTAGAAGAAGAA (2361-2384)], was labeled with fluorescein at the 3' end. For PCR, 4 mM MgCl2, 4 pmol of each hybridization probe, 20 pmol of the two PCR primers, 2 μL of LightCycler Fast Start DNA Master Hybridization probe mix (Roche Diagnostics) and 10 μL of HBV DNA samples were used. The PCR cycling program consisted of an initial denaturing step at 95°C for 10 min, followed by 45 amplification cycles at 92°C for 10 s, 50°C for 10 s, and 72°C for 10 s. To prevent carry-over contamination, 1 U of uracil N glycosylase (Roche Diagnostics, Shanghai, China) was added to the master mix. Plasma containing 4.4 ×109 HBV DNA copies/mL (HBV DNA Quantiplex assay, Chiron) was serially diluted to generate a standard curve. Serum samples with more than 108 HBV DNA copies/mL were further diluted and quantified.
Detection of PBMC Traffic from Mother to Fetus
To detect the traffic of PBMCs from mother to fetus, two polymorphic markers were used: 1) an insertion/deletion polymorphism of the glutathione S-transferase M1 (GSTM1) gene 14,19,20, and 2) an insertion/deletion polymorphism involving intron 16 of the angiotensin-converting enzyme (ACE) gene 14,21. Because the GSTM1 locus is deleted in approximately 50% of the population, mother-baby pairs informative for this locus were those in which the mother possess the GSTM1 gene and the baby carries the null allele or vice versa. Typing of the GSTM1 status has been previously described 19. Amplification of GSTM1 gene would produce a 273-bp product absent in GSTM1 null homozygotes. In addition, a heminested GSTM1 PCR was used to detect the rare maternal sequences in the blood of infants or fetal sequences in maternal blood. The expected size of the second-round PCR product was 252 bp. The primer sequences used for these assays are listed (see Supplementary Table 1) and the PCR procedures were previously described 14.
For the insertion/deletion polymorphism of intron 16 of the ACE gene, mother-baby pairs were informative only if the baby was homozygous for either the insertion (i.e., II) or deletion (i.e., DD) alleles while mother was heterozygous (ID). Fetomaternal genotyping was performed using a low-sensitivity PCR with the primers ACE-P1 and ACE-P2 (see Supplementary Table 1) and conditions that were previously described 21. PCR fragments of 479 bps and 191 bps represent insertion and deletion alleles, respectively.
Criteria for neonatus HBV infection and definition of index cases
In our study, the diagnostic criteria of neonatus HBV infection was defined for newborn infants found positive for HBsAg, HBV DNA in serum, or HBV DNA in PBMCs in their venous blood specimens collected within 24 h after birth. Index cases were defined as mother-baby pairs based on the genetic polymorphism biomarkers of GSTM1 and ACE genes as mentioned above.
Statistical Analysis
Data entry and statistical analyses were performed using the statistical package SPSS version 16.0 (SPSS Inc., Chicago, IL). The corrected chi-square test or two-sided Fisher's exact test was used to compare categorical data. The Student's t test or one-way analysis of variance was used for group comparisons of parametric quantitative data. Results are presented as the means ± standard deviation or median (range) when appropriate. In all cases, tests were all two-sided, and a p value of <0.05 was considered statistically significant.
Results
Infant HBV infection
A total of 312 HBsAg-positive mothers and 312 neonates were recruited in this study. Serum HBsAg, HBV DNA and PBMC HBV DNA were detected in 7.7% (24/312), 19.6% (61/312) and 26.0% (81/312) of the infants, respectively. A total of 142 (45.5%, 142/312) neonates were infected with HBV in utero, including 15 neonates positive for both HBsAg and HBV DNA, and 9 neonates positive for three biomarkers: HBsAg, HBV DNA and PBMC HBV DNA (Table 1). In addition, 30.8% (24/78) of the neonates born to HBsAg and HBeAg positive mothers were infected with HBV.
Table 1.
Distribution of HBV infection from 312 newborn infants
| Characteristic | N | % |
|---|---|---|
| HBsAg(+) | 24 | 7.7 |
| HBV DNA(+) | 61 | 19.6 |
| PBMC HBV DNA (+) | 81 | 26.0 |
| HBsAg(+) and HBV DNA(+) | 15 | 4.8 |
| All positive | 9 | 2.9 |
| Newbone infants HBV infection | 142 | 45.5 |
Risk factors associated with neonatal HBV infection
The demographic data and detection of HBV biomarkers for mothers and newborns are listed in Table 2. No significant differences were observed in the gender and weight of the newborns or in the maternal ages at delivery and maternal educational levels between the newborns infected with HBV (n=142) and those not infected (n=170). In addition, there were no significant differences between the two groups in terms of other risk factors, such as history of threatened preterm delivery or abortion and delivery methods.
Table 2.
Demographic data and risk factors for mother-to-infant HBV infection
| Characteristic | Newborn infants with HBV Infection (n=142) |
Newborn infants without HBV Infection (n=170) | p value |
|---|---|---|---|
| Maternal PBMC HBV DNA status | <0.001a | ||
| Positive | 67 | 33 | |
| Negative | 75 | 137 | |
| Maternal HBeAg status | <0.001a | ||
| Positive | 24 | 54 | |
| Negative | 0 | 234 | |
| Maternal serum HBV DNA status | 0.0147a | ||
| Positive | 73 | 64 | |
| Negative | 69 | 106 | |
| Gender of newborn infants | 0.5628a | ||
| Female | 68 | 87 | |
| Male | 74 | 83 | |
| Maternal age at delivery, mean±standard deviation | 26.58±2.67 | 27.06±2.31 | 0.0897b |
| Maternal educational level (years), mean±standard deviation | 10.26±2.18 | 10.43±2.45 | 0.5217b |
| Neonatal weight(g), mean±standard deviation | 3247.25±316.45 | 3184.18±389.55 | 0.1224b |
| History of threatened preterm delivery | 0.6287a | ||
| Yes | 1 | 3 | |
| No | 141 | 167 | |
| History of threatened abortion | 0.0829a | ||
| Yes | 5 | 14 | |
| No | 137 | 156 | |
| Mode of delivery | 0.1444a | ||
| Caesarean section | 83 | 113 | |
| Vaginal delivery | 59 | 57 |
Note: ap and bp values indicate comparisons of newborn infants without HBV infection with those with HBV infection using the chi-square test and Student's t test, respectively.
Abbreviations: HBV, hepatitis B virus; PBMC, peripheral blood mononuclear cells; HBeAg, hepatitis B e antigen
Of 312 mothers, 100 (32.1%) were positive for PBMC HBV DNA, 78 (25.0%) positive for serum HBeAg and 137 (43.9%) positive for serum HBV DNA. The mother-to-infant transmission of HBV was significantly associated with the status of maternal PBMC HBV DNA (p<0.001), serum HBeAg (p<0.001) and serum HBV DNA (p=0.0147) (Table 2). Furthermore, infants HBV infection was apparently associated with the level of maternal HBV DNA in serum. The higher the titers of maternal HBV DNA, the higher the risk of newborns infected with HBV (Table 3).
Table 3.
Association of serum HBV DNA content in pregnant women with the risk of HBV infection in newborn infants
| Serum HBV DNA content | N | Newborn infants HBV Infection (Yes) |
Newborn infants HBV Infection (No) |
OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| <103copy/mL | 175 | 17 | 9.71 | 158 | 90.29 | 1.00 | - |
| 103~106copy/mL | 41 | 9 | 21.95 | 32 | 78.05 | 2.61 | 1.07-6.38 |
| 106~109copy/mL | 96 | 35 | 36.46 | 61 | 63.54 | 5.33 | 2.78-10.22 |
Note: serum HBV DNA content <103copy/mL was defined as negative for HBV DNA. Results of logistic regression analysis (χ2=28.24, p<0.001).
Abbreviations: OR, odds ratio; CI, confidence interval; HBV, hepatitis B virus.
A total of 100 mothers were HBV DNA-positive in PBMCs while 67% (67/100) of their neonates were infected with HBV (Table 4). For mothers who were HBV DNA-negative in serum but HBV DNA-positive in PBMCs, their infants showed significantly higher risk of HBV infection (about five-fold) than those born to mothers with HBV DNA-negative PBMCs (OR, 4.75; 95% CI, 2.38-9.49; p<0.001). In contrast, the infants born to mothers who were HBV DNA-positive in serum, but HBV DNA-negative in PBMCs showed only two-fold higher risk of HBV infection. These results indicated that maternal PBMCs represent an important factor leading to MTCT of HBV infection.
Table 4.
Relationship between maternal HBV DNA status, PBMC HBV infection and newborn infant HBV infection
| Maternal HBV DNA status | Newborn infant HBV infection | OR | 95% CI | |
|---|---|---|---|---|
| Yes | No | |||
| Serum HBV DNA/PBMC | N (%) | N (%) | ||
| -/- | 35/123 (28.5) | 88/123 (71.5) | 1.000 | - |
| +/- | 40/89 (44.9) | 49/89 (55.1) | 2.05 | 1.16-3.64 |
| -/+ | 34/52 (65.4) | 18/52 (34.6) | 4.75 | 2.38-9.49 |
| +/+ | 33/48 (68.7) | 15/48 (31.3) | 5.53 | 2.68-11.42 |
Test of homogeneity χ2=33.07, P=0.0000; Score test for trend of odds χ2=18.78, P=0.0000;
Note: HBV DNA positive and HBV DNA negative denote mothers who were positive or negative for HBV DNA in serum, respectively; PBMC HBV DNA (+) denotes mothers who were HBV-infected in PBMCs; PBMC HBV DNA (-) denotes mothers who did not have HBV infection in PBMCs.
Abbreviations: OR, odds ratio; CI, confidence interval; PBMC, peripheral blood mononuclear cell; HBV, hepatitis B virus.
Role of PBMC traffic from mother to fetus in infant HBV infection
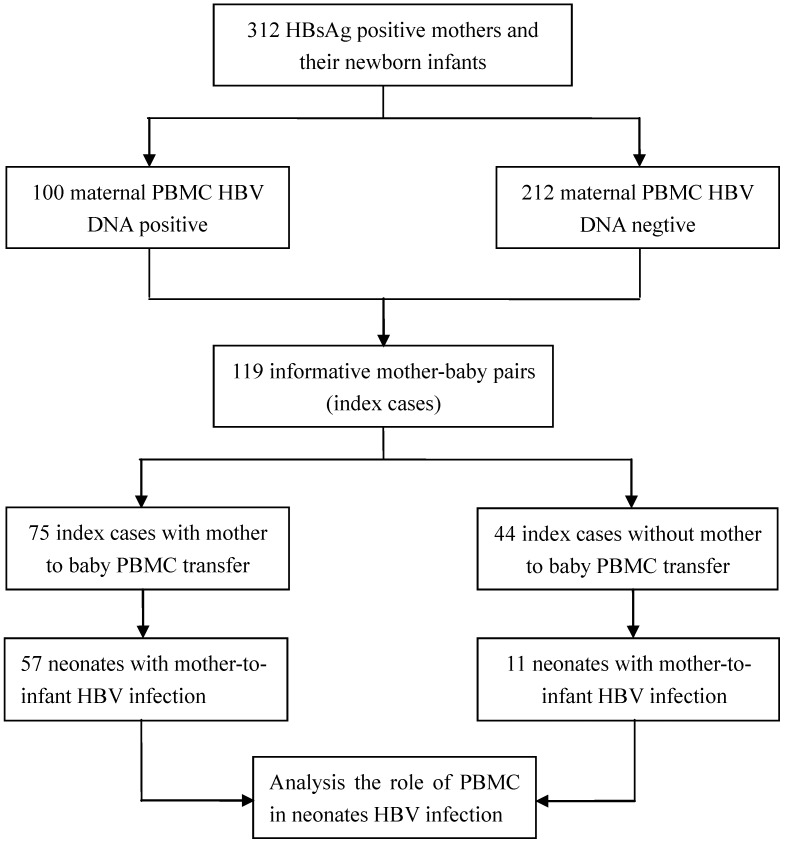
Genotyping of GSTM1 and ACE polymorphisms was performed using PBMC DNA specimens from the 312 mother-baby pairs. Using GSTM1-PCR to screen mother-baby pairs, we identified 77 mother-baby pairs as index cases in which the newborns were GSTM1 null (-/-) and the mothers were GSTM1 positive (+/+ or +/-). ACE genotyping was also performed, and revealed 42 mother-baby informative pairs in which the fetus was homozygous for the deletion allele (DD) or the insertion allele (II), while the mother was heterozygous (ID). Therefore, a total of 119 mother-baby pairs were identified as index cases to determine the role of mother-to-infant PBMC transfer in infant HBV infection.
Among the 119 informative mother-baby pairs, 75 (63.0%) showed mother-to-fetus PBMC transfer and 85 (71.4%) of the infants were infected with HBV. Sixty-eight (57.1%) of the 119 infants had HBV DNA-positive PBMCs. Furthermore, HBV infection was observed in 76.0% (57/75) of the infants born to mothers with mother-to-fetus PBMC transfer while only 25.0% (11/44) of the infants were infected with HBV when mother-to-fetus PBMC transfer was not detected. The flow chat of patients in this cohort is shown in Figure 1. We also noted that mother-to-infant PBMC traffic increased the risk of newborn infants' PBMC HBV infection by 9.50 fold (OR, 9.50; 95% CI, 3.71-24.91; P<0.001). The results of these detections were summarized in Table 5. In contrast, no significant association could be established between mother-to-infant PBMC transmission and the newborn infants' serum positivity of HBsAg and/or HBV DNA (OR, 0.90; 95% CI, 0.36-2.33; P=0.816) (Table 6). We conducted a stratified analysis of mother-to-infant HBV transmission, the results show that there was no interaction in factor of mother-to-infant PBMC transmission and mother PBMC HBV DNA. Meanwhile, PBMC transmission from mother-to-infant was association with neonatus HBV infection (OR, 3.39; 95% CI, 0.86-12.84; P=0.037) (Table 6).These results are clearly supportive of the conclusion that the increased neonatus HBV infection is largely due to HBV-infected PBMC traffic from mother to infant.
Figure 1.
Study design for PBMC traffic from mother to fetus and cohort patient flow chart.
Table 5.
Univariate analyses of the risk of PBMC traffic from mother-to-fetus in 119 mother-baby pairs
| Mode of neonates HBV infection | Mother-to-infant PBMC transmission |
Total (N=119) |
OR (95% CI) | p value | |
|---|---|---|---|---|---|
| Yes (n=75) | No (n=44) | ||||
| PBMC HBV DNA | |||||
| + | 57 | 11 | 68 | ||
| - | 18 | 33 | 51 | 9.50 (3.71-24.91) | <0.001 |
| Serum HBsAg and/or HBV DNA | |||||
| + | 19 | 12 | 31 | ||
| - | 56 | 32 | 88 | 0.90 (0.36-2.33) | 0.816 |
| Serum HBsAg and HBV DNA or PBMC HBV DNA | |||||
| + | 64 | 21 | 85 | ||
| - | 11 | 23 | 34 | 6.37 (2.46-16.84) | <0.001 |
Abbreviations: OR, odds ratio; CI, confidence interval; PBMC, peripheral blood mononuclear cell; HBV, hepatitis B virus.
Table 6.
Stratified analysis of PBMC transmission on the risk of HBV infection in 119 mother-baby pairs
| Mother-to-infant PBMC transmission | Maternal PBMC HBV DNA | Mother-to-infant HBV infection | OR (95%CI) | p value | ||
|---|---|---|---|---|---|---|
| Yes | No | Total | ||||
| Yes | positive | 48 | 9 | 57 | 3.39 (0.86-12.84) | 0.037 |
| negative | 11 | 7 | 18 | |||
| total | 59 | 16 | 75 | |||
| No | positive | 11 | 22 | 33 | 0.88 (0.17-4.99) | 0.854 |
| negative | 4 | 7 | 11 | |||
| total | 15 | 29 | 44 | |||
Abbreviations: OR, odds ratio; CI, confidence interval; PBMC, peripheral blood mononuclear cell; HBV, hepatitis B virus.
Discussion
Shi et al reported a large meta-analysis study including 37 random case-control studies and 5900 newborns of HBV carrier mothers, that serum HBsAg and HBV DNA were detected in 22.9% (595/2598) and 29.8% (208/698) of newborns respectively 22. In our study, serum HBsAg, HBV DNA and PBMC HBV DNA were detected in 7.7% (24/312), 19.6% (61/312) and 26.0% (81/312) of the infants. The infant HBV infection rate was thus 45.5%. Apparently, PBMCs infected with HBV contributed significantly to the relatively higher HBV intrauterine infection rate observed in our study. This may partly explain the differences in HBV intrauterine infection rates reported to date by various investigators because some studies only detected HBsAg and/or HBV DNA in the serum of infants 12, 23, 24, but not HBV DNA in the PBMCs.
In this study, we examined the PBMC HBV DNA as a biomarker of HBV intrauterine infection based on the following considerations: 1) HBV can infect and replicate in PBMCs. HBV DNA can be detected in PBMCs by PCR, immunohistochemistry and in situ hybridization 6,16. In addition, both covalently closed circular DNA (cccDNA) and pregenomic RNA (pgRNA) have been found in PBMCs 16,25. The transcription of pgRNA from cccDNA is a key event in HBV replication. Lu et al reported that HBV DNA and pgRNA in PBMCs correlated with the level of serum HBV DNA, and antiviral treatment with lamivudine could reduce the viral loads in PBMCs, but showed less efficacy in PBMCs than in serum 16. These results indicate that PBMCs could be a reservoir of HBV and also an extrahepatic site for virus replication. 2) HBV-infected PBMCs of mothers may serve as a source of HBV intrauterine infection. Although the fetal and maternal circulations are separated by the villous trophoblastic layers in the placenta, two-way cell traffic between mother and fetus has been well documented, and may cause intrauterine infection. Shimizu et al 26 reported that two of four infants born to three HBsAg-positive mothers who were HBV DNA negative in the plasma but HBV DNA positive in the PBMCs, developed acute or fulminant hepatitis within three months after birth. Bai et al 6 reported that 16 (26.7%) out of 60 infants born to mothers who were HBV DNA positive in the PBMCs were infected with HBV. Our study showed that the overall HBV intrauterine infection rate was 45.5% for neonates born to HBsAg-positive mothers. However, 57.0% (81/142) of the neonates born to mothers with HBV DNA-positive PBMCs were infected with HBV. In addition, 63% of the mothers were shown to have mother-to-infant PBMC traffic activity. Of note, among these infants, 76% were found to be infected with HBV. In other words, infants born to mothers with cell traffic were at an almost 10-fold higher risk of HBV infection in utero than those born to mothers without cell transfer. Taken together, these findings suggest that HBV DNA in PBMCs may serve as a critical marker for the diagnoses of HBV intrauterine infection. Therefore, when HBsAg and/or serum HBV DNA in newborn infants are used as markers for HBV intrauterine infection, the infection rate may be underestimated. Based on these findings, the 45% HBV intrauterine infection rate identified in this study may be more accurate to indicate the real infection rate among HBsAg-positive mothers. A recent study showed that the mother's PBMCs infected with HIV-1 contribute to mother-to-infant HIV-1 transmission in utero 27. Our study suggested that PBMCs played a key role in mother-to-infant HBV transmission. Together, these observations support the conclusion that virus-infected mother's PBMCs could contribute significantly to multiple viral mother-to-infant transmission diseases.
It was very difficult to conduct follow-up studies for the infants infected with HBV in utero. This limited the assessment of the clinical significance of the role PBMC infection plays in MTCT HBV infection. However, to the best of our knowledge, this is the first large-scale study to evaluate the role of maternal PBMCs in mother-to-infant HBV transmission. The traffic of HBV-infected PBMCs from mother to infant represents an alternative route by which HBV intrauterine infection occurs. Our findings provide an additional approach to evaluate HBV intrauterine infection in newborn infants born to mothers positive for HBV DNA in PBMCs. The findings may therefore provide novel strategies for the control and prevention of HBV intrauterine infection by reducing the HBV viral loads in PBMCs of pregnant women and/or by inhibiting the PBMC trafficking from mother to infant.
Supplementary Material
Acknowledgments
The authors are grateful to all of the individuals who participated in collecting the specimens, including Prof John D. Klena (United States Centers for Disease Control and Prevention, International Emerging Infections Program, Beijing, China). This work was supported by grants from the Beijing Natural Science Foundation (No.7122126) and the National Natural Science Foundation of China (No. 81072501), and partially supported by the Gillson Longenbaugh Foundation.
Abbreviations
- ACE
angiotensin-converting enzyme
- AS-PCR
allele-specific polymerase chain reaction
- bp
base pairs
- cccDNA
covalently closed circular DNA
- CI
confidence interval
- GSTM1
glutathione S transferase M1
- HBeAg
hepatitis B e antigen
- HBIG
hepatitis B immunoglobulin
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- hemi-nPCR
hemi nested polymerase chain reaction
- MTCT
mother-to-child transmission
- n-PCR
nested polymerase chain reaction
- OR
odds ratio
- PBMCs
peripheral blood mononuclear cells.
References
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. Journal of viral hepatitis. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Liu H, Wang Y, Hao R, Li Z, Song H. The next step in controlling HBV in China. Bmj. 2013;347:f4503. doi: 10.1136/bmj.f4503. [DOI] [PubMed] [Google Scholar]
- 3.Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006;43:s173–s181. doi: 10.1002/hep.20956. [DOI] [PubMed] [Google Scholar]
- 4.Carman WF, Elder AG, Wallace LA. et al. Effects of influenza vaccination of health-care workers on mortality of elderly people in long-term care: a randomised controlled trial. Lancet. 2000;355:93–97. doi: 10.1016/S0140-6736(99)05190-9. [DOI] [PubMed] [Google Scholar]
- 5.Wei JN, Wang SP, Shuang JY. [Study on the relationship between fetomaternal cellular traffic and hepatitis B virus intrauterine infection] Zhonghua liu xing bing xue za zhi = Zhonghua liuxingbingxue zazhi. 2005;26:240–244. [PubMed] [Google Scholar]
- 6.Bai GQ, Li SH, Yue YF. et al. The study on role of peripheral blood mononuclear cell in HBV intrauterine infection. Arch Gynecol Obstet. 2011;283:317–321. doi: 10.1007/s00404-010-1366-8. [DOI] [PubMed] [Google Scholar]
- 7.Shi L, Yue YF, Zhang SL, Li SH, Shi ZY, Lei CM. [Peripheral blood mononuclear cell of neonates infected with hepatitis B virus] Zhonghua Er Ke Za Zhi. 2006;44:855–858. [PubMed] [Google Scholar]
- 8.Li XM, Shi MF, Yang YB. et al. Effect of hepatitis B immunoglobulin on interruption of HBV intrauterine infection. World J Gastroenterol. 2004;10:3215–3217. doi: 10.3748/wjg.v10.i21.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Liu J, Meng L. et al. Survey of HBsAg-positive pregnant women and their infants regarding measures to prevent maternal-infantile transmission. BMC Infect Dis. 2010;10:26. doi: 10.1186/1471-2334-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin HH, Ohto H, Etoh T. et al. Studies on the risk factors of intrauterine infection of hepatitis B virus. Nippon Sanka Fujinka Gakkai zasshi. 1985;37:2393–2400. [PubMed] [Google Scholar]
- 11.Ohto H, Lin HH, Kawana T. et al. Intrauterine transmission of hepatitis B virus is closely related to placental leakage. Journal of medical virology. 1987;21:1–6. doi: 10.1002/jmv.1890210102. [DOI] [PubMed] [Google Scholar]
- 12.Xu DZ, Yan YP, Choi BC. et al. Risk factors and mechanism of transplacental transmission of hepatitis B virus: a case-control study. Journal of medical virology. 2002;67:20–26. doi: 10.1002/jmv.2187. [DOI] [PubMed] [Google Scholar]
- 13.Li F, Wang X, Men K. et al. Receptivity of human choriocarcinoma JEGIII cells and isolated trophoblast cells to hepatitis B virus infection and enhancement by tumor necrosis factor alpha. Japanese journal of infectious diseases. 2007;60:167–172. [PubMed] [Google Scholar]
- 14.Lo YM, Lo ES, Watson N. et al. Two-way cell traffic between mother and fetus: biologic and clinical implications. Blood. 1996;88:4390–4395. [PubMed] [Google Scholar]
- 15.Murakami Y, Minami M, Daimon Y. et al. Hepatitis B virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis B virus surface antigen. Journal of medical virology. 2004;72:203–214. doi: 10.1002/jmv.10547. [DOI] [PubMed] [Google Scholar]
- 16.Lu L, Zhang HY, Yueng YH. et al. Intracellular levels of hepatitis B virus DNA and pregenomic RNA in peripheral blood mononuclear cells of chronically infected patients. Journal of viral hepatitis. 2009;16:104–112. doi: 10.1111/j.1365-2893.2008.01054.x. [DOI] [PubMed] [Google Scholar]
- 17.Cabrerizo M, Bartolome J, Caramelo C. et al. Molecular analysis of hepatitis B virus DNA in serum and peripheral blood mononuclear cells from hepatitis B surface antigen-negative cases. Hepatology. 2000;32:116–123. doi: 10.1053/jhep.2000.8541. [DOI] [PubMed] [Google Scholar]
- 18.Lo YM, Chan KC. Setting up a polymerase chain reaction laboratory. Methods Mol Biol. 2006;336:11–18. doi: 10.1385/1-59745-074-X:11. [DOI] [PubMed] [Google Scholar]
- 19.Davies MH, Elias E, Acharya S. et al. GSTM1 null polymorphism at the glutathione S-transferase M1 locus: phenotype and genotype studies in patients with primary biliary cirrhosis. Gut. 1993;34:549–553. doi: 10.1136/gut.34.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fryer AA, Zhao L, Alldersea J. et al. Use of site-directed mutagenesis of allele-specific PCR primers to identify the GSTM1 A, GSTM1 B, GSTM1 A,B and GSTM1 null polymorphisms at the glutathione S-transferase, GSTM1 locus. The Biochemical journal. 1993;295:313–315. doi: 10.1042/bj2950313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigat B, Hubert C, Corvol P. et al. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidyl carboxypeptidase 1) Nucleic acids research. 1992;20:1433. doi: 10.1093/nar/20.6.1433-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Z, Li X, Ma L. et al. Hepatitis B immunoglobulin injection in pregnancy to interrupt hepatitis B virus mother-to-child transmission-a meta-analysis. Int J Infect Dis. 2010;14:e622–e634. doi: 10.1016/j.ijid.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Zhang J, Yang H. et al. Quantitative analysis of HBV DNA level and HBeAg titer in hepatitis B surface antigen positive mothers and their babies: HBeAg passage through the placenta and the rate of decay in babies. Journal of medical virology. 2003;71:360–366. doi: 10.1002/jmv.10493. [DOI] [PubMed] [Google Scholar]
- 24.Shao ZJ, Xu DZ, Xu JQ, et al.Maternal hepatitis B virus (HBV) DNA positivity, sexual intercourse are associated with HBV intrauterine transmission in China. a prospective case-control study. Journal of gastroenterology and hepatology. 2007;22:165–170. doi: 10.1111/j.1440-1746.2006.04462.x. [DOI] [PubMed] [Google Scholar]
- 25.Stoll-Becker S, Repp R, Glebe D. et al. Transcription of hepatitis B virus in peripheral blood mononuclear cells from persistently infected patients. J Virol. 1997;71:5399–5407. doi: 10.1128/jvi.71.7.5399-5407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu H, Mitsuda T, Fujita S. et al. Perinatal hepatitis B virus infection caused by antihepatitis Be positive maternal mononuclear cells. Arch Dis Child. 1991;66:718–721. doi: 10.1136/adc.66.6.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourtis AP, Amedee AM, Bulterys M. et al. Various viral compartments in HIV-1-infected mothers contribute to in utero transmission of HIV-1. AIDS research and human retroviruses. 2011;27:421–427. doi: 10.1089/aid.2010.0160. [DOI] [PubMed] [Google Scholar]
- 28.Chen SQ, Zhu QR, Wang JS. Study on the S region gene mutation of hepatitis B virus during prevention of HBV transmission in uterus with hepatitis B immunoglobulin. Zhonghua Liu Xing Bing Xue Za Zhi. 2006;27:522–525. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.