Abstract
The CFTR (cystic fibrosis transmembrane conductance regulator) gene shows a complex temporal and spatial pattern of expression that is controlled by multiple cis-acting elements interacting with the basal promoter. Although significant progress has been made towards understanding these genomic elements, there have been no reports of post-transcriptional regulation of CFTR by miRNAs (microRNAs). In the present study, we identify two miRNAs, hsa-miR-145 and hsa-miR-494, which regulate CFTR expression by directly targeting discrete sites in the CFTR 3′ UTR (untranslated region). We show that at least 12 miRNAs are capable of repressing endogenous CFTR mRNA expression in the Caco-2 cell line. Ten of these also inhibit expression of a reporter construct containing the CFTR 3′ UTR in one or more cell lines, and five repress endogenous CFTR protein expression in Caco-2 cells. Moreover, at least three are expressed in primary human airway epithelial cells, where CFTR expression is maintained at low levels in comparison with intestinal cell lines. Three of the miRNAs that target CFTR, hsa-miR-384, hsa-miR-494 and hsamiR-1246, also inhibit expression of a reporter carrying the Na+ – K+ –Cl− co-transporter SLC12A2 [solute carrier family 12 (Na+ – K+ –Cl− transporters), member 2] 3′ UTR, suggesting that these miRNAs may play a more general role in regulating chloride transport in epithelial cells.
Keywords: cystic fibrosis transmembrane conductance regulator (CFTR), microRNA (miRNA), solute carrier family 12 (Na+ -K+ -Cl− transporters), member 2 (SLC12A2), post-transcriptional regulation
INTRODUCTION
CF (cystic fibrosis), the most common lethal autosomal recessive disorder in Caucasians, results from mutation of the CFTR (CF transmembrane conductance regulator) gene [1]. The CFTR locus spans 189 kb of DNA and includes 27 exons, which encode a 6.2 kb mRNA transcript, including a 1.5 kb 3′ UTR (untranslated region). The 1480-amino-acid CFTR protein is a member of the ABC (ATP-binding cassette) family of transporters and functions primarily as a chloride channel. The gene is expressed at low levels in specialized epithelial cells of the airway, and at much higher levels in pancreatic duct, intestinal and male genital duct epithelium. The gene is also expressed in some non-epithelial cells, and may be involved in a wide range of cellular processes. CFTR expression is subject to temporal regulation during development of the airway, with high expression in fetal lung and low expression in adult airway, except in the serous portion of submucosal glands and some ciliated cells of the surface epithelium. Despite this complex regulation of expression (reviewed in [2]), the CFTR promoter lacks key cell-type-specific control elements, implicating elements elsewhere in temporal and tissue-specific regulation.
Indeed, several regulatory elements have been identified both flanking the locus and within introns (reviewed in [2,3]). Enhancer-blocking insulators were found both proximal and distal to the coding region of CFTR [4,5], and several cell-type-specific enhancers were characterized within introns that co-ordinate looping of the active locus [6–9]. Although these cis-acting genomic elements are likely to be the primary regulators of CFTR expression, post-transcriptional regulation of the CFTR mRNA may also be important. In addition to alternative splicing, which is known to influence CFTR expression levels (reviewed in [10]), targeting of the CFTR transcript by miRNAs (microRNAs) may have a post-transcriptional regulatory role. These short (~22 nt) endogenously expressed non-coding RNAs generally suppress gene expression in mammals by binding to the 3′ UTR of transcripts and blocking translation or promoting degradation of the transcript by decapping or deadenylation (reviewed in [11]). In addition to suppression, miRNAs may also up-regulate expression, although this mechanism appears to involve binding in the 5′ UTR rather than the 3′ UTR of the transcript. miRNAs are predicted to regulate more than 50% of mammalian mRNAs post-transcriptionally [11], and there is evidence that 3′ UTR length strongly correlates with miRNA regulation [12]. In this context, it may be relevant that CFTR has a long (1.5 kb) 3′ UTR in comparison with the 740 bp average for human genes.
To date, several groups have examined the potential role of miRNAs in molecular pathways involved in the pathogenesis of CF, including lung inflammation [13,14]; however, despite its potential value as a therapeutic target, miRNA regulation of CFTR itself has not been investigated. An hypoxia-induced increase in expression of a group of miRNAs predicted to target CFTR correlates with hypoxia-induced down-regulation of CFTR expression, but no direct targeting of CFTR has been reported [15]. In the present paper, we describe the identification of several miRNAs which directly repress CFTR expression by binding to the 3′ UTR of the transcript and are expressed in relevant primary human epithelial cells. Moreover, we demonstrate that three of these miRNAs also target the Na+ –K+ –Cl− co-transporter SLC12A2 [solute carrier family 12 (Na+ –K+ –Cl− transporters), member 2], implying a more general role in co-ordinating chloride transport in epithelial cells.
EXPERIMENTAL
Cell culture
The human colon carcinoma cell line Caco-2 [16], bronchial epithelial cell line 16HBE14o- [17] and pancreatic adenocarcinoma cell line PANC-1 [18] were grown by standard methods. Primary HTEs (human tracheal cells) and NHBEs (normal human bronchial and tracheal epithelial cells; Lonza) were grown as described previously [19].
qRT-PCR (quantitative reverse transcription PCR)
Total RNA for qRT-PCR was isolated using TRIzol® (Invitrogen). CFTR expression was assayed using a TaqMan probe and primer set spanning exons 5 and 6 (TAQEX5/6) [7]. SLC12A2 expression was measured using a primer set spanning exons 13 and 14 [20]. miRNA expression was assayed using TaqMan miRNA assays (Applied Biosystems) according to the manufacturer’s protocols. 18S rRNA (CFTR/SLC12A2) and U6 snRNA (small nuclear RNA) (miRNAs) were used as endogenous normalization controls.
Western blot analysis
CFTR expression was assayed by Western blot analysis using the anti-CFTR antibody 570 [21]. Cells were lysed in RIPA buffer with 1% Triton X-100, using previously published protocols [22]. Cell lysates were separated by SDS/PAGE (3%/6% gels) and transferred on to Immobilon membranes (Millipore). Western blot quantification was performed using ImageJ software (NIH; http://rsb.info.nih.gov/ij/).
Luciferase reporter constructs
The CFTR 3′ UTR (chr7:117307180–117308839; hg19) was amplified by PCR from pCOF [24] and cloned into the MCS (multiple cloning site) of the 3′ UTR luciferase reporter vector pMIR-REPORT luciferase (Ambion) to create pMIR-CFTR. Mutant variants of pMIR-CFTR were created using the QuikChange® Lightning Site-Directed Mutagenesis Kit or Lightning Multi Site-Directed Mutagenesis Kit (Agilent). The SLC12A2 3′ UTR (chr5:127522279–127525419; hg19) was amplified from human genomic DNA using Pfu DNA polymerase (Agilent) and cloned into the MCS of pMIR-REPORT luciferase to create pMIR-SLC12A2.
Transient transfections
For evaluating the effects of miRNAs on endogenous CFTR mRNA or protein expression, cells were reverse-transfected with 20 nM (final concentration) of Pre-miR™ miRNA mimics (Ambion) using Lipofectamine™ RNAiMAX (Invitrogen). For luciferase reporter assays, cells were co-transfected 48 h after plating with 200 ng/ml luciferase reporter (e.g. pMIR-CFTR), 200 ng/ml pMIR-REPORT β-galactosidase (Ambion) and 20 nM (final concentration) Pre-miRs using Lipofectamine™ 2000 (Invitrogen). Assays were performed 48 h post-transfection. All data were normalized to transfectionwith the Pre-miR™ Negative Control #2 (Ambion).
Primer sequences
Primer sequences and genomic locations used for cloning and mutagenesis are shown in Supplementary Table S1 at http://www.BiochemJ.org/bj/438/bj4380025add.htm
RESULTS
The 3′ UTR of the CFTR transcript is approximately 1.5 kb and so is a plausible candidate for regulation by miRNAs. We first used an in silico approach to search for predicted miRNA seed sites within the CFTR 3′ UTR.
Multiple miRNAs are predicted to target the CFTR 3′ UTR
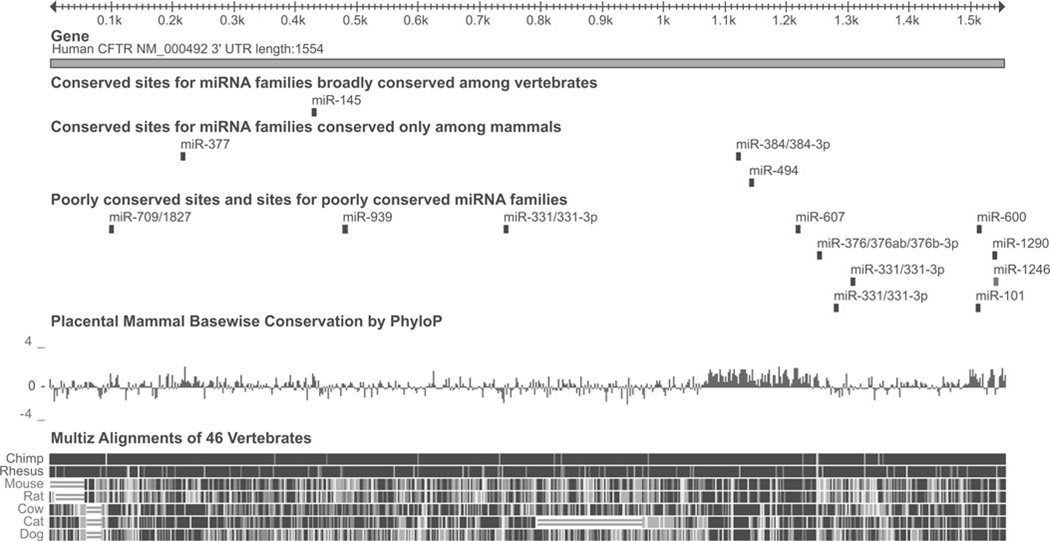
TargetScan (version 5.0) [25] was used to predict miRNAs that bind to the CFTR 3′ UTR since it is reported to be the most accurate algorithm currently available for in silico prediction of miRNAs that may bind to a gene, and does not rely exclusively on site conservation [26]. The 106 miRNAs predicted to target the CFTR 3′ UTR were ranked according to context score (the TargetScan raw binding affinity score, a measure of the affinity of a miRNA for a specific DNA sequence), and context score percentile (a measure of the context score of a predicted miRNA target site in CFTR relative to that of other predicted targets). The top 11 candidate miRNAs by context score are shown in Table 1 and the location of the predicted binding sites in the CFTR 3′ UTR for each are shown in Figure 1. Also shown are hsa-miR-101, the only highly conserved miRNA with a conserved target predicted by a previous version of TargetScan (version 4.0), and hsa-miR-939, for which sites are predicted in the 3′ and 5′ UTRs of CFTR, and thus may form a miRNA bridge [27].
Table 1. miRNAs predicted to target CFTR (TargetScan 5.1).
The top 11 miRNAs (sorted by context score) predicted to target the human CFTR 3′ UTR are listed, along with published expression data [28–34]. Two additional miRNAs predicted to target the human CFTR 3′ UTR are also included (see the text for details). CS, context score; N/A, not applicable.
| miRNA | CS | CS rank | CS percentile | Number of sites | Expression |
|---|---|---|---|---|---|
| hsa-miR-600 | −0.57 | 1 | 99 | 1 | Normal colonic epithelium |
| hsa-miR-607 | −0.53 | 2 | 99 | 1 | Caco-2 (colon carcinoma) |
| hsa-miR-494 | −0.53 | 2 | 99 | 1 | Multiple cell types |
| hsa-miR-384 | −0.5 | 4 | 99 | 1 | N/A |
| hsa-miR-376a/b | −0.49 | 5 | 97 | 1 | Multiple cell types |
| hsa-miR-1290 | −0.47 | 6 | 97 | 1 | Human embryonic stem cells |
| hsa-miR-1246 | −0.45 | 7 | 98 | 1 | Human embryonic stem cells |
| hsa-miR-377 | −0.45 | 7 | 97 | 1 | Various cell types |
| hsa-miR-145 | −0.43 | 9 | 95 | 1 | Multiple cell types, highest in prostate, uterus and epididymis |
| hsa-miR-1827 | −0.42 | 10 | 99 | 1 | HeLa (cervical carcinoma) |
| hsa-miR-331-3p* | −0.39 | 11 | 39, 40, 93* | 3 | Multiple cell types |
| hsa-miR-101 | −0.27 | 27 | 80 | 1 | Multiple cell types, highest in lymphocytes |
| hsa-miR-939 | −0.11 | 75 | 65 | 1 | Multiple cell types |
hsa-miR-331-3p has three predicted target sites in the CFTR 3′ UTR.
Figure 1. The CFTR 3′ UTR and predicted miRNAs.
The CFTR 3′ UTR, and the location of predicted miRNA seed sites, mammalian conservation and sequence alignment with relevant mammals. ‘Broadly conserved’ miRNAs are conserved across most vertebrates; ‘conserved’ miRNAs are conserved across most mammals. ‘Placental Mammal Basewise Conservation’ is based on sequence alignment of 33 mammalian genomes, including those appearing individually below [46–48]. The Figure is based on the output from http://genome.ucsc.edu (hg19 assembly) [49] and http://www.targetscan.org [25].
Since CFTR shows a complex pattern of cell-type-specific expression, with high expression in intestinal, pancreatic duct and epididymis epithelial cells, and low expression in the airway epithelium, we next evaluated previously published human expression data for the 13 candidates [28–34] (Table 1). Two candidate miRNAs, hsa-miR-600 and hsa-miR-607, were identified in colorectal epithelial cells, either the Caco-2 colon carcinoma cell line (hsa-miR-607) or primary normal colonic epithelium (hsa-miR-600) [32], hsa-miR-1246 and hsa-miR-1290 were identified in human embryonic stem cells [28], and hsa-miR-1827 was identified in HeLa cervical carcinoma cells [29]. hsa-miR-101, hsa-miR-145, hsa-miR-331-3p, hsa-miR-376a/b, hsa-miR-377, hsa-miR-494 and hsa-miR-939 are expressed in a wide range of cell types, with hsa-miR-101 expression highest in lymphocytes and hsa-miR-145 expression highest in the prostate, uterus and epididymis [30,31,33]. No human expression data are available for hsa-miR-384. This is likely to be due to the exceptionally low melting temperature of this miRNA making detection difficult [35].
miRNAs predicted by TargetScan repress endogenous CFTR expression
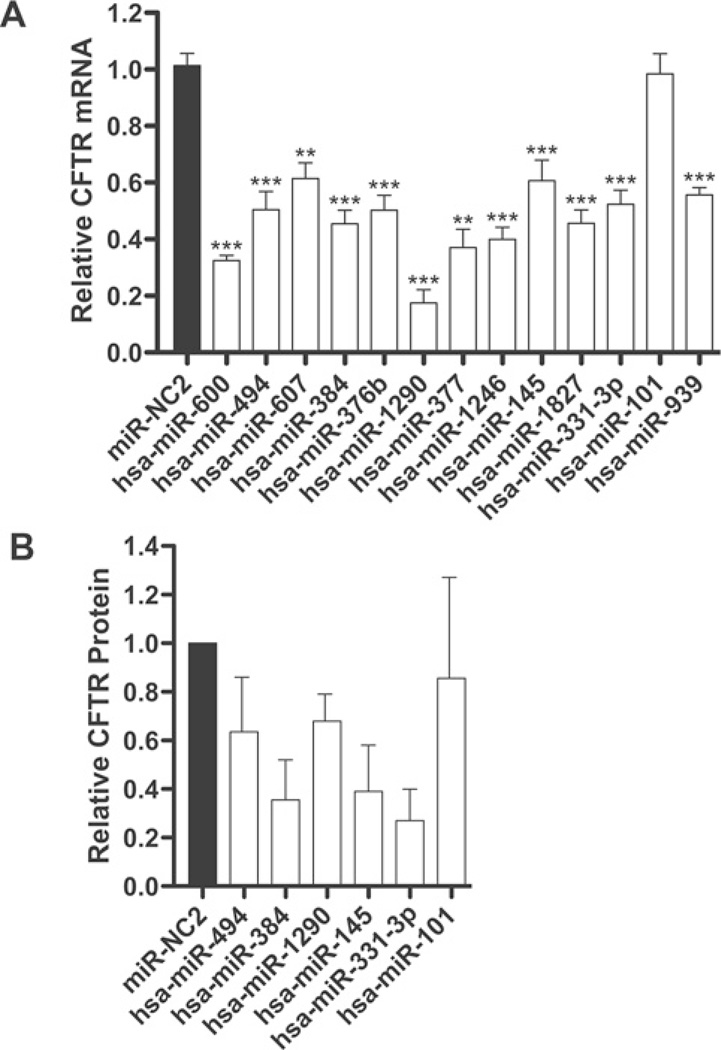
To investigate whether the TargetScan-predicted miRNAs influence CFTR mRNA and/or CFTR protein expression, miRNA mimics [dsRNAs (double-stranded RNAs) which are processed like endogenous pre-miRNAs] were transfected into Caco-2 cells. After 48 h, total RNA and protein were isolated in separate experiments and the effect of the miRNAs on CFTR mRNA and protein levels were assayed by qRT-PCR (Figure 2A) and Western blot analysis with anti-CFTR antibody 570 (Figure 2B) respectively. Twelve out of the 13 predicted miRNAs repressed CFTR mRNA expression by at least 40% relative to a negative control miRNA. The sole exception was hsa-miR-101, which has the second weakest TargetScan context score among the miRNAs assayed. Only hsa-miR-939 has a lower context score, but this does not account for the predicted site in the 5′ UTR, which may make a functional contribution [27]. Consistent with the qRT-PCR data, hsa-miR-145, hsa-miR-331-3p, hsa-miR-384, hsa-miR-494 and hsa-miR-1290 individually decreased CFTR protein levels by 30–60%, whereas hsa-miR-101 had no effect (Figure 2B). A representative Western blot is shown in Supplementary Figure S1 at http://www.BiochemJ.org/bj/438/bj4380025add.htm. Transfection of multiple miRNAs did not enhance knockdown of either protein ormRNA in Caco-2 cells relative to the transfection of individual miRNAs (results not shown).
Figure 2. Repression of CFTR expression in Caco-2 cells by miRNAs predicted with TargetScan.
(A) Expression of CFTR mRNA in Caco-2 cells, measured by TaqMan qRT-PCR, after transient transfection of the indicated miRNA mimics. miRNAs are ordered by TargetScan context score, with the strongest predictions on the left-hand side. (B) ImageJ quantification of CFTR protein expression in Caco-2 cells, measured by Western blot analysis, after transient transfection of the indicated miRNA mimics (average of two biological replicates). *P < 0.05, **P < 0.01 and ***P < 0.001. Values are means ± S.E.M.
miRNAs that repress CFTR expression target the 3′ UTR
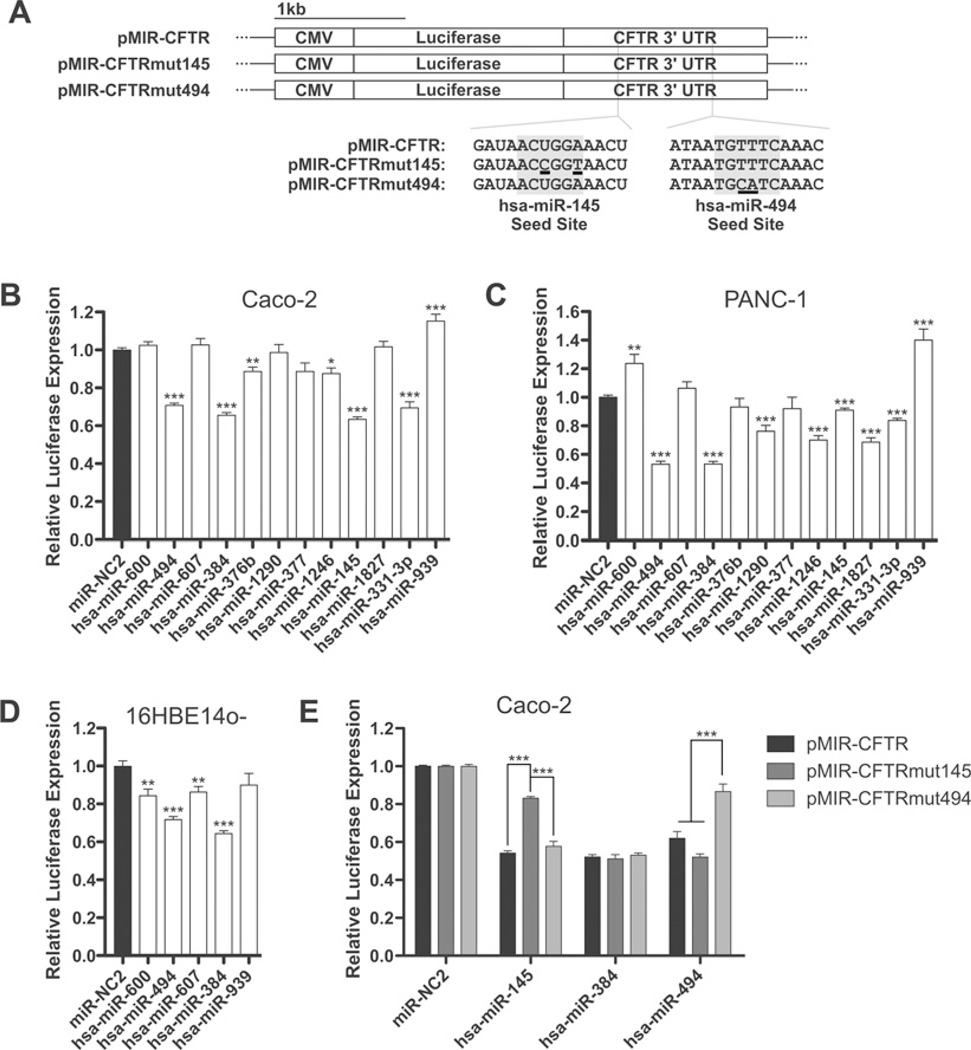
To determine whether miRNAs that repress endogenous CFTR expression act directly through the CFTR 3′ UTR, a series of luciferase reporters were generated in pMIR REPORT (Figure 3A). pMIR-CFTR contains the complete wild-type 1.5 kb CFTR 3′ UTR; pMIR-CFTRmut145 and pMIR-CFTRmut494 carry mutations in the predicted hsa-miR-145 and hsa-miR-494 target sites respectively within this 1.5 kb (Supplementary Table S1). pMIR-CFTR, pMIR-REPORT β-galactosidase [CMV (cytomegalovirus) β-galactosidase transfection control] and miRNA mimics were co-transfected into Caco-2 colon carcinoma cells and PANC-1 pancreatic adenocarcinoma cells. Several miRNAs acted directly on the CFTR 3′ UTR in this assay (Figures 3B and 3C). hsa-miR-384 and hsa-miR-494 each repressed luciferase expression by >30% in both cell lines (P < 0.001). hsa-miR-145 and hsa-miR-331-3p also significantly inhibited reporter expression in both cell lines (P < 0.001), but more effectively in Caco-2 (>30% repression) than PANC-1 (10–20%) cells. hsa-miR-1246 produced significant knockdown in PANC-1 cells (P < 0.001), but was less effective in Caco-2 cells (P < 0.05). hsa-miR-1290- and hsa-miR-1827-dependent repression was significant in PANC-1 (P < 0.001), but not in Caco-2, cells, whereas hsa-miR-376b showed the opposite result. Five out of the 11 miRNAs that caused CFTR mRNA inhibition were also assayed in 16HBE14o- bronchial epithelial cells (Figure 3D) and hsa-miR-384 and hsa-miR-494 again caused significant knockdown (P < 0.001). hsa-miR-600 and hsa-miR-607 targeted CFTR in 16HBE14o- cells, but were ineffective in the other cell lines.
Figure 3. miRNA-mediated CFTR repression acts through the 3′ UTR.
(A) The pMIR-REPORT-based luciferase reporter constructs used in these assays. (B–D) Luciferase expression from pMIR-CFTR (full-length CFTR 3′ UTR) after co-transfection with pMIR-REPORT β-galactosidase and the indicated miRNA mimics in Caco-2 (B), PANC-1 (C) and 16HBE14o- (D) cells. miRNAs are ordered by TargetScan context score, with the strongest predictions on the left-hand side. (E) Luciferase expression from pMIR-CFTR, pMIR-CFTRmut145 and pMIR-CFTRmut494 after co-transfection with pMIR-REPORT β-galactosidase and the indicated miRNA mimics in Caco-2 cells. *P < 0.05, **P < 0.01 and ***P <0.001. Values are means ± S.E.M. (n ≥ 9).
To confirm that several of these miRNAs were acting directly through their predicted seed sites in the CFTR 3′ UTR, we next evaluated the effect of mutation of the TargetScan-predicted binding sites for hsa-miR-145 and hsa-miR-494. pMIR-CFTRmut145 and pMIR-CFTRmut494 were co-transfected with pMIR-REPORT β-galactosidase and miRNA mimics into Caco-2 and PANC-1 cells. Mutation of the miR-145 and miR-494 seed sites significantly reduced repression of reporter expression in comparison with the wild-type sequence in Caco-2 cells (P ≤ 0.001) (Figure 3E), thus confirming the functional importance of these sites. Repression by another miRNA (hsa-mIR-384) was unaffected in pMIR-CFTRmut145 and pMIR-CFTRmut494.
miRNA-145 and miRNA-331-3p are differentially expressed in Caco-2, 16HBE14o-, PANC-1 and primary human airway epithelial cells
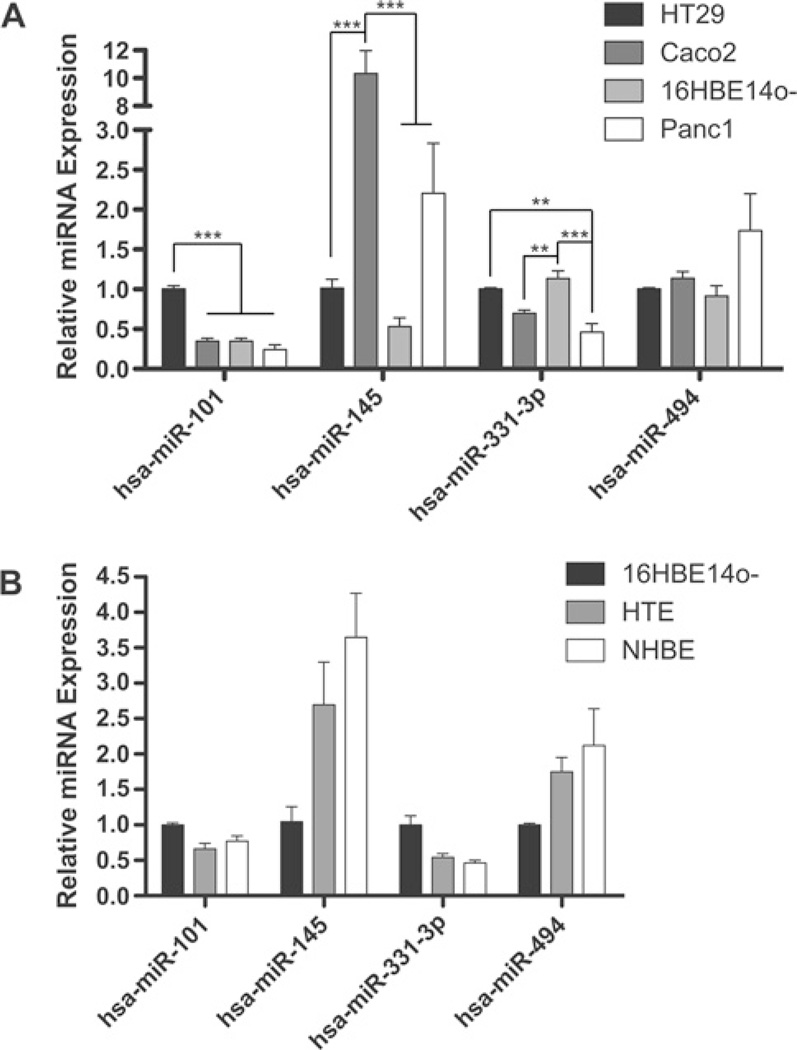
To investigate the relative expression levels of each miRNA in the cell lines employed in the present study, we used TaqMan miRNA assays for hsa-miR-101, hsa-miR-145, hsa-miR-331-3p, hsa-miR-384 and hsa-miR-494. Expression profiles for Caco-2, 16HBE14o- and PANC-1 cells are shown in Figure 4(A), together with HT29, a human colorectal carcinoma cell line included in the NCI-60 cancer cell panel, for comparison with published NCI-60 miRNA expression data [36,37]. There was no significant difference between Caco-2, PANC-1 and 16HBE14o- cells in the expression levels of hsa-miR-101 or hsa-miR-494, but expression of hsa-miR-101 was significantly higher in HT29 (P < 0.001) than in any other cell line. Levels of hsa-miR-145 were significantly higher in Caco-2 cells (P < 0.001) than in the other cell lines, whereas hsa-miR-331-3p levels were significantly higher in 16HBE14o- than in Caco-2 (P < 0.01) or PANC-1 (P < 0.001) cells. No hsa-miR-384 expression was detected in any of the cell lines tested (including K562, T47D, MCF7 and B549; results not shown), consistent with the lack of any published hsa-miR-384 expression data. We next measured expression of hsa-miR-101, hsa-miR-145, hsa-miR-331-3p and hsa-miR-494 in primary HTE cells and cultures of NHBEs in comparison with 16HBE14o- cells (Figure 4B). Three primary cultures of each cell type were used to account for individual variation.
Figure 4. Expression of miRNA-101, miRNA-145, miRNA-331-3p and miRNA-494 in (A) cell lines and (B) primary human tracheal and bronchial cells (HTEs and NHBEs).
Expression of hsa-miR-101, hsa-miR-145, hsa-miR-331-3p and hsa-miR-494, measured by qRT-PCR with TaqMan miRNA assays. *P < 0.05, **P < 0.01 and ***P < 0.001. Values are means ± S.E.M. Data from three cultures of HTEs and NHBEs are combined to account for variation between individuals.
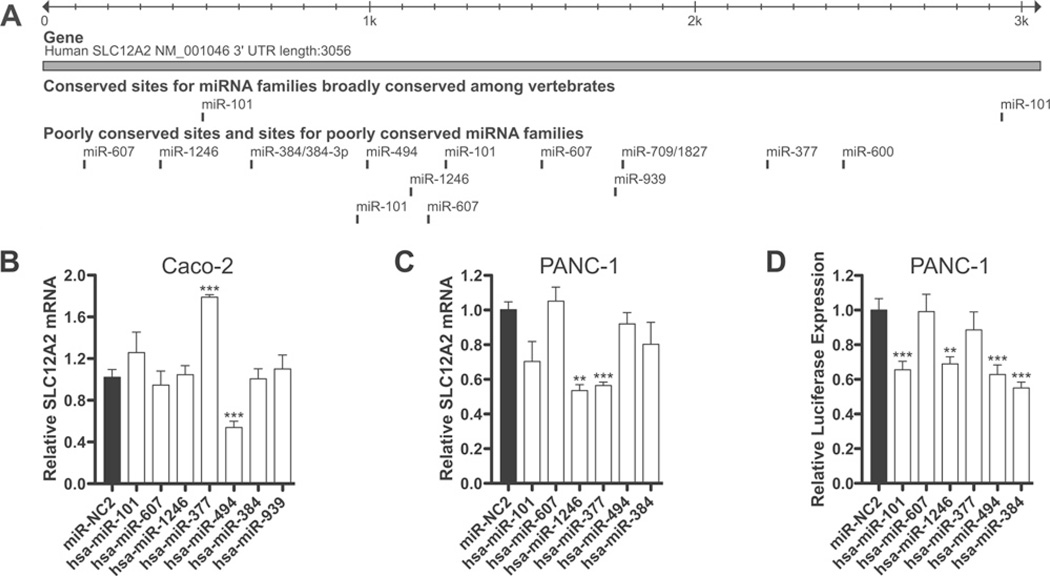
hsa-miR-384, hsa-miR-494 and hsa-miR-1246 also target the SLC12A2 3′ UTR
SLC12A2 (NKCC1) is among the group of ion channels, ion exchangers and transporters that interact directly with CFTR and influence its activity [38]. We examined the 3.1 kb 3′ UTR of SLC12A2 for predicted miRNA seed sites and identified sites for seven of the 12 miRNAs predicted to repress CFTR expression, together with a site for hsa-miR-101 (Figure 5A). To determine whether SLC12A2 is also functionally targeted by these miRNAs, we transfected miRNA mimics into Caco-2 and PANC-1 cells and assayed SLC12A2 expression by qRT-PCR (Figures 5B and 5C). All seven miRNAs predicted to target SLC12A2 were assayed in Caco-2 cells (Figure 5B), and six of these (excluding hsa-miR-939, which has a context score of 0.0) were evaluated in PANC-1 cells (Figure 5C). In contrast with miRNA-mediated CFTR mRNA repression, only three miRNAs significantly repressed SLC12A2 mRNA: hsa-miR-494 in Caco-2 cells (P < 0.001), and hsa-miR-377 and hsa-miR-1246 in PANC-1 cells (P < 0.01). To assay for direct targeting of the SLC12A2 3′ UTR by these miRNAs, a pMIR-REPORT construct was generated that encompassed the full 3′ UTR of the transcript. pMIR-SLC12A2, pMIR-REPORT β-galactosidase and miRNA mimics were co-transfected into PANC-1 cells (Figure 5D). Four miRNAs repressed reporter expression by >35% (P < 0.01), including three that also functionally target the CFTR 3′ UTR, hsa-miR-384, hsa-miR-494 and hsa-miR-1246.
Figure 5. Repression of SLC12A2 expression by miRNAs that also target CFTR.
(A) The SLC12A2 3′ UTR, and the location of predicted miRNA seed sites. The Figure is based on the output from http://genome.ucsc.edu (hg19 assembly) [49] and http://www.targetscan.org [25]. (B–C) Expression of SLC12A2 mRNA in Caco-2 (B) and PANC-1 (C) cells, measured by TaqMan qRT-PCR, after transient transfection of the indicated miRNA mimics. miRNAs are ordered by TargetScan context score, with the strongest predictions on the left-hand side. (D) Luciferase expression from pMIR-SLC12A2 (full-length SLC12A2 3′ UTR) after co-transfection with pMIR-REPORT β-galactosidase and the indicated miRNA mimics in PANC-1 cells. miRNAs are ordered by TargetScan context score, with the strongest predictions on the left-hand side. *P <0.05, **P < 0.01 and ***P < 0.001. Values are means ± S.E.M. (n ≥ 9).
DISCUSSION
It is well known that miRNAs contribute to the pathogenesis of a number of important human diseases, including cancer. The Bcl2 (B-cell lymphoma 2) oncogene, an anti-apoptotic protein that is down-regulated in several cancers, is a proven target of hsa-miR-15 and hsa-miR-16. Both of these miRNAs are able to down-regulate Bcl2 expression and induce apoptosis when expressed in cell lines, and both are down-regulated in chronic lymphocytic leukaemias [39,40]. In contrast, the ‘oncomiR’ hsa-miR-21 is overexpressed in a variety of tumours, where it acts by repressing PTEN (phosphatase and tensin homologue) and PDCD4 (programmed cell death 4) expression [41,42]. miRNAs have also been implicated in cardiac and neurodegenerative diseases, diabetes, inflammation and diseases of the immune system (reviewed in [43]). Hence they show great promise as therapeutic targets, largely due to their potential for inhibition by artificial antisense miRNAs (antagomirs or anti-miRs). LNA (locked nucleic acid) anti-miRs are exceptionally stable, non-toxic and soluble in aqueous solution. Delivery of LNA anti-miRs directed against miR-122 in monkeys results in long-lasting and reversible knockdown of the targets of miR-122 with no evidence of toxicity [44]. This ease of in vivo inhibition highlights the importance of identifying miRNA-target interactions relevant to disease states.
In the present study we demonstrate that miRNAs post-transcriptionally regulate expression of the CFTR gene. Moreover, we show that at least three of these miRNAs (hsa-miR-145, hsa-miR-331-3p and hsa-miR-494) are expressed in primary human airway epithelial cells, which have very low levels of CFTR expression in comparison with intestinal epithelial cells. Of the 12 miRNAs that repress expression of CFTR mRNA in Caco-2 cells (Figure 2A), ten of these directly target the CFTR 3′ UTR in Caco-2, PANC-1 or 16HBE14o- cells (Figures 3B–3D). For hsa-miR-145 and hsa-miR-494, this targeting occurs at least partially through the sites predicted by TargetScan. All of the top 11 TargetScan-predicted miRNAs are able to repress endogenous CFTR mRNA expression by >40% in Caco-2 cells, but only four repress 3′ UTR reporter expression by >30%in the same cell line. This may be due to miRNA regulation of upstream (transcription) factors involved in CFTR expression, or these miRNAs binding to sites outside of the 3′ UTR. The fact that the luciferase reporter is repressed by all but one of the top 11 miRNAs (hsa-miR-377) in Caco-2, PANC-1 or 16HBE14o- cells, but not necessarily in all of the cell lines, warrants further discussion. Although it is possible that target site utilization is cell-type-dependent, or that there are cell-type-specific cofactors required for repression with some miRNAs, these results may reflect significant differences in miRNA transfection efficiency between cell lines and reagents. Although Lipofectamine™ 2000 (used in luciferase experiments) was the most efficient reagent available for co-transfection of small dsRNA and plasmid DNA [measured by hsa-miR-1 knockdown of TWF1 (twinfilin-1) mRNA; results not shown], it was significantly less efficient than Lipofectamine™ RNAiMAX (used in qRT-PCR experiments) for miRNA delivery in Caco-2 cells. This difference was less evident in PANC-1 cells, where both reagents were as efficient as Lipofectamine™ RNAiMAX in Caco-2 cells. Thus, in Caco-2 cells, the functional miRNA concentrations in experiments that measured direct effects on the CFTR 3′ UTR were likely to be lower than in those with Lipofectamine™ RNAiMAX or in PANC-1 cells with either reagent.
It is of interest that the transient transfections using the pMIR-CFTRmut145 and pMIR-CFTRmut494 mutant reporters confirmed the functional relevance of the binding sites predicted by TargetScan. The fact that mutation of these sites fail to relieve repression completely suggests that there may be additional functional binding sites in the CFTR 3′ UTR. Indeed, while TargetScan predicts only a single target site for hsa-miR-494, the CFTR 3′ UTR contains two sequences complementary to the miR-494 seed site (GAAACA). The second site, located 292 bp downstream of the TargetScan-recognized site, is not predicted by this program because it has neither an ‘A’ opposite base 1 nor pairing at base 8 of the miRNA sequence. RNAHybrid, another tool for modelling RNA–RNA interactions, predicts slightly stronger pairing between the mRNA and miRNA at this 3′ site, based on calculated minimum free energy of hybridization. There is only one sequence complementary to the hsa-miR-145 seed site in the CFTR 3′ UTR, suggesting a different mechanism for maximal hsa-miR-145-mediated repression.
The physiological levels of miRNAs that are required to regulate CFTR expression in normal cells in vivo are of interest, particularly given the large variation in endogenous expression of the miRNAs seen in the different cell lines utilized in these experiments; however, there are currently few studies that directly address this question in primary human epithelial cells. The results of the present study show that the relatively high level of hsa-miR-145 expression in Caco-2 cells is not sufficient to maximally repress CFTR, as the introduction of exogenous hsa-miR-145 further represses expression of endogenous CFTR and reporters carrying the CFTR 3′ UTR. However, miRNA inhibition experiments would be necessary to definitively establish the effect of endogenous hsa-miR-145 on CFTR expression in Caco-2 cells.
Of the ten miRNAs that target the CFTR 3′ UTR in the luciferase reporter assay, three also target the SLC12A2 3′ UTR in PANC-1 cells: hsa-miR-384, hsa-miR-494 and hsa-miR-1246. Of these, only hsa-miR-494 has a validated target, PTEN [45], and none have yet been implicated in the regulation of epithelial chloride channels and transporters. However, the results of the present study show that they are able to functionally regulate two of the primary chloride transporters in epithelial cells, and thus they may have a wider role in co-ordinating epithelial ion transport.
The dominant mechanisms controlling CFTR expression levels in vivo are driven by cis-acting regulatory elements (reviewed in [2]). However, miRNAs could also play an important role in directing changes in the amounts of mature CFTR protein, which in turn could have a significant effect on cell physiology. Thus the identification of multiple miRNAs that target the CFTR gene may have a direct therapeutic application. This would be particularly relevant to certain classes of mutations in the CFTR gene that result in low and insufficient expression of a functional CFTR protein. Among others, these include mutations that reduce transcriptional activity, generate an unstable mRNA (e.g. stop mutations) and affect splicing (reviewed in [10]). In patients with these disease-causing mutations, inhibition (by antagomirs or anti-miRs) of miRNAs targeting CFTR in relevant tissues has the potential to elevate CFTR expression levels to within the normal range.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr J. Rommens (Hospital for Sick Children, Toronto, Canada) for the pCOF plasmid, Dr M. Gentzsch (University of North Carolina, Chapel Hill, NC, U.S.A.) for advice on CFTR Western blots and E. Seftor (CBE Program, Children’s Memorial Research Center, Chicago, IL, U.S.A.) for reagents.
FUNDING
This work was supported by the Cystic Fibrosis Foundation USA [grant number Harris1010]; and the National Institutes of Health [grant number R01HL094585 (to A.H.)]. A.E.G. and N.G. are in the Northwestern University Integrated Graduate Programme.
Abbreviations used
- Bcl2
B-cell lymphoma 2
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- dsRNA
double-stranded RNA
- HTE
human tracheal epithelial cell
- LNA
locked nucleic acid
- MCS
multiple cloning site
- miRNA
microRNA
- NHBE
human bronchial and tracheal epithelial cell
- PTEN
phosphatase and tensin homologue
- qRT-PCR
quantitative reverse transcription PCR
- SLC12A2
solute carrier family 12 (Na+ –K+ – Cl− transporters), member 2
- UTR
untranslated region
Footnotes
AUTHOR CONTRIBUTION
Austin Gillen and Ann Harris designed the experiments and analysed the data. Austin Gillen, Nehal Gosalia and Shih-Hsing Leir performed the experiments. Austin Gillen and Ann Harris wrote the paper.
REFERENCES
- 1.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 2.Gillen AE, Harris A. Transcriptional regulation of CFTR gene expression. Front. Biosci. 2011 doi: 10.2741/401. in the press. [DOI] [PubMed] [Google Scholar]
- 3.Ott CJ, Harris A. Genomic approaches for the discovery of CFTR regulatory elements. Transcription. 2010;2:23–27. doi: 10.4161/trns.2.1.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 7.Mouchel N, Henstra SA, McCarthy VA, Williams SH, Phylactides M, Harris A. HNF1α is involved in tissue-specific regulation of CFTR gene expression. Biochem. J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowntree RK, Vassaux G, McDowell TL, Howe S, McGuigan A, Phylactides M, Huxley C, Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001;10:1455–1464. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
- 10.Rowntree RK, Harris A. The phenotypic consequences of CFTR mutations. Ann. Hum. Genet. 2003;67:471–485. doi: 10.1046/j.1469-1809.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- 11.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′ UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am. J. Respir. Cell Mol. Biol. 2010;42:506–513. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation, but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guimbellot JS, Erickson SW, Mehta T, Wen H, Page GP, Sorscher EJ, Hong JS. Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis. BMC Med. Genomics. 2009;2:15. doi: 10.1186/1755-8794-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J. Natl. Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 17.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 18.Lieber M, Mazzetta J, Nelson-Rees W, Kaplan M, Todaro G. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int. J. Cancer. 1975;15:741–747. doi: 10.1002/ijc.2910150505. [DOI] [PubMed] [Google Scholar]
- 19.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell. Mol. Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright PK, May FE, Darby S, Saif R, Lennard TW, Westley BR. Estrogen regulates vesicle trafficking gene expression in EFF-3, EFM-19 and MCF-7 breast cancer cells. Int. J. Clin. Exp. Pathol. 2009;2:463–475. [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR. Domain interdependence in the biosynthetic assembly of CFTR. J. Mol. Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 22.Farinha CM, Penque D, Roxo-Rosa M, Lukacs G, Dormer R, McPherson M, Pereira M, Bot AG, Jorna H, Willemsen R, et al. Biochemical methods to assess CFTR expression and membrane localization. J. Cystic Fibrosis. 2004;3(Suppl. 2):73–77. doi: 10.1016/j.jcf.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 23. Reference deleted. [Google Scholar]
- 24.Rommens JM, Dho S, Bear CE, Kartner N, Kennedy D, Riordan JR, Tsui LC, Foskett JK. cAMP-inducible chloride conductance in mouse fibroblast lines stably expressing the human cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7500–7504. doi: 10.1073/pnas.88.17.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 27.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 30.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 31.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 34.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, D’Andrade P, Fulmer-Smentek S, Lorenzi P, Kohn KW, Weinstein JN, Pommier Y, Reinhold WC. mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol. Cancer Ther. 2010;9:1080–1091. doi: 10.1158/1535-7163.MCT-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blower PE, Verducci JS, Lin S, Zhou J, Chung JH, Dai Z, Liu CG, Reinhold W, Lorenzi PL, Kaldjian EP, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol. Cancer Ther. 2007;6:1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 38.Shumaker H, Soleimani M. CFTR upregulates the expression of the basolateral Na+ -K+ -2Cl− cotransporter in cultured pancreatic duct cells. Am. J. Physiol. 1999;277:C1100–C1110. doi: 10.1152/ajpcell.1999.277.6.C1100. [DOI] [PubMed] [Google Scholar]
- 39.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D’Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 43.Tsai LM, Yu D. MicroRNAs in common diseases and potential therapeutic applications. Clin. Exp. Pharmacol. Physiol. 2010;37:102–107. doi: 10.1111/j.1440-1681.2009.05269.x. [DOI] [PubMed] [Google Scholar]
- 44.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Jiang Y, Zhang H, Greenlee AR, Han Z. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life Sci. 2010;86:192–198. doi: 10.1016/j.lfs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.